Abstract
Fecal coliforms and enterococci are indicator organisms used worldwide to monitor water quality. These bacteria are used in microbial source tracking (MST) studies, which attempt to assess the contribution of various host species to fecal pollution in water. Ideally, all strains of a given indicator organism (IO) would experience equal persistence (maintenance of culturable populations) in water; however, some strains may have comparatively extended persistence outside the host, while others may persist very poorly in environmental waters. Assessment of the relative contribution of host species to fecal pollution would be confounded by differential persistence of strains. Here, freshwater and saltwater mesocosms, including sediments, were inoculated with dog feces, sewage, or contaminated soil and were incubated under conditions that included natural stressors such as microbial predators, radiation, and temperature fluctuations. Persistence of IOs was measured by decay rates (change in culturable counts over time). Decay rates were influenced by IO, inoculum, water type, sediment versus water column location, and Escherichia coli strain. Fecal coliform decay rates were significantly lower than those of enterococci in freshwater but were not significantly different in saltwater. IO persistence according to mesocosm treatment followed the trend: contaminated soil > wastewater > dog feces. E. coli ribotyping demonstrated that certain strains were more persistent than others in freshwater mesocosms, and the distribution of ribotypes sampled from mesocosm waters was dissimilar from the distribution in fecal material. These results have implications for the accuracy of MST methods, modeling of microbial populations in water, and efficacy of regulatory standards for protection of water quality.
Indicator organisms (IOs), which reside in the gastrointestinal tracts of humans and animals, are used in the United States and throughout the world to assess the microbiological safety of drinking water, recreational waters, and shellfish waters. The U.S. Environmental Protection Agency recommends the use of Escherichia coli, a member of the fecal coliform group, as an indicator organism for recreational waters in freshwater bodies and members of the genus Enterococcus (the enterococci) for both freshwater and saltwater (34). The state of Florida utilizes both the fecal coliform group and the enterococci for monitoring recreational water (http://esetappsdoh.doh.state.fl.us/irm006eachwater/default.aspx); however, fecal coliforms are the sole criterion for classification of shellfish waters in Florida (http://www.floridaaquaculture.com/SEAS/SEAS_intro.htm). Growth of these organisms in natural waters would compromise their use as indicators of fecal contamination, and evidence from a number of studies suggests that E. coli and enterococci may multiply in warm, subtropical waters (4, 9, 31). Many more studies have demonstrated extended persistence of culturable indicator bacteria in the sediments of environmental waters (8, 11, 29).
Indicator bacteria are frequently used as the source identifier in microbial source tracking (MST) methods, which are designed to determine the source(s) of fecal pollution and/or indicator organisms in water. A group of methods termed “library-based” methods rely on a common approach: indicator bacteria from various fecal sources are isolated and phylotyped by a genotypic (5, 10, 27) or phenotypic (18, 20, 36, 40) technique. The database, or library, of phylotype patterns from known sources then forms the basis for classifying bacteria isolated from water into a source category (39). One of the important assumptions of these methods is that the distribution of indicator organism phylotypes in water samples is roughly proportional to the population distribution of phylotypes in fecal samples; this relationship is much less probable if differential persistence of phylotypes occurs in environmental waters. Indicator organisms that are adapted to living in the gastrointestinal tract but which pass to a different habitat, such as a septic system or wastewater collection system, may be leaving a primary habitat and entering a secondary habitat (14). Typing of E. coli populations in primary and secondary habitats has shown substantial shifts in the dominant members over time, suggesting that only certain members of the population in the primary habitat remain culturable in the secondary habitat (14, 32, 38). To date, no studies have been published in which population shifts over time have been determined for water bodies contaminated with fecal indicator organisms, although this is essential information for the improvement of MST approaches.
Many of the studies published on the survival of culturable indicator bacteria in environmental waters relied on laboratory microcosm experiments. While a wealth of information was obtained from such studies, including the negative relationship between salinity and E. coli survival (2), the persistence of fecal indicator bacteria in environmental waters is affected by a complex array of physical, chemical, and biological factors (28) that are difficult to simulate in the laboratory (24). Experiments described in this work were therefore designed to simulate environmental conditions as closely as possible, including construction of the mesocosms with nonsterile water and sediments. Persistence over incubation times as long as several weeks was assessed because of the possible role of soil and sediments as a repository and reservoir of indicator bacteria (8, 15, 26).
This study utilized mesocosms designed to simulate natural conditions, which were inoculated with fecal material, to test the hypothesis that certain E. coli phylotypes exhibit greater persistence than others in aquatic environments. Dog feces, untreated wastewater, and sediment from a chronically contaminated stream bank were chosen because they represent fecal sources that are frequently implicated in contamination of water and because microorganisms contained in these types of samples have experienced various degrees of previous exposure to environmental stress at the time of inoculation. Decay rates of fecal coliforms (FC) and enterococci (ENT) in the mesocosm water column and sediment in both freshwater and saltwater environments were calculated to compare the change in culturable indicator organism concentrations (persistence) over time. Ribotyping of E. coli genomic DNA was used to monitor the persistence of E. coli phylotypes inoculated as fecal material into freshwater mesocosms.
MATERIALS AND METHODS
Establishment of mesocosms.
Mesocosms were designed to simulate environmental conditions by allowing exposure to radiation, ambient temperatures, and inclusion of sediments and nonsterile water (which would include predators). Mesocosms were created by adding one of two natural water types to sterile, uncovered, polypropylene containers with dimensions of 10 in. by 7.5 in. by 16 in. high. Either freshwater from the Hillsborough River or saltwater from the Gulf of Mexico (∼30 ppt salinity) was utilized. Each container was filled to a depth of 5 cm with the sediment type associated with the water (wet weight, ∼2.5 to 3 kg, depending upon sediment sample), and 5 liters of water was added. All mesocosms were incubated outdoors in a roofed greenhouse without sides, providing conditions of variable environmental stresses such as temperature and UV radiation. Each mesocosm was aerated using aquarium air pumps, which served both to maintain dissolved oxygen concentration and to keep the water column mixed. Mesocosms were placed in several large, round containers (plastic pools) filled with water to moderate the temperature. Three replicate mesocosms containing each water type were inoculated with one of three different inoculum types: feces from a single dog, untreated wastewater collected from a sewer line receiving waste directly from a hospital, or soil collected from the banks of a tidally influenced stream that was known to be contaminated with indicator bacteria (22). Initial concentrations of fecal coliforms in the freshwater and sediment were ∼102 CFU · 100 ml−1 and ∼5 × 104 CFU · 100 g−1, respectively. Initial concentrations of enterococci in the freshwater and sediment were 23 CFU · 100 ml−1 and <66 CFU · 100 g−1, respectively. In the saltwater column and sediment, no culturable indicator organisms were detected prior to inoculation.
Mesocosms were allowed to equilibrate for 24 h prior to inoculation. The concentrations of indicator organisms in each inoculum type were determined prior to addition into mesocosms. Inoculum volumes were adjusted to provide a final concentration of approximately 105 CFU · 100 ml−1 fecal coliforms. Dog feces were prepared for inoculation by suspending 2 g fecal material in 10 ml buffered water, which was homogenized by vortexing and pipetted into the overlying waters of three mesocosms. Two hundred milliliters of wastewater was pipetted into each of three mesocosms, and 1 kg soil was weighed into a sterile plastic bag and added to each of three mesocosms with stirring. In each case, vigorous stirring was employed to ensure even distribution of the inoculum.
Measurements for dissolved oxygen, pH, salinity, and temperature were taken daily using YSI model 600QS (YSI, Yellow Springs, OH). Averages over the course of the experiment are reported.
Sampling of mesocosm populations.
Both the sediment and the water column of each mesocosm (one water column and one sediment sample per mesocosm per time point) were sampled daily for the first week and weekly for three following weeks. Mesocosms were sampled within 15 min after inoculation (D0). Core samplers were made from sterile, plastic 25-ml pipettes whose ends had been aseptically removed. The sediment was minimally disturbed during sampling to avoid artificially elevated fecal indicator concentrations in the water column. Procedures for enumeration of fecal coliforms and Enterococcus spp. were carried out according to standardized membrane filtration protocols (1, 35). Water samples were filtered directly, and sediment samples were processed by adding 10 g (wet weight) of sediment to 100 ml of buffered water followed by sonication of the mixture using an ultrasonic dismembrator (model 100; Fisher Scientific) for 30 s at 14 W. Preliminary experiments were conducted to determine optimal dissociation of fecal coliform and enterococcal cells from particles while maintaining culturability. The sample was allowed to settle for 10 min, and the supernatant was pipetted to a membrane filtration apparatus. After this point, water column and sediment samples were processed identically.
After 24 h, fecal coliform colonies were transferred to microtiter dishes containing EC broth amended with 50 μg · ml−1 4-methylumbelliferyl-β-d-glucuronide, which were incubated at 37°C for 24 h. Cultures that fluoresced under UV were designated 4-methylumbelliferyl-β-d-glucuronide positive and considered putative E. coli. Ten percent of putative E. coli isolates were subjected to biochemical confirmation using the API 20E system. One hundred percent of these isolates were confirmed as E. coli. E. coli strains from each treatment were randomly selected for ribotyping at each time point. Between 14 and 21 E. coli strains per time point were ribotyped from the mesocosm. The DNA of some isolates could not be digested, and toward the end of the experiments, fewer colonies were recovered.
Ribotyping.
The E. coli ribotyping protocol was modified from a previously published method (27) by altering the probe synthesis protocol. The 16S rRNA gene probe was synthesized by PCR using 5 μl purified DNA from E. coli ATCC 9637 as a template. Each PCR mixture contained 25 mM MgCl2, 5 μl 10× buffer B, 1.25 U Taq polymerase (Fisher, Pittsburgh, PA), 10 mM concentrations of deoxynucleoside triphosphates (Promega, Madison, WI), 2 mM concentrations of digoxigenin-labeled deoxynucleoside triphosphates (Roche Diagnostics, Indianapolis, IN), and 10 mM concentrations of each primer (IDT, Coralville, IA). Universal primers targeting the 16S rRNA gene (33), U1 (5′-AGAGTTTGATCMTGGCTCAG-3′) and U2 (5′-GGTTACCTTGTTACGACTT-3′), were used, producing a 1,484-bp amplicon. DNA fragments that hybridized with the probe were detected using a digoxigenin nucleic acid detection kit (Roche Diagnostics, Indianapolis, IN).
Dialysis tube microcosms.
Microcosms of dialysis tubing were incubated in a pond to test the stability of E. coli ribotypes. Spectra/Por 7 membrane dialysis tubing (MWCO 50000; Spectrum Laboratories, Inc., Rancho Dominguez, CA) was cut into 30-cm strips, soaked in sterile nanopure water overnight, and washed prior to inoculation. Six replicate dialysis tube microcosms were made for each E. coli source (dog feces, wastewater, contaminated soil), and each was filled with 30 ml pond water that had been sterilized by autoclaving. Three E. coli isolates from each source were randomly selected to inoculate the microcosms and were grown in pure culture in 1 ml tryptic soy broth at 37°C overnight. Cells were centrifuged, washed three times, and then diluted 10-fold by resuspension in 10 ml sterile pond water. Thirty-microliter aliquots of cultures were transferred to the microcosms, and each microcosm was inoculated with cells from three E. coli cultures derived from one of the sources.
Dialysis tube microcosms were incubated in a freshwater pond on the University of South Florida campus. Microcosms were placed in a metal cage and suspended 0.15 m below the water surface. One microcosm per host source was processed upon inoculation (T0). Microcosms were subsequently sampled 1 hour after inoculation (T1), and at ∼24, 48, 72, and 96 h (T24 to T96). Culturable E. coli concentrations were obtained by membrane filtration as described above, and ribotyping was conducted as described above. This study was conducted during three consecutive weeks to obtain replicates for each treatment.
Decay rate calculations.
Decay rates of culturable indicator organism concentrations were calculated using the following equation, which is a standard exponential growth/decline equation:
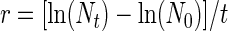 |
where r = decay rate, Nt = log10(CFU 100 ml−1) at time t, N0 = log10(CFU 100 ml−1) at time zero, and t = time (in days). Time (t) was determined by the days between the first sampling event and either the last sampling event (28 days) or when culturable cells could no longer be detected. The absolute value of the decay rate reflects the magnitude of change in culturable concentrations. A negative r represents a decrease in CFU (net cell die off), while a positive r represents an increase in CFU (growth). The decay rate of enterococci in the water column of the dog feces treatment was estimated because no enterococci were detected postinoculation. The estimate was accomplished by assuming D0 numbers equal to that in the freshwater treatment, and assuming 1 CFU on day 1.
Statistical analysis.
Decay rates were compared using two-way analysis of variance and pairwise t tests (JMP, version 3.0; SAS Institute, Inc.). Alpha was set at 0.05.
RESULTS
Physical and chemical parameters in the mesocosms varied by less than 10% over the course of the experiment. The average salinity of the freshwater mesocosms was 0.15‰ in the dog feces treatment, 0.16‰ in the wastewater treatment, and 0.52‰ in the contaminated-soil treatment. Average salinities for all saltwater treatments were 24.6‰. Average pH values for freshwater mesocosms were 8.2 in the dog feces treatment, 8.0 in the wastewater treatment, and 8.2 in the contaminated-soil treatment. Average pH values in all saltwater mesocosms were 7.9. Dissolved oxygen in freshwater mesocosms was an average 7.6 mg liter−1 in the dog feces treatment, 7.5 mg liter−1 in the wastewater treatment, and 7.3 mg liter−1 in the contaminated-soil treatment. Corresponding dissolved oxygen values for saltwater mesocosms were 6.2 mg liter−1, 6.1 mg liter−1, and 6.2 mg liter−1.
Persistence of indicator organisms in mesocosms.
Culturable FC and ENT from dog feces, sewage, and contaminated soil were enumerated over the course of 4 weeks to assess the persistence of the indicator organisms. At all time points in all treatments, at least 90% of fecal coliforms were E. coli isolates. Average decay rates in culturable counts allowed direct comparison of the persistence of indicator organisms that were inoculated at various concentrations and which became undetectable at various time points (Table 1). A negative value represents a net decrease in culturable cells, and the larger the value, the greater the departure from initial (D0) concentrations. No positive decay rates, which would indicate an increase in culturable counts, were noted during the experiments.
TABLE 1.
Decay rates for fecal coliforms and Enterococcus spp. from various sources in fresh water and saltwater mesocosms
| Water type | Location | Decay rate froma:
|
Overall decay rate
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Dog feces
|
Wastewater
|
Soil inoculum
|
|||||||
| FC | ENT | FC | ENT | FC | ENT | FC | ENT | ||
| Fresh | Water | −0.37 | −1.49 | −0.27 | −0.31 | −0.08 | −0.39 | −0.24 | −0.73 |
| Sediment | −0.03 | −0.29 | −0.03 | −0.21 | −0.02 | −0.16 | −0.02 | −0.22 | |
| Salt | Water | −3.8 | ≤−4.2b | −4.2 | −1.05 | −0.83 | −0.99 | −2.9 | ≤−2.1b |
| Sediment | −0.65 | −3.1 | −3.1 | −0.22 | −0.28 | −0.01 | −1.3 | −1.1 | |
Decay rates are calculated as log10 (CFU 100 ml−1) per day (for water columns) and log10 (CFU 100 g−1) per day (for sediments).
Decay rate estimated; see Materials and Methods.
The decay rates of fecal coliforms under various physicochemical conditions of location (water column versus sediment) and water type (fresh versus saltwater) were generally in line with those of previous studies. Saltwater had a negative effect on FC persistence, as the decay rates of FC (all inoculum sources combined) in saltwater sediments and water column were greater than those in freshwater. Saltwater also significantly increased enterococcal decay rates compared to freshwater.
IO persistence tended to be greater in sediments than in the water column. The average decay rate of FC in sediments of freshwater mesocosms was significantly less than those in the water column, and the difference was nearly significantly at the α = 0.05 level in saltwater (P = 0.083). Although decay rates of enterococci tended to be greater in the water column than in sediments, the difference was not significant in freshwater or saltwater mesocosms.
In some treatments, the type of material used to inoculate mesocosms had a significant effect on IO decay rates; however, these differences should be interpreted cautiously because not only were inoculum sources different but the amount of inoculum material and, therefore, organic carbon differed between treatments. The trend was for the lowest decay rates (greatest persistence) in mesocosms inoculated with contaminated soil, followed by wastewater and, finally, dog feces. Differences were generally not significant, with the exception of FC in the water column of freshwater and saltwater mesocosms and ENT in saltwater mesocosms.
The persistence of FC and ENT was also compared. FC decay rates in freshwater were significantly lower than ENT decay rates when all data were combined. FC and ENT decay rates were not significantly different in saltwater mesocosms.
Ribotype distribution in freshwater mesocosms.
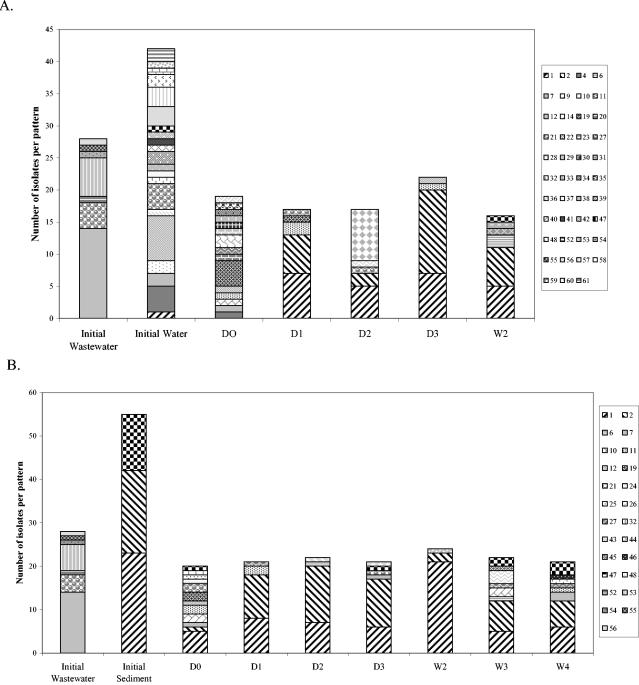
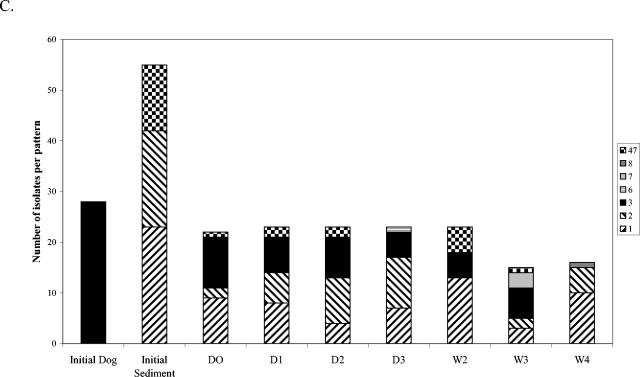
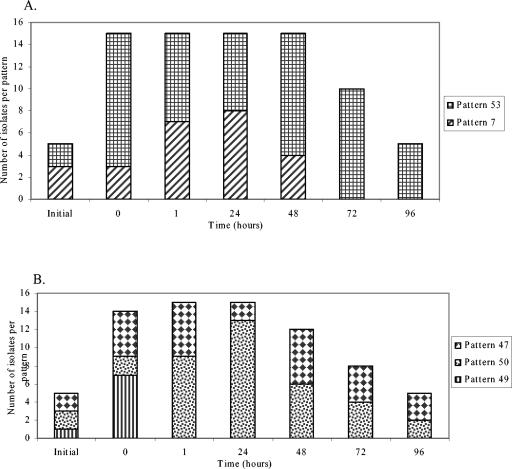
The change over time in distribution of E. coli phylotypes in freshwater mesocosms was investigated by ribotyping (Fig. 1 and 2). E. coli isolates were isolated and ribotyped from water (n = 42) and sediment (n = 55) to characterize the preexisting populations, and isolates from inoculum samples were also typed before inoculation (Fig. 1). Generally 23 to 24 isolates per treatment per day were typed (minimum, 15). E. coli ribotype diversity was very high in the uninoculated (initial) river water, high in the wastewater, lower in the uninoculated sediments, and still lower (apparently clonal) in the dog feces inoculum (Fig. 1A to C). Ribotype diversity in mesocosms inoculated with wastewater (Fig. 1A and 1B) was quite high on day 0 (D0), which was not surprising considering the relatively diverse E. coli population in wastewater. Ribotypes 1 and 2, which were detected in the initial water and sediment samples but not in the wastewater inoculum, increased in relative abundance over time in water and sediment samples, while most of the other ribotypes became undetectable over time. Fecal coliforms were not detected in the water column after week 2 but could be cultured from the sediment through week 4.
FIG. 1.
E. coli ribotype distribution in freshwater mesocosms inoculated with wastewater or dog feces. Each column pattern represents a different ribotype, and patterns that appear in separate graphs represent the same ribotype. Ribotypes from the initial inoculum sample are represented by the first bar on each graph, and ribotypes from the water sample or sediment before inoculation are also included. (A) Water column of wastewater mesocosms; (B) sediment of wastewater mesocosms; (C) sediment of dog feces mesocosms. D, day; W, week.
FIG. 2.
Stability of ribotypes over time in dialysis tube microcosms containing sterile water inoculated with wastewater (A) or contaminated soil (B).
In contrast to the wastewater inoculum, ribotype diversity in the dog feces inoculum was extremely low (Fig. 1C). In the water column of mesocosms inoculated with dog feces (data not shown), all isolates sampled on D0 shared the ribotypes of the initial dog feces isolates. Ribotypes 1 and 2 came to dominate the water column population by week 2, after which E. coli was no longer detected in the water column. The sediments of dog feces mesocosms (Fig. 1C) harbored the initial dog feces isolate and ribotypes 1 and 2 through week 3, but the dog feces isolate was not observed in week 4, indicating its great decrease in relative abundance over time. The initial ribotype diversity in the contaminated sediment inoculum was intermediate between those of wastewater and the dog feces inoculum, and the trends observed for these mesocosms were very similar to those described above (data not shown). Ribotypes 1 and 2 were observed through week 2 in the water column and through week 4 in the sediments.
The differential survival of a randomly selected group of E. coli strains was also tested in dialysis tube microcosms incubated in situ in a freshwater pond for 5 days. Pure cultures of E. coli isolated from the same source material that was used to inoculate the mesocosms (dog feces, wastewater, and contaminated soil) were used to inoculate dialysis tube microcosms made with sterile pond water. Although each microcosm was inoculated with cultures derived from three randomly chosen isolates, all of the initial (inoculum) isolates for the dog treatment were identical due to the low-diversity E. coli population (data not shown) and only two of the three wastewater strains were different. Differential survival of the E. coli strains was observed, i.e., ribotype 7 from wastewater (Fig. 2A) could not be detected after T48 and ribotype 49 from contaminated soil (Fig. 2B) became undetectable within 1 hour after inoculation.
DISCUSSION
The wisdom of reliance on water quality standards that are based on indicator bacterial concentrations has been widely discussed for decades; e.g., Wolf (41) commented that the predictive relationship between fecal coliforms and nonbacterial pathogens in water was uncertain. These concerns have grown over time and are outlined in recent reviews (17, 23). A major concern has been the possibility that E. coli (3, 31, 37) and enterococci (4, 11, 12) can multiply in environmental waters, sediments, and associated soils. The growth of indicator organisms in water and sediments would obviously jeopardize the connection between indicator organism concentration and human health risk. Differential survival of certain E. coli or ENT strains or of indicator bacteria from different pollution sources would also present a major challenge to MST methods that utilize these indicator organisms to determine the contributions of various sources to fecal pollution in water.
This study was designed to investigate the relative persistence of E. coli phylotypes in a setting that included environmental stresses characteristic of subtropical environments. Because the deliberate addition of fecal material to natural waters could be hazardous to human health and because assessment of differential survival would be very difficult in a natural system with continued inputs of indicator organisms, these experiments were conducted in a mesocosm setting. Differential survival of Enterococcus spp. phylotypes and of E. coli in saltwater will be investigated in future work.
Persistence of indicator bacteria.
The population trends of FC and ENT in freshwater mesocosms did not yield evidence of growth of the organisms, unlike some other studies (9, 31); however, prolonged persistence (assessed by decay rates of culturable bacteria) was characteristic of IOs in many treatment groups. FC had greater persistence in freshwater than ENT, which was reported under some conditions by Sinton et al. (30). Other findings that were in general agreement with previous studies were that FC decay rates were lower in freshwater than in saltwater (2) and that FC decay rates in sediments were lower than in the water column (7, 11, 13). Interestingly, ENT decay rates (all inoculum treatments combined) were not significantly lower than FC decay rates in saltwater mesocosms, although the trend was evident. The extremely high decay rate for the dog feces inoculum no doubt influenced this result, which was somewhat unexpected based on the literature. Greater persistence of enterococci than of fecal coliforms or E. coli in saline waters has been documented (8). In contrast, one study demonstrated that enterococci from waste stabilization pond effluent had a greater inactivation rate in seawater than fecal coliforms or E. coli (30), but the opposite was true for IOs from untreated wastewater. The results of the preceding study and this work suggest that persistence characteristics of enterococci (whether in terms of species or phylotypes) are heterogeneous and that applying extensive generalizations to IO persistence in secondary habitats is not valid given our present, incomplete understanding.
Higher variability between replicate sediment samples was observed compared to water samples and probably has multiple contributing factors, including patchy distribution of organisms in sediments and difficulty in dissociating bacteria from sediment particles. Therefore, comparisons of sediment samples in which significant differences were not found must be interpreted cautiously, i.e., real differences in persistence may exist that were not detected in these experiments. These factors are not unique to this study and should be considered when designing sampling strategies for environmental studies.
Effect of inoculum type on survival.
In some cases, IO populations from different sources that were subjected to the same environmental conditions had significantly different decay rates. The relative persistence of FC and ENT from different sources in saltwater and freshwater was contaminated sediment > wastewater > dog feces. This trend not only reflects the relative time period for exposure of the IO populations to selective pressure outside the host gastrointestinal tract but it also reflects the amount of material added to the mesocosms to achieve the desired IO concentrations. Dissolved organic carbon (DOC) measurements were attempted for these experiments, but DOC values in many mesocosms were lower than in uninoculated mesocosms, leading to the conclusion that the results were not reliable. We believe that the filtration step necessary for DOC measurement removed aggregates of organic material, contributing to artificially low numbers. Because the availability of organic carbon and other nutrients doubtless varied between inoculum treatments, comparison of decay rates between these treatments must be interpreted cautiously.
Differential survival of specific E. coli phylotypes.
E. coli phylotypes (strains) exhibited differential survival. Ribotypes 1 and 2 were observed in the sediment and water column of all mesocosms by day 1, even though both fecal source and differences caused by different inoculum sizes (e.g., nutrient availability) would be expected to influence persistence. Ribotypes 1 and 2 eventually became dominant members of the E. coli populations in the sediments and water columns of all mesocosms. Differential survival of E. coli strains was also noted in the microcosm experiments, i.e., ribotype 7 from wastewater and ribotype 49 from contaminated soil could be cultured for only a portion of the 4-day study. Other studies have shown that E. coli survivorship in habitats outside the gastrointestinal tract, such as soil and poultry litter, is strain dependent (32, 38).
This study demonstrated a high degree of variability in the response of fecal indicator organisms to stresses in aquatic environments on all levels investigated. Responses to water type (saline versus fresh), location (sediment versus water column), and inoculum type all varied within and between indicator bacterial groups (FC and ENT). The discrepant results emphasize the difficulties encountered in attempting to regulate diverse types of water bodies by one regulatory standard. Also cautionary is the persistence of indicator organisms in sediments, which leads to elevation of their densities and a false indication of recent pollution in the water column after events such as rain storms, construction, or recreational use. Differential survival of IOs has profound implications for MST methods that rely on these organisms, particularly those that estimate IO loadings from various possible contamination sources (for examples, see references 6, 16, 19-21, 25, 36, and 40). An assumption of library-based MST methods is that the population distribution of IO phylotypes in feces or wastewater reflects the population distribution in water contaminated by those sources. The feasibility of making semiquantitative assessments of relative source contributions to water will be doubtful if future studies confirm that IO source and/or phylotype significantly influence persistence in environmental waters (14).
Acknowledgments
This study was funded by STAR grant no. R828829 from the United States Environmental Protection Agency to V.J.H.
We thank the University of South Florida Botanical Gardens and Laurie Walker, director, for the use of their facilities. We also thank Gordon Fox (USF Department of Biology) for help with decay rate equations and the use of greenhouse space.
REFERENCES
- 1.American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Inc., Washington, D.C.
- 2.Anderson, I. C., M. Rhodes, and H. Kator. 1979. Sublethal stress in Escherichia coli: a function of salinity. Appl. Environ. Microbiol. 38:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byappanahalli, M., D. A. Shively, M. B. Nevers, M. J. Sadowsky, and R. L. Whitman. 2003. Growth and survival of Escherichia coli and enterococci populations in the macro-alga Cladophora (Chlorophyta). FEMS Microbiol. Ecol. 46:203-211. [DOI] [PubMed] [Google Scholar]
- 4.Byappanahalli, M. N., and R. S. Fujioka. 1998. Evidence that tropical soil can support the growth of Escherichia coli. Water Sci. Technol. 38:171-174. [Google Scholar]
- 5.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, S., W. Chu, J. Brown, S. J. Becker, V. J. Harwood, and S. C. Jiang. 2003. Application of enterococci antibiotic resistance patterns for contamination source identification at Huntington Beach, California. Mar. Pollut. Bull. 46:748-755. [DOI] [PubMed] [Google Scholar]
- 7.Craig, D. L., H. J. Fallowfield, and N. J. Cromar. 2004. Use of microcosms to determine persistence of Escherichia coli in recreational coastal water and sediment and validation with in situ measurements. J. Appl. Microbiol. 96:922-930. [DOI] [PubMed] [Google Scholar]
- 8.Davies, C. M., J. A. Long, M. Donald, and N. J. Ashbolt. 1995. Survival of fecal microorganisms in marine and freshwater sediments. Appl. Environ. Microbiol. 61:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desmarais, T. R., H. M. Solo-Gabriele, and C. J. Palmer. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fish, J. T., and G. W. Pettibone. 1995. Influence of freshwater sediment on the survival of Escherichia coli and Salmonella sp. as measured by three methods of enumeration. Lett. Appl. Microbiol. 20:277-281. [DOI] [PubMed] [Google Scholar]
- 12.Fujioka, R. S., C. Sian-Denton, M. Borja, J. Castro, and K. Morphew. 1999. Soil: the environmental source of Escherichia coli and enterococci in Guam's streams. J. Appl. Microbiol. Symp. Suppl. 85:83S-89S. [DOI] [PubMed] [Google Scholar]
- 13.Gerba, C. P., and J. S. McLeod. 1976. Effect of sediments on the survival of Escherichia coli in marine waters. Appl. Environ. Microbiol. 32:114-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 15.Goyal, S. M., C. P. Gerba, and J. L. Melnick. 1977. Occurrence and distribution of bacterial indicators and pathogens in canal communities along the Texas coast. Appl. Environ. Microbiol. 34:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graves, A. K., C. Hagedorn, A. Teetor, M. Mahal, A. M. Booth, and R. B. Reneau, Jr. 2002. Antibiotic resistance profiles to determine sources of fecal contamination in a rural Virginia watershed. J. Environ. Qual. 31:1300-1308. [DOI] [PubMed] [Google Scholar]
- 17.Griffin, D. W., E. K. Lipp, M. R. McLaughlin, and J. B. Rose. 2001. Marine recreation and public health microbiology: quest for the ideal indicator. Bioscience 51:817-825. [Google Scholar]
- 18.Hagedorn, C., J. B. Crozier, K. A. Mentz, A. M. Booth, A. K. Graves, N. J. Nelson, and R. B. Reneau. 2003. Carbon source utilization profiles as a method to identify sources of faecal pollution in water. J Appl. Microbiol. 94:792-799. [DOI] [PubMed] [Google Scholar]
- 19.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau, Jr. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harwood, V. J., B. Wiggins, C. Hagedorn, R. D. Ellender, J. Gooch, J. Kern, M. Samadpour, A. C. H. Chapman, B. J. Robinson, and B. C. Thompson. 2003. Phenotypic library-based microbial source tracking methods: efficacy in the California collaborative study. J. Water Health 01:153-166. [PubMed] [Google Scholar]
- 22.Hood, K. L., J. E. Whitlock, M. R. McLaughlin, J. B. Rose, and V. J. Harwood. 2002. Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. Q-455.
- 23.Leclerc, H., D. A. Mossel, S. C. Edberg, and C. B. Struijk. 2001. Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety. Annu. Rev. Microbiol. 55:201-234. [DOI] [PubMed] [Google Scholar]
- 24.McFeters, G. A., and S. I. Terzieva. 1991. Survival of Escherichia coli and Yersinia enterocolitica in stream water: comparison of field and laboratory exposure. Microb. Ecol. 22:65-74. [DOI] [PubMed] [Google Scholar]
- 25.Myoda, S. P., C. A. Carson, J. J. Fuhrmann, B.-K. Hahm, P. G. Hartel, H. Yampara-Iquise, L. Johnson, R. L. Kuntz, C. H. Nakatsu, M. J. Sadowsky, and M. Samadpour. 2003. Comparison of genotypic-based microbial source tracking methods requiring a host origin database. J. Water Health 01:167-180. [PubMed] [Google Scholar]
- 26.Obiri-Danso, K., and K. Jones. 2000. Intertidal sediments as reservoirs for hippurate negative campylobacters, salmonellae and faecal indicators in three EU recognized bathing waters in north west England. Water Res. 23:519-527. [Google Scholar]
- 27.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes, M. W., and H. Kator. 1988. Survival of Escherichia coli and Salmonella spp. in estuarine environments. Appl. Environ. Microbiol. 54:2902-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherer, B. M., J. R. Miner, J. A. Moore, and J. D. Buckhouse. 1992. Indicator bacterial survival in stream sediments. J. Environ. Qual. 21:591-595. [Google Scholar]
- 30.Sinton, L. W., C. H. Hall, P. A. Lynch, and R. J. Davies-Colley. 2002. Sunlight inactivation of fecal indicator bacteria and bacteriophages from waste stabilization pond effluent in fresh and saline waters. Appl. Environ. Microbiol. 68:1122-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Topp, E., M. Welsh, Y.-C. Tien, A. Dang, G. Lazarovits, K. Conn, and H. Zhu. 2003. Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microbiol. Ecol. 44:303-308. [DOI] [PubMed] [Google Scholar]
- 33.Urakawa, H., K. Kita-Tsukamoto, and K. Ohwada. 1999. 16S rDNA restriction fragment length polymorphism analysis of psychrotrophic vibrios from Japanese coastal water. Can. J. Microbiol. 45:1001-1007. [PubMed] [Google Scholar]
- 34.U.S. Environmental Protection Agency. 2000. Improved enumeration methods for the recreational water quality indicators: enterococci and Escherichia coli EPA-821/R-97/004. U.S. Environmental Protection Agency, Washington, D.C.
- 35.U.S. Environmental Protection Agency. 1997. Method 1600: membrane filter test method for enterococci in water EPA 821-R-97-004. Office of Water and Hazardous Materials, U.S. Environmental Protection Agency, Washington, D.C.
- 36.Whitlock, J. E., D. T. Jones, and V. J. Harwood. 2002. Identification of the sources of fecal coliforms in an urban watershed using antibiotic resistance analysis. Water Res. 36:4273-4282. [DOI] [PubMed] [Google Scholar]
- 37.Whitman, R. L., and M. B. Nevers. 2003. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl. Environ. Microbiol. 69:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittam, T. S. 1989. Clonal dynamics of Escherichia coli in its natural habitat. Antonie Leeuwenhoek 55:23-32. [DOI] [PubMed] [Google Scholar]
- 39.Wiggins, B. A. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiggins, B. A., R. W. Andrews, R. A. Conway, C. L. Corr, E. J. Dobratz, D. P. Dougherty, J. R. Eppard, S. R. Knupp, M. C. Limjoco, J. M. Mettenburg, J. M. Rinehardt, J. Sonsino, R. L. Torrijos, and M. E. Zimmerman. 1999. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl. Environ. Microbiol. 65:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf, H. W. 1972. The coliform count as a measure of water quality. In R. Mitchell (ed.), Water pollution microbiology. Wiley Interscience, New York, N.Y.