Abstract
The human short interspersed repeated element (SINE), Alu, amplifies through a poorly understood RNA-mediated mechanism, termed retroposition. There are over one million copies of Alu per haploid human genome. The copies show some internal variations in sequence and are very heterogeneous in chromosomal environment. However, very few Alu elements actively amplify. The amplification rate has decreased greatly in the last 40 million years. Factors influencing Alu transcription would directly affect an element’s retroposition capability. Therefore, we evaluated several features that might influence expression from individual Alu elements. The influence of various internal sequence variations and 3′ unique flanks on full-length Alu RNA steady-state levels was determined. Alu subfamily diagnostic mutations do not significantly alter the amount of Alu RNA observed. However, sequences containing random mutations throughout the right half of selected genomic Alu elements altered Alu RNA steady-state levels in cultured cells. In addition, sequence variations at the 3′ unique end of the transcript also significantly altered the Alu RNA levels. In general, sequence mutations and 3′ end sequences contribute to Alu RNA levels, suggesting that the master Alu element(s) have a multitude of individual differences that collectively gives them a selective advantage over other Alu elements.
INTRODUCTION
Alu belongs to a family of mobile, repetitive elements that have amplified to over one million copies per haploid human genome (~10% of the genome) (1). Alu elements have amplified over the past 65 million years through an RNA-mediated process, termed retroposition (2,3). Alu elements can be organized into subfamilies based on a series of diagnostic positions (4). These subfamilies amplified sequentially during primate evolution, with a decrease in amplification rate by as much as 100-fold for the more recent subfamilies (5). Currently, only a few low copy, young subfamilies of Alu elements are retropositionally active in the human genome (6).
Retroposition first involves transcription from the internal RNA polymerase III promoter (7), which is influenced by upstream flanking sequences (8,9). The transcript is then subjected to reverse transcription, and integration into a new genomic site in an as yet uncharacterized process (10). Despite their high copy number, Alu elements are generally expressed at very low concentrations (11–14), but are subject to stimulation by a number of factors (15–19). Several studies have demonstrated the importance of Alu upstream flanking sequences to the transcription from individual sites (8,20). Mutations that alter the internal core promoter are also known to influence Alu expression (21). Although Alu elements have high sequence identity to one another, there are a number of different types of sequence variations within the individual Alu elements that may influence Alu RNA steady-state levels, either through transcription or stability. These include the subfamily diagnostic mutations, since the subfamilies have amplified to widely different numbers (5). Random mutations within the Alu element, in addition to those in the core promoter, may influence protein binding to DNA–RNA, or RNA secondary structures. Any of these factors could influence the steady-state RNA level from an individual Alu element and may contribute to the observed changes in Alu subfamily amplification rate.
Another source of variability is found in the 3′ region of Alu transcripts. Alu elements do not encode an RNA polymerase III terminator (22). Instead, the polymerase continues past the Alu sequence into the 3′ flanking sequence until it finds a termination sequence. Therefore, each individual Alu element will have a unique sequence at the 3′ end of its RNA. These unique sequences may affect the amount of RNA accumulated from the different Alu elements, possibly accounting for some of the variation in individual Alu expression levels. For example, the termination signal has been suggested to influence the recycling of the polymerase (23), potentially influencing transcription rates. Furthermore, either the primary sequence or the secondary structure present at the 3′ end may be important for RNA stability.
We have created a number of mutant Alu constructs in order to study the effects of diagnostic mutations, random mutations and various RNA polymerase III terminators on Alu RNA steady-state concentrations and presumably their amplification potential. These data will allow us to further characterize the features necessary for an active Alu element.
MATERIALS AND METHODS
Construction of plasmids
The subfamily constructs were generated using the Bio-Rad Muta-Gene Phagemid In Vitro Mutagenesis v.2 (Hercules, CA) starting from a pBluescript clone carrying the consensus Ya5 element, pPD39 (24). The oligonucleotides used to create the mutations are presented in Table 1A. Mutations were created successively by making the older subfamilies from the younger subfamilies.
Table 1. Mutagenic oligonucleotides and primers used to make plasmid constructs containing each Alu subfamily sequence (A), the different Alu right-half (B) and altered RNA polymerase III terminators (C).
| Oligo name | Oligonucleotide sequence (5′–3′) |
|---|---|
| A | |
| Y-1 | ACGGGGTTTCACCGTGTTAGCCAGGATGGTCTCGATCT |
| Y-2 | ACAGGCGCCCGCCACCACGCCCGGCTAATTT |
| Y-3 | CTGCCTCAGCCTCCCGAGTAGCTGGGACTAC |
| Y-4 | TGGAGTGCAGTGGCGCGATCTCGGCTCACTG |
| Sg-1 | CGAACTCCTGACCTCGGTGATCCGCCCGCCT |
| Sx-1 | AGACGGGGTTTCACCATGTTGGCCAGGCTGGTCTCGAACCCTGACCTCAGGT |
| Sx-2 | ACCACGCCCGGCTAATTTTTGTATTTTTAGTAGAGA |
| Sx-3 | TGGGACTACAGGCGCGCGCCACCACGCCCGG |
| Sx-4 | TCTCGGCTCACTGCAACCTCCGCCTCCCGGGTTCAAGCGATTTCCTGCCTCA |
| ERE | TTTCACCATGTTGGCTAGGCTGGTCTCGAAC |
| B | |
| Full3959-5′ | CTTGAATGATCTTTACTTTGAGAAA |
| Full3959-3′ | CACCTTACAATTAGGTGAGACCCAT |
| Full0115-5′ | CACAGGTAACTTTGTAGATGT |
| Full0115-3′ | TTGAGAGATTGGTGTGGCTCT |
| Full5368-5′ | GCAGAACAGAGAGATAAGGGTCCAG |
| Full5368-3′ | GTCCTCTGAGACAACAGAACCCTAG |
| Full98047-5′ | ACAGTGTGTGGGTTTTAATCAGATG |
| Full98047-3′ | AACCAAGGAGTGTCTTTCTCAAGGG |
| Full84472-5′ | TCCCGAGGCTGTCACCAGGTGAGGT |
| Full84472-3′ | GGGCTGCCATTTGCTAGCACTCTGT |
| Full0453-5′ | CTTGAATGATCTTTACTTTGAGAAA |
| Full0453-3′ | CACCTTACAATTAGGTGAGACCCAT |
| Full31274-5′ | TTATTCCATTGGTCCTTTCCACCAG |
| Full31274-3′ | CAGGCAGGGAGGTACTTGTCTCTTG |
| Alu-midA5′ | TGGTGAAACCCCGTCTCTACT |
| 5368Right5′ | GAGACGAAGTCTCACTCTGTTGCCT |
| 5368Right3′ | TTAGCCGAGTGAGGTGATGGGCGCC |
| 3959Right3′ | TTTTTTTTTTGAGACGGAGTCTCGCTCTGTCGCCCAGGC |
| 0115Right3′ | TTTTTTTTTTGAGATGAAGTCTCGCTCTTGTCCCCCAG |
| 0453Right3′ | TTTTTTTTTTGAGACGGAGTCTCGCTCTGTCGCCCAGGC |
| 98047Right3′ | GAGACGGAGTCTCGCTCTGTCACCCAGGCTGG |
| 84472Right3′ | GAGACAGAGTTTCGCTCGTCACCCAGGCTGGA |
| C | |
| F-BC1 3′n | CACACAACCTTTTTCATTTTCAAAGACCCCCAAGGGCATTTTCA |
| R-BC1 3′n | TGAAAATGCCCTTGGGGGTCTTTGAAAATGAAAAAGGTTGTGTG |
| F-BC1 3′B1 | CACACAAAATTTTTAATTTTCAAAGACCCCCAAGGGCATTTTCA |
| R-BC1 3′B1 | TGAAAATGCCCTTGGGGGTCTTTGAAAATTAAAAATTTTGTGTG |
| F-BC1 3′5S | CACACAAGCTTTTTGCTTTTCAAAGACCCCCAAGGGCATTTTCA |
| R-BC1 3′5S | TGAAAATGCCCTTGGGGGTCTTTGAAAAGCAAAAAGCTTGTGTG |
| R-AluTs | GGTCTTGAAAATGAAAAAGAGACGGAGTCTC |
Human DNA was isolated from peripheral lymphocytes as described previously (1). Specific Alu elements were amplified to provide authentic right-half sequences. Primers for PCR amplification of genomic Alu elements were designed from the upstream and downstream flanking sequences of random Alu elements selected from the database (Table 1B). Amplification of DNA samples was carried out in 10 µl reactions using 50 ng of target DNA, 50 pmol of each oligonucleotide primer, 2 mM dNTPs in 2.5 mM MgCl2 and 2.5 U AmpliTaq DNA polymerase (Perkin Elemer, Branchburg, NJ). Each sample was subjected to the following amplification conditions: 2 min at 95°C (denaturation). The reaction was then cycled for: 0.5 min at 95°C, 0.5 min at 58°C (annealing) and 1.5 min at 72°C (extension) for 35 cycles. A final extension period of 3 min at 72°C was added. A second round PCR reaction was performed to amplify only the right half of the specific Alu element. The PCR reaction was the same as above except that the annealing temperature was 55°C. The Alu-mid A region primer was used as the 5′ primer for Alu3959, Alu0115, Alu0453, Alu98047 and Alu84472 and their corresponding 3′ primer (Table 1B).
The PCR products were used as mutagenic primers in the QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA) to change the right half of p7SLSxBC1. The extension reaction was as follows: 95°C for 0.5 min (denaturation), 55°C for 1.0 min (annealing) and 72°C for 14.0 min (extension) for 17 cycles. DNA from mutagenized colonies was extracted using the Wizard Plus SV Minipreps DNA purification system (Promega, Madison, WI). Positive clones were confirmed by DNA sequencing with ThermoSequenase Radiolabeled Terminator Cycle Sequencing kit (USB, Cleveland, OH), following the manufacturer’s protocol.
To make the constructs with the different RNA polymerase III terminator variants, the primers shown in Table 1C were used. The forward and reverse primers were used in the extension reaction using the QuikChange Mutagenesis Kit (Stratagene) as above.
Large scale DNA preparations of the constructs were performed with the Qiagen Plasmid Maxi Kit (Qiagen, Valencia, CA) using the manufacturer’s recommended protocol. After final resuspension, the DNA samples were clarified by centrifugation for 10 min at maximum speed in an Eppendorf tabletop centrifuge 5415C (Brinkman Instruments, Inc., Westbury, NY). The supernatant was filtered through a Millex-GS 0.2 µm filter unit with protein-binding capacity (Millipore, Molsheim, France). DNA was quantitated by absorption at 260 nm and DNA quality and concentrations confirmed by agarose gel electrophoresis.
Transcription in cell lines
Transient transfections were carried out in NIH 3T3 (ATCC CRL1658), 293 (ATCC CRL 1573), H1299 (human lung carcinoma) and HeLa (ATCC CCL2) cell lines. Cell lines were grown in Dulbecco’s modified essential medium plus glutamine and 10% calf serum (Gibco BRL Life Technologies, Rockville, MD). T25 flasks were seeded with 5 × 105 cells and co-transfected the following day with 2 or 3 µg of the construct and 1 µg of control vector (pBC1) using Lipofectamine Plus system (Gibco BRL Life Technologies) following the manufacturer’s recommended protocol. Total RNA was isolated 16–20 h post-transfection.
RNA extraction was performed using the Trizol Reagent (Gibco BRL Life Technologies) according to the manufacturer’s protocol. RNA samples were electrophoresed and transferred to Hybond-N Nylon membrane (Amersham Pharmacia Biotech). Hybridization using a probe (unique-1: 5′-TGTGTGTGCCAGTTACCTTG-3′) complementary to the unique region of BC1 RNA, or (aYa5-1: 5′-ACCGTTTTAGCCGGGATGGTC-3′) complementary to Alu Ya5 RNA was carried out at 42°C overnight in 5× SSC, 5× Denhardt’s, 1% SDS and 100 µg/ml herring sperm DNA. The oligonucleotide was end-labeled with [γ-32P]ATP (Amersham Pharmacia Biotech) using T4 polynucleotide kinase (NEB, Beverly, MA), and purified by filtration through a Sephadex G-50 column. The filter was washed with 1× SSPE, 0.1% SDS for 1 h intervals. Quantitative analysis was performed using a FujiFilm FLA-2000 fluorescent image analyzer (Fuji Photo Film Co., Ltd, Tokyo, Japan).
Statistical analysis was performed using a one-way analysis of variance (ANOVA) followed where appropriate by Dunnett’s post-hoc test. A P-value <0.05 was considered to be statistically significant. All statistics were performed using Jandel Sigma Stat for Windows, v.2.0 (SPSS, Inc., Chicago, IL).
RESULTS
Diagnostic mutations do not affect Alu RNA steady-state levels
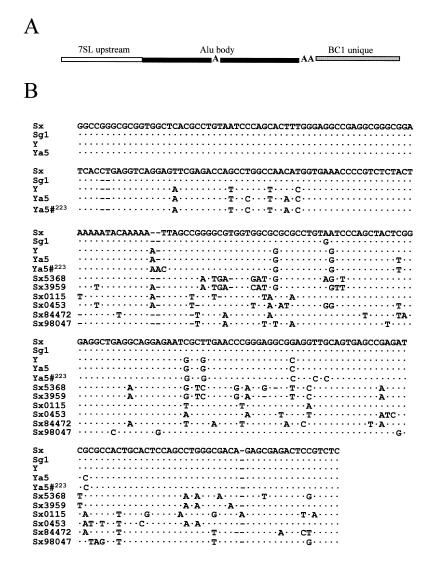
To determine whether diagnostic mutations that classify different Alu subfamilies affect Alu transcript levels, several constructs were made to represent the different subfamily consensus sequences. The basic backbone of the constructs contains the 7SL upstream region to allow active transcription (9), an Alu body and the 3′ unique region from the highly transcribed rodent short interspersed repeated element (SINE), BC1, downstream of the Alu body (Fig. 1A). Each of the major Alu subfamilies (4,5) was evaluated by transient transfections in NIH 3T3 cells, using a construct expressing the rodent SINE, BC1 (p7SLBC1BC1), as an internal control (Fig. 2). The sequences are shown in Figure 1B. The constructs contain the consensus of the currently active Ya5 subfamily, the Y, the Sg1 and our Sx (previously PS) that differs from the Sx consensus (A→G) at position 163 (4).
Figure 1.
Alu construct used for transfections and sequence alignment of mutations made. (A) Schematic diagram of the basic construct used for the transient transfections in NIH 3T3 cells. The 7SL upstream (117 bases of the human 7SL RNA gene accession no. M20910) contains cis-acting enhancer elements (white). The Alu body sequence (black) is divided into the left- and right-half by the middle A-rich region. The RNA polymerase III promoter is in the left half of Alu. The BC1 unique region from the BC1 master gene (47) (gray) contains the sequence complementary to the oligo used for RNA detection. The Alu body sequence was changed to represent the different subfamilies and the different right half sequences obtained from the GenBank database (B). Residues were changed in the BC1 unique region to represent the different 3′ end flanking sequences (C). (B) Sequence alignment of four Alu subfamilies (Sx, Sg1, Y and Ya5) that show the diagnostic mutations and the Alu body sequence from elements found in the GenBank databases (Sx5368, Sx3959, Sx0115, Sx0453, Sx84472 and Sx98047). Ya5#223 represents clone pYa5-31274223). The initial 5′ sequences of Sx5369, Sx3959, Sx0115, Sx0453, Sx84472 and Sx98047 is not shown since they are identical to their consensus counterpart. (C) The mutations incorporated into the BC1 unique region of the construct are shown. The sequence alignment contains the 3′ end flank sequences from the B1 gene, 5S RNA gene, SxBC1–tm (BC1 tm), Ya5-31274223 (Ya5#223) and Ya5-31274TTT (Ya5#TTT). Dots represent sequence identity, dashes represent lack of sequence and letters represent the nucleotide change from the consensus sequence on the top. The asterisk represents the rest of the sequence downstream of the terminator sequence.
Figure 2.
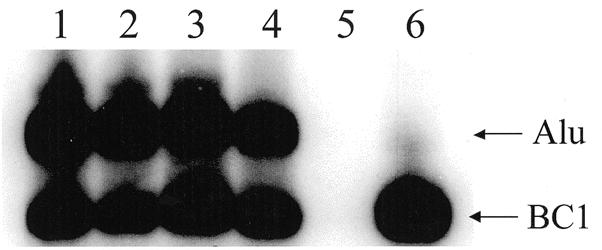
Northern blot of Alu subfamily RNA expression. Comparison of the expression from transient transfection in NIH 3T3 of the different subfamilies: lane 1, p7SLSxBC1–tm; lane 2, p7SLSg1BC1–tm; lane 3, p7SLYBC1–tm; lane 4, p7SLYa5BC1–tm; lane 5, pTAblue only (empty vector control); lane 6, p7SLBC1BC1 only. Arrows indicate positions of Alu RNA and BC1 RNA (internal control). No bands were detected in the mock transfection with no DNA (data not shown).
The northern blot shown in Figure 2 displays the steady-state of full-length RNA of each subfamily construct. Endogenous BC1 RNA levels in NIH 3T3 cells are very low and do not interfere with the control plasmid transcript levels, as shown in the vector control lane (Fig. 2, lane 5). One-way ANOVA failed to show any significant differences between the four Alu subfamilies steady-state RNA amounts (P = 0.102). Because no difference was observed, it is likely that neither transcription nor stability were affected by the diagnostic mutations. In vitro transcription with HeLa nuclear extracts and transient transfections in the human cell lines (H1299 and HeLa) also presented no significant difference in RNA levels between the four subfamilies (data not shown).
Random mutations of the right half affect Alu RNA steady-state levels
The A and B boxes of the internal RNA polymerase III promoter are necessary for transcription (7). We wanted to determine whether random mutations in non-promoter regions also influence Alu RNA levels. To avoid affecting the internal promoter, we chose to leave the left half of the Alu as the Sx consensus sequence, while only altering the right half sequences. Screening of the GenBank non-redundant (nr) database to identify Sx Alu elements was performed using the Advanced Basic Local Alignment Search Tool 2.0 (BLAST) (25) available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). The right halves of the selected Alu elements (Fig. 1B) were incorporated into the p7SLSxBC1 base construct (Fig. 1A). The nomenclature used for the new constructs include the accession numbers of the Alu elements found in the database: p7SLSx3959ABC1, p7SLSx0453BC1, p7SLSx0115BC1, p7SLSx5368BC1, p7SLSx98047DBC1 and p7SLSx84472BC1.
These constructs were tested in parallel transfections on NIH 3T3 cells using p7SLBC1BC1 as an internal control. Northern blot analysis was performed to determine RNA levels (Table 2). Only two of the constructs evaluated show a statistically significant effect in the amounts of Alu RNA detected. Construct Sx0453 consistently has an expression level that is ∼50% of the Sx construct, whereas construct Sx98047D has an expression level that is consistently ∼150% of the Sx construct. The sequence divergence between the Sx construct and the Sx0453 and Sx98047D constructs is 15.3 and 10.4%, respectively (Table 2). Because the promoter portions and 3′ terminator containing flanking sequence are identical, these data suggest that the stability of these two transcripts may have been affected. Interestingly, most of the mutations in the constructs have no real effect on the amount of Alu RNA detected. In addition, no correlation between the RNA levels and the estimated thermostability values (ΔG) of the transcripts (Table 2) was observed.
Table 2. Comparison of right half divergence, stability and expression of Alu Sx constructs with mutations in the right half.
| Right half construct | Accession numbera | Divergence from Sx right half (%) | Stability ΔGb | Steady-state level ratio Alu/BC1 ± SEM (PI unit)c |
|---|---|---|---|---|
| p7SLSxBC1 | – | 0 | –74.1 | 0.898 ± 0.118 |
| p7SLSx98047DBC1 | Z98047 | 10.4 | –65.8 | 1.498 ± 0.0835d |
| p7SLSx5368BC1 | AC005368 | 17.7 | –56.4 | 1.205 ± 0.1 |
| p7SLSx0115BC1 | AC00115 | 12.2 | –56.4 | 1.212 ± 0.0611 |
| p7SLSx0453BC1 | HSG0453 | 15.3 | –50.0 | 0.496 ± 0.0376d |
| p7SLSx3959ABC1 | AC003959 | 17.2 | –49.5 | 0.946 ± 0.0968 |
| p7SLSx84472BC1 | M84472 | 14.7 | –49.0 | 1.158 ± 0.0656 |
aThe accession number identifies the Alu locus in each individual clone.
bThe stability values (kcal/mol) are predicted from the mfold program (45,46).
cThe Alu/BC1 ratios are means ± SEM expressed in PI units. Statistical significance calculated by ANOVA was used to determine P values with n ≥ 7.
dSignificantly different to p7SLSxBC1 P < 0.05.
Sequence changes at the 3′ end affect Alu RNA steady-state levels
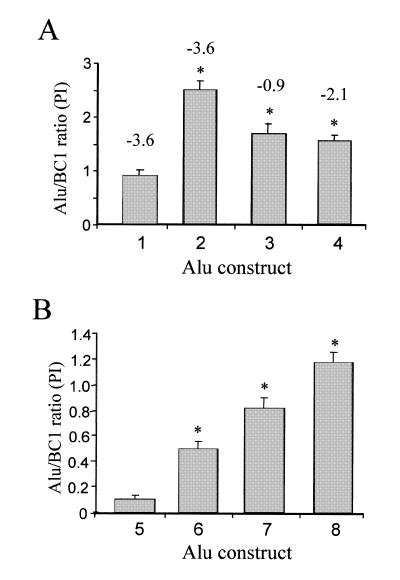
Goodier and Maraia (23) demonstrated in vitro that specific sequences affected La binding to the 3′ end of some RNA polymerase III transcripts, which in turn influenced polymerase recycling and possibly transcription rate. We created several constructs with the same 3′-end sequences (Fig. 1C) to test whether similar changes would have similar effects on Alu transcripts in vivo. The construct p7SLSxBC1–tm contains a mutation in the terminator causing transcripts to end further downstream (Fig. 1C). The ‘normal’ (SxBC1) contains the wild-type BC1 RNA gene (rodent-specific SINE) terminator. The SxBC1-5S contains the 5S rRNA terminator, and SxBC1–B1 contains a specific B1 SINE terminator. The results of the transient transfections are shown in Figure 3A. Compared to the p7SLSxBC1–tm construct, the RNA steady-state levels of p7SLSxBC1, p7SLSxBC1–B1 and p7SLSxBC1-5S are significantly higher. In addition, the transcript levels of SxBC1-5S and SxBC1–B1 are not significantly different in vivo, in contrast to the previous in vitro data (23). All the constructs presented higher transcript levels than SxBC1–tm, which has its terminator further downstream.
Figure 3.
Alu RNA expression from constructs with altered pol III terminator regions. Each experiment consists of a co-transfection of the individual plasmid with the internal control, p7SLBC1BC1. The Alu/BC1 ratios (n ≥ 7) were calculated by dividing the amount of Alu RNA by the amount of BC1 RNA detected and expressed as Phosphoimager (PI) units. The bars represent the means ± the standard error of the mean (SEM). Asterisks indicate P < 0.05 from ANOVA tests when compared to p7SLSxBC1 (1) or p–416Ya5-31274223 (5). (A) The effect of different pol III terminator sequences on Alu expression. The following constructs were evaluated: 1, p7SLSxBC1–tm (n = 12) SEM ± 0.118; 2, p7SLSxBC1 (n = 12) SEM ± 0.176; 3, p7SLSxBC1–B1 (n = 7) SEM ± 0.188; 4, p7SLSxBC1-5S (n = 9) SEM ± 0.103. The numbers on top of the bars represent the stability values (kcal/mol) as determined from mfold analyses (45,46). (B) Effect of the endogenous 3′ end on RNA expression from a specific genomic Alu element. The following constructs were evaluated: 5, p–416Ya5-31274223 (n = 8) SEM ± 0.032; 6, p–416Ya5-31274TTT (n = 7) SEM ± 0.072; 7, p7SLYa5-31274223 (n = 8) SEM ± 0.090; 8, p7SLYa5-31274TTT (n = 8) SEM ± 0.086. No secondary structure formation was detected by mfold analyses.
The thermostability values for potential structures at the 3′ ends of the RNA made by the different constructs are shown in Figure 3. No correlation between thermostability and RNA levels was observed. For example, the thermostability of SxBC1–B1 transcript is low compared to SxBC1–tm, but the transcript levels of SxBC1–B1 are significantly higher. In addition, the SxBC1–tm transcript with the highest predicted thermostability has the lowest RNA level detected.
To analyze the effect of an endogenous Alu 3′ end sequence, a specific Alu Ya5 was selected from the GenBank ‘nr’ database. Constructs of Alu Ya5-31274 (accession no. AL031274) with endogenous 5′ flank or 7SL upstream were made with its endogenous genomic 3′ flanking sequence (p–416Ya5-31274223 and p7SLYa5-31274223) and with a synthetic terminator immediately flanking the Alu element (p–416Ya5-31274TTT and p7SLYa5-31274TTT) (Fig. 1C). The constructs with the endogenous 3′ sequence (A26GAGTAACCTTTTTGT) presented lower RNA levels independent of the upstream enhancing region (Fig. 3B). Introducing a terminator immediately downstream of the Alu element increases the RNA levels between 2- and 5-fold (Fig. 3B). These data confirm that the 3′ flank of Alu elements has a major influence on the RNA steady-state levels.
DISCUSSION
Alu has amplified in primate genomes for the past 65 million years (5). As new Alu subfamilies emerged during the evolution of primates, the Alu amplification rate decreased (5). The specific amplification of subfamilies at different times in primate evolution is best explained by specific Alu ‘master genes’ having preferential retropositional activity. Any factors that influence the retroposition process may select which Alu elements may serve as master genes. Transcription of Alu RNA is the first step in the retroposition process. Thus, the accumulation of higher levels of steady-state RNA may influence retroposition rate. However, there is strong evidence that there is post-transcriptional selection of specific subsets of RNAs for retroposition (26).
We evaluated the effect of several factors including subfamily mutations, random mutations and variations around the transcription terminator on the steady-state levels of full-length Alu RNA. Our data demonstrate that the diagnostic mutations do not significantly alter full-length Alu transcript levels. This is consistent with the previously reported half-life assessments of these Alu subfamilies (27). The subfamily diagnostics were found to influence accumulation of intermediate breakdown products of Alu RNA (i.e. scAlu), but did not significantly alter levels of full-length Alu RNA. In vivo Alu RNA interacts with the RNA-binding subunit of the signal recognition particle, SRP9/14 (28) potentially influencing retropositional capability. Although the mouse SRP9/14 binds with less affinity to Alu RNA than the human counterpart (29) and increases its stability (30,31), we also obtained the same results using a human cell line (HeLa) as in NIH 3T3 (data not shown). This indicates that differential SRP9/14 binding to Alu subfamily RNA right halves in vitro (32) does not seem to influence full-length Alu RNA steady-state levels to any significant extent. Therefore, it is unlikely that the evolutionary decrease in Alu amplification rates was simply due to changes in RNA stability or promoter strength caused by the subfamily mutations. However, our assay is not able to test for epigenetic influences, such as chromatin silencing or methylation. DNA methylation of the different Alu loci can influence Alu expression (33–35). In addition, subfamily mutations may not influence RNA steady-state levels, but may still influence retroposition rate through a post-transcriptional selection process. It is worth noting that previous RNA expression studies found an ~8-fold enrichment for Ya5 Alu RNA relative to Sx Alu RNA in relation to copy number (20). Thus, the vast majority of transcripts are still from the retropositionally inactive older subfamilies due to their higher copy number. Also, the RNA levels observed in vivo represent a collection of molecules with varying degrees of sequence diversity that come from an assortment of Ya5 containing loci each affecting both individual transcription rates and stability. Our data suggest that the difference is not related to the subfamily diagnostics, but is instead a combination of the influence of the random mutations in the older Alu elements influencing the promoters on some of the elements (36,37) and the rest of the influence is likely to be from RNA stability.
Studies of expressed Alu RNAs confirm that the canonical, internal promoter elements are intact in most of the transcripts detected (20). Mutations in the internal promoter regions would influence expression rates. Since older subfamilies have accumulated more mutations throughout evolution, a higher percentage of them may have disabling promoter mutations relative to the younger subfamilies. However, we wanted to determine whether mutations outside of the promoter elements would also influence RNA expression levels. By altering only the right half of the Alu element, we were able to demonstrate that some mutations could positively or negatively change expression by a factor of two or less. Given that none of the mutations are anywhere near the promoter, plus the left halves and the 3′ unique flanks are identical in these two constructs, the right half changes are almost certainly influencing the stability of the RNA. However, our data also suggest that naturally occurring sequence divergence of the Alu right half may not have a major impact in the expression of these elements, since most of our constructs presented no statistical difference relative to the control. In this case, sequence divergence of the left half of Alu elements, either alone or in conjunction with the mutations on the right half, may contribute more efficiently in reducing Alu expression in vivo. Also, changes in the left half, besides potentially influencing the promoter could further destabilize the structure or alter protein binding sites (38) and subsequently affect retroposition (32,39). However, our data does not rule out the model proposed by Sinnett et al. (40) suggesting that there could be a post-transcriptional selection in one of the latter steps of the retroposition process resulting in the integration of only specific Alu subfamilies.
We also determined the effects of the sequence variations in the termination sequence on transcript levels. New Alu inserts will have a different 3′ flanking region with potentially a new transcription terminator. Thus, all Alu elements and their transcripts will differ in this region. Changes in the flanking sequences are likely to play a major role in transcriptional selection of one Alu over another, as previously observed (9,41) and exemplified here by the major effect that changing of the 5′ and/or 3′ flanks had on an endogenous Alu element (Fig. 3B).
Goodier and Maraia (23) demonstrated through in vitro assays that the sequences surrounding the terminator are important for recycling the polymerase via La protein. We found that similar changes influenced the expression levels of Alu RNAs in our transfection assay. However, the direction of the changes did not always agree with the in vitro findings. Thus, it seems likely that in vivo there are other factors contributing to RNA expression other than the La-influenced polymerase recycling. This could include other influences on polymerase recycling, but it could also relate to factors that influence the RNA stability. For instance, the wild-type BC1 RNA 3′ end structure forms a stable hairpin structure (42). Our studies demonstrate that even a few bases changed around the terminator influence RNA expression. The p7SLSxBC1–tm construct that presented the lowest amounts of Alu RNA levels, features a terminator which consists of only four Ts, instead of the very rich T terminator (T5NNT4) present in the other constructs. This is in agreement with previous observations (43,44), where the data indicate that a reduction in the number of T residues will reduce expression. Alternatively, the mutations around the terminator may influence the RNA structure as calculated by ΔG, which in turn may affect the RNA stability. The altered RNA structure may be exposed to endonucleases or may lose the ability to bind protein(s), which protect the 3′ end structure. Finally, the 3′ unique region may represent a critical portion of the post-transcriptional selection process that determines which Alu RNAs are capable of retroposition. One hypothesis is that this selection process is carried out by the preferential interaction with the retrotransposition machinery provided by L1 elements (6). This selection could be provided by the specific 3′ flanking ends of a small number of elements, by the subfamily diagnostic changes in the younger, active subfamilies, or by a combination of the two.
In general, variations within Alu elements and their 3′ flanking regions will have an effect on its steady-state RNA levels. The contributions of these variations between Alu elements to the RNA expression and stability are complex. Polymerase recycling by perhaps several different mechanisms, hairpin stability limiting endonuclease access and potential binding of proteins to these structures may all influence the RNA levels, as well as post-transcriptional interactions with the retroposition apparatus. Therefore it is likely that the master Alu element(s) have a multitude of individual differences that collectively give them a selective advantage over the other one million or more elements.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a student grant from the Cancer Association of Greater New Orleans (CAGNO) to C.A. and by the National Institutes of Health RO1 GM45668 and Department of the Army DAMD17-98-1-8119 to P.L.D. A.M.R. was supported by a Brown Foundation fellowship from the Tulane Cancer Center.
REFERENCES
- 1.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1996) Current Protocols In Molecular Biology. John Wiley & Sons, Inc., Canada.
- 2.Rogers J.H. and Willison,K.R. (1983) Nature, 304, 549–552. [DOI] [PubMed] [Google Scholar]
- 3.Weiner A., Deininger,P. and Efstradiatis,A. (1986) Annu. Rev. Biochem., 55, 631–661. [DOI] [PubMed] [Google Scholar]
- 4.Batzer M.A., Deininger,P.L., Hellmann-Blumberg,U., Jurka,J., Labuda,D., Rubin,C.M., Schmid,C.W., Zietkiewicz,E. and Zuckerkandl,E. (1996) J. Mol. Evol., 42, 3–6. [DOI] [PubMed] [Google Scholar]
- 5.Shen M., Batzer,M. and Deininger,P. (1991) J. Mol. Evol., 33, 311–320. [DOI] [PubMed] [Google Scholar]
- 6.Deininger P.L. and Batzer,M.A. (1999) Mol. Genet. Metab., 67, 183–193. [DOI] [PubMed] [Google Scholar]
- 7.Fuhrman S., Deininger,P.L., LaPorte,P., Friedmann,T. and Geiduschek,P. (1981) Nucleic Acids Res., 9, 6439–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesnokov I. and Schmid,C.W. (1996) J. Mol. Evol., 42, 30–36. [DOI] [PubMed] [Google Scholar]
- 9.Roy A.M., West,N.C., Rao,A., Adhikari,P., Alemán,C., Barnes,A.P. and Deininger,P.L. (2000) J. Mol. Biol., 302, 17–25. [DOI] [PubMed] [Google Scholar]
- 10.Jagadeeswaran P., Forget,B.G. and Weissman,S.M. (1981) Cell, 26, 141–142. [DOI] [PubMed] [Google Scholar]
- 11.Paulson K.E. and Schmid,C.W. (1986) Nucleic Acids Res., 14, 6145–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matera A.G., Hellmann,U. and Schmid,C.W. (1990) Mol. Cell. Biol., 10, 5424–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Englander E.W., Wolffe,A.P. and Howard,B.H. (1993) J. Biol. Chem., 268, 19565–19573. [PubMed] [Google Scholar]
- 14.Maraia R.J., Driscoll,C.T., Bilyeu,T., Hsu,K. and Darlington,G.J. (1993) Mol. Cell. Biol., 13, 4233–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid C.W. (1996) Prog. Nucleic Acid Res. Mol. Biol., 53, 283–319. [DOI] [PubMed] [Google Scholar]
- 16.Chu W.M., Liu,W.M. and Schmid,C.W. (1995) Nucleic Acids Res., 23, 1750–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chesnokov I., Chu,W.M., Botchan,M.R. and Schmid,C.W. (1996) Mol. Cell. Biol., 16, 7084–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W., Heierhorst,J., Brosius,J. and Tiedge,H. (1997) Eur. J. Cancer, 33, 288–292. [DOI] [PubMed] [Google Scholar]
- 19.Chen W., Bocker,W., Brosius,J. and Tiedge,H. (1997) J. Pathol., 183, 345–351. [DOI] [PubMed] [Google Scholar]
- 20.Shaikh T.H., Roy,A.M., Kim,J., Batzer,M.A. and Deininger,P.L. (1997) J. Mol. Biol., 271, 222–234. [DOI] [PubMed] [Google Scholar]
- 21.Schmid C.W. and Maraia,R. (1992) Curr. Opin. Genet. Dev., 2, 874–882. [DOI] [PubMed] [Google Scholar]
- 22.Deininger P. (1988) In Howe,M. and Berg,D. (eds), Mobile DNA. ASM Press, pp. 619–636.
- 23.Goodier J.L. and Maraia,R.J. (1998) J. Biol. Chem., 273, 26110–26116. [DOI] [PubMed] [Google Scholar]
- 24.Batzer M., Alegria-Hartman,M. and Deininger,P. (1994) Genet. Anal. Tech. Appl., 11, 34–38. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 26.Shaikh T.H. and Deininger,P.L. (1996) J. Mol. Evol., 42, 15–21. [DOI] [PubMed] [Google Scholar]
- 27.Sarrowa J., Chang,D.Y. and Maraia,R.J. (1997) Mol. Cell. Biol., 17, 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang D.Y., Hsu,K. and Maraia,R.J. (1996) Nucleic Acids Res., 24, 4165–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bovia F., Fornallaz,M., Leffers,H. and Strub,K. (1995) Mol. Biol. Cell, 6, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang D.Y., Sasaki-Tozawa,N., Green,L.K. and Maraia,R.J. (1995) Mol. Cell. Biol., 15, 2109–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang D.Y., Nelson,B., Bilyeu,T., Hsu,K., Darlington,G.J. and Maraia,R.J. (1994) Mol. Cell. Biol., 14, 3949–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarrowa J., Chang,D.Y. and Maraia,R.J. (1997) Mol. Cell. Biol., 17, 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W.M. and Schmid,C.W. (1993) Nucleic Acids Res., 21, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W.M., Maraia,R.J., Rubin,C.M. and Schmid,C.W. (1994) Nucleic Acids Res., 22, 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vorce R.L., Lee,B. and Howard,B.H. (1994) Biochem. Biophys. Res. Commun., 203, 845–851. [DOI] [PubMed] [Google Scholar]
- 36.Liu W.M., Leeflang,E.P. and Schmid,C.W. (1992) Biochim. Biophys. Acta, 1132, 306–308. [DOI] [PubMed] [Google Scholar]
- 37.Daniels G. and Deininger,P. (1991) Nucleic Acids Res., 19, 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bui N., Wolff,N., Cusack,S. and Strub,K. (1997) RNA, 3, 748–763. [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu K., Chang,D.Y. and Maraia,R.J. (1995) J. Biol. Chem., 270, 10179–10186. [DOI] [PubMed] [Google Scholar]
- 40.Sinnett D., Richer,C., Deragon,J.M. and Labuda,D. (1992) J. Mol. Biol., 226, 689–706. [DOI] [PubMed] [Google Scholar]
- 41.Chu W.M., Liu,W.M. and Schmid,C.W. (1995) Nucleic Acids Res., 23, 1750–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen M.R., Brosius,J. and Deininger,P.L. (1997) Nucleic Acids Res., 25, 1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu W.M., Liu,W.M. and Schmid,C.W. (1995) Nucleic Acids Res., 23, 1750–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu W.M., Ballard,R.E. and Schmid,C.W. (1997) Nucleic Acids Res., 25, 2077–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 46.Zuker M., Mathews,D.H. and Turner,D.H. (1999) In Barciszewski,J. and Clark,B.F.C. (eds), Biochemistry and Biotechnology. Kluwer Academic Publishers, New York, NY, NATO ASI series.
- 47.Kim J., Martignetti,J., Shen,M.R., Brosius,J. and Deininger,P. (1994) Proc. Natl Acad. Sci. USA, 91, 3607–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]