Abstract
We describe isolation and characterization of the gene encoding the glucosidase II alpha subunit (GIIα) of the industrially important fungus Trichoderma reesei. This subunit is the catalytic part of the glucosidase II heterodimeric enzyme involved in the structural modification within the endoplasmic reticulum (ER) of N-linked oligosaccharides present on glycoproteins. The gene encoding GIIα (gls2α) in the hypercellulolytic strain Rut-C30 contains a frameshift mutation resulting in a truncated gene product. Based on the peculiar monoglucosylated N-glycan pattern on proteins produced by the strain, we concluded that the truncated protein can still hydrolyze the first α-1,3-linked glucose residue but not the innermost α-1,3-linked glucose residue from the Glc2Man9GlcNAc2 N-glycan ER structure. Transformation of the Rut-C30 strain with a repaired T. reesei gls2α gene changed the glycosylation profile significantly, decreasing the amount of monoglucosylated structures and increasing the amount of high-mannose N-glycans. Full conversion to high-mannose carbohydrates was not obtained, and this was probably due to competition between the endogenous mutant subunit and the introduced wild-type GIIα protein. Since glucosidase II is also involved in the ER quality control of nascent polypeptide chains, its transcriptional regulation was studied in a strain producing recombinant tissue plasminogen activator (tPA) and in cultures treated with the stress agents dithiothreitol (DTT) and brefeldin A (BFA), which are known to block protein transport and to induce the unfolded protein response. While the mRNA levels were clearly upregulated upon tPA production or BFA treatment, no such enhancement was observed after DTT addition.
N glycosylation of proteins is a cotranslational process that starts in the lumen of the endoplasmic reticulum (ER) by the en bloc transfer of the precursor glycan Glc3Man9GlcNAc2 to suitable Asn-X-Ser/Thr sequons of nascent polypeptide chains (40). Shortly after transfer, the first α-1,2-linked glucose residue is removed by glucosidase I, a type II ER membrane protein with a luminal catalytic domain (36). This rapid removal probably plays an important role in the detachment of the newly formed glycoprotein from the oligosaccharyl transferase (68). The two remaining α-1,3-glucoses are hydrolyzed by glucosidase II (40). Subcellular fractionation (8, 39) and immunolocalization (42) studies determined that glucosidase II is mostly confined to the ER.
The existence of an enzyme capable of removing the two remaining α-1,3-linked glucose residues was first proposed by Ugalde and coworkers after partial purification of microsomal glucosidase activities from rat liver cells (90). Since then, glucosidase II has been partially or completely purified from a wide variety of both lower and higher eukaryotic organisms (2, 6, 7, 11, 20, 38, 78, 80, 88). Following some initial controversy, it is now generally accepted that the protein is a rather asymmetric nonglobular heterodimer consisting of a catalytic alpha subunit with a molecular mass of about 110 kDa (GIIα) and a beta subunit with a molecular mass of 60 kDa (GIIβ) (6, 15, 65, 85, 86, 88).
The sequence coding for the alpha subunit, originally cloned from rat liver cells, showed significant homology with members of glycosyl hydrolase families 9 and 31 (88). A BLAST search identified a human cDNA sequence with an unknown function (GenBank accession no. D42041), as well as a Saccharomyces cerevisiae open reading frame (ORF) from chromosome II (GenBank accession no. Z36098). Disruption of the latter ORF yielded a yeast strain devoid of glucosidase II activity (88). Shortly after this, GIIα-encoding sequences were cloned from mouse T lymphocytes (6), pig liver (20), the slime mold Dictyostelium discoideum (22), and Schizosaccharomyces pombe (15). All derived protein sequences carry a conserved WXDLNE motif, which is the typical consensus sequence for the active site of glycosidase family 31 (29).
Retrieval of the sequences encoding the beta subunit was based on the amino acid composition of the purified protein or on homology searches of available databases (6, 88). GIIβ is a hydrophilic protein with a molecular mass of about 60 kDa and an elongated conformation (86). It carries a C-terminal HDEL tag, which is known to be an ER retrieval signal (64). Since the glucosidase II alpha subunit does not carry a known ER retention signal, it is generally believed that one of the major tasks of the beta subunit is to constrain the enzymatic subunit to the ER (15). Recent data also indicate the involvement of GIIβ in the maturation of the glucosidase II heterodimer. While coexpression of the alpha and beta subunits increases total glucosidase II activity, expression of GIIα alone results in an insoluble and inactive aggregate (65, 85). However, once GIIα is fully folded, the beta subunit is no longer necessary for activity (86).
Glucosidase II has a neutral pH optimum (7, 11, 30, 38, 78), and its function does not depend on metal ions (30, 77). Its optimal substrate is Glc1-2Man9GlcNAc2, and its hydrolytic capacity diminishes as the number of mannose residues decreases (27). Incubation with compounds such as deoxynojirimycin (77) or castanospermine (38) blocks the removal of both α-1,3-linked glucoses. Bromoconduritol, on the other hand, inhibits the hydrolysis only of the innermost glucose residue (16). This was explained by the kinetic model proposed by Alonso and coworkers, who identified a low-affinity substrate binding site and a high-affinity substrate binding site (2, 3). In this model, the first α-1,3-glucose is rapidly removed by binding of the high-affinity site, while the second glucose residue is hydrolyzed after binding of the low-affinity site.
After deglucosylation by glucosidase II, the ER resident UDP-glucose:glycoprotein glucosyltransferase (GT) can add a new α-1,3-linked glucose residue to the glucose-free oligosaccharide (19, 57, 89). However, a prerequisite for this transfer is that the underlying peptide chain has not yet folded into a mature glycoprotein (87). GT seems to be a very sensitive sensor for the tertiary structure of the protein (83) and as such lies at the core of the glycoprotein quality control mechanism. Monoglucosylated carbohydrates, formed either after partial hydrolysis of the α-1,3-linked glucose residues by glucosidase II or through the action of GT on glucose-free N-glycans, can interact with the ER-resident lectins calnexin and calreticulin (28). This greatly reduces aggregation of unfolded proteins and thereby increases the possibility of their interaction with other ER chaperones, such as the binding protein BiP (60).
Filamentous fungi, such as Trichoderma reesei, have a great capacity to secrete proteins and are therefore exploited for industrial production of autologous proteins (10, 23, 24) and heterologous proteins (41, 66). However, although the yield of secreted autologous proteins can reach tens of grams per liter, the yield of proteins of mammalian origin remains within the range of a few milligrams per liter (66). The synthesis of heterologous proteins and supplementation of the medium with factors that result in ER secretion stress (such as dithiothreitol [DTT] or brefeldin A [BFA]) are known to induce the unfolded protein response (UPR). In yeast, this response is mediated by the kinase/endoribonuclease Ire1p, which when activated splices the HAC1 mRNA in an unconventional way. The resulting mRNA is translated into a functional Hac1p that acts as a positive transcription factor for genes under control of the UPR (reviewed in references 43 and 61). Recent results indicate that a similar mechanism in filamentous fungi (74) induces elevated synthesis of chaperones (18, 32, 35, 52, 70, 73, 92) and other proteins involved in different processes in the secretion pathway (75). However, with the exception of one report on the Aspergillus niger calnexin gene (91), little is known about the sensitivity of several other partners of the calnexin-related quality control system (such as glucosidase II and GT) to the accumulation of unfolded proteins and to secretion stress in general.
In this study, we cloned the gene encoding the glucosidase II alpha subunit of T. reesei Rut-C30. The glycan pattern of the extracellular hydrolases of this hypercellulolytic strain is very peculiar, in that the carbohydrates carry a terminal α-1,3-glucose residue (17, 45, 79). This could result from insufficient glucosidase II activity under cellulose-inducing conditions, or for some reason the enzyme activity could be partially inhibited in the Rut-C30 strain. As the Rut-C30 strain was generated after several rounds of mutagenesis, the aberrant glycosylation pattern could also be the result of a defective glucosidase II promoter or protein. Here we present data indicating that a frameshift mutation in the glucosidase II ORF is responsible for the observed glycan pattern and discuss the effect on the strain's phenotype when a functional version of this gene is introduced.
MATERIALS AND METHODS
Strains and growth conditions.
For preparation of genomic DNA or RNA for 5′ rapid amplification of cDNA ends (5′-RACE) cloning, T. reesei strains Rut-C30 (= ATCC 56765), Rut-NG14 (= ATCC 56765), and QM9414 (= ATCC 26921) were grown in minimal medium. This medium contained (per liter) 20 g dextrose monohydrate, 5 g (NH4)2SO4, 15 g KH2PO4, 0.3 g CaCl2, and 0.3 g MgSO4 and was supplemented with essential minerals (5 mg FeSO4 · 7H2O, 1.4 mg ZnSO4 · 7H2O, 3.7 mg CoCl2, and 1.6 mg MnSO4 · 7H2O).
T. reesei Rut-C30 and a transformant of this strain producing tissue plasminogen activator (tPA) (clone 306/36; Uusitalo, Pakula, and Penttilä, unpublished data) were cultivated in laboratory-scale bioreactors. Fungal spores were first inoculated into shake flasks (107 spores per 50 ml) in culture medium containing (per liter) 20 g lactose, 4 g peptone, 1 g yeast extract, 15 g KH2PO4, 2.8 g (NH4)2SO4, 0.6 g MgSO4 · 7H2O, and 0.8 g CaCl2 · 2H2O and supplemented with essential minerals, and the pH was adjusted to 5.2 by addition of KOH. The precultures were cultivated for 2 days at 28°C on a rotary shaker at 200 rpm, transferred into fresh medium (1:10, vol/vol), and cultivated for another day. Before the precultures were transferred to the bioreactors, the mycelium was centrifuged for 5 min at 1,500 × g and washed with culture medium. The medium used in the final fermentation was the medium described above, except that a higher concentration of lactose (40 g/liter) and a lower concentration of KH2PO4 (4 g/liter) were used. The pH of the medium in the bioreactor was adjusted to 6.1 to 6.3 by addition of NH4OH and H3PO4.
For cultivation under ER stress conditions, the Rut-C30 strain was grown on minimal medium containing (per liter) 20 g lactose, 15 g KH2PO4, 7.6 g (NH4)2SO4, 0.5 g MgSO4 · 7H2O, 0.2 g CaCl2 · 2H2O, 5 mg FeSO4 · 7H2O, 1.4 mg ZnSO4 · 7H2O, 3.7 mg CoCl2, and 1.6 mg MnSO4 · 7H2O. The pH of the medium was adjusted to 5.2 with KOH. The culture medium was inoculated with 2 × 107 spores per 200 ml of medium, and the culture was grown in conical shake flasks at 28°C with shaking at 200 rpm for 4 days. The cultures were then diluted 1:10 in fresh medium, grown for another 24 h, and treated with DTT (final concentration, 10 mM) or BFA (50 μg/ml). The same volume of solvent used in the drug stock solution (double-distilled water for cultivation with DTT and 0.5% dimethyl sulfoxide for cultivation with BFA) was added to the untreated control cultures.
Growth of T. reesei Rut-C30 in the presence of 1 mg of the α-glucosidase inhibitor castanospermine (Calbiochem-Merck Eurolab NV/SA, Leuven, Belgium) was done in 15 ml (total volume) of minimal dextrose medium using a 50-ml Falcon tube. The culture was incubated for 2 days at 30°C with shaking at 200 rpm.
Electrocompetent or chemocompetent Escherichia coli MC1061 cells [hsdR2 hsdM+ hsdS+ araD139 Δ(ara leu)7697 ΔlacX74 galE15 galK16 rpsL (Strr) mcrA mcrB1] (13) were used for DNA manipulations. Growth and transformation of E. coli were performed as described previously (76).
Nucleic acid preparation for cloning purposes.
Trichoderma genomic DNA was extracted from mycelium grown for 5 to 6 days in minimal medium at 30°C. The mycelium was separated from the growth medium, washed, dried, frozen in liquid nitrogen, and then ground. The disrupted fungal cells were resuspended in 5 to 10 ml of extraction buffer (200 mM Tris-HCl [pH 8.5], 250 mM NaCl, 0.5% sodium dodecyl sulfate [SDS]), and after phenol extraction, the aqueous phase was treated with 1 mg of RNase A (30 min at 37°C). Following chloroform extraction, the DNA was precipitated with isopropanol, and the DNA pellet was washed, dried, and resuspended in a suitable volume of H2O.
Total Trichoderma RNA for 5′-RACE was prepared from mycelium grown in minimal medium for 5 to 6 days at 30°C. The mycelium was separated from the growth medium, washed, dried, frozen in liquid nitrogen, and then ground. The disrupted cells were resuspended in extraction buffer (25 mM sodium citrate, 4 M guanidinium HCl, 100 mM sodium lauryl sarcosine, and 100 mM beta-mercaptoethanol), and the resulting suspension was extracted twice with phenol and once with chloroform. The RNA was precipitated from the aqueous phase with LiCl, and the resulting pellet was resuspended in 0.3 M sodium acetate, pH 5.7. After centrifugation, the RNA was precipitated from the supernatant with ice-cold ethanol. The RNA pellet was washed, dried, and resuspended in a suitable volume of diethyl pyrocarbonate-treated H2O.
cDNA cloning of the T. reesei Rut-C30 glucosidase II gene.
Based on an alignment of several known mammalian and yeast glucosidase II alpha subunit amino acid sequences, three homologous regions were selected for synthesis of degenerate primers. The primers were sense primer 5′-GTITATGGIATHCCIGAGCATGC-3′ (primer 1) and antisense primers 5′-GIGCGTGIGCICKGAAGAAIG-3′ (primer 2) and 5′-TGISWICCIGCGAAGAAIGCIC-3′ (primer 3) (H = A, C, or T; K = G or T; S = G or C; W = A or T). A PCR with 200 ng of total DNA from a T. reesei cDNA library (72) was performed using Taq polymerase and 50 pmol of primer 1 and primer 3. Amplification was carried out for 35 cycles of 94°C for 45 s, 55°C for 1 min, and 72°C for 1.5 min. Similar reaction conditions were used for a PCR with primers 1 and 2, except that the annealing temperature was decreased to 50°C. The amplified fragments were cloned in pCR2.1-TOPO (N.V. Invitrogen SA, Merelbeke, Belgium) for sequence analysis.
Clones containing the T. reesei glucosidase II alpha subunit were isolated using a PCR-based method as described previously (82). In brief, the cDNA library was transformed into E. coli MC1061 competent cells. The transformation mixture was diluted and transferred into a 96-well plate at about 5,000 cDNA clones per well. The microtiter plate thus contained about five times the number of cDNA clones in the cDNA library. After incubation for several hours at 37°C, PCR was performed with primers 1 and 3 using the pooled cellular suspension from each of the 12 columns and each of the eight rows of the 96-well plate. Based on the results, positive wells lying at the intersection of positive columns and positive rows could be identified by gel analysis of the obtained PCR product. One of these wells was used to inoculate a new microtiter plate at a concentration of 500 clones per well. After incubation, the PCR was repeated, and the cell suspension of one of the resulting positive wells was again inoculated into the wells of a new microtiter plate, this time at a concentration of 50 clones per well. After incubation, the cell suspension from one of the positive wells was plated on solid Luria-Bertani (LB) medium. About 200 colonies were transferred to Hybond N filters (Amersham Biosciences Benelux, Roosendaal, The Netherlands) and incubated overnight. Colony hybridization was carried out using the PCR fragment, amplified from the cDNA library with primers 1 and 3, as the probe. 32P labeling of the probe was done with a High-Prime kit (Roche Diagnostics Belgium sa/nv, Vilvoorde, Belgium), following the instructions of the manufacturer.
DNA was prepared from several positive clones and digested with EcoRI and XhoI to release the DNA insert. Southern blotting was performed to confirm the glucosidase II specificity of the fragments obtained. Furthermore, the fragments were cloned for sequence analysis, either as EcoRI/XhoI fragments into an EcoRI/SalI-opened pUC19 vector or as blunted XhoI fragments into an EcoRV-opened pBluescriptII KS(+/−) vector (Stratagene Europe, Amsterdam, The Netherlands).
5′-RACE and iPCR cloning.
To clone the missing 5′ part of the glucosidase II alpha subunit gene, 5′-RACE and inverse PCR (iPCR) were used. For the iPCR, an antisense PCR primer (ROT2TR4_AS; 5′-GTTAAACGTTTCGTCCCACC-3′) and a sense PCR primer (ROT2TR1_S; 5′-GGCTCCATCCCTTTCATGC-3′) were designed on the basis of the 5′ sequence of the cloned but incomplete gls2α cDNA fragment. The 5′ ends of the primers faced each other and hybridized to positions on the cDNA that were separated by a 229-bp region which contained an NcoI restriction site. Trichoderma genomic DNA (10 μg) was digested with BamHI, a restriction enzyme that cuts the glucosidase II alpha subunit sequence at a position 3′ to both iPCR primers. After heat inactivation of BamHI, the genomic DNA fragments were self-ligated into circular molecules with T4 DNA ligase, purified, and digested with NcoI. The iPCR primers, now with their 3′ ends facing each other, flanked the missing 5′ sequence of the glucosidase II alpha subunit. After the NcoI digestion, the DNA was purified by phenol extraction and isopropanol precipitation and dissolved in 50 μl of H2O. One microliter of the DNA suspension was used as a template in a PCR with 50 pmol of each iPCR primer. The PCR was performed with Pfu polymerase for 20 cycles of 94°C for 45 s, 55°C for 30 s, and 72°C for 1.5 min. A schematic view of the iPCR strategy is shown in Fig. 1A.
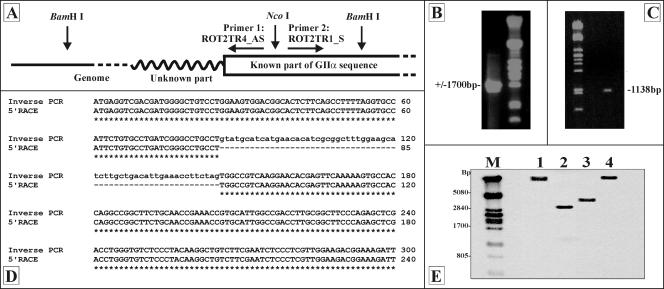
FIG. 1.
Cloning and characterization of the glucosidase II alpha subunit gene. (A) Inverse PCR strategy. Genomic DNA was digested with BamHI, and the fragments were self-ligated. After NcoI digestion, a PCR was performed using primers ROT2TR4_AS and ROT2TR1_S. (B) Gel analysis of the DNA fragment obtained by iPCR. (C) Gel analysis of the DNA fragment obtained by 5′-RACE. (D) Alignment of part of the cloned iPCR and 5′-RACE sequences, indicating the presence of a 60-bp intron sequence. (E) Southern analysis of Rut-C30 genomic DNA (treated with EcoRI [lane1], HindII [lane 2], HindIII [lane 3], and BglII [lane 4]) using a gls2α-specific 32P-labeld probe. Lane M contained a λ PstI-digested DNA reference marker.
For the 5′-RACE procedure, a First Choice RLM-RACE kit was used [Ambion (Europe) Ltd., Huntingdon, United Kingdom]. For primer design and the experimental procedure we followed the instructions of the manufacturer. The gene-specific primers used for the outer and inner PCRs were ROT2TR-RLMRACE (5′-GATATACTCGAAGACGTCGG-3′) and ROT2TR4_AS (5′-GTTAAACGTTTCGTCCCACC-3′), respectively. Annealing during the outer and inner PCRs was done at 57°C and 55°C, respectively.
For sequence analysis, the 5′-RACE and inverse PCR fragments were cloned in the pCR-blunt II-TOPO vector (N.V. Invitrogen SA, Merelbeke, Belgium).
PCR-based intron and frameshift analyses.
The intron-exon composition of the glucosidase II gene was analyzed by amplifying the whole gene from the Rut-C30 genome. Genomic DNA (1 μg) was used as the template, and the sense and antisense primers were 5′-ATGAGGTCGACGATGGGG-3′ and 5′-AGCCAGCTTGATGCTCC-3′, respectively. The fragment was amplified with Pfu polymerase using 25 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 7 min.
Frameshift analysis was done on the Rut-C30, Rut-NG14, and QM9414 genomes by PCR using 1 μg of genomic DNA as the template. The sequences of the internal gls2α-specific primers were 5′-TATCTCTGGTTTCCCGTTCTCG-3′ for the sense primer ROT2TR3_S and 5′-CTGGTCATCAATCGCCAAGCC-3′ for the antisense primer ROT2TR0_AS. PCR was performed using Pfu polymerase for 25 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min.
For sequence analysis, the PCR fragments were cloned in the pCR-blunt II-TOPO vector (N.V. Invitrogen SA, Merelbeke, Belgium).
Southern analysis.
About 15 μg of genomic DNA was digested overnight with suitable restriction enzymes. After fractionation on a 1% agarose gel, the DNA was transferred to a nylon membrane (Hybond N+; Amersham Biosciences Benelux, Roosendaal, The Netherlands) via alkali blotting. Hybridization analysis was done using a 32P-labeled 599-bp NcoI fragment of the Trichoderma glucosidase II alpha subunit gene. Nucleic acid blotting and the hybridization analysis were done using standard procedures (76). Labeling of the probe was performed with a High-Prime kit (Roche Diagnostics Belgium sa/nv, Vilvoorde, Belgium).
Northern analysis of T. reesei Rut-C30 treated with DTT or BFA or expressing tPA.
Mycelium samples (50 ml) were collected from cultures treated with either DTT or BFA and from the corresponding untreated control cultures at different times (0, 15, 30, 60, 90, 120, 240, and 360 min after addition of the drug). The first sample (time zero) was obtained immediately before DTT or BFA was added. Similarly, 50-ml samples of mycelia were collected frequently during laboratory-scale fermentation of untransformed strain Rut-C30 and the tPA-producing clone 306/36.
The mycelium was filtered, washed with an equal volume of 0.7% NaCl, frozen immediately in liquid nitrogen, and stored at −80°C. Total RNA was isolated using the Trizol reagent (Gibco BRL) according to the manufacturer's instructions. Northern blot analysis on nitrocellulose filters was carried out using standard procedures (76). A cDNA fragment (499-bp BamHI fragment) of the glucosidase II alpha subunit gene was used as a probe for the gls2α transcript. The gls2α signals were normalized using the signals of gpd1 or act1 (accession no. X75421), as indicated below. For gpd1, a 900-bp cDNA fragment (VTT Biotechnology, Espoo, Finland) was used as a probe, while for act1 a 767-bp PCR-amplified fragment was used.
Construction of the Trichoderma expression vector for a functional Trichoderma glucosidase II alpha subunit gene.
The Trichoderma expression vector for a functional Trichoderma glucosidase II alpha subunit gene was constructed in six steps, as follows. (i) The cloned glucosidase II cDNA fragment was cut out of the pAJ401 library vector as an approximately 3,000-bp EcoRI/HindIII fragment. This fragment was ligated into an EcoRI/HindIII-opened pUC19 vector to obtain plasmid pUC19Δgls2αTreesei(shift). (ii) The frameshift within the cloned Rut-C30 cDNA fragment was repaired. A PCR fragment was amplified from genomic DNA of the QM9414 strain using the Pfu polymerase and primers ROT2TR2_S (5′-ATCAATGAGCAACTCCTGGC-3′) and ROT2TR0_AS (5′-CTGGTCATCAATCGCCAAGCC-3′). A PCR was performed by using 25 cycles of 1 min at 95°C, 1 min at 60°C, and 1 min at 72°C. The amplified fragment was digested with XcmI/PflMI and ligated into the XcmI/PflMI-opened vector pUC19Δgls2αTreesei(shift) to obtain plasmid pUC19Δgls2αTreesei(repaired). (iii) The ORF of the glucosidase II alpha subunit (gls2α) was completed; the 5′-RACE fragment was digested with DraIII and MspAI and ligated to the DraIII and Klenow-blunted EcoRI sites of vector pUC19Δgls2αTreesei(repaired), giving rise to plasmid pUC19gls2αTreesei. (iv) A unique SmaI site was incorporated at the 3′ terminus of the gls2α ORF by mutagenesis using a Quick Change mutagenesis kit (Stratagene Europe, Amsterdam, The Netherlands). The two primers used to induce the silent mutation (from CGT to CGG) were the sense primer 5′-CCATGTGAAGGCCCGGGTTGGGGATGACTGG-3′ and the antisense primer 5′-CCAGTCATCCCCAACCCGGGCCTTCACATGG-3′. The resulting plasmid was designated pUC19gls2αTreesei(SmaI). (v) The plasmid was cut with EcoRI/SalI for integration of a linker at the 5′ end of the gls2α ORF. The linker consisted of two partially complementary primers (sense primer 5′-GAATTCCCGCGGTACGTAATTATGAGG-3′ and antisense primer 5′GTCGACCTCATAATTACGTACCGCGGG-3′) and was prepared by mixing equimolar amounts of the two primers, boiling the mixture, and gradually cooling it to room temperature. By inserting the linker, two new unique restriction sites (SacII and SnaBI) were integrated at the 5′ end of the glucosidase II ORF, creating plasmid pUC19(5′)gls2αTreesei(SmaI). (vi). The glucosidase II ORF was isolated from this plasmid by HindIII/SacII-T4 polymerase treatment and ligated into HindIII/NcoI-S1 nuclease-treated plasmid pFGPDGLAT3 (14). The glucosidase II alpha subunit ORF was thus placed under transcriptional control of the constitutive Aspergillus nidulans gpdA promoter. To decrease the distance between the 3′ end of the ORF and the trpC terminator, the vector was digested with MluI to remove an approximately 500-bp fragment and then self-ligated to produce plasmid pFGPDgls2αTreesei.
Transformation of T. reesei.
T. reesei was cotransformed as described by Penttila et al. (67) using the hygromycin resistance gene (plasmid pAN7.1 [69]) as a selection marker. Before transformation, the glucosidase II alpha subunit expression vector pFGPDgls2αTreesei was linearized with FspI. Transformants were selected on minimal medium containing 150 μg/ml hygromycin.
N-glycan analysis.
Trichoderma strains were grown for 6 days at 28°C in 100-ml shake flasks containing 50 ml minimal medium with glucose as the single carbon source [composition (per liter): 20 g dextrose monohydrate, 5 g (NH4)2SO4, 15 g KH2PO4, 0.3 g CaCl2, 0.3 g MgSO4, and essential minerals] or as otherwise indicated. N-glycans of the total pool of secreted proteins were prepared as described previously (56) from 1 ml of growth culture supernatant. The final glycan pellet was resuspended in 5 μl of double-distilled H2O, and 1 μl of this preparation was used for oligosaccharide analysis by laser-induced DNA sequencer-assisted fluorescence-assisted carbohydrate electrophoresis (DSA-FACE), as described recently (12). Briefly, glycoproteins were immobilized on a Multiscreen Immobilon-P plate and deglycosylated using peptide N-glycosidase F (New England Bioloabs GmbH, Frankfurt am Main, Germany). N-glycans were recovered and derivatized using 8-amino-1,3,6-pyrenetrisulfonic acid. After removal of excess label by size fractionation on Sephadex G-10 resin, the 8-amino-1,3,6-pyrenetrisulfonic acid-labeled carbohydrates were dried by evaporation and reconstituted in a 5-μl mixture. A rodamine X-labeled Genescan 500 mixture (Perkin-Elmer, Foster City, Calif.) was added as an internal standard. Electrophoresis was done with an ABI 377 DNA sequencer (Applied Biosystems, Foster City, Calif.) using a 12% polyacrylamide gel. The electrophoresis buffer consisted of 89 mM Tris, 89 mM boric acid, and 2.2 mM EDTA. A maltodextran ladder and an N-glycan mixture of RNase B, consisting of Man5-9GlcNAc2, were used as references. Data analysis was performed with the GENESCAN 3.1 software (Applied Biosystems, Foster City, Calif.).
Mild acid hydrolysis of the N-glycans was performed with 1 μl of the prepared N-glycan mixture by incubation with 9 μl of 10 mM HCl at 100°C for 30 min. α-1,2-Mannosidase and jack bean mannosidase (0.1 U; Sigma Biochemicals, Bornem, Belgium) digestion of the N-glycan mixture (1 μl) was done overnight at 37°C in 20 mM sodium acetate (pH 5.0; total volume, 10 μl). The α-1,2-mannosidase used was 1.35 μg (about 50 μU) of T. reesei α-1,2-mannosidase that was produced in house (44). Before DSA-FACE analysis, the in vitro-treated glycan samples were dried, and the pellets were resuspended in 1 μl double-distilled H2O.
Protein analysis.
Trichoderma strain Rut-C30 and transformants derived from this strain were grown in 100-ml shake flasks for 6 days in 50 ml minimal glucose medium. The cultures were incubated at 28°C with continuous shaking at 200 rpm. After growth, the mycelium was separated from the culture medium and dried overnight at 50°C. Proteins were precipitated from the medium with trichloroacetic acid. The proteins were resuspended in 2× Laemmli loading buffer, separated by conventional SDS-polyacrylamide gel electrophoresis (PAGE), and stained with Coomassie brilliant blue R-250 (Merck-VWR International BVBA, Leuven, Belgium) using standard procedures (76).
Bioinformatics.
All bioinformatics were performed using tools that are accessible via a link on the ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (5). Homology searches were done using the BLAST algorithm (4). Dual and multiple alignments were performed using the Clustal W algorithm (84) or the Align program (GENESTREAM network server; IGH, Montpellier, France) (62). General features of the protein (molecular weight, pI, amino acid composition) were assessed using the ProtParam tool, and the presence of a putative signal sequence was predicted using Signal P (version 1.1). Predictions of the presence of transmembrane helices were made using the TMHMM (version 2.0) or HMMTOP (version 2.0) program (by G. E. Tusnady).
Nucleotide sequence accession number.
The sequence data presented in this paper have been deposited in the EMBL/GenBank databases under accession number AY623904.
RESULTS
Cloning of the T. reesei glucosidase II alpha subunit gene.
The amino acid sequence of S. cerevisiae glucosidase II was aligned with the sequences of several known mammalian glucosidase II alpha subunits, using the Clustal W algorithm website. Three degenerate primers were designed on the basis of several homologous regions in order to screen a cDNA library of the T. reesei Rut-C30 strain. PCR amplification using sense primer 1 and antisense primer 3 produced a 1,170-bp fragment, and nested PCR amplification with primers 1 and 2 resulted in an approximately 970-bp fragment. Both fragments were of the expected length, and sequence analysis showed that they exhibited similarity with known glucosidase II alpha subunits.
Based on these results, cloning of the glucosidase II alpha subunit cDNA was started from the Rut-C30 cDNA library, using the technique for cDNA cloning by PCR screening (82) with sense primer 1 and antisense primer 3. Each PCR round (there were a total of three rounds) indicated that several wells in the microtiter plate (see Materials and Methods) contained at least one gls2α-specific clone. In the final PCR round, each well contained a cell suspension consisting of about 50 different cDNA clones, and two wells proved to be positive. After incubation at 37°C, a dilution of the cell suspension of one of the two positive wells was plated on solid LB medium. Two hundred of the resultant colonies were analyzed by colony hybridization using a 32P-labeled gls2α probe. Seven positive clones were identified. XhoI/EcoRI restriction fragments (1,700, 600, and 200 bp) of the plasmid DNA proved to be glucosidase II specific as determined by Southern hybridization or by sequence analysis. The isolated cDNA fragment was completely sequenced and consisted of 2,290 bp. However, similarity analysis also indicated that a substantial part of the 5′ end of the ORF was missing.
To clone the 5′ end of the gene, both an inverse PCR strategy and a 5′-RACE strategy were used. When inverse PCR was used with Rut-C30 genomic DNA, a 1,700-bp fragment was cloned (Fig. 1B). Partial sequence analysis indicated the presence of two regions showing homology to the 5′ part of the ORFs of other known glucosidase II alpha subunit genes that were separated from each other by a 60-bp putative intron sequence. The existence of an intron in the 5′ part of the Trichoderma gls2α sequence was confirmed by 5′-RACE (Fig. 1D). Using a First Choice RLM-RACE kit, we obtained an 1,138-bp fragment (Fig. 1C) missing the 60-bp intron sequence but otherwise showing 100% homology to the cloned iPCR fragment.
Southern analysis, after digestion of the T. reesei Rut-C30 genomic DNA with different restriction enzymes, indicated that a single copy of the glucosidase II alpha subunit gene was present (Fig. 1E).
Nucleotide sequence analysis. (i) Intron analysis.
To evaluate the intron-exon composition of the Trichoderma glucosidase II alpha subunit gene, PCR was performed with genomic Rut-C30 DNA using a high-fidelity polymerase and gene-specific primers. A fragment consisting of about 3,000 bp was amplified, which was approximately the length of the coding cDNA. This indicated that only a few rather small intron sequences could be present. Alignment of the PCR fragment with the cloned cDNA indicated that the 60-bp intron identified earlier at the 5′ terminus was the only intron present in the gls2α gene. The small size of this intron is consistent with the sizes of most of the introns of the other filamentous fungi that have been characterized (47, 48, 81). The intron follows the GT/AG rule for the 5′ and 3′ splice site (51). It contains a lariat sequence with the consensus sequence CTRAC (R = purine) 13 nucleotides upstream of the 3′ splice site, which is characteristic of other fungal intron sequences (33, 48, 53).
(ii) Promoter analysis.
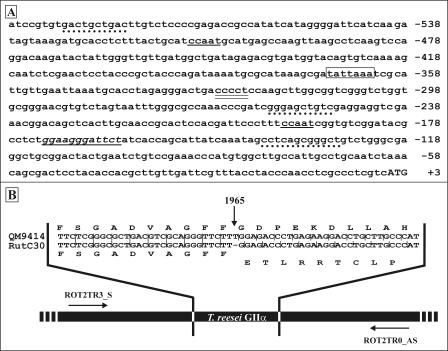
The inverse PCR strategy not only succeeded in cloning the missing 5′ part of the alpha subunit ORF but also enabled us to isolate about 600 bp immediately upstream of the initiating ATG codon (Fig. 2A). Analysis of this 5′ noncoding region revealed the presence of two putative CCAAT boxes, one at position −509 and the other at position −195. This motif has been identified in the promoter region of several biotechnologically important filamentous fungal genes (reviewed in reference 9) and is believed to be involved in Ca2+ stress signal transduction in the human binding protein promoter (71). No standard TATA box was found, but a TATTAAA sequence was identified at position −368. An identical sequence was found at positions −391 and −352 of the cre1 promoter region of T. reesei and Trichoderma harzianum, respectively (34). A putative heat shock element, resembling the consensus sequence CT-GAA-TTC-AG (63), was identified at position −171. A possible stress response element, CCCCT (46), was found at position −325. Three putative UPR-specific elements, showing 60 to 70% homology to the yeast consensus sequence (50), were found either on the forward strand or on the reverse strand at positions −139/−128, −249/−258, and −580/−589.
FIG. 2.
(A) Analysis of the sequence 5′ of the initiating ATG codon of the cloned glucosidase II alpha subunit. CCAAT boxes are underlined; the putative TATA box is enclosed in a box; the putative heat shock element is underlined and in italics; the possible general stress response element CCCCT is double underlined; putative UPR elements are indicated by dotted lines; and the beginning of the coding sequence is indicated by uppercase letters. (B) Frameshift analysis of the Rut-C30, Rut-NG14, and QM9414 genomic DNA: strategy used to analyze the genomic DNA. Using primers ROT2TR3_S and ROT2TR0_AS, fragments consisting of about 320 bp were amplified from all three genomes. The alignment (a portion of the alignment is shown) indicates that a T is missing at position 1965 of the ORF (indicated by the vertical arrow). Above and below the alignment are the amino acid sequences encoded by the corresponding nucleotide sequences of QM9414 and Rut-C30, respectively. The data for Rut-NG14 are identical to those for Rut-C30 (data not shown).
(iii) Frameshift analysis.
The 5′-RACE fragment and the cloned cDNA sequence obtained from Rut-C30 were ligated in silico, resulting in a 3,621-bp fragment. Translation and BLAST analysis indicated the presence of an ORF encoding an 807-amino-acid polypeptide showing homology to the N-terminal regions of known glucosidase II alpha subunits. In contrast to the first 655 amino acids, the C-terminal 152-amino-acid sequence did not show any sequence homology to other known glucosidase II alpha subunits. Moreover, the Trichoderma GIIα polypeptide sequence is significantly shorter than the yeast or mammalian homologues. Detailed sequence comparison indicated the presence of a frameshift within the cloned cDNA causing premature termination of translation. Indeed, computer analysis of the 3′ 1,500 bp of the 3,621-bp fragment identified a 927-bp out-of-frame sequence encoding a 309-amino-acid polypeptide that shows high homology to the C terminus of known glucosidase II alpha subunits.
Using the gls2α internal primers ROT2TR3_S and ROT2TR0_AS, a fragment consisting of about 320 bp was amplified from the genomic DNA of T. reesei strains Rut-C30, QM9414, and Rut-NG14 (Rut-NG14 is a cellulose-hypersecreting precursor strain of Rut-C30) (49). Based on BLAST homology searches, the annealing sites of the two primers were chosen so that the amplified fragment would contain the site of the frameshift. Sequence alignment of the PCR fragments generated clearly indicated the presence of a frameshift in the Rut-C30 and Rut-NG14 genomes (but not in the QM9414 genome); at position 1965 of the glucosidase II alpha subunit ORF a T was missing (Fig. 2B). This generated a premature stop codon 456 nucleotides 3′ of the position of the frameshift, resulting in the synthesis of a truncated protein with 152 C-terminal amino acids that were not specific for the glucosidase II alpha subunit.
Analysis of the deduced amino acid sequence.
A full-size nonmutant version of the gls2α ORF (total length, 2,892 bp) encoded a 964-amino-acid polypeptide, whose length was similar to the lengths of other known glucosidase II alpha subunits. The protein had a calculated molecular mass of 109,858 Da and a theoretical pI of 5.6. Analysis using Signal P (version 1.1) indicated the presence of a putative eukaryotic N-terminal signal sequence consisting of 30 amino acids. A signal cleavage site was predicted after Leu29Ala30. The polypeptide sequence seemed to lack any known ER retention signals, such as an HDEL tag or putative transmembrane helices. These data are in agreement with the general model for the GIIα protein (6, 88). The T. reesei glucosidase II alpha subunit exhibited 64.2% sequence identity with its Neurospora crassa counterpart (SP-TrEMBL accession no. Q8NIY3) but only 37.9% sequence identity with the S. cerevisiae homologue (88) (GenBank accession no. Z36098). Low sequence identities, ranging from 40 to 43%, were also found for S. pombe (15) (SP-TrEMBL accession no. Q9US55) and higher eukaryotic organisms, such as the pig (20) (SP-TrEMBL accession no. P79403), humans (88) (SP-TrEMBL accession no. Q9P0X0), and Arabidopsis thaliana (SP-TrEMBL accession no. Q9FN05). Figure 3 shows an alignment of some known glucosidase II alpha subunits with the deduced amino acid sequence of the cloned T. reesei homologue. Similar to all known glucosidase II alpha subunits, the Trichoderma homologue contains the conserved WXDMNE motif of the glycosyl hydrolase family 31 that is essential for enzymatic activity (29).
FIG. 3.
Alignment of the deduced amino acid sequence of the cloned T. reesei glucosidase II alpha subunit with the sequences of other known alpha subunits, including those of N. crassa, Sus scrofa (pig), Homo sapiens (human), A. thaliana, S. pombe, and S. cerevisiae. The solid bar at position 635 of the alignment indicates the conserved motif of family 31 glycosyl hydrolases. The horizontal arrows with numbers indicate the amino acid sequences on which degenerate primers 1, 2, and 3 were based (resulting from alignment of the human, pig, and yeast sequences). The vertical arrow indicates the position where the frameshift mutation in the Rut-C30 gene results in termination of the alpha subunit-specific polypeptide sequence and the starting point of the 152 nonspecific C-terminal amino acids.
Transcriptional response of the glucosidase II alpha subunit gene in a T. reesei strain producing a heterologous protein.
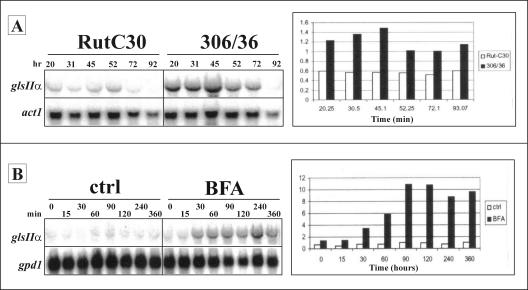
In fungi, the production of heterologous proteins sometimes induces a UPR (52, 70, 91, 92). As glucosidase II is involved in the quality control of protein folding in the ER, it might be required at elevated levels under such conditions. Hence, we analyzed the expression levels of the gls2α gene in bioreactor cultures of a T. reesei strain (clone 306/36) expressing human tPA and the parental Rut-C30 strain. Northern analysis using a gls2α-specific probe revealed a band corresponding to a 3.0-kb transcript, which is in accordance with the estimated length of the gls2α mRNA consisting of the 2,891-bp ORF and the 3′ and 5′ flanking regions. Throughout cultivation, the observed gls2α-specific signals were approximately 2- to 2.5-fold greater for the strain expressing tPA than for the untransformed Rut-C30 strain (Fig. 4A).
FIG. 4.
Analysis of the transcription levels of the glucosidase II alpha subunit by Northern blotting. (A) Effect of tPA production on mRNA levels. Total RNA was prepared from mycelial samples taken at different times after the start of fermentation (20, 31, 45, 52, 72, and 92 h) of the Rut-C30 strain and the Rut-C30-derived tPA-producing transformant 306/36 and was analyzed by Northern blotting. The signals were quantified with a PhosphorImager and were normalized against those obtained for the act1 gene. (B) Effect of BFA treatment on the mRNA levels. Total RNA was prepared from mycelial samples taken at different times after addition of the drug (0, 15, 30, 60, 90, 120, 240, and 360 min) and subjected to Northern analysis. The signals were normalized against those obtained for the gpd1 gene. ctrl, control.
Transcriptional response of the glucosidase II alpha subunit gene in cultures treated with DTT and BFA.
Treatment of T. reesei cultures with DTT or brefeldin A has been shown to efficiently block transport and to induce UPR in the cultures (55). To study further whether the expression of the glucosidase II alpha subunit is sensitive to such stress conditions, Northern blot analyses were carried out on filters containing RNA samples from Rut-C30 cultures treated with DTT (10 mM) or BFA (50 μg).
Hybridization of the BFA stress filters with a gls2α-specific probe and normalization of the signals with gpd1 indicated that there was moderate induction of the gene encoding Trichoderma GIIα. The response was detectable about 30 min after addition of the stress factors and significantly increased around 90 min, after which the mRNA signals remained more or less stable for the next 270 min. The maximum upregulation was about 10-fold (Fig. 4B).
No upregulation of the mRNA levels was observed when DTT was used as a stress factor. Depending on the experimental conditions, the transcript levels either remained more or less stable or seemed to decrease (results not shown). This indicates that although stress conditions such as tPA production and BFA treatment enhance transcription of the alpha subunit gene, the expression of glucosidase II activity does not behave in exactly the same way as the expression of many other UPR-regulated genes.
Expression of a fully active Trichoderma glucosidase II alpha subunit in the Rut-C30 strain.
An expression vector encoding a predicted functional variant of the T. reesei glucosidase II alpha subunit was constructed. The frameshift within the cloned cDNA fragment was repaired, and then the 5′-RACE fragment and the repaired cDNA were ligated to obtain a full-length ORF encoding a full-size GIIα subunit. Finally, the ORF was placed under the transcriptional control of the constitutive gpdA promoter and the trpC terminator. The resultant plasmid (pFGPDgls2αTreesei) was transformed into T. reesei Rut-C30.
Transformants were first analyzed for functionality of the expressed glucosidase II alpha subunit. Following 6 days of culture in glucose minimal medium, the N-glycans were prepared from 0.25 to 1 ml of culture supernatant as described by Papac and coworkers (56) and were analyzed by DSA-FACE (12). The N-glycan profiles of the transformants were compared with those of the QM9414 strain, which does not carry monoglucosylated N-glycans (25), and the Rut-C30 untransformed strain.
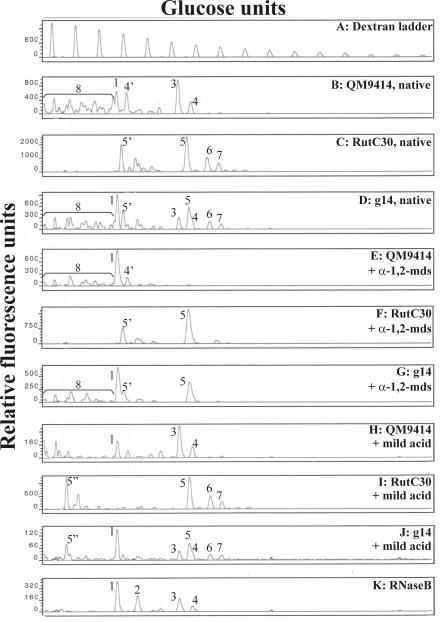
Based on the previously published structural data for the most predominant oligosaccharides synthesized on secreted cellobiohydrolase I (17, 45, 79), the profile of the Rut-C30 strain appeared to be relatively easy to interpret. In vitro α-1,2-mannosidase digestion was used to characterize the peaks representing monoglucosylated high-mannose glycans. Since α-1,3-linked glucose blocks the hydrolysis of the two underlying α-1,2-linked mannose residues, a maximum of two mannoses can be released from the glycan substrate, resulting in GlcMan7GlcNAc2. Mild acid treatment, which hydrolyzes phosphodiester linkages, was used to characterize the peaks representing phosphorylated carbohydrates. Release of the phosphate-coupled mannose resulted in a phosphomonoester glycan, which carried an extra negative charge and therefore had greater electrophoretic mobility. Peaks representing these glycans were shifted to the left side of the DSA-FACE profile. Using a combination of α-1,2-mannosidase digestion and mild acid hydrolysis, the most predominant peaks in the DSA-FACE glycan pattern of the Rut-C30 strain could be assigned to GlcMan7-8GlcNAc2 and the charged counterparts ManPGlcMan7-8GlcNAc2 (Fig. 5C, F, and I). Similar results were obtained for the Rut-NG14 strain (not shown).
FIG. 5.
Glycosylation profiles of the Rut-C30, QM9414, and g14 transformants, including the native profiles, the profiles after α-1,2-mannosidase digestion, and the profiles after mild acid hydrolysis. In all three cases it is clear that the g14 transformant has a glycosylation profile that has characteristics of the profiles of both the Rut-C30 and QM9414 strains. The deduced N-glycans are indicated as follows: 1, Man5GlcNAc2; 2, Man6GlcNAc2; 3, Man8GlcNAc2; 4, Man9GlcNAc2; 5, GlcMan7GlcNAc2; 6, GlcMan8GlcNAc2; 7, GlcMan9GlcNAc2; 4′, ManPMan9GlcNAc2; 5′, ManPGlcMan7GlcNAc2; 5", PGlcMan7GlcNAc2. In the g14 native and mild acid-treated patterns, Man9GlcNAc2 appears as a shoulder peak on the GlcMan7GlcNAc2 peak. α-1,2-mds, N-glycan pattern after treatment with α-1,2-mannosidase; mild acid, N-glycan pattern after mild acid hydrolysis of the carbohydrates; RNaseB, standard high-mannose N-glycan mixture derived from RNase B. The glycan profile of the Rut-NG14 strain is identical to that of Rut-C30 (data not shown).
The glycan pattern of QM9414 was analyzed in the same way. Initially, the QM9414 DSA-FACE profile looked far more complex. However, comparison with the standard RNase B profile indicated that a significant fraction of the glycan pool consisted of Man5-9GlcNAc2. This was confirmed by in vitro digestion with α-1,2-mannosidase. Moreover, mild acid hydrolysis of the carbohydrates indicated that most of the peaks on the left side of the Man5GlcNAc2 signal represented glycans containing one or more phosphodiester linkages. The QM9414 glycan peaks could thus be assigned to neutral and phosphorylated high-mannose N-glycans. The distribution of the phosphorylated N-glycans was not significantly changed after α-1,2-mannosidase digestion, because the phosphodiester linkages sterically hindered the enzyme or blocked access to the underlying α-1,2-linked mannose residues (Fig. 5B, E, and H).
Next, the N-glycan profiles of several hygromycin-resistant transformants were analyzed. Only one of the transformants analyzed (g14) had a clearly different N-glycan pattern compared to the Rut-C30 wild-type strain. The g14 transformant glycan pool appeared to be more heterogeneous, and closer examination indicated that it consisted of a combination of the Rut-C30 and QM9414 carbohydrate profiles (Fig. 5D, G, and J). Especially on the left side of the Man5GlcNAc2 peak, many new peaks, representing fast-migrating oligosaccharides, emerged. Since most of these molecules are susceptible to mild acid hydrolysis, we believe that they represent the structural diversity of phosphorylated high-mannose glycans, analogous to the situation in QM9414. In addition to these charged high-mannose N-glycans, some peaks representing neutral unglucosylated carbohydrates also emerged in the DSA-FACE profile. The presence of these structures was further confirmed by in vitro α-1,2-mannosidase digestion. Comparison of the glycan profile of the g14 transformant with that of the Rut-C30 untransformed strain, however, clearly indicated that a significant amount of monoglucosylated glycans (neutral and charged GlcMan7-9GlcNAc2) was still synthesized on the proteins of the transformed strain.
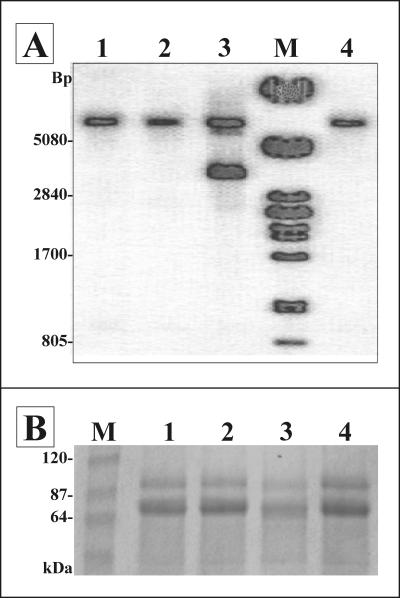
A number of hygromycin-resistant transformants, including g14, and the wild-type Rut-C30 strain were analyzed at the genomic level. To clearly discriminate between the autologous mutant alpha subunit locus and the repaired cDNA from the integrated expression vector, genomic DNA was digested with KpnI/NheI (NheI cut within the gpdA promoter of the expression vector) and analyzed by Southern blot using a gls2α-specific probe. Two bands at more than 5,000 bp and approximately 3,400 bp were visualized for the g14 transformant, and the latter resulted from random integration of the alpha subunit expression cassette into the Rut-C30 genome. For all other transformants, which exhibited no change in the N-glycan profile, only the fragment of more than 5,000 bp was identified. This corresponded to the band obtained for the untransformed Rut-C30 strain and as such could only have resulted from hybridization to the endogenous locus encoding the truncated GIIα (Fig. 6A).
FIG. 6.
(A) Genomic analysis by Southern hybridization to check integration of the T. reesei gls2α expression plasmid. Lanes 1 and 2 contained hygromycin-resistant clones that showed a typical Rut-C30 N-glycan profile; lane 3 shows the results for transformant g14; and lane 4 contained the untransformed Rut-C30 strain. Lane M contained a PstI-digested λ DNA reference marker. (B) SDS-PAGE analysis of proteins present in 1 ml growth medium. Lanes 1 and 2, two hygromycin-resistant transformants having a typical Rut-C30 N-glycan profile; lane 3, transformant g14; lane 4, untransformed Rut-C30 strain; lane M, protein reference marker.
Finally, the effect of gls2α overexpression on the secretion level of autologous proteins was analyzed. All strains grew equally well, based on the dry weight of the isolated mycelium. Hence, proteins were precipitated from an equal volume of growth medium and separated by SDS-PAGE. Coomassie blue staining indicated that the g14 transformant, expressing the repaired variant of the Trichoderma gls2α ORF, had a reduced capacity to secrete proteins into the medium (Fig. 6B). Repetitive experiments confirmed this result.
T. reesei produces Glc3Man9GlcNAc2 N-glycan precursor structures.
Most eukaryotic organisms produce a lipid-linked tetradecasaccharide Glc3Man9GlcNAc2 precursor structure that is transferred within the lumen of the ER onto suitable asparagine residues of a nascent polypeptide chain. After transfer to the protein, the terminal α-1,2-linked glucose is rapidly removed by glucosidase I, while the two innermost α-1,3-linked glucose residues are cleaved off via the action of glucosidase II (40). However, some exceptions occur in nature, as some protozoans do not add glucose structures to the N-glycan precursor (58, 59).
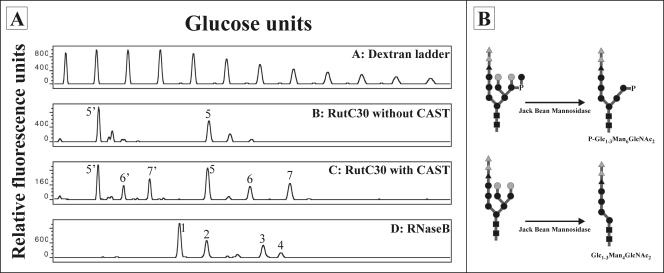
To check whether a triglucosylated precursor structure is initially transferred to the T. reesei N-glycans, the fungus was grown for 2 days in the presence of an α-glucosidase inhibitor, castanospermine. N-glycans from the secreted proteins were isolated from 1 ml of the growth medium and analyzed by DSA-FACE (12). To allow easier interpretation of the glycan profile obtained, carbohydrates were treated with jack bean mannosidase. This mannosidase removes all alpha-linked mannose residues that are not capped by other components (such as terminal glucose residues or phosphates), including the terminal mannose residue in a phosphodiester linkage. The profiles of the jack bean mannosidase-treated N-glycans, obtained from the Rut-C30 strain grown with and without castanospermine, are shown in Fig. 7A. A schematic representation of the expected N-glycans and the corresponding products after jack bean mannosidase digestion is shown in Fig. 7B. Treatment of T. reesei Rut-C30 with an α-glucosidase inhibitor clearly resulted in the appearance of carbohydrates with one or two extra monosaccharide residues.
FIG. 7.
(A) DSA-FACE profiles of the Rut-C30 strain grown with (lane C) and without (lane B) castanospermine (CAST) after in vitro treatment of the isolated N-glycans with jack bean mannosidase. The deduced N-glycans are indicated as follows: 1, Man5GlcNAc2; 2, Man6GlcNAc2; 3, Man8GlcNAc2; 4, Man9GlcNAc2; 5, GlcMan4GlcNAc2; 6, Glc2Man4GlcNAc2; 7, Glc3Man4GlcNAc2; 5′, P-GlcMan6GlcNAc2; 6′, P-Glc2Man6GlcNAc2; 7′, P-Glc3Man6GlcNAc2. RNaseB, standard high-mannose N-glycan mixture derived from RNase B. (B) Schematic representation of the expected N-glycan structures before and after in vitro treatment. ▪, GlcNAc; •, mannose; ▴, glucose; P, phosphate. The grey dots represent α-1,2-mannose residues that are potentially removed from the N-glycans due to intra- and/or extracellular Trichoderma α-1,2-mannosidase activity. The grey triangles represent glucose residues that are potentially still present after T. reesei is grown in the presence of castanospermine.
DISCUSSION
The T. reesei Rut-C30 strain, which secretes large amounts of cellulolytic enzymes, synthesizes unusual N-glycan structures on its secreted proteins. Thorough analyses have indicated that most of the oligosaccharides still carry an α-1,3-glucose residue (17, 45, 79). This capping structure, which prevents further trimming of the glycans by α-1,2-mannosidase, is probably the result of an inefficient glucosidase II activity. Several factors might be at the root of this problem. First, oligosaccharide analysis of cellobiohydrolase I was previously carried out after the fungus was grown in cellulose-inducing conditions. Since the Rut-C30 strain is known to hypersecrete this cellulase, inefficient glucose trimming could be due to the inability of the glucosidase II to handle the heavy load of glycoproteins passing through the secretion pathway. Second, analysis of the N-glycan pool revealed the presence of an α-1,2-mannosidase activity that results in the generation of GlcMan7GlcNAc2 (44, 45). This compound is known to be a poor substrate for glucosidase II (27). Hence, the glucosidase II activity could be partially inhibited by competition with the α-1,2-mannosidase for the same glucosylated Man9GlcNAc2 substrate. This would imply that there is little or no difference in the localization of the two enzymes. Since subcellular localization revealed the presence of some Golgi-like enzymes in the proliferated ER after the Rut-C30 strain was grown in cellulose-inducing conditions (26), this possibility was also worth considering. However, oligosaccharide analysis after the Trichoderma Rut-C30 strain was grown in cellulase-inducing and noninducing conditions revealed similar large amounts of monoglucosylated glycans (results not shown), ruling out these first two explanations. Finally, we considered the possibility that during the consecutive rounds of mutagenesis leading to this hypersecreting strain, one or more mutations might have occurred within the glucosidase II gene or its transcriptional elements.
In order to evaluate this hypothesis, we cloned the Rut-C30 gene encoding the catalytic glucosidase II alpha subunit, using a combination of rapid cDNA cloning by PCR screening (82), inverse PCR, and 5′-RACE. Analysis of the sequence data, however, identified a frameshift in the cloned Rut-C30 cDNA. Based on PCR amplification of the Rut-C30/NG14 and QM9414 gls2α genes, we concluded that a T was missing at position 1965 of the Rut-C30/NG14 alpha subunit sequence. The presence of the frameshift resulted in a premature translation stop and consequently a truncated protein lacking about one-third of the complete polypeptide chain. However, this premature stop codon is situated more than 450 nucleotides 3′ of the position of the frameshift, resulting in a peptide stretch of 152 extra nonspecific amino acids added to the first 654 glucosidase II-specific amino acids. This was rather unexpected, since statistically 5% of out-of-frame codons encode a translational stop. Indeed, in silico introduction of a similar frameshift mutation into the glucosidase II alpha subunit genes of other organisms resulted in elongation of the deduced truncated polypeptide product by only a few amino acids before an in-frame stop codon was encountered (results not shown). At this moment, we can only speculate on the effect, if any, of this additional peptide stretch of 152 amino acids on the truncated Rut-C30 GIIα protein.
Growth of T. reesei Rut-C30 in the presence of the α-glucosidase inhibitor castanospermine clearly indicated that Glc3Man9GlcNAc2 is initially transferred to nascent polypeptide chains. Hence, the activities of both glucosidase I and glucosidase II are necessary for hydrolysis of the terminal triglucose cap. Translation of the mutant locus of the Rut-C30 glucosidase II alpha subunit resulted in a protein that still contained the conserved WXDMNE motif typical of the catalytic site of the family 31 glycosyl hydrolases (29). A few years ago, a kinetic model was proposed for the glucosidase II alpha subunit (1, 3). In this model, the protein consists of a catalytic site and two substrate binding sites, one recognizing the Glc-α1,3-Glc and the other one recognizing the Glc-α1,3-Man linkage of the ER precursor oligosaccharide. The Rut-C30 glycan profile clearly indicates that the first α-1,3-linked glucose residue is removed by glucosidase II. However, since most of the glycans still contain the innermost glucose, we believe that the truncated GIIα subunit still contains a functional Glc-α1,3-Glc substrate binding site and catalytic center but no Glc-α1,3-Man binding site. Hence, we propose that the latter is encoded in the 3′ part of the alpha subunit gene and is either severely damaged or absent in the truncated form of the Rut-C30 GIIα protein. To our knowledge, this is the first report speculating on the position of the substrate binding sites in the primary sequence of a glucosidase II alpha subunit. The fact that the first α-1,3-glucose residue is removed from the N-glycans indicates that the truncated protein is not unstable and is probably still localized within the ER. This also suggests that the extra 152 nonspecific amino acids do not have a big influence on the protein's stability (but they potentially have a stabilizing effect on the truncated protein product) and do not significantly interfere with its subcellular localization.
The nonmutated T. reesei gls2α gene encodes a protein consisting of 964 amino acids, and it exhibits the highest level of homology with the N. crassa homologue. Interestingly, Trichoderma GIIα showed more sequence homology to the alpha subunits of higher eukaryotic organisms than to the glucosidase II of S. cerevisiae. Similar to its mammalian (6, 88) and S. pombe (15) counterparts, the T. reesei GIIα protein is probably retained within the ER through interaction with a GIIβ subunit, as no ER retention signals or transmembrane regions could be predicted from its amino acid sequence.
Analysis of the 5′ noncoding sequence revealed the presence of several putative stress elements, including possible UPR elements, indicating that the level of gls2α expression can vary as a result of different growth and/or physiological conditions. The sensitivity of the gls2α transcriptional regulation to accumulation of unfolded proteins was investigated further. For this, the gls2α mRNA levels were analyzed in a Rut-C30 strain either producing a heterologous protein (tPA) or cultivated in the presence of DTT or BFA, two compounds known to interfere with protein folding and secretion. As expected for a UPR-sensitive gene, the transcript levels of the glucosidase II alpha subunit were significantly increased upon tPA secretion or BFA treatment. In both cases, the induction was similar to that of pdi1 but not as strong as that of bip1 (55; Uusitalo, Pakula, and Penttila, unpublished). However, DTT treatment did not result in transcriptional upregulation; rather, the mRNA levels remained stable or even decreased after addition of the drug (data not shown). A similar effect has been observed in the Rut-C30 strain for two other secretion-related Trichoderma genes, sar1 and ypt1, since a significant increase in transcription was observed for these genes after the fungus was grown in the presence of BFA but not after it was grown in the presence of DTT (75). Although both drugs efficiently block protein transport, their different modes of action could perhaps explain the Northern data obtained for sar1 and ypt1, as well as gls2α. It has been shown that in higher eukaryotic cell lines accumulation of unfolded proteins by BFA treatment also induces NF-kappa B DNA binding and kappa B-dependent gene expression, but this is not the case during DTT treatment (54). Hence, alternative regulatory systems in T. reesei could perhaps also account for the higher sensitivity of several fungal genes to BFA-related secretion stress than to DTT-induced secretion stress (75). Furthermore, it is quite possible that the expression of several secretion-related genes increases only when a certain threshold value of accumulated proteins is exceeded. Indeed, in vivo labeling studies with T. reesei have shown that the intracellular accumulation of proteins is significantly higher after BFA treatment than after DTT treatment (55).
A plasmid for constitutive expression of a repaired variant of the gls2α gene was constructed to evaluate the effect of its overexpression on the glycan profile of T. reesei Rut-C30. Since this profile is dominated by monoglucosylated oligosaccharides, we expected a significant change after successful gls2α gene expression. Indeed, we isolated one transformant that had a clearly different N-glycan composition. The expression of the full-size GIIα subunit did not result in complete hydrolysis of the innermost α-1,3-linked glucose residue; rather, the oligosaccharide profile proved to be a mixture of the QM9414 and untransformed Rut-C30 glycan patterns and thus consisted of a wide variety of charged and neutral high-mannose and monoglucosylated carbohydrates. This moderate efficiency in glucose hydrolysis could indicate the presence of one or several extra mutations within the Rut-C30 coding sequence, apart from the repaired frameshift. However, comparison of the gls2α genes of the Rut-C30 and QM9414 strains did not reveal any other sequence differences (results not shown). Possibly, the repaired functional and endogenous mutant versions of the GIIα protein, which are expressed from different promoter sequences, compete with each other in one or more ways. On the one hand, they could compete for the same substrate. This would indicate that the truncated glucosidase II protein is somehow still able to interact with or shield the Glc-α1,3-Man substrate of the glycan core without cleaving and/or releasing it. Another and perhaps more plausible explanation is that both forms of the alpha subunit compete with each other for interaction with the beta subunit. Thus, not all full-size glucosidase II alpha subunits are retained within the ER. Hence, the lack of a normal level of fully functional glucosidase II heterodimers could very well explain the incomplete trimming of the Glc-α1,3-Man substrate. Work is in progress to obtain complete conversion of the monoglucosylated oligosaccharides into glucose-free high-mannose N-glycans by actually replacing the mutant Rut-C30 gls2α gene with its repaired variant.
A striking feature is that even partial removal of the innermost glucose residues in the g14 transformant results in a very heterogeneous glycan pattern compared to that of the untransformed Rut-C30 strain. Especially on the left side of the Man5GlcNAc2 peak, a lot of new peaks emerge in the profile, which is indicative of fast-migrating oligosaccharides. Since most of these oligosaccharides are susceptible to mild acid hydrolysis, we believe that these peaks represent structural diversity of charged high-mannose N-glycans carrying one or more phosphodiester linkages. In the untransformed Rut-C30 strain, addition of a phosphodiester linkage is restricted to the α-1,6 arm (position C-1) of the N-glycan (17). Analysis of the high-mannose N-glycans of S. cerevisiae indicated that position A-3 of the α-1,3 arm could also serve as an acceptor site for a phosphodiester linkage (31). However, in the slime mold D. discoideum, the formation of a phosphodiester linkage at position A-3 was not possible in a glucosidase II (modA) mutant (21, 22), indicating that terminal glucose residues selectively blocked the occurrence of this glycan modification in the α-1,3 arm. Hence, steric hindrance as a result of a terminal α-1,3-linked glucose residue on the T. reesei Rut-C30 carbohydrates could also account for the lack of N-glycan phosphorylation at position A-3. As a result of the partial hydrolysis of this terminal glucose residue in transformant g14, the underlying α-1,2-mannobiose becomes accessible as a substrate for the mannosephosphate transferase (37, 93), as well as endogenous α-1,2-mannosidases. Competition between the two glycan processing activities for the high-mannose structures obtained can result in trimming of one to four α-1,2-mannose residues and/or addition of one or two phosphodiester linkages. This would explain the great heterogeneity of neutral and charged oligomannose structures synthesized on the secreted proteins of transformant g14 compared to those of the untransformed Rut-C30 strain.
In a final experiment, the secretion capacity of the g14 transformant was compared to that of the Rut-C30 strain and some hygromycin resistant transformants with an unchanged glycosylation profile. Southern analysis indicated that the latter had not integrated the gls2α expression construct. Although no differences in growth capacity were observed between the strains analyzed, transformant g14 continuously seemed to secrete less protein into the medium. Together with the genomic and N-glycan data for T. reesei Rut-C30 and Rut-NG14, the hypersecreting parent strain of Rut-C30 (49), these results suggest a possible link between the secretion capacity of T. reesei on the one hand and the glucosidase II mutation on the other hand. Experiments are in progress to further evaluate this putative link.
To conclude, we cloned a mutant gene coding for the Rut-C30 glucosidase II alpha subunit. After the cre1 mutation (34), this was the second mutation identified in the genome of the hypercellulolytic strain. To our knowledge, this represents the first report of cloning of a glycosyl hydrolase involved in the processing of protein-linked N-glycans in the ER of filamentous fungi. Moreover, our results provide genetic evidence for the kinetic model proposed by Alonso and coworkers for the substrate binding specificity of the glucosidase II alpha subunit.
Acknowledgments
This work was funded by an EC grant to the Eurofung project (contract no. QLK3-CT1999-00729) and by GOA grant 12051099 from Ghent University. S.G. held an IWT fellowship.
We thank Annelies Van Hecke for DSA-FACE and Rudy Cocquyt for DNA sequence analysis. Fruitful discussions were held with Ingeborg Stals, Wouter Vervecken, and Wouter Laroy. We thank Amin Bredan for critical reading of the manuscript.
REFERENCES
- 1.Alonso, J. M., A. Santa-Cecilia, and P. Calvo. 1993. Effect of bromoconduritol on glucosidase II from rat liver. A new kinetic model for the binding and hydrolysis of the substrate. Eur. J. Biochem. 215:37-42. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, J. M., A. Santa-Cecilia, and P. Calvo. 1991. Glucosidase II from rat liver microsomes. Kinetic model for binding and hydrolysis. Biochem. J. 278:721-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, J. M., A. Santa-Cecilia, M. A. Chinchetru, and P. Calvo. 1993. Characterization of the maltase activity of glucosidase II from rat liver. Kinetic model. Biol. Chem. Hoppe-Seyler 374:977-982. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Appel, R. D., A. Bairoch, and D. F. Hochstrasser. 1994. A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem. Sci. 19:258-260. [DOI] [PubMed] [Google Scholar]
- 6.Arendt, C. W., and H. L. Ostergaard. 1997. Identification of the CD45-associated 116-kDa and 80-kDa proteins as the alpha- and beta-subunits of alpha-glucosidase II. J. Biol. Chem. 272:13117-13125. [DOI] [PubMed] [Google Scholar]
- 7.Brada, D., and U. C. Dubach. 1984. Isolation of a homogeneous glucosidase II from pig kidney microsomes. Eur. J. Biochem. 141:149-156. [DOI] [PubMed] [Google Scholar]
- 8.Brada, D., D. Kerjaschki, and J. Roth. 1990. Cell type-specific post-Golgi apparatus localization of a “resident” endoplasmic reticulum glycoprotein, glucosidase II. J. Cell Biol. 110:309-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brakhage, A. A., A. Andrianopoulos, M. Kato, S. Steidl, M. A. Davis, N. Tsukagoshi, and M. J. Hynes. 1999. HAP-Like CCAAT-binding complexes in filamentous fungi: implications for biotechnology. Fungal Genet. Biol. 27:243-252. [DOI] [PubMed] [Google Scholar]
- 10.Buchert, J., T. Oksanen, J. Pere, M. Sikka-Aho, A. Suurnakki, and L. Viikari. 1998. Application of Trichoderma reesei enzymes in the pulp and paper industry, p. 343-363. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma & Gliocladium, vol. 2. Taylor & Francis, London, United Kingdom. [Google Scholar]
- 11.Burns, D. M., and O. Touster. 1982. Purification and characterization of glucosidase II, an endoplasmic reticulum hydrolase involved in glycoprotein biosynthesis. J. Biol. Chem. 257:9990-10000. [PubMed] [Google Scholar]
- 12.Callewaert, N., S. Geysens, F. Molemans, and R. Contreras. 2001. Ultrasensitive profiling and sequencing of N-linked oligosaccharides using standard DNA-sequencing equipment. Glycobiology 11:275-281. [DOI] [PubMed] [Google Scholar]
- 13.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 14.Contreras, R., D. Carrez, J. R. Kinghorn, C. A. van den Hondel, and W. Fiers. 1991. Efficient KEX2-like processing of a glucoamylase-interleukin-6 fusion protein by Aspergillus nidulans and secretion of mature interleukin-6. Bio/Technology 9:378-381. [DOI] [PubMed] [Google Scholar]
- 15.D'Alessio, C., F. Fernandez, E. S. Trombetta, and A. J. Parodi. 1999. Genetic evidence for the heterodimeric structure of glucosidase II. The effect of disrupting the subunit-encoding genes on glycoprotein folding. J. Biol. Chem. 274:25899-25905. [DOI] [PubMed] [Google Scholar]
- 16.Datema, R., P. A. Romero, G. Legler, and R. T. Schwarz. 1982. Inhibition of formation of complex oligosaccharides by the glucosidase inhibitor bromoconduritol. Proc. Natl. Acad. Sci. USA 79:6787-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Bruyn, A., M. Maras, J. Schraml, P. Herdewijn, and R. Contreras. 1997. NMR evidence for a novel asparagine-linked oligosaccharide on cellobiohydrolase I from Trichoderma reesei RUTC 30. FEBS Lett. 405:111-113. [DOI] [PubMed] [Google Scholar]
- 18.Derkx, P. M., and S. M. Madrid. 2001. The foldase CYPB is a component of the secretory pathway of Aspergillus niger and contains the endoplasmic reticulum retention signal HEEL. Mol. Genet. Genomics 266:537-545. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez, F. S., S. E. Trombetta, U. Hellman, and A. J. Parodi. 1994. Purification to homogeneity of UDP-glucose:glycoprotein glucosyltransferase from Schizosaccharomyces pombe and apparent absence of the enzyme from Saccharomyces cerevisiae. J. Biol. Chem. 269:30701-30706. [PubMed] [Google Scholar]
- 20.Flura, T., D. Brada, M. Ziak, and J. Roth. 1997. Expression of a cDNA encoding the glucose trimming enzyme glucosidase II in CHO cells and molecular characterization of the enzyme deficiency in a mutant mouse lymphoma cell line. Glycobiology 7:617-624. [DOI] [PubMed] [Google Scholar]
- 21.Freeze, H. H. 1985. Interaction of Dictyostelium discoideum lysosomal enzymes with the mammalian phosphomannosyl receptor. The importance of oligosaccharides which contain phosphodiesters. J. Biol. Chem. 260:8857-8864. [PubMed] [Google Scholar]
- 22.Freeze, H. H., M. Lammertz, N. Iranfar, D. Fuller, K. Panneerselvam, and W. F. Loomis. 1997. Consequences of disrupting the gene that encodes alpha-glucosidase II in the N-linked oligosaccharide biosynthesis pathway of Dictyostelium discoideum. Dev. Genet. 21:177-186. [DOI] [PubMed] [Google Scholar]
- 23.Galante, Y. M., A. De Conti, and R. Monteverdi. 1998. Application of Trichoderma enzymes in the food and feed industries, p. 327-342. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma & Gliocladium, vol. 2. Taylor & Francis, London, United Kingdom. [Google Scholar]
- 24.Galante, Y. M., A. De Conti, and R. Monteverdi. 1998. Application of Trichoderma enzymes in the textile industry, p. 311-325. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma & Gliocladium, vol. 2. Taylor & Francis, London, United Kingdom. [Google Scholar]
- 25.Garcia, R., J. A. Cremata, O. Quintero, R. Montesino, K. Benkestock, and J. Stahlberg. 2001. Characterization of protein glycoforms with N-linked neutral and phosphorylated oligosaccharides: studies on the glycosylation of endoglucanase 1 (Cel7B) from Trichoderma reesei. Biotechnol. Appl. Biochem. 33:141-152. [DOI] [PubMed] [Google Scholar]
- 26.Glenn, M., A. Ghosh, and B. K. Ghosh. 1985. Subcellular fractionation of a hypercellulolytic mutant, Trichoderma reesei Rut-C30: localization of endoglucanase in microsomal fraction. Appl. Environ. Microbiol. 50:1137-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grinna, L. S., and P. W. Robbins. 1980. Substrate specificities of rat liver microsomal glucosidases which process glycoproteins. J. Biol. Chem. 255:2255-2258. [PubMed] [Google Scholar]
- 28.Helenius, A., E. S. Trombetta, D. N. Hebert, and J. F. Simons. 1997. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 7:193-200. [DOI] [PubMed] [Google Scholar]
- 29.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hentges, A., and E. Bause. 1997. Affinity purification and characterization of glucosidase II from pig liver. Biol. Chem. 378:1031-1038. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez, L. M., I. Olivero, E. Alvarado, and G. Larriba. 1992. Oligosaccharide structures of the major exoglucanase secreted by Saccharomyces cerevisiae. Biochemistry 31:9823-9831. [DOI] [PubMed] [Google Scholar]
- 32.Hijarrubia, M. J., J. Casqueiro, S. Gutierrez, F. J. Fernandez, and J. F. Martin. 1997. Characterization of the bip gene of Aspergillus awamori encoding a protein with an HDEL retention signal homologous to the mammalian BiP involved in polypeptide secretion. Curr. Genet. 32:139-146. [DOI] [PubMed] [Google Scholar]
- 33.Hiraoka, Y., T. Toda, and M. Yanagida. 1984. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell 39:349-358. [DOI] [PubMed] [Google Scholar]
- 34.Ilmen, M., C. Thrane, and M. Penttila. 1996. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol. Gen. Genet. 251:451-460. [DOI] [PubMed] [Google Scholar]
- 35.Jeenes, D. J., R. Pfaller, and D. B. Archer. 1997. Isolation and characterisation of a novel stress-inducible PDI-family gene from Aspergillus niger. Gene 193:151-156. [DOI] [PubMed] [Google Scholar]
- 36.Kalz-Fuller, B., E. Bieberich, and E. Bause. 1995. Cloning and expression of glucosidase I from human hippocampus. Eur. J. Biochem. 231:344-351. [DOI] [PubMed] [Google Scholar]
- 37.Karson, E. M., and C. E. Ballou. 1978. Biosynthesis of yeast mannan. Properties of a mannosylphosphate transferase in Saccharomyces cerevisiae. J. Biol. Chem. 253:6484-6492. [PubMed] [Google Scholar]
- 38.Kaushal, G. P., I. Pastuszak, K. Hatanaka, and A. D. Elbein. 1990. Purification to homogeneity and properties of glucosidase II from mung bean seedlings and suspension-cultured soybean cells. J. Biol. Chem. 265:16271-16279. [PubMed] [Google Scholar]
- 39.Kaushal, G. P., Y. Zeng, and A. D. Elbein. 1993. Biosynthesis of glucosidase II in suspension-cultured soybean cells. J. Biol. Chem. 268:14536-14542. [PubMed] [Google Scholar]
- 40.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-664. [DOI] [PubMed] [Google Scholar]
- 41.Kruszewska, J. S. 1999. Heterologous expression of genes in filamentous fungi. Acta Biochim. Pol. 46:181-195. [PubMed] [Google Scholar]
- 42.Lucocq, J. M., D. Brada, and J. Roth. 1986. Immunolocalization of the oligosaccharide trimming enzyme glucosidase II. J. Cell Biol. 102:2137-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma, Y., and L. M. Hendershot. 2001. The unfolding tale of the unfolded protein response. Cell 107:827-830. [DOI] [PubMed] [Google Scholar]
- 44.Maras, M., N. Callewaert, K. Piens, M. Claeyssens, W. Martinet, S. Dewaele, H. Contreras, I. Dewerte, M. Penttila, and R. Contreras. 2000. Molecular cloning and enzymatic characterization of a Trichoderma reesei 1,2-alpha-d-mannosidase. J. Biotechnol. 77:255-263. [DOI] [PubMed] [Google Scholar]
- 45.Maras, M., A. De Bruyn, J. Schraml, P. Herdewijn, M. Claeyssens, W. Fiers, and R. Contreras. 1997. Structural characterization of N-linked oligosaccharides from cellobiohydrolase I secreted by the filamentous fungus Trichoderma reesei RUTC 30. Eur. J. Biochem. 245:617-625. [DOI] [PubMed] [Google Scholar]
- 46.Marchler, G., C. Schuller, G. Adam, and H. Ruis. 1993. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 12:1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Blanco, H., M. Orejas, A. Reglero, J. M. Luengo, and M. A. Penalva. 1993. Characterisation of the gene encoding acetyl-CoA synthetase in Penicillium chrysogenum: conservation of intron position in plectomycetes. Gene 130:265-270. [DOI] [PubMed] [Google Scholar]
- 48.May, G. S., and N. R. Morris. 1987. The unique histone H2A gene of Aspergillus nidulans contains three introns. Gene 58:59-66. [DOI] [PubMed] [Google Scholar]
- 49.Montenecourt, B. S., and D. E. Eveleigh. 1979. Selective screening methods for the isolation of high yielding cellulase mutants of Trichoderma reesei. Adv. Chem. Ser. 181:289-301. [Google Scholar]
- 50.Mori, K., A. Sant, K. Kohno, K. Normington, M. J. Gething, and J. F. Sambrook. 1992. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 11:2583-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mount, S. M. 1982. A catalogue of splice junction sequences. Nucleic Acids Res. 10:459-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ngiam, C., D. J. Jeenes, P. J. Punt, C. A. Van Den Hondel, and D. B. Archer. 2000. Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl. Environ. Microbiol. 66:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orbach, M. J., E. B. Porro, and C. Yanofsky. 1986. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol. Cell. Biol. 6:2452-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pahl, H. L., and P. A. Baeuerle. 1995. A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-kappa B. EMBO J. 14:2580-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pakula, T. M., M. Laxell, A. Huuskonen, J. Uusitalo, M. Saloheimo, and M. Penttila. 2003. The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei. Evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J. Biol. Chem. 278:45011-45020. [DOI] [PubMed] [Google Scholar]
- 56.Papac, D. I., J. B. Briggs, E. T. Chin, and A. J. Jones. 1998. A high-throughput microscale method to release N-linked oligosaccharides from glycoproteins for matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis. Glycobiology 8:445-454. [DOI] [PubMed] [Google Scholar]
- 57.Parker, C. G., L. I. Fessler, R. E. Nelson, and J. H. Fessler. 1995. Drosophila UDP-glucose:glycoprotein glucosyltransferase: sequence and characterization of an enzyme that distinguishes between denatured and native proteins. EMBO J. 14:1294-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parodi, A. J. 1993. N-glycosylation in trypanosomatid protozoa. Glycobiology 3:193-199. [DOI] [PubMed] [Google Scholar]
- 59.Parodi, A. J. 1993. Protein N-glycosylation in trypanosomatids. A pathway with odd enzymes at both ends. Biol. Res. 26:69-75. [PubMed] [Google Scholar]
- 60.Parodi, A. J. 2000. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem. J. 348:1-13. [PMC free article] [PubMed] [Google Scholar]
- 61.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349-355. [DOI] [PubMed] [Google Scholar]
- 62.Pearson, W. R., T. Wood, Z. Zhang, and W. Miller. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24-36. [DOI] [PubMed] [Google Scholar]
- 63.Pelham, H. R., and M. Bienz. 1982. A synthetic heat-shock promoter element confers heat-inducibility on the herpes simplex virus thymidine kinase gene. EMBO J. 1:1473-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pelham, H. R., K. G. Hardwick, and M. J. Lewis. 1988. Sorting of soluble ER proteins in yeast. EMBO J. 7:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pelletier, M. F., A. Marcil, G. Sevigny, C. A. Jakob, D. C. Tessier, E. Chevet, R. Menard, J. J. Bergeron, and D. Y. Thomas. 2000. The heterodimeric structure of glucosidase II is required for its activity, solubility, and localization in vivo. Glycobiology 10:815-827. [DOI] [PubMed] [Google Scholar]
- 66.Penttila, M. 1998. Heterologous protein production in Trichoderma, p. 365-379. In G. Harman and C. Kubicek (ed.), Trichoderma & Gliocladium, vol. 2. Taylor & Francis, London, United Kingdom. [Google Scholar]
- 67.Penttila, M., H. Nevalainen, M. Ratto, E. Salminen, and J. Knowles. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155-164. [DOI] [PubMed] [Google Scholar]
- 68.Petrescu, A. J., T. D. Butters, G. Reinkensmeier, S. Petrescu, F. M. Platt, R. A. Dwek, and M. R. Wormald. 1997. The solution NMR structure of glucosylated N-glycans involved in the early stages of glycoprotein biosynthesis and folding. EMBO J. 16:4302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 70.Punt, P. J., I. A. van Gemeren, J. Drint-Kuijvenhoven, J. G. Hessing, G. M. van Muijlwijk-Harteveld, A. Beijersbergen, C. T. Verrips, and C. A. van den Hondel. 1998. Analysis of the role of the gene bipA, encoding the major endoplasmic reticulum chaperone protein in the secretion of homologous and heterologous proteins in black aspergilli. Appl. Microbiol. Biotechnol. 50:447-454. [DOI] [PubMed] [Google Scholar]
- 71.Roy, B., and A. S. Lee. 1995. Transduction of calcium stress through interaction of the human transcription factor CBF with the proximal CCAAT regulatory element of the grp78/BiP promoter. Mol. Cell. Biol. 15:2263-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saloheimo, A., B. Henrissat, A. M. Hoffren, O. Teleman, and M. Penttila. 1994. A novel, small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Mol. Microbiol. 13:219-228. [DOI] [PubMed] [Google Scholar]
- 73.Saloheimo, M., M. Lund, and M. E. Penttila. 1999. The protein disulphide isomerase gene of the fungus Trichoderma reesei is induced by endoplasmic reticulum stress and regulated by the carbon source. Mol. Gen. Genet. 262:35-45. [DOI] [PubMed] [Google Scholar]
- 74.Saloheimo, M., M. Valkonen, and M. Penttila. 2003. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 47:1149-1161. [DOI] [PubMed] [Google Scholar]
- 75.Saloheimo, M., H. Wang, M. Valkonen, T. Vasara, A. Huuskonen, M. Riikonen, T. Pakula, M. Ward, and M. Penttila. 2004. Characterization of secretory genes ypt1/yptA and nsf1/nsfA from two filamentous fungi: induction of secretory pathway genes of Trichoderma reesei under secretion stress conditions. Appl. Environ. Microbiol. 70:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 77.Saunier, B., R. D. Kilker, Jr., J. S. Tkacz, A. Quaroni, and A. Herscovics. 1982. Inhibition of N-linked complex oligosaccharide formation by 1-deoxynojirimycin, an inhibitor of processing glucosidases. J. Biol. Chem. 257:14155-14161. [PubMed] [Google Scholar]
- 78.Saxena, S., K. Shailubhai, B. Dong-Yu, and I. K. Vijay. 1987. Purification and characterization of glucosidase II involved in N-linked glycoprotein processing in bovine mammary gland. Biochem. J. 247:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stals, I., K. Sandra, S. Geysens, R. Contreras, J. Van Beeumen, and M. Claeyssens. 2004. Factors influencing glycosylation of Trichoderma reesei cellulases. Part one. Post-secretorial changes of the O- and N-glycosylation pattern of Cel7A. Glycobiology 14:1-12. [DOI] [PubMed] [Google Scholar]
- 80.Strous, G. J., P. Van Kerkhof, R. Brok, J. Roth, and D. Brada. 1987. Glucosidase II, a protein of the endoplasmic reticulum with high mannose oligosaccharide chains and a rapid turnover. J. Biol. Chem. 262:3620-3625. [PubMed] [Google Scholar]
- 81.Takaya, N., K. Yanai, H. Horiuchi, A. Ohta, and M. Takagi. 1994. Cloning and characterization of two 3-phosphoglycerate kinase genes of Rhizopus niveus and heterologous gene expression using their promoters. Curr. Genet. 25:524-530. [DOI] [PubMed] [Google Scholar]
- 82.Takumi, T., and H. F. Lodish. 1994. Rapid cDNA cloning by PCR screening. BioTechniques 17:443-444. [PubMed] [Google Scholar]
- 83.Taylor, S. C., A. D. Ferguson, J. J. Bergeron, and D. Y. Thomas. 2004. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat. Struct. Mol. Biol. 11:128-134. [DOI] [PubMed] [Google Scholar]
- 84.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Treml, K., D. Meimaroglou, A. Hentges, and E. Bause. 2000. The alpha- and beta-subunits are required for expression of catalytic activity in the hetero-dimeric glucosidase II complex from human liver. Glycobiology 10:493-502. [DOI] [PubMed] [Google Scholar]
- 86.Trombetta, E. S., K. G. Fleming, and A. Helenius. 2001. Quaternary and domain structure of glycoprotein processing glucosidase II. Biochemistry 40:10717-10722. [DOI] [PubMed] [Google Scholar]
- 87.Trombetta, E. S., and A. Helenius. 2000. Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J. Cell Biol. 148:1123-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trombetta, E. S., J. F. Simons, and A. Helenius. 1996. Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J. Biol. Chem. 271:27509-27516. [DOI] [PubMed] [Google Scholar]
- 89.Trombetta, S. E., and A. J. Parodi. 1992. Purification to apparent homogeneity and partial characterization of rat liver UDP-glucose:glycoprotein glucosyltransferase. J. Biol. Chem. 267:9236-9240. [PubMed] [Google Scholar]
- 90.Ugalde, R. A., R. J. Staneloni, and L. F. Leloir. 1980. Microsomal glucosidases of rat liver. Partial purification and inhibition by disaccharides. Eur. J. Biochem. 113:97-103. [DOI] [PubMed] [Google Scholar]
- 91.Wang, H., J. Entwistle, E. Morlon, D. B. Archer, J. F. Peberdy, M. Ward, and D. J. Jeenes. 2003. Isolation and characterisation of a calnexin homologue, clxA, from Aspergillus niger. Mol. Genet. Genomics 268:684-691. [DOI] [PubMed] [Google Scholar]
- 92.Wang, H., and M. Ward. 2000. Molecular characterization of a PDI-related gene prpA in Aspergillus niger var. awamori. Curr. Genet. 37:57-64. [DOI] [PubMed] [Google Scholar]
- 93.Wang, X. H., K. Nakayama, Y. Shimma, A. Tanaka, and Y. Jigami. 1997. MNN6, a member of the KRE2/MNT1 family, is the gene for mannosylphosphate transfer in Saccharomyces cerevisiae. J. Biol. Chem. 272:18117-18124. [DOI] [PubMed] [Google Scholar]