Abstract
DNA-based methods are increasingly important for bacterial typing. The high number of polymorphic sites present among closely related bacterial genomes is the basis for the presented method. The method identifies multilocus genomic polymorphisms in intergenic regions termed AILP (amplified intergenic locus polymorphism). For each locus, a pair of unique PCR primers was designed to amplify an intergenic sequence from one open reading frame (ORF) to the adjacent ORF. Presence, absence, and size variation of the amplification products were identified and used as genetic markers for rapidly differentiating among strains. Polymorphism was evaluated using 18 AILP sites among 28 strains of Listeria monocytogenes and 6 strains of Listeria spp. and 30 AILP markers among 27 strains of Escherichia coli. Up to four alleles per locus were identified among Listeria strains, and up to six were identified among E. coli strains. In both species, more than half of the AILP sites revealed intraspecies polymorphism. The AILP data were applied to phylogenetic analysis among Listeria and E. coli strains. A clear distinction between L. monocytogenes and Listeria spp. was demonstrated. In addition, the method separated L. monocytogenes into the three known lineages and discriminated the most common virulent serotypic group, 4b. In E. coli, AILP analysis separated the known groups as well as the virulent O157:H7 isolates. These findings for both Listeria and E. coli are in agreement with other phylogenetic studies using molecular markers. The AILP method was found to be rapid, simple, reproducible, and a low-cost method for initial bacterial typing that could serve as a basis for epidemiological investigation.
Bacterial strain typing has several important applications in microbiology. In clinical practice, strain typing is useful for diagnosis and determining treatment strategy and is essential for rapid identification of disease outbreaks and new virulent strains. In the food industry, strain typing is necessary to ensure food safety and for linking cases of food-borne infections to suspected items in the food chain. Classical bacterial identification is based on selective enrichment, followed by plating on selective media. Species identification is mainly by biochemical characterization, and strain identification is primarily based on serology. These methods do not meet the requirement for rapid identification and typing in clinical, epidemiological, and food industry applications. Recent advances in biotechnology have resulted in the development of numerous methods for detection and typing of microorganisms (11-13, 19, 25) which differ in their sensitivity, rapidity, labor intensiveness, complexity, discriminatory power, reproducibility, and cost (5, 32, 43, 49). In principle, by screening a large number of polymorphic sites, genomic methods should be able to provide very accurate discrimination among closely related strains. The total multilocus output of these methods is often termed “DNA fingerprints” or a “DNA bar code.”
In the present study, we present a new method (amplified intergenic locus polymorphism [AILP]), based on the above principles, which is specifically useful for generating DNA bar codes for discrimination among bacterial strains. The method is based on the finding that whole-genome sequence comparisons within and between closely related bacterial species show the presence of numerous single nucleotide polymorphic sites (SNPs) and genome rearrangements (e.g., see references 15, 17, 20, 28, 33, and 36). This implies that a pair of PCR primers designed to amplify a randomly selected genomic fragment in one strain will often produce different fragment sizes in other strains or may fail to amplify the genomic fragment altogether due to sequence mismatch, insertion/deletions, and other variation at the priming site. A major advantage of the proposed method is that, given complete or partial genome sequences, no additional prior information is required to identify informative AILP markers. The experiments described here were carried out to evaluate the potential of this new strain typing methodology for representative gram-positive and gram-negative bacterial species. Listeria spp. are gram-positive bacteria that include seven classified species, among which only Listeria monocytogenes is pathogenic to humans and responsible for listeriosis (14, 39). Escherichia coli is a gram-negative bacterium composed of numerous strains and serotypes. The species includes commensal strains and a variety of pathogenic strains, such as enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), and enterotoxigenic E. coli (ETEC) (1, 30, 31, 34, 42, 47). The availability of the complete genome sequences for E. coli K-12 (4) and L. monocytogenes (17) provided the basis to designing primers for PCR amplification of random genomic targets in these organisms. The results showed the ability of the AILP method to discriminate within and between species in Listeria spp. and E. coli. The uncovered genetic variation data were used for phylogeny analysis in both species.
MATERIALS AND METHODS
Bacterial strains.
The study included a set of 34 L. monocytogenes strains, six strains of the remaining species of the Listeria genus (Table 1), and two sets of E. coli strains: a set of 27 pathogenic and nonpathogenic strains of E. coli (Table 2) (12) and a set of 72 wild-type E. coli strains from a reference collection (30).
TABLE 1.
Listeria sp. strains screened in the present study
| Organism | Description and source |
|---|---|
| Listeria monocytogenes | |
| LM6 DA3 | Serotype 4ba |
| LM8 Scott A | Serotype 4ba |
| LM26H | Serotype 4b, ATCC 19115 |
| LM28/2 | Serotype 4b, food isolateb |
| LM54 | Serotype 4b, human isolateb |
| LM21/1 | Serotype 1/2b, human isolateb |
| LM25/2 | Serotype 1/2b, food isolateb |
| LM31 | Serotype 1/2b, human isolateb |
| LM1 | Serotype 1/2b, human isolateb |
| LM14 EGD | Serotype 1/2aa |
| LM16 | Serotype 1/2a, SLCC5764 |
| LM17/3 | Serotype 1/2a, food isolateb |
| LM19/1 | Serotype 1/2a, food isolateb |
| LM10 | Serotype unknowna |
| LM11 | Serotype unknowna |
| LM17 | Serotype 4ca |
| LM25H | Serotype 4a, ATCC 19114 |
| LM15 LO28 | Serotype 1/2ca |
| LM24H | Serotype 1/2c,bATCC 7644 |
| WHO/1 | Serotype 3ab |
| WHO/16 | Serotype 3ab |
| WHO/28 | Serotype 3ab |
| WHO/11 | Serotype 3bb |
| WHO/14 | Serotype 3bb |
| WHO/19 | Serotype 3bb |
| WHO/33 | Serotype 3cb |
| WHO/52 | Serotype 3cb |
| WHO/60 | Serotype 3cb |
| Listeria spp. | |
| L. innocua | ATCC 33090 |
| L. ivanovii | ATCC 19119 |
| L. seeligeri | ATCC 35967 |
| L. welshimeri | ATCC 35897 |
| L. grayi | ATCC 19120 |
| L. murrayi | ATCC 25401 |
Wayne State University Food Microbiology Laboratory, Detroit, Mich.
Central Laboratories, Ministry of Health, Jerusalem, Israel.
TABLE 2.
E. coli strains screened in the present study
| E. coli group and strain serotype | Description and sourcea |
|---|---|
| EHEC | |
| O22:H8 | E. coli Reference Center, 90.0327 |
| O42:H2 | E. coli Reference Center, 88.0501 |
| O111:NM | E. coli Reference Center, 88.0015 |
| O113:H2 | E. coli Reference Center, 88.0632 |
| O26:H11 | Centers for Disease Control and Prevention CDC 2239-69 |
| O157:NM | USDA-FSIS, MF7123A |
| O157:H7 | USFDA, SEA13B88, Odwalla cider outbreak strain |
| O157:H7 HER phage type 1057 | Ontario Public Health Laboratory; 1 |
| O157:H7 HER phage type 1058 | |
| O157:H7 HER phage type 1261 | |
| O157:H7 HER phage type 1265 | |
| O157:H7 HER phage type 1266 | |
| ETEC | |
| O78:NM | Haifa Public Health Department, Rowe no. E10407 |
| O8:H9 | Central Laboratories, Ministry of Health, Jerusalem, Israel |
| O9:H33 | |
| O86:H10 | |
| O86:H18 | |
| O153:H | |
| EPEC | |
| O111ac:NM | Haifa Public Health Department, Rowe no. E639616 |
| O26:H | Central Laboratories, Ministry of Health, Jerusalem, Israel |
| O55:H7 | |
| O127:H21 | |
| K-12 | |
| DH5α | Technion Faculty of Food Engineering & Biotechnology collection |
| W3110 | |
| W4100 | |
| B | |
| SR9b, SR9c | |
| Wild type | |
| Reference collection strains 1-72 | Michigan State University; 30 |
USDA, U.S. Department of Agriculture; FSIS, Food Safety and Inspection Service; USFDA, U.S. Food and Drug Administration.
DNA preparation.
A modified procedure of Jersek et al. (22) was used for DNA extraction from pure cultures. Cultures of listeriae and E. coli were grown for 24 h at 37°C on brain heart infusion and Luria agar plates, respectively. A loop was transferred from the plate to a microcentrifuge tube containing 1 ml of SSC buffer (0.15 M NaCl, 15 mM sodium citrate, pH 8.0) and vortexed thoroughly. The suspension was centrifuged for 1 min at 21,000 × g.
For listeriae, the pellet was resuspended in 100 μl lysozyme solution (4 mg/ml in 20% sucrose-1 mM sodium phosphate) and incubated for 1 h at 37°C. To this were added 200 μl TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 8.0), 100 μl N-lauryl-sarcosine solution (5% in TE), and 100 μl proteinase K solution (20 mg/ml in TE). The mixture was incubated overnight at 50°C, 500 μl EZ-DNA solution (Biological Industries, Beit-Haemek, Israel) was added, followed by incubation at 60 °C for 1 h and ethanol precipitation according to the manufacturer's instructions. The DNA extract was treated with 0.1 mg/ml RNase, followed by extraction with phenol chloroform and ethanol precipitation. The DNA was stored at −20°C.
For E. coli, the pellet was resuspended in 200 μl TE buffer (10 mM Tris-Cl, 1 mM EDTA, pH 8.0)-100 μl proteinase K solution (20 mg/ml in TE) and incubated overnight at 50°C. Two percent sodium dodecyl sulfate was added, and the mixture was incubated at 55 °C for 1 h, followed by extraction with phenol chloroform and ethanol precipitation. The DNA was stored at −20°C.
In addition, standard boiling for rapid DNA purification was used with equal success for both E. coli and Listeria.
Locus selection and primer construction.
The complete genomic sequence of L. monocytogenes (EGD-e, serotype 1/2a) was obtained from http:/www.ncbi.nlm.nih.gov/, and that of E. coli (K-12) was obtained from http://mol.genes.nig.ac.jp/ecoli/. With the exceptions noted below, genomic loci were randomly selected along the bacterial chromosome. PCR primers were usually selected to amplify a specific intergenic locus, from an open reading frame (ORF) to the adjacent ORF. Primers were selected using the Gene Runner (version 3.05) with up to 3°C melting temperature (Tm) difference between the primers. In the case of Listeria, the study began before the complete sequence was available. Consequently, the first nine markers were based on various GenBank sequences of L. monocytogenes; two markers were derived from the unfinished genome sequence of L. monocytogenes (serotype 4b [The Institute for Genomic Research]), six were from the published genome sequence (EGD-e, serotype 1/2a) (17), and one was from the published genome sequence of L. innocua (Clip11262) (17; Table S1 in the supplemental material). All loci, except abc and Lin0694, were found in the published genome of EGD-e, serotype 1/2a. The E. coli markers were based on the finished K-12 genome sequence (GenBank accession no. U00096) (4; Table S2 in the supplemental material). Loci were named after the downstream ORF.
PCR amplification.
The PCR mixture contained 0.2 mM deoxynucleoside triphosphates, 0.4 μM each forward and reverse primer, 0.5 U Taq polymerase (Super Nova; JMR Holding, Kent, England), 1× buffer (1.5 mM MgCl2), and 50 ng template DNA in a total volume of 25 μl. The reaction was carried out in a PCR thermocycler (HYBAID Omn-E; Hybaid, Ashford, United Kingdom) as follows: 95°C for 5 min; five cycles of 45 s at 95°C, 45 s at the Tm (Tables S1 and S2 in the supplemental material), and 45 s at 72°C; 20 cycles of 45 s at 95°C, 45 s at the Tm minus 5°C, and 45 s at 72°C; and a final step of 72°C for 7 min. PCR amplification products were analyzed by agarose gel (2%) electrophoresis and observed by UV fluorescence.
Data analysis.
Two classes of polymorphism were observed: class I, presence or absence of amplification product, and class II, presence of a product different in size from the expected product, based on the published sequence. In some instances, class II polymorphisms included two bands, the expected product and an additional product. Allele designations for each locus were as follows: allele 1, the obtained product with the expected length according to the GenBank sequence (Tables S1 and S2 in the supplemental material); allele 2, absence of product; allele 3, two amplification products, the expected product and an additional product different in size; alleles 4 to 7, all products present different in length from the expected product and from each other. For further analysis, fragment data for all genotypes of a specific locus were scored as 1 (present) or 0 (absent) for each of the alleles. Where two fragments were obtained (allele 3), each was scored as 1. Using SAS version 8.02 (38), the data were used to calculate the simple matching coefficients of association (41) and to generate two corresponding genetic distance matrices, one for the 34 Listeria isolates and the other for the 27 E. coli isolates. These matrices were used to determine the relationships among strains. The dendrograms were constructed by means of the unweighted-pair group method using average linkages (UPGMA) with MEGA version 2.1 (24).
RESULTS
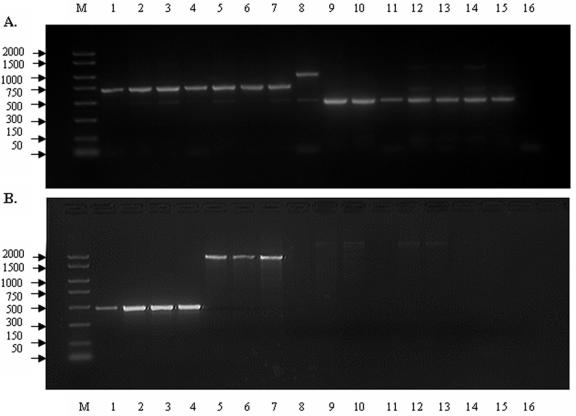
A set of distributed intergenic loci were selected along the published genomes of L. monocytogenes and E. coli for AILP analysis. Eighteen sites were chosen along the genome of L. monocytogenes (EGD-e 1/2a), and 30 sites were chosen along the genome of E. coli (K-12) (Tables S1 and S2, respectively, in the supplemental material). Analysis of the PCR amplification products by gel electrophoresis showed considerable polymorphism among the tested strains of Listeria and E. coli (Fig. 1). To ensure reproducibility, only the major bands were considered as products. Results were verified by at least three independent PCRs for each strain. In addition, identical results were obtained in our lab by different personnel/staff using various thermal cyclers, as well as in other labs (e.g., David Walt, Tufts, University, Boston, Mass.) (40). Figure 1 depicts the intraspecies polymorphism at both the abc and gbuA loci among 15 strains of L. monocytogenes.
FIG. 1.
AILP analysis of 15 L. monocytogenes strains at the gbuA (a) and abc (b) loci. Amplification products were separated on a 2% agarose gel. Lanes were as follows: M, size standards (bp); 1, LM8:4b; 2, LM26H:4b; 3, LM6:4b; 4, LM54:4b; 5, LM21/1:1/2b; 6, LM25/2:1/2b; 7, LM31:1/2b; 8, LM25H:4a; 9, LM15:1/2c; 10, LM14:1/2a; 11, LM16:1/2a; 12, LM17/3:1/2a; 13, LM19/1:1/2a; 14, WHO/1:3a; 15, WHO/33:3c; 16, no DNA (for details, see Table 1).
AILP in Listeria spp.
Eighteen loci were used to assess the variation among 28 L. monocytogenes strains (including human pathogenic isolates) and 6 strains of other Listeria spp. (Table 3). The other Listeria species served as control isolates presenting the ability of the method to distinguish between the pathogenic L. monocytogenes isolates and the other Listeria species. All loci were polymorphic, having 2 to 4 alleles with an average of 2.61 alleles per locus. Nine (50%) of the loci showed class I polymorphism, possessing two alleles (product or no product); the remaining loci showed class II polymorphism, seven loci showing three alleles, and two showing four alleles. Using the information from all tested loci, the AILP method can assign the 34 Listeria strains to 18 different AILP types (Table 3). Five of the loci (clpE, ATTM, cheR, Lmo0196, and gid), clearly differentiated between L. monocytogenes and other Listeria species, presented the same allele in all L. monocytogenes strains. Ten (56%) loci were polymorphic within L. monocytogenes, thereby facilitating differentiation among strains. Furthermore, a combination of two loci, abc and gbuA, enabled the identification of L. monocytogenes from the other Listeria spp., discriminated the three known L. monocytogenes lineages, and differentiated the 4b serotypic group from the other serotypic groups in lineage I (with the exception of strain LM17; Table 3).
TABLE 3.
AILP types based on electrophoretic profiles of PCR amplification products for 28 L. monocytogenes strains and 6 Listeria spp. at 18 sites
| Strain or species | Allelea at following locus:
|
AILP type | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lmo1430 | clpE | BetL | ATTM | ltrC | cheR | lisR | Lmo0672 | Lmo0196 | Lmo0075 | Lin0694 | Lmo0023 | gid | fhuB | Lmo0042 | Lmo0176 | abc | gbuA | ||
| LM6-4b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 |
| LM8-4b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 4 | 2 |
| LM26H-4b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 4 | 2 |
| LM28/2-4b | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 2 | 3 |
| LM54-4b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 |
| LM21/1-1/2b | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 4 | 4 |
| LM25/2-1/2b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 4 | 5 |
| LM31-1/2b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 4 | 5 |
| LM1-1/2b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 2 | 6 |
| LM14-1/2a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| LM16-1/2a | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 8 |
| LM17/3-1/2a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| LM19/1-1/2a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| LM10 | 2 | 1 | 2 | 1 | 2 | 1 | 1 | 4 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 3 | 9 |
| LM11 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 10 |
| LM17-4c | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 4 | 1 | 4 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 11 |
| LM25H-4a | 2 | 1 | 2 | 1 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 2 | 3 | 12 |
| LM15-1/2c | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 13 |
| LM24H-1/2c | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| WHO/1-3a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| WHO/16-3a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| WHO/28-3a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| WHO/11-3b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 4 | 4 | 14 |
| WHO/14-3b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 4 | 4 | 14 |
| WHO/19-3b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 4 | 4 | 14 |
| WHO/33-3c | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| WHO/52-3c | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| WHO/60-3c | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| L. innocua | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 3 | 2 | 1 | 1 | 4 | 1 | 2 | 1 | 2 | 2 | 15 |
| L. ivanovii | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 4 | 2 | 2 | 16 |
| L. seeligeri | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 3 | 2 | 1 | 1 | 4 | 1 | 2 | 1 | 2 | 2 | 15 |
| L. welshimeri | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 17 |
| L. grayi | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 18 |
| L. murrayi | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 18 |
Allele designations: 1, amplification product at the expected size according to GenBank sequence; 2, absence of product; 3, one product in addition to the expected product; 4 to 7, products differing in length from one another and from the expected product.
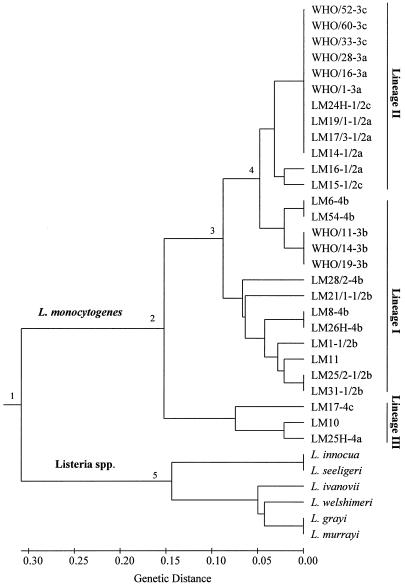
The AILP data were also used for analysis of phylogenetic relationships among the 34 Listeria isolates. A genetic-distance matrix was generated based on 47 polymorphic points (18 loci by the number of alleles in each locus), followed by cluster analysis using the UPGMA. The resulting dendrogram showed five main branching nodes (Fig. 2). There was clear and deep separation of the L. monocytogenes strains from the other six Listeria species (node 1; average genetic distance, 0.615 ± 0.099). Genetic distances among L. monocytogenes strains ranged from 0.000 to 0.383 (average genetic distance, 0.150 ± 0.10), with strains from serotypes 4a and 4c being the most distant (average genetic distance, 0.325 ± 0.079). Strains of L. monocytogenes were clustered in the three known lineages. Strains of serotypes 4a and 4c comprised a separate node (lineage III, node 2). Strains from serotypes 1/2a, 1/2c, 3a, and 3c were grouped together (lineage II, node 4), separately from strains belonging to serotypes 1/2b, 4b, and 3b (lineage I). Lineage I serotypes, in turn, were divided into two nodes (nodes 3 and 4). Although some of the lineage I strains clustered closely to lineage II, the genetic distance between lineage II and lineage I was significant (average of 0.155 ± 0.056). Two pairs of Listeria species gave identical AILP fingerprint patterns: L. grayi-L. murrayi and L. innocua-L. seeligeri. The latter pair was separated from the other Listeria species (node 5).
FIG. 2.
Dendrogram presenting the genetic relationships among Listeria isolates using UPGMA cluster analysis of the AILP data (for details, see Table 1).
AILP in E. coli.
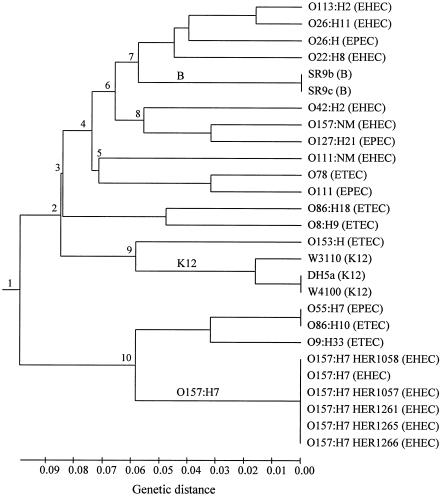
Thirty loci were used to assess the variation among a set of 27 E. coli isolates (Tables 2 and 4). Sixteen loci (53%) were polymorphic, having 2 to 6 alleles (with an average of 2.62 alleles per locus); 14 loci were monomorphic, presenting only a single allele. Eleven (69%) of the polymorphic loci showed class I polymorphism, possessing two alleles; five loci showed class II polymorphism, presenting two to six alleles. Using the 16 informative loci, the AILP method can assign the 27 strains to 19 AILP types. The AILP data were used for analysis of genetic relationships among the 27 isolates. A genetic-distance matrix was generated based on 64 polymorphic points (total number of alleles in the 30 loci). Genetic distances among E. coli isolates ranged from 0.000 for the very close isolates to 0.27 for the most distant isolates, with a mean genetic distance of 0.162 ± 0.06. Cluster analysis of the distance matrix was performed using the UPGMA. The resulting dendrogram presented in Fig. 3, shows 10 main branching nodes. As expected, all O157:H7 isolates exhibited similar patterns and clustered together (node 10). Similarly, isolates SR9b and SR9c of E. coli group B (node 7) had the same AILP pattern. Isolates from E. coli K-12 were grouped together (node 9).
TABLE 4.
AILP types based on electrophoretic profiles of PCR amplification products of 27 E. coli strains at 30 sites
| Group and strain | Allelea at following locus:
|
AILP type | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hisC | viaB | ftsZ | b1688 | aidB | b1284 | yaiN | ycgW | caiF | yacA | serW | dsrB | yaaH | molR-1 | yjiD | b0829 | yibA | b1031 | mhpR | folA | pyrD | b2345 | ykgE | osmB | gutP | b1248 | galS | uvrB | yafY | pepD | ||
| EHEC | |||||||||||||||||||||||||||||||
| O22:H8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 2 | 1 | 1 |
| O42:H2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 2 | 2 | 1 | 2 |
| O111:NM | 2 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 3 |
| O113:H2 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 4 |
| O26:H11 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 5 |
| O157:NM | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 6 |
| O157:H7 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| O157:H7-1057 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| O157:H7-1058 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| O157:H7-1261 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| O157:H7-1265 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| O157:H7-1266 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 7 |
| ETEC | |||||||||||||||||||||||||||||||
| O78 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 8 |
| O8:H9 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 5 | 1 | 1 | 1 | 1 | 2 | 1 | 7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 9 |
| O9:H33 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 10 |
| O86:H10 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 11 |
| O86:H18 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 6 | 1 | 1 | 1 | 2 | 2 | 1 | 7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 12 |
| O153:H | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 13 |
| EPEC | |||||||||||||||||||||||||||||||
| O111 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 14 |
| O26:H | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 2 | 1 | 15 |
| O55:H7 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 11 |
| O127:H21 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 16 |
| K-12 | |||||||||||||||||||||||||||||||
| DH5a | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 17 |
| W3110 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 18 |
| W4100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 17 |
| B | |||||||||||||||||||||||||||||||
| SR9b | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 19 |
| SR9c | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 19 |
Allele designations: 1, amplification product at the expected size according to GenBank sequence; 2, absence of product; 3, one product in addition to the expected product; 4 to 7, products differing in length from one another and from the expected product.
FIG. 3.
Dendrogram presenting the genetic relationships among E. coli isolates using UPGMA cluster analysis of the AILP data (for details, see Table 2).
In addition, the E. coli reference collection of 72 strains (30) was analyzed at a subset of five AILP markers (yaiN, ycgW, serW, b2345, and ykgE), yielding 15 polymorphic points (total number of alleles across the five loci). The genetic distance between the most distant isolates (ECOR23 and ECOR66) was 0.73, and the average genetic distance was 0.24 ± 0.18. Cluster analysis, presented in Fig. 4, revealed that all B2 group ECOR isolates (21) were grouped to a distinct node (node 1), followed by a cluster of most of the group D ECOR isolates (node 2) and a cluster consisting of most of the group A and B1 isolates (node 3).
FIG. 4.
Dendrogram presenting the genetic relationships among 72 strains of an E. coli reference collection using UPGMA cluster analysis of five AILP markers (yaiN, ycgW, serW, b2345, and ykgE).
DISCUSSION
This study presents a new, simple, DNA-based bacterial typing method, AILP. The method is based on PCR amplification of a randomly chosen intergenic locus. The typing is determined by presence, absence, or size variation in the amplified products. Specific strain typing is achieved by multilocus analysis. The power of the method derives from the large number of polymorphic sites that are found across the whole genome (2, 15, 20, 33, 35, 36). These numerous polymorphic sites make it likely that a pair of PCR primers that amplify a specific, randomly chosen intergenic locus will produce polymorphic products when screened across a set of closely related species or strains within a species. Yet, prior information of variation among strains is not required, only the primary genome sequence. The observed polymorphisms may be caused by SNPs at the primer sites, as well as insertions or deletions (20). Recent findings of serotype- or strain-specific genes and many SNPs among three L. monocytogenes 4b and 1/2a strains (28) support the basis for the AILP typing approach. Similar approaches of presence and absence variations have been used in binary typing based on probe hybridization (44, 45). Recently, Nekrutenko et al. (27) discussed the possible use of an in silico screen for the identification of presence/absence variation among strains, as well as the use of PCR amplification.
In the present study, we found that more than half of the randomly chosen loci in both L. monocytogenes and E. coli were polymorphic within a representative set of strains, revealing intraspecies polymorphism. Interspecies polymorphism was found in Listeria as well. Up to four alleles for a given locus were identified among 34 Listeria strains and up to six alleles were identified among 27 E. coli strains.
Reliability of a typing method is crucial for accurate distinction among different bacterial isolates (e.g., see references 32 and 49). The AILP method uses unique primers for PCR amplification under high-stringency conditions, providing reliable and reproducible results. In order to compare the results obtained by AILP analysis with those obtained using other typing methods, sets of AILP data were applied to phylogenetic analyses of a set of Listeria and E. coli strains. Phylogenetic analyses using AILP data were consistent with other studies. In Listeria, five of the loci clearly differentiated between the pathogenic species L. monocytogenes and the other Listeria species. Ten (56%) loci were polymorphic within L. monocytogenes, thereby facilitating assignment of the 28 L. monocytogenes strains to 14 different AILP types (Table 3). AILP phylogenetic analysis divided L. monocytogenes strains into three distinct genetic lineages (Fig. 2, nodes 2, 3, and 4). This accords closely with other DNA subtyping methods, including pulsed-field gel electrophoresis (PFGE), ribotyping, mixed-genome microarray, and multilocus sequence typing (MLST) (e.g., see references 6, 10, 26, 37, and 48). Serotypes 4a and 4c (lineage III) were found to be the most genetically distant strains (average genetic distance of 0.325 ± 0.079), in accordance with previous studies suggesting that lineage III represents a unique subset of L. monocytogenes characterized by reduced virulence for humans and by other genetic features (48). Similar lineage clustering but higher discrimination was achieved by MLST analysis of simple sequence repeat (SSR) loci (19 sequence types) and by PFGE (24 PFGE profiles) in the same set of 28 L. monocytogenes strains (L. Somer, Y. Danin-Poleg, L. Valinsky, and Y. Kashi, unpublished data). In conclusion, AILP analysis separated L. monocytogenes from the other Listeria species, divided L. monocytogenes isolates among the three known lineages comparable to the serological division, and discriminated the most virulent 4b serotypic group. These results support the efficiency of the AILP method for rapid strain typing of Listeria.
In E. coli, the 16 informative loci facilitated the assignment of the 27 strains to 19 AILP types. Similar discrimination ability was achieved (17 sequence types) with the same bacterial set using MLST analysis of SSR loci (12). Phylogenetic analysis of AILP data separated the B, K-12, and O157:H7 serological groups into distinct clusters. O157:H7 isolates were clustered together with the O55:H7 isolate (Fig. 3, node 10), supporting the findings that O55:H7 and O157:H7 have recently evolved from a common ancestor (46). The six O157:H7 isolates exhibited the same pattern, indicating close genetic relations and low diversity, in agreement with other methods (23, 29). However, due to wide genome rearrangements reported between O157:H7 and K-12 (20, 33), it is likely that AILP analysis with primers designed on the basis of the O157:H7 genome (rather than the K-12 genome) would facilitate discrimination between O157:H7 isolates. Indeed, in silico variation was found in the studied serW AILP site between the two published genomes of O157:H7 (33). In addition, analysis of a second E. coli set consisting of the E. coli reference collection strains (30) was performed at five AILP markers. The analysis showed increased genetic distance among strains (average distance of 0.24 ± 0.18 compared to 0.162 ± 0.06 in the first set of 27 strains) as a result of the higher genetic diversity in the wild-type ECOR collection. The phylogenetic analysis clearly separated B2 strains from the D strains and from A and B1 strains (Fig. 4) (21). Parallel clustering was achieved with the same bacterial set using MLST analysis of SSR loci (12).
Comparing the AILP results for Listeria and E. coli revealed that in L. monocytogenes a high correlation was found between serologic profiles and other genetic typing methods (3), providing initial typing comparable to serology (48). In contrast, in E. coli, only a low correlation was found between the serologic profiles or pathogenic groups (e.g., EPEC, ETEC, and EHEC) and AILP typing. This is expected as in general there is a low correlation between serological profiles and genetic relationships in E. coli (3, 42, 46). The higher rates of horizontal gene transfer reported in E. coli (46) compared to the clonal structure characterization of L. monocytogenes (23, 50) could explain these differences. This is further documented by the whole-genome comparison between L. monocytogenes and L. innocua showing that no large genome rearrangements have occurred between these species (8, 17, 28). In contrast, genome-wide rearrangements were described between E. coli strains K-12 and O157:H7 (20, 33).
The AILP typing method was found to be more efficient in bacterial species with a clonal structure, such as Listeria species. Its efficiency is demonstrated by the fact that using only two AILP sites, abc and gbuA, enabled the discrimination of the three known L. monocytogenes lineages (Fig. 2; Table 3) and distinguished the most common virulent serotypic group, 4b, from the other serotypic groups of lineage I. Similar differentiation was achieved using infrequent restriction site PCR (16). Furthermore, similar to the AILP analysis, only 4 probes (out of 29) of the mixed-genome microarray were needed for the same differentiation (10).
Compared to other typing methods, AILP is a rapid, low-cost, and simple typing method. It provides discrimination power comparable to serology but lower than MLST (12, 37) and PFGE (7, 18). However, AILP is much faster (a few hours compared to a few days) and less expensive, provides results that are simple to analyze, and requires distinctly less experienced manpower. Equipment and infrastructure requirements for AILP are minimal, consisting of basic laboratory equipment, thermocycler and minigel apparatus. Thus, this method is suitable for initial rapid bacterial typing scanning. Following this, detailed strain typing can be done as a second step by using typing methods such as MLST (9, 37) or PFGE (7, 18), which provide higher discrimination but are labor intensive and expensive.
Due to the high stringency of the reaction conditions and the unique set of primers, the efficiency of AILP analysis can be increased by multiplexing a number of loci in the same reaction mixture. A rapid DNA preparation (such as standard boiling) is suitable for routine high-throughput identification, as similar results were obtained using rapid DNA purification methods (data not shown). However, the same DNA purification method should be applied to all isolates. High-throughput strain identification with the AILP method could be achieved by technologies such as microarrays (40) or real-time PCR. AILP analysis can be applied to any microorganism with prior knowledge of part or all of its genome sequence but does not require prior knowledge of sequence variation among species or strains. In cases where genomes of different strains are available, in silico selection of AILP primers directed to variable (multiallelic) chromosomal sites is possible.
In conclusion, the AILP method provides rapid and simple initial identification of isolates as a basis for epidemiological investigation, clearly discriminating between different strains or revealing similarities that can be further tested using high discriminatory power typing methods. Thus, the AILP method should be a useful addition to the available methodologies for rapid initial microbial strain typing.
Supplementary Material
Acknowledgments
We thank Thomas and Beth Whittam (Michigan State University), Leora Shelef (Wayne State University), and the Ministry of Health Central Laboratories Jerusalem and Haifa Department, Israel, for providing the samples of the bacterial strains and serotyping and The Institute for Genomic Research for the unfinished genomic sequence data of L. monocytogenes serotype 4b. We are grateful to Leora Shelef for fruitful discussions.
This research was supported by the Grand Water Research Institute, the Israeli Water Commission, and the Mitchel Soref Innovation Awards program, Technion. R. Gur-Arie was supported by the Food Control Administration in the Israel Ministry of Health.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahmed, R., C. Bopp, A. Borczyk, and S. Kasatiya. 1987. Phage-typing scheme for Escherichia coli O157:H7. J. Infect. Dis. 155:806-809. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, W. F., B. G. Gellin, R. Weaver, B. Schwartz, B. D. Plikaytis, M. W. Reeves, R. W. Pinner, and C. V. Broome. 1990. Analysis of clinical and food-borne isolates of Listeria monocytogenes in the United States by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Appl. Environ. Microbiol. 56:2133-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Boerlin, P., E. Bannerman, F. Ischer, J. Rocourt, and J. Bille. 1995. Typing Listeria monocytogenes: a comparison of random amplification of polymorphic DNA with 5 other methods. Res. Microbiol. 146:35-49. [DOI] [PubMed] [Google Scholar]
- 6.Borucki, M. K., M. J. Krug, W. T. Muraoka, and D. R. Call. 2003. Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92:351-362. [DOI] [PubMed] [Google Scholar]
- 7.Buchrieser, C., R. Brosch, B. Catimel, and J. Rocourt. 1993. Pulsed-field gel electrophoresis applied for comparing Listeria monocytogenes strains involved in outbreaks. Can. J. Microbiol. 39:395-401. [DOI] [PubMed] [Google Scholar]
- 8.Buchrieser, C., C. Rusniok, F. Kunst, P. Cossart, and P. Glaser. 2003. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 35:207-213. [DOI] [PubMed] [Google Scholar]
- 9.Cai, S., D. Y. Kabuki, A. Y. Kuaye, T. G. Cargioli, M. S. Chung, R. Nielsen, and M. Wiedmann. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J. Clin. Microbiol. 40:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Call, D. R., M. K. Borucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype- and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke, S. C. 2002. Nucleotide sequence-based typing of bacteria and the impact of automation. Bioessays 24:858-862. [DOI] [PubMed] [Google Scholar]
- 12.Diamant, E., Y. Palti, R. Gur-Arie, H. Cohen, E. M. Hallerman, and Y. Kashi. 2004. Phylogeny and strain typing of Escherichia coli, inferred from variation at mononucleotide repeat loci. Appl. Environ. Microbiol. 70:2464-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duim, B., C. W. Ang, A. van Belkum, A. Rigter, N. W. J. van Leeuwen, H. P. Endtz, and J. A. Wagenaar. 2000. Amplified fragment length polymorphism analysis of Campylobacter jejuni strains isolated from chickens and from patients with gastroenteritis or Guillain-Barre or Miller Fisher syndrome. Appl. Environ. Microbiol. 66:3917-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, J. W. J. Jacobs, J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franciosa, G., S. Tartaro, C. Wedell-Neergaard, and P. Aureli. 2001. Characterization of Listeria monocytogenes strains involved in invasive and noninvasive listeriosis outbreaks by PCR-based fingerprinting techniques. Appl. Environ. Microbiol. 67:1793-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 18.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 19.Gur-Arie, R., C. J. Cohen, Y. Eitan, L. Shelef, E. M. Hallerman, and Y. Kashi. 2000. Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res. 10:62-71. [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 21.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jersek, B., P. Gilot, M. Gubina, N. Klun, J. Mehle, E. Tcherneva, N. Rijpens, and L. Herman. 1999. Typing of Listeria monocytogenes strains by repetitive element sequence-based PCR. J. Clin. Microbiol. 37:103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudva, I. T., P. S. Evans, N. T. Perna, T. J. Barrett, G. J. DeCastro, F. M. Ausubel, F. R. Blattner, and S. B. Calderwood. 2002. Polymorphic amplified typing sequences provide a novel approach to Escherichia coli O157:H7 strain typing. J. Clin. Microbiol. 40:1152-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 25.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nekrutenko, A., K. D. Makova, and R. J. Baker. 2000. Isolation of binary species-specific PCR-based markers and their value for diagnostic applications. Gene 249:47-51. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. G. J. Morris, D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochman, H., T. S. Whittam, D. A. Caugant, and R. K. Selander. 1983. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J. Gen. Microbiol. 129(Pt. 9):2715-2726. [DOI] [PubMed] [Google Scholar]
- 32.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 34.Pupo, G. M., D. K. Karaolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 37.Salcedo, C., L. Arreaza, B. Alcala, L. De La Fuente, and J. A. Vazquez. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.SAS. 2003. The SAS system for Windows, version 8.02. SAS Institute Inc., Cary, N.C.
- 39.Schuchat, A., B. Swaminathan, and C. V. Broome. 1991. Epidemiology of human listeriosis. Clin. Microbiol. Rev. 4:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shepard, J. R. E., Y. Danin-Poleg, Y. Kashi, and D. R. Walt. 2005. Array-based binary analysis for bacterial typing. Anal. Chem. 77:319-326. [DOI] [PubMed]
- 41.Sokal, R. R., and P. H. A. Sneath. 1963. Principles of numerical taxonomy. W. H. Freeman & Co., San Francisco, Calif.
- 42.Strockbine, N. A., J. G. Wells, C. A. Bopp, and T. J. Barrett. 1998. Overview of detection and subtyping methods, p. 331-356. In J. B. Kapper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 43.van Belkum, A., M. Struelens, A. de Visser, H. Verbrugh, and M. Tibayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14:547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Leeuwen, W., C. Libregts, M. Schalk, J. Veuskens, H. Verbrugh, and A. van Belkum. 2001. Binary typing of Staphylococcus aureus strains through reversed hybridization using digoxigenin-universal linkage system-labeled bacterial genomic DNA. J. Clin. Microbiol. 39:328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Leeuwen, W. B., S. Snoeijers, C. Werken-Libregts, A. Tuip, A. van der Zee, D. Egberink, M. de Proost, E. Bik, B. Lunter, J. Kluytmans, E. Gits, I. van Duyn, M. Heck, K. van der Zwaluw, W. Wannet, G. T. Noordhoek, S. Mulder, N. Renders, M. Boers, S. Zaat, D. van der Riet, M. Kooistra, A. Talens, L. Dijkshoorn, T. van der Reyden, D. Veenendaal, N. Bakker, B. Cookson, A. Lynch, W. Witte, C. Cuny, D. Blanc, I. Vernez, W. Hryniewicz, J. Fiett, M. Struelens, A. Deplano, J. Landegent, H. A. Verbrugh, and A. van Belkum. 2002. Intercenter reproducibility of binary typing for Staphylococcus aureus. J. Microbiol. Methods 51:19-28. [DOI] [PubMed] [Google Scholar]
- 46.Whittam, T. S. 1996. Genetic variation and evolutionary processes in natural populations of Escherichia coli, p. 2708-2720. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 47.Whittam, T. S., H. Ochman, and R. K. Selander. 1983. Multilocus genetic structure in natural populations of Escherichia coli. Proc. Natl. Acad. Sci. USA 80:1751-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-531. [PubMed] [Google Scholar]
- 49.Wiedmann, M. 2002. Subtyping of bacterial foodborne pathogens. Nutr. Rev. 60:201-208. [DOI] [PubMed] [Google Scholar]
- 50.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.