Abstract
Xylanase B1 (XlnB1) from Streptomyces lividans is a protein consisting of two discrete structural and functional units, an N-terminal catalytic domain and a C-terminal substrate binding domain. In the culture medium, two forms of xylanase B are present, namely, XlnB1 and XlnB2, the latter of which corresponds to the catalytic domain of XlnB1 deprived of the substrate binding domain. Both forms of the xylanase have the same activity on xylan. The enzyme is secreted through the Sec-dependent pathway with a better yield of XlnB1 than XlnB2. Interestingly, XlnB2 exhibits 80% identity with XlnC which is secreted exclusively through the Tat-dependent pathway. To demonstrate whether XlnB1 and XlnB2 could also be secreted through the Tat-dependent pathway, the Tat-targeting xlnC signal sequence was fused to the structural genes of xlnB1 and xlnB2. Both XlnB1 and XlnB2 were secreted through the Tat-dependent pathway, but XlnB2 was better produced than XlnB1. As XlnB1 and XlnB2 could be better secreted through the Sec- and Tat-dependent systems, respectively, a copy of the structural gene of xlnB1 fused to a Sec signal sequence and a copy of the structural gene of xlnB2 fused to a Tat signal sequence were inserted into the same plasmid under the control of the xlnA promoter. The transformant produced xylanase activity which corresponded approximately to the sum of activities of the individual strain producing xylanase by either the Sec- or Tat-dependent secretion system. This indicated that both secretion systems are functional and independent of each other in the recombinant strain. This is the first report on the efficient secretion of a protein using two different secretion systems at the same time. Assuming that the protein to be secreted could be properly folded prior to and after translocation via the Tat- and Sec-dependent pathways, respectively, the simultaneous use of the Sec- and Tat-dependent pathways provides an efficient means to increase the production of a given protein.
Streptomyces species are gram-positive soil-living bacteria known to secrete large quantities of proteins (10). Most of the secreted proteins are enzymes which are well suited to hydrolyze complex organic material, thus permitting the bacteria to survive in their environment. In the past, we reported the homologous cloning of three xylanase genes (30) and purified the corresponding enzymes XlnA, XlnB, and XlnC from Streptomyces lividans (16, 17, 19). Xylanases degrade xylan, one of the major components of hemicellulose, which is a β-1,4-linked polymer of xylose substituted with uronic acid, hexoses, and side chains of other pentoses. These enzymes have attracted considerable attention for their potential use as bleaching agents in pulp and paper, biomass energy, and chemical industries (14, 34).
The vast majority of bacterial proteins are exported across the cytoplasmic membrane by the well-known Sec system, which acts on unfolded polypeptide chains (7). On the contrary, a novel alternate translocation system recently identified in bacteria and thylakoids, i.e., the Tat (twin-arginine translocation) pathway in bacteria or ΔpH pathway in thylakoids, functions to transport folded or even oligomerized preproteins (2, 6, 23, 24, 28, 29). The Tat-dependent proteins possess a signal peptide that preserves the tripartite structural organization of Sec signal peptides but contains an S/T-R-R-X-F-L-K consensus motif at the boundary of its n and h regions (2). In most cases, precursors bind redox cofactors, but some cofactorless precursors may also be transported by the Tat pathway, probably because they either require cytoplasmic factors for folding or fold too rapidly or too tightly to be transported by the Sec system (3).
XlnA and XlnB are exported by the Sec system (21), while XlnC is exported through the Tat-dependent secretion pathway (9). Since the catalytic domains of XlnB1 and XlnC exhibit 80% identity (30), we surmised that XlnB1 and XlnB2 could also be secreted efficiently through the Tat-dependent pathway. XlnA is not very well secreted through the Tat secretion system. Fusion of the xlnC signal sequence to the xlnA structural gene resulted in a yield that was 10% of that obtained with the wild-type signal sequence (21).
So far, to our knowledge, the simultaneous use of the Tat- and Sec-dependent secretion systems for protein export has not been investigated. Herein, we show for the first time that the Sec-dependent xylanase B can be efficiently translocated by the two secretion systems simultaneously. This represents an additional way, together with the existing methods, to increase the production of a given protein by replacements of signal peptides, promoters, codons, and additional ribosome-binding sites (4, 22, 31).
MATERIALS AND METHODS
Bacterial strains and media.
Streptomyces lividans 10-164 is a cellulase- and xylanase-negative mutant of S. lividans 1326 which was interrupted in the msiK gene required for generation of energy to disaccharide transport systems (13). This strain was used as the host for recombinant plasmids. Spores were prepared according to a method described previously by Hopwood et al. (11). The transformants were grown in M14 minimal medium containing xylose as the sole carbon source (21). Thiostrepton was added where required up to a final concentration of 50 μg/ml. Protoplasting and transformation of S. lividans were performed as described previously by Hopwood et al. (11).
Phagemid pTZ19U was from Pharmacia. The screening of transformants of Escherichia coli was performed on 2× YT broth plates containing ampicillin (100 μg/ml). DNA manipulations were performed according to methods described previously by Sambrook et al. (25).
Plasmid construction.
Plasmids used in this study are listed in Table 1. The sequence encoding XlnB1 and XlnB2 without its signal sequence was amplified using pIAF42 which contained xlnB as the template (17). To allow further cloning of the amplified fragments, the restriction sites were introduced at the 5′ and 3′ ends using the oligonucleotide XBfor (5′-CCGGTACCGACACGGTCGTCACGA-3′ [the KpnI site is underlined]) in combination with XB1rev (5′-CCGAGCTCTCAGCCCGCGCTGCA-3′ [the SacI site is underlined]) for the xlnB1 coding sequence and XB2rev (5′-CCCGAGCTCTCACCCGCCGACGTTGATGCT-3′ [the SacI site is underlined]) for the xlnB2 coding sequence. The fragments were amplified by PCRs under the following conditions: 5 min at 94°C followed by 30 cycles of 1 min at 94°C, 2 min at 50°C, and 3 min at 72°C. The reaction mixture was electrophoresed on a 1% agarose gel, and the amplified sequences were isolated with a QIAEX gel extraction kit according to the manufacturer's instructions (QIAGEN Inc., Chatsworth, CA). The 901-bp and 593-bp purified fragments containing the coding sequence of xlnB1 and xlnB2, respectively, were digested with KpnI-SacI and were then cloned at the same sites in pIAF815 and pIAF816 (21). The insertion of these xylanase sequences generated plasmids pIAF815B1 and pIAF815B2 (Sec signal sequence from xlnB1) and pIAF816B1 and pIAF816B2 (Tat signal sequence from xlnC). The expected sequences of the clones were confirmed by the dideoxy chain termination method (27). After digestion of the plasmids by HindIII-SacI, the resulting fragments harboring a signal sequence fused to a xylanase gene were cloned into pIAF906 (21) at the same restriction sites in replacement of the xlnA sequence. This generated plasmids pIAF915B1, pIAF915B2, pIAF916B1, and pIAF916B2. The transcription of xlnB1 and xlnB2 in these constructs was then under the control of the xlnA promoter. These plasmids were used to transform S. lividans 10-164 protoplasts.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. lividans | ||
| pIJ702 | Vector containing tsr and mel markers | 11 |
| pIAF42 | pIJ702 derivative containing xlnB1 from S. lividans | 17 |
| pIAF906 | pIJ702 derivative containing xlnA from S. lividans as a 2-kb SphI-SacI fragment where a HindIII and KpnI restriction site were added on each side of the signal sequence | 21 |
| pIAF907 | pIAF906 derivative in which the KpnI, PstI, SphI, and BglII sites were removed and the xlnA coding sequence was eliminated and replaced by a cloning cassette of HindIII, EcoRI, XbaI, and SacI | 21 |
| pIAF911A.8 | pIAF906 derivative in which the xlnA signal sequence was replaced by the celA signal sequence plus 57 nt upstream of it, containing a sequence complementary to the 16S rRNA and two translation initiation codons | 15 |
| pIAF916C | pIAF906 derivative containing the xlnC signal sequence and the xlnC coding sequence | 9 |
| pIAF916C-40 | pIAF916C derivative in which the xlnC signal sequence was deleted of 30 nt from its 5′ end | 9 |
| pIAF907-SB1 | pIAF911A.8 derivative in which the xlnA coding sequence was replaced by the xlnB1 coding sequence | This work |
| pIAF907-TB2 | pIAF916C-40 derivative in which the xlnC coding sequence was replaced by the xlnB2 coding sequence and the 57 nt upstream of the celA signal sequence was added in front of the truncated xlnC signalsequence | This work |
| pIAF907-SB1 TB2 | pIAF907-SB1 derivative in which a fragment containing the signal sequence and xlnB2 present in pIAF907-TB2 was inserted downstream of xlnB1 | This work |
| pIAF907-SB1 pATB2 | pIAF907-SB1 derivative in which the fragment containing the xlnA promoter, the signal sequence, and xlnB2 present in pIAF907-TB2 was inserted downstream of xlnB1 | This work |
| pIAF907-SB1pASB1 | pIAF907-SB1 derivative in which the fragment containing the xlnA promoter, the signal sequence, and xlnB1 present in pIAF907-SB1 was inserted downstream of xlnB1 | This work |
| pIAF907-TB2pATB2 | pIAF907-TB2 derivative in which the fragment containing the xlnA promoter, the signal sequence, and xlnB2 present in pIAF907-TB2 was inserted downstream of xlnB2 | This work |
| pIAF915 | pIAF906 derivative containing the xlnB1 signal sequence | 21 |
| pIAF915B1 | pIAF915 derivative in which the xlnA coding sequence was replaced by the xlnB1 coding sequence | This work |
| pIAF915B2 | pIAF915 derivative in which the xlnA coding sequence was replaced by the xlnB2 coding sequence | This work |
| pIAF916 | pIAF906 derivative containing the xlnC signal sequence | 21 |
| pIAF916B1 | pIAF916 derivative in which the xlnA coding sequence was replaced by the xlnB1 coding sequence | This work |
| pIAF916B2 | pIAF916 derivative in which the xlnA coding sequence was replaced by the xlnB2 coding sequence | This work |
| E. coli | ||
| pTZ19 | Phagemid carrying bla and part of lacZ | Pharmacia |
| pIAF807 | pTZ19 derivative containing promotorless xlnA with a HindIII and KpnI restriction site on each side of the signal sequence | 21 |
| pIAF811-A.8 | pIAF807 derivative in which the xlnA signal sequence was replaced by the celA signal sequence plus 57 nt upstream of it, containing a sequence complementary to the 16S rRNA and two initiation codons | 15 |
| pIAF815 | pIAF807 derivative in which the xlnA signal sequence was replaced by the xlnB1 signal sequence | 21 |
| pIAF815B1 | pIAF815 derivative in which the xlnA coding sequence was replaced by the xlnB1 coding sequence | This work |
| pIAF815B2 | pIAF815 derivative in which the xlnA coding sequence was replaced by the xlnB2 coding sequence | This work |
| pIAF816 | pIAF807 derivative in which the xlnA signal sequence was replaced by the xlnC signal sequence | 21 |
| pIAF816B1 | pIAF816 derivative in which the xlnA coding sequence was replaced by the xlnB1 coding sequence | This work |
| pIAF816B2 | pIAF816 derivative in which the xlnA coding sequence was replaced by the xlnB2 coding sequence | This work |
For the double translation initiation construct involving a Sec-dependent signal peptide, pIAF811-A.8, which contains the celA signal sequence plus the 57 nucleotides (nt) upstream of it (15), was used. The KpnI-SacI fragment of the xlnB1 coding sequence was cloned into pIAF811-A.8 at the same sites after elimination of the xlnA coding sequence, generating plasmid pIAF811-A.8-SB1. The plasmid was digested with HindIII-SacI, and the fragment was cloned into pIAF907 at the same sites, generating plasmid pIAF907-SB1.
For the double translation initiation construct involving a Tat-dependent signal peptide, the 57-nt sequence upstream of the celA signal sequence was cloned in front of the xlnC signal sequence which was deleted of 30 nt in the plasmid pIAF916C-40 (9). The deleted xlnC signal sequence was used to reduce the size of the precursor for better processing. The cloning was achieved by using overlap extension PCR (12). The DNA fragment upstream of the joint was amplified using pIAF811-A.8 as the template. The two oligonucleotides used were Afor (5′-TGCTCACATGTTCTTTCCTGCGT-3′ [the AflIII site is underlined]), located around the AflIII restriction site in pTZ19, and Jrev (5′-AGGGGCGGGACTCTGCTTCATAGCGTTCCTCCTCGTGCGA-3′), containing the 19 nt from the 3′ end of the 57-nt celA sequence and the 21 nt from the 5′ end of the truncated xlnC signal sequence. A fragment of 488 bp was generated. The DNA fragment downstream of the joint was amplified using pIAF816B2 as the template. The two oligonucleotides used were Jfor (5′-TCGCACGAGGAGGAACGCTATGAAGCAGAGTCCCGCCCCT-3′), which is the complementary sequence of Jrev, and XB2rev, corresponding to the 3′ end of xlnB2. A 748-bp fragment was obtained. Both amplified fragments were mixed, and a fragment of 1,196 bp was amplified with primers Afor and XB2rev. After digestion with HindIII-SacI (The HindIII site is located just upstream of the ribosome binding site of the 57-nt celA sequence), the resulting fragment of 800 bp was inserted at the same sites in pIAF907, resulting in plasmid pIAF907-TB2.
For the construction of the Sec/Tat secretion chimera, the plasmid pIAF907-SB1 was digested with SacI, and a copy of xlnB2 fused to the xlnC signal sequence having a double translation initiation was cloned at the same site. The fragment was obtained by PCR with pIAF907-TB2 as the template using the oligonucleotides GAU1 (5′-GGCCGAGCTCAAGCTTGGAGGCACAGTC-3′ [the SacI site is underlined]) and XB2rev. The amplified fragment of 808 bp was digested with SacI and inserted into pIAF907-SB1 at the same site, resulting in plasmid pIAF907-SB1TB2. Both genes were arranged in a bicistronic operon under the control of the xlnA promoter.
To have xlnB2 directly under the control of the xlnA promoter, a similar construct was achieved as described above, except that the amplified xlnB2 fragment started at the 5′ end with the xlnA promoter located 91 nt upstream of the first translation initiation codon. An 887-bp fragment was amplified with primers GAU2 (5′-GGCCGAGCTCTTGACCGAGCATCGTCAA-3′ [the SacI site is underlined]) and XB2rev while using pIAF907-TB2 as the template. After digestion with SacI, the fragment was inserted at the same site in pIAF907-SB1, yielding plasmid pIAF907-SB1pATB2. In this plasmid, xlnB1 and xlnB2 were then each under the control of the xlnA promoter.
For the construction of the Sec/Sec and Tat/Tat secretion chimeras, a strategy similar to that for pIAF907-SB1pATB2 was used. Starting from the plasmids pIAF907-SB1 and pIAF907-TB2, plasmids pIAF907-SB1pASB1 and pIAF907-TB2pATB2 were constructed, respectively.
Culture conditions and sampling procedures.
Transformants of S. lividans 10-164 were grown in shake flasks containing M14 medium supplemented with 1% xylose and 50 μg/ml of thiostrepton. The medium was inoculated with 2 × 106 spores per ml and incubated at 34°C. One-milliliter samples were removed at intervals. Supernatant fractions were recovered after centrifugation of 1 ml of culture for 5 min in a microcentrifuge and used for the measurement of extracellular xylanase activity. Growth was assessed by determining the DNA content of the mycelium pellet (5).
Enzyme assay.
Xylanase activity was measured in supernatants by the dinitrosalicylic acid method (17). One international unit (IU) of enzyme activity is defined as the amount of enzyme required to liberate 1 μmol of reducing sugars (expressed as xylose) in 1 min at 57°C.
Proteins and Western blot analysis.
Proteins from the culture supernatants were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (18) followed by Western blotting (33). The identification of xylanase-specific polypeptides was carried out with anti-XlnB antibodies and alkaline phosphatase-labeled goat anti-rabbit immunoglobulin (Roche).
Pulse-chase experiments.
Pulse-chase labeling experiments with S. lividans, immunoprecipitations, and SDS-PAGE were performed as described previously (20), but the mycelia were broken for 30 s with a glass bead shaker (FastPrep FP120; Thermo Savant, Holbrook, NY) instead of sonication. The gels were fixed in a solution of methanol-acetic acid and soaked for 30 min in an Amplify fluorographic reagent (Amersham Life Science), dried down on filter paper, exposed to a phosphor screen (Molecular Dynamics), and analyzed with a PhosphorImager SI (Molecular Dynamics).
RESULTS AND DISCUSSION
XlnB1 and XlnB2 production through the Sec-dependent pathway.
In these experiments, S. lividans 10-164 was used as the host for recombinant plasmids to make sure that the xylanase activity measured in the culture supernatants originated from the different constructs only (13). XlnB1, which contains both the catalytic domain and the xylan binding domain, and XlnB2, which contains the catalytic domain only, have the same specific activity on xylan (unpublished results). As we are interested in increasing xylanase production, it was necessary to show which form of the enzyme is more efficiently secreted through the Sec-dependent pathway in S. lividans. Therefore, the structural genes of xlnB1 and xlnB2 were inserted, respectively, downstream of the xlnB signal sequence in the multicopy plasmid pIAF915 in replacement of the structural gene of xlnA. In these constructs, the structural genes of xlnB1 and xlnB2 are under the control of the xlnA promoter. This promoter was chosen because it was used successfully to express xylanase genes and allowed us to compare the present data to previous results (21). As shown in Table 2, the xylanase activity in the IAF915B1 supernatant was about 20 to 30% higher than that of IAF915B2. This indicated that in XlnB1, the xylan binding domain contributes to secretion.
TABLE 2.
Xylanase activity in the extracellular fraction of S. lividans 10-164 and ΔtatC mutant transformed with different plasmids
| Transformant | Signal peptide | Xylanase activity (IU/ml)a |
|---|---|---|
| IAF915B1 | Sec dependent | 90 ± 6 |
| IAF915B2 | Sec dependent | 76 ± 5 |
| IAF916B1 | Tat dependent | 32 ± 4 |
| IAF916B2 | Tat dependent | 52 ± 5 |
| IAF907-SB1 | Sec dependent | 256 ± 10 |
| IAF907-SB1ΔtatC | Sec dependent | 207 ± 8 |
| IAF907-TB2 | Tat dependent | 173 ± 6 |
| IAF907-TB2ΔtatC | Tat dependent | 18 ± 5 |
| IAF907-SB1TB2 | Sec and Tat dependent | 350 ± 12 |
| IAF907-SB1TB2ΔtatC | Sec and Tat dependent | 172 ± 4 |
| IAF907-SB1pATB2 | Sec and Tat dependent | 360 ± 15 |
| IAF907-SB1pATB2ΔtatC | Sec and Tat dependent | 167 ± 3 |
| IAF907-TB2pATB2 | Tat dependent | 251 ± 14 |
The activity was measured in the culture supernatants along the growth of the transformants. The activity after 144 h of incubation is reported. Activity is the average of three independent cultures. Growth was similar for all transformants of S. lividans 10-164 and for all transformants of the ΔtatC mutant as determined by the DNA content of the cultures (data not shown). However, in these conditions, the growth of S. lividans 10-164 transformants was 30% higher than that of ΔtatC transformants.
We have reported previously that the presence of two ribosome-binding sites and two in-frame initiation codons increased XlnA production (22). These changes were introduced into the XlnA signal sequence, resulting in the synthesis of two precursors of different lengths. As the signal peptidase cleavage site was the same for both precursors, a mature XlnA that originated from the maturation of both precursors was produced. The signal sequence of the cellulase A (CelA) gene from S. lividans preceded by a 57-nt upstream sequence was used. The short and long signal sequences encode 27-amino-acid (aa) and 46-aa residues, respectively. In addition, an 8-nt sequence complementary to the 16S rRNA was introduced into the upstream sequence of celA, giving rise to enhanced translation (Fig. 1). Altogether, these modifications stimulated the XlnA production by 10-fold compared to that of wild-type xlnA (15). This construction was used to express xlnB1 since XlnB1 was produced to a higher level than XlnB2 in the Sec-dependent system. The results are shown in Table 2. The growth and the enzymatic activity of culture supernatants were determined at different times, but only the results after 144 h of cultivation were reported since after this time, no increase of xylanase activity was observed and the growth of all transformants was similar (data not shown). The transformant IAF907-SB1 produced 2.8-fold more enzyme than wild-type IAF915B1. This contrasted to the results obtained with XlnA, where a 10-fold increase was observed for a similar construct. This suggested that XlnB1 is probably less stable than XlnA. It should be noted that the XlnA catalytic domain folds into an (αβ)8 barrel motif (8), while XlnB1 is formed mostly of β-sheets (32) which might be more susceptible to proteolytic degradation. However, in 96-h culture supernatants containing XlnA, XlnB1, and XlnB2, there was no loss of activity after an incubation of up to 5 days at 30°C, thus ruling out the possibility of proteolytic degradation of these enzymes once secreted (data not shown). As the structural genes of xlnA, xlnB1, and xlnB2 are under the control of the same promoter, the difference is probably not at the transcriptional level but more likely at the translational level.
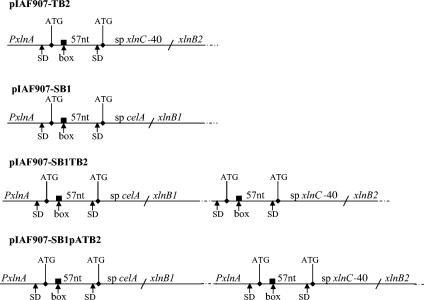
FIG. 1.
Scheme of different constructs for the secretion of XlnB1 and XlnB2 through either the Sec- or the Tat-dependent secretion system. For the Sec-targeted signal sequence, the celA signal sequence (sp celA) plus a 57-nt upstream sequence next to the ATG codon was fused to xlnB1 (pIAF907-SB1). For the Tat-targeted signal sequence, the xlnC signal sequence deleted from the 5′ end (sp xlnC-40), to which the 57 nt upstream of the celA signal sequence were added, was fused to xlnB2 (pIAF907-TB2). PxlnA, xlnA promoter; SD, Shine-Dalgarno sequence; dark box, sequence complementary to the 16S rRNA; ATG, translation initiation codon. The xlnB1 and xlnB2 genes were arranged either in bicistronic operon (pIAF907-SB1TB2) under the control of the xlnA promoter or separately, each preceded by the xlnA promoter (pIAF907-SB1pATB2).
XlnB1 and XlnB2 production through the Tat-dependent pathway.
Since XlnB2, which represents the XlnB1 catalytic domain only, exhibits 80% identity with XlnC, a xylanase secreted exclusively through the Tat-dependent pathway (9), we determined whether active XlnB2 could be secreted through the Tat system. The structural genes of xlnB1 and xlnB2 were inserted into plasmid pIAF916 (21) downstream of the Tat-targeting xlnC signal sequence in replacement of the structural gene of xlnA. As shown in Table 2, the activity in the supernatant of IAF916B2 was about 60% higher than that of IAF916B1. A 30% lower yield of XlnB2 was obtained when secreted through the Tat-dependent pathway compared to that of the Sec-dependent pathway. This result was surprising if we consider the half-life of a Sec precursor, which is 2 min (15), compared to the 12-min half-life for a Tat precursor (9). When this large difference in the precursor processing rate is taken into account, a lower yield will be expected for XlnB2 secreted through the Tat pathway.
However, the XlnB1 yield was threefold lower when the Tat secretion system was used than when the Sec one was used. This indicated that the Tat machinery has some difficulty in properly folding the two independent domains of XlnB1 to secrete an active enzyme.
To increase XlnB2 production through the Tat pathway as it was done for the Sec-dependent system, the xlnC signal sequence encoding 49 aa was reduced in size. Indeed, if 19 aa were added to the XlnC signal peptide, the precursor would probably be too long to be processed efficiently by the Tat machinery (22). It was observed that a 10-aa deletion of the N domain of the XlnC signal peptide did not affect the XlnC secretion (9). Therefore, this truncated xlnC signal sequence was fused to the 57 nt upstream of the celA signal sequence (Fig. 1). The resulting precursors will have signal peptides of 39 aa and 58 aa, respectively. With this construct, the XlnB2 yield increased 3.3-fold compared with that of IAF916B2 (Table 2). As shown above, comparable results were obtained with IAF907-SB1 containing the Sec signal sequence with a double ribosome binding site, a double translation initiation site, and the complementary sequence to the 16S rRNA.
Xylanase production by a recombinant strain containing a plasmid harboring both the Tat- and Sec-targeting signal sequences.
As shown above, the independent use of the modified Sec- and Tat-targeting signal sequences increased xylanase production. We investigated whether a further increase in xylanase yield could be obtained by using both secretion systems simultaneously in the same strain. To do so, plasmid pIAF907-SB1 containing the Sec signal sequence was digested with SacI, and a copy of the Tat signal sequence fused with xlnB2 was inserted at the same site. Only 24 nt separated the stop codon of xlnB1 from the first translation initiation codon of xlnB2. Moreover, this region contained a ribosome binding site. This construct constituted a bicistronic operon under the control of the xlnA promoter (Fig. 1). A total of 350 IU/ml of xylanase was produced by strain IAF907-SB1TB2. However, it is well known that in a bicistronic operon, the second gene is generally less expressed than the first one. For this reason, a second xlnA promoter was introduced upstream of the xlnB2 construct by using the plasmid pIAF907-SB1 digested with SacI, in which a copy of xlnB2 fused to the xlnA promoter (91 nt) was inserted (Fig. 1). The resulting recombinant strain, IAF907-SB1pATB2, produced around 360 IU/ml, independently of the orientation of the two promoters. This implied that both genes in the construct under the control of a single promoter (IAF907-SB1TB2) were expressed at the same level. This value of 360 IU/ml represented (not exactly) the sum of the production of the two secretion pathways taken individually, which was expected to be around 430 IU/ml. The 70-IU/ml deficit (16% less than expected) in the xylanase production is difficult to explain when both secretion systems function simultaneously. It is not clear if either both secretion systems are equally affected or only is one preferentially affected.
As two copies of the xylanase gene present in pIAF907-SB1pATB2 increased the xylanase yield via the two secretion pathways, it was of interest to investigate whether similar results would be obtained with two copies of the gene containing either the Sec or Tat signal sequence in the same plasmid. To determine this possibility, plasmids pIAF907-SB1pASB1 and pIAF907-TB2pATB2 were constructed and used to transform S. lividans. In the Sec-dependent system, the xylanase production could not be measured because plasmid pIAF907-SB1pASB1 was found to be unstable in the strain. Plasmid pIAF907-TB2pATB2 was stable, and XlnB2 production increased by 45% compared with that of IAF907-TB2 containing a single copy of the gene (Table 2). The fact that the XlnB2 yield didn't double but increased only 45% indicated that the Tat translocation machinery might be saturated in this case when the Tat-dependent xlnB2 gene was overexpressed. Thus, using a plasmid containing two copies of the same gene with a Tat signature was less efficient than a plasmid bearing two copies of the gene, one with a Tat signal sequence and the other with a Sec signal sequence.
Importance of the Tat-dependent secretion pathway in the different recombinant strains.
TatC is a key component of the Tat-dependent pathway (3). A ΔtatC mutant of S. lividans 10-164 was shown to impair secretion of XlnC, which is exclusively secreted through the Tat-dependent pathway (9). To analyze the contribution of the Tat-dependent pathway to xylanase production, plasmids were used to transform protoplasts of the ΔtatC mutant. It should be noted that at their stationary phase, the ΔtatC mutant strain and S. lividans 10-164 showed an average optical density difference (OD595 − OD700) of 0.35 and 0.5, respectively, according to their DNA content. This indicated that the growth of the strains was not comparable and that the ΔtatC mutation reduced the growth of the strain by 30% in the minimal medium. Therefore, the data were analyzed qualitatively, as a strict comparison was impossible. Only the results after 144 h of cultivation are shown, which represented approximately the middle of stationary phase. In the ΔtatC mutant, all the transformants having the Tat-targeting xlnC signal sequence showed a loss of xylanase production, corresponding to the contribution of the Tat-dependent pathway (Table 2). The xylanase yield of IAF907-SB1 containing a Sec-dependent signal sequence was about 20% higher than that obtained with the ΔtatC mutant transformed with the same plasmid. This was likely due to the inferior growth of the ΔtatC mutant as shown above, rather than a negative effect on the Sec-dependent pathway. However, with pIAF907-TB2, 18 IU/ml was produced by the ΔtatC mutant, representing 10% of the yield of IAF907-TB2 when the Tat-dependent pathway was not affected. This might indicate that XlnB2, even though it possesses a Tat-targeting signal peptide, could be partially secreted through the Sec-dependent pathway when the Tat-dependent pathway is defective. A similar event also occurred in E. coli, where a protein fused to a Tat-dependent signal can be translocated through the Sec-dependent pathway (26). For pIAF907-SB1TB2 and pIAF907-SB1pATB2 introduced into the ΔtatC mutant, there was a decrease of xylanase production of about 50% which represented approximately the loss due to the defective Tat-dependent secretion system. Considering the differential growth of both strains, the results are consistent and clearly indicate that the Tat pathway is functional in S. lividans 10-164 harboring a plasmid containing both a Sec- and a Tat-targeting signal sequence fused to the structural xylanase genes.
XlnB1 and XlnB2 precursor processing in the recombinant strains.
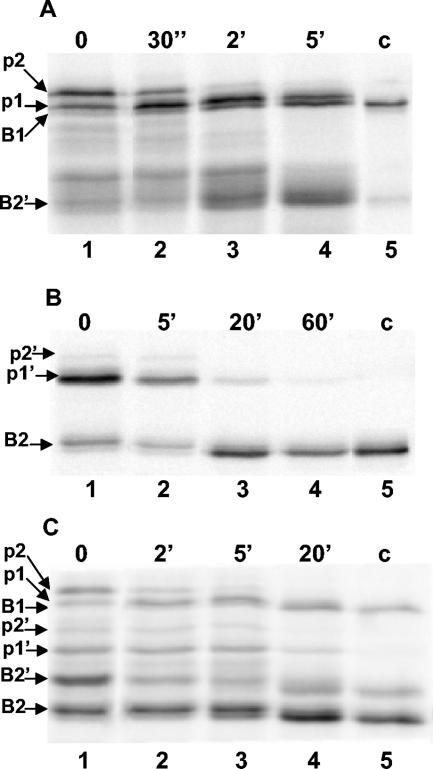
In order to show the presence of the different precursors and follow their processing in the recombinant strains, the transformants were grown in an M14 minimal medium containing xylose as the carbon source. After 48 h of growth, the mycelia were collected and submitted to pulse-chase experiments. The mycelia were labeled with [35S]methionine and chased. Samples were withdrawn at intervals, cells were broken, and xylanase-specific polypeptides were immunoprecipitated with anti-XlnB1 antibodies. The results of pulse-chase labeling experiments are shown in Fig. 2. In the Sec-dependent pathway with IAF907-SB1, the 34-kDa and 36-kDa precursors were labeled and disappeared during the chase with a half-life of 2.5 min, generating the 31-kDa mature XlnB1 (Fig. 2A). This processing rate is typical for a Sec-dependent precursor in S. lividans (20). As shown previously (17), there was also secretion of a 23-kDa xylanase called XlnB2′. This truncated xylanase shares the same N-terminal sequence as XlnB1, indicating that XlnB2′ has lost a part of the xylan binding domain (unpublished results). XlnB2′ probably resulted from the degradation of XlnB1 precursors. The origin of these precursors could be the result of the premature translation termination because there is a relatively strong secondary structure in the XlnB1 mRNA just upstream of the coding sequence of the xylan binding domain. It might be also due to the proteolytic degradation of XlnB1 in the culture medium.
FIG. 2.
Processing of pre-XlnB1 and pre-XlnB2 in S. lividans. S. lividans recombinant strains were grown in a minimal medium containing xylose as carbon source. Mycelia were pulse-labeled for 2 min with [35S]methionine and chased with nonradioactive methionine for the indicated times. Proteins were precipitated with trichloroacetic acid and recovered with the mycelia by centrifugation. After disrupting the mycelia, xylanase-like polypeptides were precipitated with anti-xylanase antibodies, and the products were analyzed by SDS-PAGE and autoradiography. Lanes 1 to 4, xylanase processing; c, control xylanase immunoprecipitated from the culture supernatant after labeling experiment (lane 5); p1 and p2, precursor forms of XlnB1; p1′and p2′, precursor forms of XlnB2; B1, XlnB1; B2, XlnB2; B2′, XlnB2′. A) IAF907-SB1 transformant carrying the plasmid pIAF907-SB1 encoding XlnB1 fused to a Sec-dependent signal sequence. B) IAF907-TB2 transformant carrying the plasmid pIAF907-TB2 encoding XlnB2 fused to a Tat-dependent signal sequence. C) IAF907-SB1TB2 transformant carrying the plasmid pIAF907-SB1TB2 encoding a bicistronic operon with XlnB1 and XlnB2 coding sequences fused, respectively, to a Sec- and Tat-targeting sequence.
The experiment with IAF907-TB2 showed two XlnB2 precursors of 25 kDa and 27 kDa. The long precursor was less abundant, indicating that translation at the first initiation codon was less efficient than translation at the internal one (Fig. 2B). The half-life of the short precursor was around 12 min, which was the same as what was determined for XlnC, a typical Tat-dependent substrate (9). As the long precursor was present in low amounts, its half-life was difficult to establish, but it was similar to that of the short precursor. XlnB2 has a calculated molecular weight of 20 kDa, which is lower than that of XlnB2′. The two enzymes differ by a few amino acids at the C-terminal end, between the catalytic and the xylan binding domains.
Strain IAF907-SB1TB2, harboring the plasmid containing both the Sec and Tat signal sequences, demonstrated the same pattern and a similar processing rate of different precursors to those observed in the strain harboring plasmids containing either Sec- or Tat-dependent signal sequences alone (Fig. 2C). In this strain, XlnB1, XlnB2, and XlnB2′ were produced and were secreted in the culture supernatant as shown by Western blot analysis (Fig. 3).
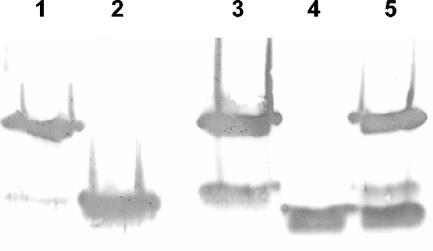
FIG. 3.
Western blot analysis of extracellular xylanase produced by S. lividans transformants. Samples of culture supernatant (10 μl) after 72 h of cultivation were separated by SDS-PAGE and then transferred onto nitrocellulose membrane and probed with anti-XlnB1 antibodies coupled with alkaline phosphatase-labeled goat anti-rabbit immunoglobulin. Lane 1, control XlnB1; lane 2, control XlnB2′; lane 3, IAF907-SB1; lane 4, IAF907-TB2; lane 5, IAF907-SB1TB2.
Although the cloning experiments were carried out in S. lividans 10-164 (a cellulase- and xylanase-negative mutant), it was nevertheless important to show that the xylanase activity measured in the supernatants of these transformants correlated with the presence of XlnB1, XlnB2, and XlnB2′. Western blot analyses with anti-XlnB1 serum were performed on proteins secreted in the culture supernatants of the different transformants after 72 h of growth (Fig. 3). XlnB1 and small amounts of XlnB2′ were detected in the supernatants of all transformants expressing only xlnB1. It is in agreement with what was found in the supernatant of the wild-type strain expressing chromosomal xlnB1 (17). In IAF907-SB1TB2, apart from the production of XlnB1 and XlnB2′, there was also production of XlnB2, which was secreted through the Tat-dependent system, as in IAF907-TB2.
In the present work, we have shown that XlnB2 of S. lividans was secreted efficiently through the Tat-dependent pathway, a recently described export system which transports folded proteins across the cytoplasmic membrane (2, 3). This suggests that XlnB2 folded after synthesis and was translocated as such through the Tat secretion system. XlnB2 was also equally well secreted through the Sec-dependent pathway and folded after secretion. Hence, for this enzyme, folding either prior to or after secretion led to production of an active product. This is in contrast to XlnC, which was active only when secreted through the Tat-dependent pathway (9). The results show that two related enzymes with the same substrate target are secreted by distinct protein export systems in S. lividans. Similarly, the alkaline phosphatase of Thermus thermophilus cloned in E. coli was Tat dependent, while the endogenous enzyme of E. coli was dependent on the Sec pathway for secretion (1). The distinct performance of XlnB2 and XlnC precursors in the secretion process might be due to differences in their primary structure. Two cysteine residues forming a disulfide bridge in XlnC are absent in XlnB2 (30). It is possible that the XlnC precursor may require some folding and that these two cysteine residues could form a disulfide bridge during the Tat-dependent maturation process.
In our experiments, different xylanases were produced, namely, XlnB1, XlnB2, and XlnB2′, and all of them exhibited activity. In order to obtain a homogenous enzyme preparation, it would have been preferable to fuse the structural gene of xlnB2 instead of xlnB1 to the Sec-dependent signal sequence to produce XlnB2 exclusively. However, the difference in the molecular weight of XlnB1 and XlnB2 allowed us to clearly demonstrate the presence of Sec and Tat precursors in vivo. We have shown that the production of xylanase could be increased through the simultaneous use of the Tat- and Sec-dependent secretion pathways in S. lividans. These results support the potential use of this system as an efficient method for producing secreted proteins. However, it remains to be determined if this secretion technology will be useful for expression and secretion of other recombinant proteins.
Acknowledgments
We thank J. Lemay for technical assistance and C. M. Dozois and D. Cossar for critical reading of the manuscript.
This research was supported by grants from the Natural Sciences and Engineering Research Council of Canada and Cangene Corporation.
REFERENCES
- 1.Angelini, S., R. Moreno, K. Gouffi, C.-L. Santini, A. Yamagishi, J. Berenger, and L.-F. Wu. 2001. Export of Thermus thermophilus alkaline phosphatase via the twin-arginine translocation pathway in Escherichia coli. FEBS Lett. 506:103-107. [DOI] [PubMed] [Google Scholar]
- 2.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 3.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 4.Binnie, C., J. D. Cossar, and D. I. Stewart. 1997. Heterologous biopharmaceutical protein expression in Streptomyces. Trends Biotechnol. 15:315-320. [DOI] [PubMed] [Google Scholar]
- 5.Burton, K. 1956. A study on the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem. J. 62:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaddock, A. M., A. Mant, I. Karnauchov, S. Brink, R. G. Herrmann, R. B. Klösgen, and C. Robinson. 1995. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the delta pH-dependent thylakoidal protein translocase. EMBO J. 14:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danese, P. N., and T. J. Silhavy. 1998. Targeting and assembly of periplasmic and outer membrane proteins in Escherichia coli. Annu. Rev. Genet. 32:59-94. [DOI] [PubMed] [Google Scholar]
- 8.Derewenda, U., L. Swenson, R. Green, Y. Wei, R. Morosoli, F. Shareck, D. Kluepfel, and Z. S. Derewenda. 1994. Crystal structure, at 2.6-Å resolution, of the Streptomyces lividans xylanase A, a member of the F family of beta-1,4-D-glycanases. J. Biol. Chem. 269:20811-20814. [PubMed] [Google Scholar]
- 9.Faury, D., S. Saidane, H. Li, and R. Morosoli. 2004. Secretion of active xylanase C from Streptomyces lividans is exclusively mediated by the Tat protein export system. Biochim. Biophys. Acta 1699:155-162. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, M., R. Morosoli, F. Shareck, and D. Kluepfel. 1995. Production and secretion of proteins by streptomycetes. Crit. Rev. Biotechnol. 15:13-39. [DOI] [PubMed] [Google Scholar]
- 11.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Keiser, C. J. Bruton, H. M. Keiser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 12.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 13.Hurtubise, Y., F. Shareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol. Microbiol. 17:367-377. [DOI] [PubMed] [Google Scholar]
- 14.Jeffries, T. W. 1992. Enzymatic treatment of pulp. ACS Symp. Ser. 476:313-329. [Google Scholar]
- 15.Kébir, H., C. Dupont, and R. Morosoli. 2000. Increased xylanase production in Streptomyces lividans after replacement of the signal peptide: dependence on box and inverted repeat sequence. Biochim. Biophys. Acta 1491:177-184. [DOI] [PubMed] [Google Scholar]
- 16.Kluepfel, D., N. Daigneault, R. Morosoli, and F. Shareck. 1992. Purification of a new xylanase (xylanase C) produced by Streptomyces lividans. Appl. Microbiol. Biotechnol. 36:626-631. [Google Scholar]
- 17.Kluepfel, D., S. Vats-Mehta, F. Aumont, F. Shareck, and R. Morosoli. 1990. Purification and characterization of a new xylanase (xylanase B) produced by Streptomyces lividans 66. Biochem. J. 267:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Morosoli, R., J. L. Bertrand, F. Mondou, F. Shareck, and D. Kluepfel. 1986. Purification and properties of a xylanase from Streptomyces lividans. Biochem. J. 239:587-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morosoli, R., and C. Dupont. 1999. Secretion of xylanase A2 in Streptomyces lividans: dependence on signal peptides length, number and composition. FEMS Microbiol. Lett. 179:437-445. [DOI] [PubMed] [Google Scholar]
- 21.Pagé, N., D. Kluepfel, F. Shareck, and R. Morosoli. 1996. Effect of signal peptide alterations and replacement on export of xylanase A in Streptomyces lividans. Appl. Environ. Microbiol. 62:109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pagé, N., D. Kluepfel, F. Shareck, and R. Morosoli. 1996. Increased xylanase yield in Streptomyces lividans: dependence on number of ribosome-binding sites. Nat. Biotechnol. 14:756-759. [DOI] [PubMed] [Google Scholar]
- 23.Robinson, C., and A. Bolhuis. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2:350-356. [DOI] [PubMed] [Google Scholar]
- 24.Robinson, C., D. Cai, A. Hulford, I. W. Brock, D. Michl, L. Hazell, I. Schmidt, R. G. Herrmann, and R. B. Klösgen. 1994. The presequence of a chimeric construct dictates which of two mechanisms are utilized for translocation across the thylakoid membrane: evidence for the existence of two distinct translocation systems. EMBO J. 13:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sanders, C., N. Wethkamp, and H. Lill. 2001. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol. Microbiol. 41:241-246. [DOI] [PubMed] [Google Scholar]
- 27.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santini, C.-L., B. Ize, A. Chanal, M. Müller, G. Giordano, and L.-F. Wu. 1998. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 17:101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shareck, F., C. Roy, M. Yaguchi, R. Morosoli, and D. Kluepfel. 1991. Sequences of three genes specifying xylanases in Streptomyces lividans. Gene 107:75-82. [DOI] [PubMed] [Google Scholar]
- 31.Te'o, V. S., A. E. Cziferszky, P. L. Bergquist, and K. M. Nevalainen. 2000. Codon optimization of xylanase gene xynB from the thermophilic bacterium Dictyoglomus thermophilum for expression in the filamentous fungus Trichoderma reesei. FEMS Microbiol. Lett. 190:13-19. [DOI] [PubMed] [Google Scholar]
- 32.Törronen, A., and J. Rouvinen. 1995. Structural comparison of two major endo-1,4-xylanases from Trichoderma reesei. Biochemistry 34:847-856. [DOI] [PubMed] [Google Scholar]
- 33.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viikari, L., A. Kantelinen, J. Sundquist, and M. Linko. 1994. Xylanases in bleaching from an idea to the industry. FEMS Microbiol. Rev. 13:335-350. [Google Scholar]