Abstract
The glutamate decarboxylase (GAD) system is critical to the survival of Listeria monocytogenes LO28 at low-pH stress (<pH 4.0). The GAD system classically involves two proteins, a glutamate decarboxylase enzyme coupled to a glutamate/γ-aminobutyrate antiporter, which results in the consumption of an intracellular proton for each glutamate entering the system. Uniquely among prokaryotes, some strains of L. monocytogenes, including strain LO28, possess genes encoding three decarboxylases (gadD1, gadD2, and gadD3) and two antiporters (gadT1 and gadT2). These are organized in two pairs (gadD1T1 and gadD2T2) and a distinct gadD3. While the creation of a gadD3 mutant has not been possible, analysis of 15 isogenic mutants has confirmed previous observations that GadD2/T2 are primarily responsible for surviving severe acid challenge (pH 2.8). However, we have now established that GadD1 plays a major role in growth at mildly acidic pHs (pH 5.1). When strain variation studies revealed that a large number of L. monocytogenes strains (including all serotype 4 strains) lack the gadD1 gadT1 pair, low-pH growth assays were carried out. It was found that the majority of strains that grew poorly at pH 5.1 lacked these genes. The strain-variable ability to grow in mildly acidic conditions may explain why non-serotype 4 strains of L. monocytogenes predominate in foods.
During its pathogenic life cycle, the gram-positive intracellular food pathogen Listeria monocytogenes frequently encounters low-pH environments. In order to cause an infection, L. monocytogenes requires robust acid resistance mechanisms to overcome the acidic stress presented by fermented foods, gastric juice, volatile fatty acids in the intestine, and even the low pH of the macrophage phagosome (10). Of the mechanisms characterized thus far (6, 7, 11, 33), L. monocytogenes is most dependent on the glutamate decarboxylase (GAD) system (8). Such is the role of this system that in the presence of glutamate, survival of wild-type Listeria in acidified skim milk can be up to 4 logs greater than that in its absence (9), while supplementation of porcine gastric juice with glutamate enhances survival by up to 6 logs (8). It has also been demonstrated that variations in GAD activity between L. monocytogenes strains correlates significantly with their degree of tolerance to gastric fluid (8). Furthermore, the percentage of strains with high GAD activity at pH 4 is greater among human carriage and clinical isolates than among food and environmental isolates (23).
The GAD system operates by converting a molecule of glutamate to γ-aminobutyrate (GABA), thus consuming an intracellular proton and alleviating acidification of the cytoplasm (Fig. 1A; for reviews, see references 26 and 27). The intracellular GABA is then exchanged for an extracellular glutamate via an antiporter, and the system is thus primed to consume another intracellular proton. Thus, at least two proteins are usually involved in the GAD system: a cytoplasmic glutamate decarboxylase, usually designated GadA or GadB, and a putative glutamate/GABA antiporter at the cell membrane, usually designated GadC. Previous investigations have shown that L. monocytogenes LO28 (serotype 1/2c) possesses at least two GAD enzymes (previously designated GadA and GadB) and one transporter (previously designated GadC) (8). In this study, we demonstrate that strain LO28 possesses two additional gad genes not identified in the previous study, giving a total of three glutamate decarboxylase homologs and two transporters, as is the case with the sequenced L. monocytogenes EGDe (serotype 1/2a) (12). By using a set of isogenic mutants, the role of four of these genes was investigated and the consequences of the strain-variable nature of two gad genes was determined.
FIG. 1.
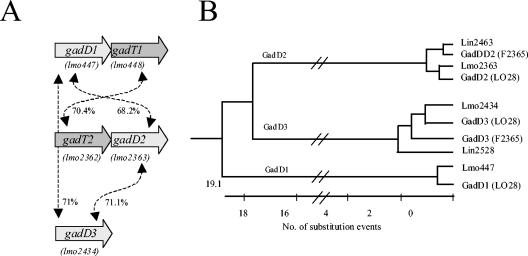
A. Relative orientation of gene homologs in LO28 (corresponding EGDe designations in brackets) and percent identity of the predicted protein products. B. The GadD proteins of Listeria can be divided into three subgroups on the basis of homology. The three subgroups correspond to three different chromosomal locations.
MATERIALS AND METHODS
Strains, plasmids, and media.
Table 1 includes a list of L. monocytogenes strains used in this study. The culture medium was tryptone soya broth (Merck, Darmstadt, Germany) supplemented with 0.6% yeast extract (Difco, Detroit, MI) (TSB-YE). Plasmid pKSV7 was a gift from K. Boor, Cornell University, New York, N.Y. Plasmid pKSV7gadT2SOE (pKSV7gadCSOE) was created previously (8). Plasmid pKSV7gadT1SOE was generated during the course of this study. Plasmid pTopo2.1a and Top 10 competent cells were purchased from Invitrogen (Carlsbad, CA) and used in accordance with the manufacturer's instructions.
TABLE 1.
Strains used in this study
| Strain | Ribotypea | gadD1T1 PCRb | gtcA PCRc | Serotyped | Source or referencee |
|---|---|---|---|---|---|
| LO28 | ND | + | − | 1/2c | Clinical |
| LO28ΔgadD1 | − | 8 (LO28ΔgadA) | |||
| LO28ΔgadD2 | + | 8 (LO28ΔgadB) | |||
| LO28ΔgadT2 | + | 8 (LO28ΔgadC) | |||
| LO28ΔgadD1D2 | − | 8 (LO28ΔgadAB) | |||
| LO28ΔgadD1T2 | − | This study | |||
| LO28ΔgadD2T2 | + | This study | |||
| LO28ΔgadD1D2T2 | − | This study | |||
| LO28ΔgadT1 | + | This study | |||
| LO28ΔgadD1T1 | − | This study | |||
| LO28ΔgadD2T1 | + | This study | |||
| LO28ΔgadT1T2 | + | This study | |||
| LO28ΔgadD1D2T1 | − | This study | |||
| LO28ΔgadD1T1T2 | − | This study | |||
| LO28ΔgadD2T1T2 | + | This study | |||
| LO28ΔgadD1D2T1T2 | − | This study | |||
| NCTC7973 | ND | + | − | 1/2a | Clinical |
| Mackaness | ND | + | − | 1/2a | Clinical |
| DPC 4594 | ND | + | − | 1/2c | ATCC 15313* (rabbit) |
| DPC 4605 | ND | + | − | 3c | SLCC 2479* (unknown) |
| DPC 4608 | ND | + | − | 1/2b | SLCC 1694* (unknown) |
| DPC 4604 | ND | + | − | 3b | Unknown* |
| ScottA | ND | − | + | 4b | Clinical |
| F5817 | ND | − | + | 4b | Clinical |
| ATCC 19118(DPC 4591) | ND | − | + | 4c | ATCC 19118T (chicken) |
| ATCC 19117(DPC 4593) | ND | − | + | 4d | ATCC 19117* (sheep) |
| 147 | 1-916-1 C | − | + | 4 (A) | Dairy enrichment** |
| 168 | 1-916-1 C | − | − | 1/2 (AS) | Aborted calf fetus** |
| 246 | 1-919-3 C | − | + | 4 (A) | Silage |
| 878 | 1-916-1 C | − | + | 4 (A) | Clinical** |
| 1038 | 1-907-1 C | + | − | Non-4 (A) | Pork sausage** |
| 1061 | 1-923-1 C | + | − | Non-4 (A) | Pork sausage** |
| 1059 | 1-907-1 C | − | − | 1/2 (AS) | Pork sausage** |
| 1078 | 1-916-1 C | − | − | 1/2 (AS) | Chicken** |
| 1198 | 1-923-1 C | + | − | Non-4 (A) | Turkey ground** |
| 1742 | 1-923-1 C | + | − | Non-4 (A) | Pork sausage** |
| 2088 | 1-916-1 C | − | − | 1/2 (AS) | Mercier clinical** |
| 83 | 1-907-5 NC | − | + | 4 (A) | Silage** |
| 241 | 1-909-2 NC | − | − | 3 (AS) | Silage** |
| 243 | 5-418-3 NC | − | − | 1/2 (A) | Silage** |
| 748 | 2-862-3 NC | + | − | Non-4 (A) | Ground beef** |
| 749 | 5-418-4 NC | + | − | Non-4 (A) | Ground beef** |
| 1028 | 5-413-2 NC | + | − | Non-4 (A) | Pork sausage** |
| 1032 | 5-408-1 NC | + | − | Non-4 (A) | Pork sausage** |
| 1066 | 5-413-5 NC | + | − | Non-4 (A) | Pork sausage** |
| 1121 | 5-408-1 NC | + | − | Non-4 (A) | Ground beef** |
| 1191 | 1-915-7 NC | + | − | Non-4 (A) | Ground turkey** |
| 1205 | 1-919-2 NC | + | − | Non-4 (A) | Ground turkey** |
| 1223 | 2-864-7 NC | + | − | Non-4 (A) | Pork sausage** |
| 1520 | 1-923-6 NC | + | − | Non-4 (A) | Ground turkey** |
| 30 | 1-910-6 O | + | − | Non-4 (A) | Brie** |
| 104 | 1-916-7 O | − | + | 4 (A) | Milk** |
| 169 | 1-910-3 O | − | − | 1/2 (AS) | Mercier clinical** |
| 179 | 1-907-5 O | − | + | 4 (A) | Dead cow brain** |
ND, not determined; C, clinical ribotype; NC, nonclinical ribotype; O, other.
Results of gadD1 and gadT1 PCRs.
The gene is present only in type 4 strains.
A, serotype determined by agglutination assay; S, serotype analysis by the Public Health Laboratory Service (PHLS).
In addition to the original source of the strains, the laboratory to which they were sent is indicated (*Teagasc, Moorepark; **Catherine Donnelly).
DNA manipulations.
PCR amplification with degenerate primers was carried out with the primer pairs gadDF1-gadTF2, gadDF2-gadTF1, gadDF1-tribestF, and gadDF2-tribestF (Table 2), respectively, using 50 pmol of each primer. These PCR products were cloned into pTopo2.1a and sequenced.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| gadDF1 | GGCAGATCGCTTACAAATGAAAGGNTGGCARGT |
| gadDF2 | GCGAGACAAAATCTAGCTACGTTYTGYCARAC |
| gadTF1 | AAAGCCGGAAGTGGCAAATGTNGGRTAYTCRTA |
| gadTF2 | CACTTTAACAATTTTGCCACCNGGDATYTGRTA |
| TribestF | ACTTTTATAGCTGGTCATTCCATGGGNGGNTA |
| gadT1SOEA | ACTTCTAGATTTTCACTGGTATTC (XbaI)a |
| gadT1SOEB | CGTCAGTCCCCAGAAAAT |
| gadT1SOEC | ATTTTCTGGGGACTGACGGCTTCCGTTGTGCCATCCAb |
| gadT1SOED | ACCCCCGGGGCCGAATGTCAATAC (XmaI)a |
| gadD3SOEA | GATCTAGAAAATCGGTGCGTC (XbaI)a |
| gadT3SOEB | CCCAATTGTATACTCATC |
| gadD3SOEC | GATGAGTATACAATTGGGACGCACTAAAAAAGAAGAb |
| gadD3SOED | GAGAATTCCAGTACGCGAGCC (EcoRI)a |
| gtcA For | TCCGTGGTACACAGATCA |
| gtcA Rev | GCAATACGATAATCCCAG |
| gadD1SOEA | AATCTAGATACGTTCTGTC |
| gadD1SOEB | CCAACAAACTTGATAGCC |
| gadD1probe1 | TTCTGTGTGTACTGGGAT |
| gadD1probe2 | GTGCCCTGATGTATTAATCG |
Base changes made to incorporate cut site (change in boldface type). The relevant restriction enzyme is in parentheses.
Overhang complementary to the corresponding SOEB primer is underlined.
SOE PCR.
Deletion mutants were created using a splicing by overlap extension (SOE) procedure (14) as described previously (8). This involved the cloning of spliced DNA fragments generated by PCR into the temperature-sensitive vector pKSV7 to facilitate allelic exchange resulting in the generation of nonpolar deletion mutations. Primers gadT1SOEA, gadT1SOEB, gadT1SOEC, and gadT1SOED were used to generate the gadT1SOE insert in pKSV7gadT1SOE, while gadD3SOEA, gadD3SOEB, gadD3SOEC, and gadD3SOED were used to generate the gadD3SOE insert in pKSV7gadD3SOE (Table 2).
Growth curves.
To determine the ability of strains to grow in mildly acidic environments, the cultures were grown overnight in TSB-YE and inoculated (2%) into TSB-YE adjusted to pH 5.1 with HCl. All cultures were grown with shaking (50 rpm) at 37°C. Growth was determined by measuring optical density at 600 nm (OD600) with a Spectra max 340 spectrophotometer (Molecular Devices).
Kill curve procedure.
To determine the ability of strains to survive in extremely acidic environments, the cultures were grown overnight with shaking (50 rpm) in TSB-YE at 37°C. Cultures were centrifuged at 12,000 × g for 10 min and then washed once in one-quarter-strength Ringers solution (LABM, Lancashire, United Kingdom) and resuspended in an equivalent volume of TSB-YE which had been adjusted to pH 2.8 with HCl. The number of cells present at time zero and 60 min was determined by serial dilution in one-quarter-strength Ringers solution and plating onto TSA-YE.
Agglutination assays.
To differentiate serotype 4 and non-serotype 4 strains, an agglutination assay was carried out using Listeria O-antigen type 4 (Difco) in accordance with the manufacturer's instructions. Additional serotyping was carried out at the Food Safety Microbiology Laboratory, London, United Kingdom.
Serotype 4-specific PCR.
The serotype 4-specific PCR primer pair gtcA For and gtcA Rev was used at an annealing temperature of 51°C to confirm the serotype of L. monocytogenes strains.
Statistical analysis.
Statistical analysis was carried out using SPSS, version 11.0. Analyses were preceded by Kolmogorov-Smirnov tests for normality and Levene's test for homogeneity of variances. Where variances showed significant heterogeneity, the data were transformed using square root or ln transformation. One-way analysis of variance (ANOVA) and post hoc Tukey tests were used to determine significant differences.
RESULTS
Identification of additional gad genes in L. monocytogenes LO28.
Analysis of the genome sequence of L. monocytogenes EGDe revealed putative genetic determinants for three glutamate decarboxylase enzymes and two glutamate:γ-aminobutyrate antiporters. These are organized as a single decarboxylase gene and two apparent enzyme-antiporter operons (Fig. 1; 12). The decarboxylases are hereby designated EGDe gadD1 (decarboxylase 1), gadD2, and gadD3 (corresponding to lmo447, lmo2363, and lmo2434, respectively), while the putative antiporters are designated EGDe gadT1 (transporter 1) and gadT2 (corresponding to lmo448 and lmo2362, respectively) (Fig. 1). We suggest that these new designations supplant those used in previous publications, which will help eliminate any confusion regarding the naming of two different open reading frames as gadA in different reports (5, 8).
Listeria monocytogenes LO28 has previously been shown to possess two glutamate decarboxylase-encoding genes and a transporter determinant, which are equivalent to gadD1, gadD2, and gadT2 (formerly gadA, gadB and gadC, respectively) (8). The use of two PCR primer pairs (gadDF1-gadTF1 and gadDF2-gadTF2) and subsequent sequencing of the products generated confirmed that LO28 also possesses a gadT1 equivalent downstream from gadD1, while the primer pair (gadDF1-TibestF) designed to amplify from regions conserved among gadD genes and among genes encoding tributyrin esterases (the gene adjacent to gadD3 in EGDe), respectively, was used to confirm that LO28 possesses gadD3 in a genetic locus similar to that of EGDe. In contrast to strains EGDe and LO28, analysis of the genomes of L. monocytogenes F2365 (ATCC 19115, serotype 4b), F6854 (serotype 1/2a), and H7858 (serotype 4b) (22) as well as that of L. innocua CLIP11262 (12) reveals that they lack gadD1 and gadT1 homologs.
The five individual Gad proteins encoded in the LO28 genome are highly similar to their homologs in EGDe (Fig. 1), with an average score of ∼99% identity between the corresponding proteins in both strains. However, the similarity between homologs produced by genes at different loci, e.g., the three gadD homologs in LO28, is lower (Fig. 1). This situation contrasts with the situation in Escherichia coli, where two glutamate decarboxylase enzymes produced by genes located on the same chromosome are virtually identical (28). Nonetheless, the relatively low rate of substitution among the GadD homologs of Listeria (Fig. 1) suggests that all three protein families evolved from a common ancestor. In addition, given that the codon bias for gadD1T1 (as for the other gad genes) does not vary significantly from that of the EGDe genome (data not shown), it is unlikely that gadD1 and gadT1 have been acquired from a foreign source.
Creation of a set of gadD1, gadD2, gadT1, and gadT2 mutants and characterization at bactericidal pH.
A 333-bp in-frame deletion was created in LO28 gadT1 using the same allelic exchange strategy previously used to generate ΔgadD1, ΔgadD2, ΔgadT2, and ΔgadD1D2 mutants (8). This strategy is particularly useful, as it creates nonpolar mutations and can be repeated in successive cycles to create double, triple, and quadruple mutants. By using plasmids pKSV7gadT1SOE and pKSV7gadT2SOE, the creation of 15 deletion mutants lacking every combination of gadD1, gadD2, gadT1, and gadT2 was possible. In contrast, numerous attempts to mutate gadD3 using either this strategy or one involving gene disruption by single-crossover plasmid insertion (19) failed.
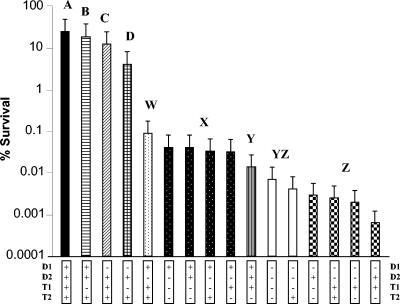
A kill curve was carried out to determine the consequence of these mutations in the survival of the LO28 derivatives. It was found that following growth (16 h) in TSB-YE (pH 7.0) overnight, the final optical density (OD600) of the various strains did not differ significantly (data not shown). Such cultures grown overnight were assayed with respect to their ability to withstand extreme pH stress, i.e., exposure for 1 h to complex broth (TSB-YE) adjusted to pH 2.8 with HCl (Fig. 2). This challenge was used since it caused a significant reduction in the numbers of parent and mutant LO28 derivatives but permitted the recovery of some survivors in all cases, such that their relative acid sensitivity could be determined. As indicated in Fig. 2, strains LO28, ΔgadT1, ΔgadD1, and ΔgadD1T1 all demonstrated dramatically higher survival capabilities (>4%) than the remaining strains (<0.1%). The survival of these four strains differs significantly from one another according to one-way ANOVA (F3,8 = 79.23; P < 0.001) and post hoc Tukey test (Fig. 2). It is thus apparent that gadT1 and gadD1 play a slight though not inconsequential role in survival at extreme acidic pHs, which is most apparent in the ΔgadD1T1 mutant. One-way ANOVA was also used to test the difference in survival between the 12 most sensitive strains (F11,24 = 163.69; P < 0.001), and Tukey test results are given in Fig. 2. In line with previous reports (5), elimination of either gadD2 or gadT2, either in isolation or in combination with any other gene, resulted in an extremely acid-sensitive phenotype (<0.1% survival in all cases), indicating that low-pH survival of LO28 is primarily dependent on GadD2/T2. It was determined that even these very sensitive strains could be divided into four significantly different statistical groups (W to Z) (Fig. 2). Of these, the group of mutants lacking GadD1 in combination with GadD2 and/or GadT2 were most sensitive (YZ/Z).
FIG. 2.
Percent survival of strains after 1 h of growth in TSB-YE, pH 2.8. Bars are arranged in order of decreasing survival. Percent survival values that do not differ significantly at the 0.05 level (Tukey tests) are noted with the same letter: A to D for the relatively more acid-resistant strains and W to Z for the relatively more acid-sensitive strains.
Contribution of the gad loci to growth at mildly acidic pH.
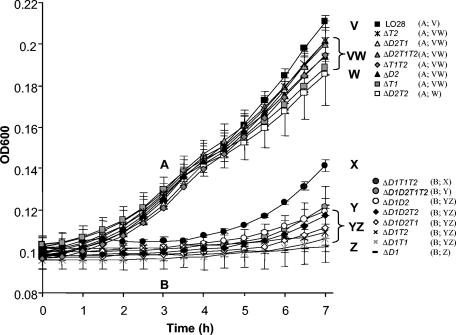
The contribution of the various gad loci to growth in acidic environments has not been examined previously. When grown at mildly acidic pHs (TSB-YE adjusted to pH 5.1 with HCl), it was found that the mutants could be divided into two distinct categories, i.e., those possessing GadD1, which grow well, and those lacking GadD1, which grow poorly (Fig. 3). In contrast, there was not a significant difference between the growth of LO28 and ΔgadD1 when grown in buffered TSB-YE (TSB-YE, 55 mM β-glycerophosphate) (data not shown). The significance of the growth differences at pH 5.1 was calculated with respect to the values at 3 and 7 h, respectively. It was determined that after 3 h, the strains could be divided into two significantly different groups (one-way ANOVA, F15,32 = 51.07; P < 0.001 following ln transformation); i.e., all strains possessing GadD1 display enhanced growth relative to those that lack the enzyme. Significant differences were again apparent after 7 h (one-way ANOVA, F15,32 = 124.39; P < 0.001 following square root transformation) (Tukey test results for both ANOVAs are presented in Fig. 3). While it is still apparent at this time point that strains possessing GadD1 have an advantage, additional variations are obvious. Of the GadD1-positive strains, only ΔgadD2T2 showed a significant difference in growth relative to LO28. The observation that the ΔgadT1T2 and ΔgadD2 mutants continued to grow relatively well demonstrates that GadD1 can function without the aid of GadD2 and even without specific GAD antiporters under these circumstances.
FIG. 3.
Growth in TSB-YE, pH 5.1, as determined by OD600. Values that do not differ significantly at the 0.05 level (Tukey tests) are noted with the same letter. Tukey test results are presented after 3 h (categories A and B) and 7 h (categories V, W, X, Y, and Z).
Impact of the strain-variable gadD1T1 genes on growth of L. monocytogenes strains at pH 5.1.
To determine whether the presence/absence of gadD1T1 genes among L. monocytogenes strains followed a discernible pattern, a multistrain PCR-based screen, using various pairs of gadD1-specific (gadD1SOEA-gadDSOEB/gadD1probe1-gadD1probe2) and gadT1-specific (gadT1SOEA-gadT1SOEB/gadT1SOEC-gadT1SOED) PCR primers, was carried out. Initially, only 10 strains of known serotype, NCTC7973 (1/2a), Mackaness (1/2a), DPC 4594 (1/2c), DPC 4605 (3c), DPC 4608 (1/2b), DPC 4604 (3b), ScottA (4b), F5817 (4b), ATCC 19118 (4c), and ATCC 19117 (4d), were tested. In all cases, it was found that strains lacking gadD1 also lacked gadT1 (Table 1). Low-stringency Southern analysis of a number of representative strains using a probe generated using the PCR primers gadD1probe1 and gadD1probe2 confirmed the presence of three glutamate decarboxylase homologs in strains LO28, Mackaness, and DPC 4608 and the presence of two glutamate decarboxylase homologs in ScottA, ATCC 19118, and ATCC 19117 (data not shown). It was noted that the serotype 4 strains tested lacked gadD1T1. gadD1- and gadT1-specific PCR analysis was extended to a further 28 strains of unknown serotype. Serotype 4-specific PCR primers (gtcA For and gtcA Rev) for gtcA (24) and Bacto-Listeria O-antiserum type 4 (Difco, Detroit, MI) were used to determine that six of these were serotype 4 strains (Table 1). All six lacked gadD1 and gadT1. The absence of these genes in the predicted locus was confirmed by PCR using primers based on flanking regions in L. innocua CLIP11262 and L. monocytogenes F2365 (data not shown). Fifteen of the remaining 22 non-serotype 4 strains possessed the gadD1T1 genes. When the serotypes of the remaining seven gadD1T1-negative strains were determined, it was found that six were of serotype 1/2 while one was of serotype 3. It would thus seem that while all serotype 4 strains tested to date lack gadD1 and gadT1, this trait is not unique to this serotype. We observed that the manner in which the presence of the gadD1T1 genes varies between strains closely corresponds to trends identified previously by Call et al. (3). Here, mixed-genome array analysis was used to identify a gadD gene that varies between clusters and serotypes of L. monocytogenes (3). In this case, clusters IIa (serotype 1/2a), IIc (1/2a and 1/2c), and Ib (mixed) were all found to possess the gene, and clusters Ic (4b) and Id (4b) do not, while its presence varies in strains of clusters IIb (1/2a) and Ia (1/2b) (3). However, in contrast to our findings that all serotype 4 strains lack gadD1, it was suggested that strains of cluster Ib (serotypes 1/2b, 4b, and 4c) possess this gene. However, as it is acknowledged that the polyphyletic composition of this cluster could be an artifact and that the positioning of 4b and 4c within this cluster is in contrast with some other cluster designations (15, 25, 32), we contend that serotype 4 strains are indeed gadD1 (and gadT1) negative.
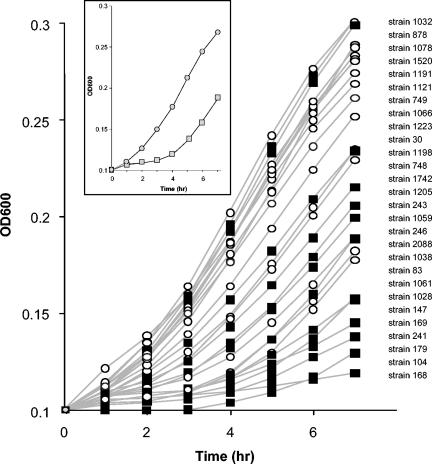
Given that the growth deficiency at pH 5.1 of the LO28 ΔgadD1 and LO28 ΔgadD1T1 mutants is so pronounced relative to LO28, a strain variation study was carried out to determine whether the natural presence or absence of gadD1 and gadT1 influenced the growth at low pH of the wild-type strains listed in Table 1 (Fig. 4). Laboratory strains were not included, as strains that have been subjected to repeated culturing frequently present atypical phenotypes (1, 4, 18). It was found that, in general, the link between rapid growth at mildly acidic pHs was indeed linked to the presence of gadD1 and gadT1, although this link is not absolute. The obvious exceptions are strains 878 and 1078, which grew quite well despite lacking gadD1T1, while strains 1038, 1061, and 1028 grew relatively poorly despite possessing these genes. Nonetheless, the mean growth of the GadD1T1-positive strains was significantly greater than that of the GadD1T1-negative strains at each time point, i.e., 1 to 7 h (one-way ANOVA, P < 0.001). The existence of individual strains that do not follow the trend is unsurprising, as unlike the LO28 mutants, these strains are not isogenic, and it is thus likely that some other factors associated with growth at low pH are altered.
FIG. 4.
Plot of growth of GadD1-positive (circles) and GadD1-negative (squares) strains in TSB-YE, pH 5.1, as determined by OD600. The strains are listed in order of descending OD600 after 7 h. (Inset) The mean growth of the GadD1-positive (grey circles) and GadD1-negative (grey squares) strains is presented as trend lines.
DISCUSSION
L. monocytogenes is thus far the only bacterium known to possess three gad genes (the presence of two corresponding antiporters is also unparalleled). GadD2 plays a major role in the ability of this species to survive low-pH stress while, where present, GadD1 facilitates growth at relatively milder acidic pHs. The reason why these enzymes behave differently is at least partially due to the expression profiles of the corresponding genes. It has been shown in strain LO28 that levels of gadD1 transcription are low and are apparent primarily in log phase, while transcription of gadD2 and gadT2 is relatively higher and is induced in response to acid induction and entry into stationary phase (8). In other strains of L. monocytogenes, this trend was also observed, and it has also been established that gadT1 and gadD3 are induced by acid adaptation (31), with gadD3 also being induced by entry into stationary phase and osmotic stress (17). In an EGDe mutant lacking σB, a stress sigma factor, acid induction of gadD2, gadT2, and gadD3 was negatively affected and induction of gadT1 was unaffected, while induction of gadD1 was positively affected (possibly to compensate for reduced levels of the other GadD enzymes) (31). However, there is obviously more to consider than transcriptional regulation alone when phenotypic results in a complex system are studied. Another obvious difference between the three genes is their sequences. When compared to the gad genes in E. coli, the various homologs of L. monocytogenes display relatively lower levels of conservation. While members of the gadD1, gadD2, and gadD3 subfamilies show a minimum of 96% similarity within the same subfamily, the levels are lower, 69 to 73%, when compared across subfamilies. It remains to be determined to what extent the differences between the proteins contribute to their roles. For example, we could speculate that the various GadD subfamilies may vary in the pH at which they display optimal activity.
The creation of a complete set of gadD1, gadT1, gadD2, and gadT2 mutants has allowed us to better understand the contribution of individual genes in different environments. At pH 2.8, the absence of either gadD2 or gadT2 has a major impact in all cases. These results are in agreement with those observed when gadD2 and gadT2 mutants were exposed to lactic acid at pH 3.5 (8). Mutants which possess both of these genes are relatively fit in this environment, as the absence of gadD1 and/or gadT1 has a relatively smaller, yet significant, impact. Further analysis of results generated indicates that the four proteins studied form discrete decarboxylase-transporter pairs, as the presence of intact but “nonmatching” pairs in ΔgadD1T2 and ΔgadD2T1 does not offer a high level of protection at pH 2.8. With respect to the two pairs, however, the degree to which the individual decarboxylases depend on the corresponding transporter varies. Under conditions (pH 2.8) where LO28 is more reliant on GadD2 than GadD1, deletion of GadT2 results in an acid-sensitive phenotype. However, at pH 5.1, where GadD1 plays the greater role, deletion of GadT1 does not have dramatic consequences, suggesting that GadD1 can provide a protective effect in the absence of GadT1. For GadD1 to act in isolation, decarboxylation would depend on intracellular levels of glutamate being sufficiently high. In such circumstances, the synthesis of glutamate by, for example, glutamate synthase (lmo1733 and lmo1734) and glutamate dehydrogenase (lmo0560), which have been found to be present in strain EGDe (12), may be significant. Such metabolic flux has also been suggested with respect to the glutamate decarboxylase-mediated response of L. monocytogenes to CO2 stress (16). While it has not yet been established whether glutamate decarboxylation can be facilitated by glutamate synthesis, it is intriguing that microarray-generated data have demonstrated that overexpression of the E. coli regulator GadE brings about increased expression of genes involved in glutamate-mediated acid resistance and of gltD, involved in glutamate synthesis (13). For a decarboxylase to be effective in the absence of an associated transporter, GABA would need to be metabolized/removed by an alternative means. Based on genes present in the EGDe genome, GABA could be converted by the product of lmo0913, succinate semialdehyde dehydrogenase, to succinyl-coenzyme A or by the products of lmo1006 and lmo1005, putatively encoding an aminotransferase and a 3-hydroxyisobutyrate dehydrogenase, respectively, to 4-hydroxybutyrate. The polymer of this compound has been shown to accumulate in L. monocytogenes, where it acts as a reserve pool (2). It is interesting that while ΔgadD1D2T1T2 is among the most sensitive mutants at pH 2.8, it is not the most sensitive strain. It may be that the presence of gadD2, gadT1, or gadT2 alone or in some combinations may further disadvantage the cell due to futile production of these proteins or even the catalysis of reactions which are counterproductive; e.g., the importation of glutamic acid in the absence of decarboxylation may serve to further reduce the internal pH. Our inability to mutate GadD3 is interesting in that it suggests that neither GadD1 nor GadD2 can compensate for its absence. It is unlikely that it plays a role in the survival of LO28 at low pH, given the efficiency with which GadD1, GadD2, GadT1, and GadT2 perform their roles, the previous observation that a ΔgadD1D2 mutant displays negligible GAD activity (8), and the exquisite sensitivity of the ΔgadD1D2 mutant in bacteriostatic and bactericidal low-pH environments. However, as it is regulated by σB and induced in response to a variety of stresses (17, 31), it is likely that it is involved in some aspect of the stress response.
The duplication/deletion of bacterial amino acid decarboxylases is undoubtedly significant. In E. coli gadA, a number of its regulators and other acid-induced genes are present on a “fitness island” that contributes significantly to acid adaptation (13, 29). Given the apparent advantage of gadD1T1+ strains in mildly acidic conditions, akin to those that Listeria encounters in low-pH foods, the absence of these genes in serogroup 4 strains may at least partly explain why they are less frequently identified in foods (21, 30). The absence of gadD1T1 from serotype 4b L. monocytogenes, the causative agent of epidemic listeriosis, indicates that its presence is not required for the organism to bring about this disease. However, it has not been established whether there is an association between the absence of these genes and virulence in the same way that the loss of lysine decarboxylase genes during evolution of Shigella species from E. coli has been demonstrated as being pathoadaptive (20). While it is apparent that possessing GadD1 benefits L. monocytogenes strains in mildly acidic conditions, it remains to be established whether its absence from a number of strains may under certain circumstances enhance pathogenic potential.
Acknowledgments
We acknowledge Catherine W. Donnelly (University of Vermont) and Teagasc, Moorepark, Fermoy, Ireland, for the kind gift of L. monocytogenes strains. Thanks also to Vina Mithani and Jim McLauchlin of the Food Safety Microbiology Laboratory, PHLS Central Public Health Laboratory, London, United Kingdom, for the serotyping of strains and to Elizabeth Cotter for carrying out statistical analysis.
We acknowledge the funding received from the Irish Government under the National Development Plan 2000-2006 and through funding of the Alimentary Pharmabiotic Centre by the Science Foundation of Ireland Centres for Science Engineering and Technology (CSET) scheme. S.R. is a recipient of a postgraduate fellowship from safefood, Food Safety Promotion Board, Ireland.
REFERENCES
- 1.Blevins, J. S., M. O. Elasri, S. D. Allmendinger, K. E. Beenken, R. A. Skinner, J. R. Thomas, and M. S. Smeltzer. 2003. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect. Immun. 71:516-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buzoleva, L. S., and A. D. Chumak. 2000. Use of poly-beta-oxybutyric acid by Yersinia pseudotuberculosis and Listeria monocytogenes bacteria at various temperatures. Mikrobiologiia 69:770-773. [PubMed] [Google Scholar]
- 3.Call, D. R., M. K. Borucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, G., C. L. Patten, and H. E. Schellhorn. 2004. Positive selection for loss of RpoS function in Escherichia coli. Mutat. Res. 554:193-203. [DOI] [PubMed] [Google Scholar]
- 5.Conte, M. P., G. Petrone, A. M. Di Biase, C. Longhi, M. Penta, A. Tinari, F. Superti, G. Fabozzi, P. Visca, and L. Seganti. 2002. Effect of acid adaptation on the fate of Listeria monocytogenes in THP-1 human macrophages activated by gamma interferon. Infect. Immun. 70:4369-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotter, P. D., N. Emerson, C. G. M. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, P. D., C. G. M. Gahan, and C. Hill. 2000. Analysis of the role of the Listeria monocytogenes F0F1-ATPase operon in the acid tolerance response. Int. J. Food Microbiol. 60:137-146. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. D., C. G. M. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. D., K. O'Reilly, and C. Hill. 2001. Role of the glutamate decarboxylase acid resistance system in the survival of Listeria monocytogenes LO28 in low pH foods. J. Food Prot. 64:1362-1368. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, et al. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 13.Hommais, F., E. Krin, J. Y. Coppee, C. Lacroix, E. Yeramian, A. Danchin, and P. Bertin. 2004. GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150:61-72. [DOI] [PubMed] [Google Scholar]
- 14.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 15.Jinneman, K. C., and W. E. Hill. 2001. Listeria monocytogenes lineage group classification by MAMA-PCR of the listeriolysin gene. Curr. Microbiol. 43:129-133. [DOI] [PubMed] [Google Scholar]
- 16.Jydegaard-Axelsen, A. M., P. E. Hoiby, K. Holmstrom, N. Russell, and S. Knochel. 2004. CO2- and anaerobiosis-induced changes in physiology and gene expression of different Listeria monocytogenes strains. Appl. Environ. Microbiol. 70:4111-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 19.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muraoka, W., C. Gay, D. Knowles, and M. Borucki. 2003. Prevalence of Listeria monocytogenes subtypes in bulk milk of the Pacific Northwest. J. Food Prot. 66:1413-1419. [DOI] [PubMed] [Google Scholar]
- 22.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olier, M., S. Rousseaux, P. Piveteau, J. P. Lemaitre, A. Rousset, and J. Guzzo. 2004. Screening of glutamate decarboxylase activity and bile salt resistance of human asymptomatic carriage, clinical, food, and environmental isolates of Listeria monocytogenes. Int. J. Food Microbiol. 93:87-99. [DOI] [PubMed] [Google Scholar]
- 24.Promadej, N., F. Fiedler, P. Cossart, S. Dramsi, and S. Kathariou. 1999. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J. Bacteriol. 181:418-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 26.Richard, H. T., and J. W. Foster. 2003. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 52:167-186. [DOI] [PubMed] [Google Scholar]
- 27.Small, P. L., and S. R. Waterman. 1998. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol. 6:214-216. [DOI] [PubMed] [Google Scholar]
- 28.Smith, D. K., T. Kassam, B. Singh, and J. F. Elliott. 1992. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 174:5820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitas, A. I., and V. A. Garcia-Jalon. 2004. Occurrence of Listeria monocytogenes in fresh and processed foods in Navarra (Spain). Int. J. Food Microbiol. 90:349-556. [DOI] [PubMed] [Google Scholar]
- 31.Wemekamp-Kamphuis, H. H., J. A. Wouters, P. P. L. A. de Leeuw, T. Hain, T. Chakraborty, and T. Abee. 2004. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70:3457-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]