Abstract
The aims of the present investigation were to develop and test a sensitive and reproducible method for the study of gene expression in the porcine lung pathogen Actinobacillus pleuropneumoniae by real-time quantitative reverse transcription (RT)-PCR and to evaluate a number of suitable internal controls, as such controls have not been defined yet for this bacterium. Bacterial gene expression was studied during in vitro exponential and early stationary growth in medium with and without sufficient iron, respectively. First, the stability of expression of five genes, the glyA, tpiA, pykA, recF, and rhoAP genes involved in basic housekeeping, was evaluated on the basis of the mean pairwise variation. All the housekeeping genes included were stably expressed under the conditions investigated and consequently were included in the normalization procedure. Next, the geometric mean of the internal control genes was used to correct five genes of interest. These genes were three genes involved in iron acquisition (tbpA, exbB, and fhuD), the heat shock protein gene groEL, and a putative quorum-sensing gene (luxS). The level of tbpA, exbB, and fhuD expression in A. pleuropneumoniae showed significant up-regulation under iron-restricted conditions compared to bacteria grown in medium with sufficient iron. The observed expression patterns of the genes of interest were consistent with previous observations. This study therefore lends further support to the use of real-time quantitative RT-PCR, with the glyA, tpiA, pykA, recF, and rhoAP genes as internal controls, for future similar gene expression studies in A. pleuropneumoniae.
An infected host represents a complex and dynamic environment to which the invading pathogen constantly needs to respond. Understanding how bacteria survive and multiply in this hostile environment and which genes are involved in this process is a prerequisite for the rational design of novel approaches for the treatment and prevention of disease.
Porcine pleuropneumonia caused by Actinobacillus pleuropneumoniae is one of the important bacterial diseases of the respiratory tract of the pig; it is distributed worldwide and results in severe losses in the pig-rearing industry (8, 23). The pathogenesis of this disease, which is complex and still not completely understood, was chosen as a model for the study of the interrelated host and pathogen gene expression patterns during an infection. In this context, real-time quantitative reverse transcription-PCR (qRT-PCR) is the method of choice, both as an independent method for expression analysis and as a method to confirm results of differential gene regulation obtained by microarray analysis.
The accuracy of a relative comparison by qRT-PCR depends largely on the normalization of raw values by means of stably expressed internal reference genes, which are used to eliminate sample-to-sample variation of RNA isolation and reverse transcription. However, the “housekeeping” metabolism of prokaryotes has been reported to be highly variable depending on the experimental procedure (26). Identification of candidate genes that are at least minimally regulated under the conditions investigated and preferably the inclusion of more than one reference gene in the analysis are therefore important for the accuracy of the qRT-PCR test (20, 22, 27).
In order to validate internal reference genes for a qRT-PCR assay and to test the reproducibility of the method, the expression of five housekeeping genes and five genes of interest in response to low-iron conditions was examined during in vitro exponential and early stationary growth in the porcine lung pathogen A. pleuropneumoniae.
Iron is essential for bacterial growth and replication, and the ability to acquire this micronutrient from the very limited free iron sources in the mammalian host is an important component of bacterial virulence (11). The five genes of interest that were analyzed by qRT-PCR included transferrin binding protein gene A (tbpA) (5, 7) and inner membrane protein complex gene B (exbB), both encoding proteins involved in iron binding and transport (25) and considered to be important for A. pleuropneumoniae virulence (1, 2). Furthermore, previous studies of A. pleuropneumoniae expression by polyacrylamide gel electrophoresis and immunoblotting have demonstrated that proteins encoded by tpbA and exbB are expressed only under iron-limiting growth conditions (5, 25). Another A. pleuropneumoniae iron transport system, via ferrichrome, was represented in the analysis by a gene coding for ferric hydroxamate uptake protein (fhuD), which is part of the fhuCDBA operon (14). Previous results indicated that the regulation of fhuA appears to be independent of the iron supply (15).
Also included in the analysis were the heat shock protein-encoding gene groEL (28) and a putative quorum-sensing protein-encoding gene (luxS) responsible for the production of autoinducer 2, which has been reported to be important for aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation conditions (6).
The putative reference genes included in the analysis were genes encoding recombinase F (recF) (12), glycine/serine hydroxymethyl transferase A (glyA), rho factor (rhoAP) (17), triosephosphate isomerase A (tpiA), and pyruvate kinase A (pykA) (13). The expression stabilities of these genes were measured as proposed by Vandesompele et al. (27), and the resulting normalization factor was used to correct the raw values for the five genes of interest.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
Type strain L20 (serotype 5b) of A. pleuropneumoniae was used in this investigation. For the iron depletion study, an overnight culture was diluted 1:10 in brain heart infusion supplemented with 0.03% NAD (Sigma-Aldrich, Copenhagen, Denmark) and incubated until the optical density at 660 nm was 0.7. The culture was split into two 50-ml portions. Iron-depleted conditions were established in one of the flasks by addition of the iron chelator 2,2′-dipyridyl (Sigma-Aldrich) (300 μM). The cultures were grown in a shaking incubator at 37°C. Samples (0.5 ml) were removed from both cultures (control and sample) 0, 10, 20, 30, 40, and 60 min after addition of the iron chelator, and these samples represented the exponential and early stationary growth phases (the optical density changed from 0.7 at time zero to 1.3 after 1 h of growth). Samples were harvested by centrifugation, immediately resuspended in RNAlater RNA stabilization reagent as described by the manufacturer (Ambion, Cambridgeshire, United Kingdom), and stored at 4°C until they were used; however, they were not stored for more than 1 month.
RNA isolation and reverse transcription.
Total RNA was isolated from 0.5-ml portions of bacterial samples by using an RNeasy mini kit (QIAGEN, Hilden, Germany) as described by the manufacturer. Genomic DNA was eliminated by RNase-free DNase I treatment during the isolation procedure. Reverse transcription was performed at 42°C for 120 min using random hexamer primers and 100 U of Moloney murine leukemia virus reverse transcriptase (Ambion). For each time extraction was performed in duplicate. Furthermore, serial dilutions containing from approximately 1.8 μg to 0.6 ng of total RNA extracted from a control sample at an optical density at 660 nm of 0.58 were reverse transcribed.
Quantitative real-time PCR.
Gene quantification was performed with a Rotor-Gene 3000 (Corbett Research, Sydney, Australia). Primers were designed from GenBank (http://www.ncbi.nlm.nih.gov/) sequences with the aid of DNASIS MAX V1.0 (Hitachi Software Engineering Company, Ltd., San Francisco, CA). The sequences of the primers are shown in Table 1. Each PCR was performed in a 20-μl reaction mixture containing SYBR Green I (stock diluted 1:20,000; BMA, Rockland, ME) and 0.5 U of AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA). The final concentrations of primers and Mg2+ were 0.5 μM and 4 mM, respectively. The thermal cycling conditions were as follows: 6 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Data collection was performed during each extension phase. Positive controls (DNA), a negative control (distilled water), and RT-negative controls (total RNA sample) were included in each run. For each of the RNA extractions, measurements of gene expression were obtained in triplicate, and the mean of these values was used for further analysis.
TABLE 1.
Primers used in real-time qRT-PCR study
| Target | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| recF | TATGCCGAGATTCTTGCTCA | AATTTAAGCTGCCCACGAGA |
| glyA | CAAGCGAATGCAGCTGTTTA | CTGTGATGCCGTAGAGGACA |
| rhoAP | AATACCGTGACGCCTGTTTC | ACTAATGCCGTCGCGATAAT |
| tpi | CTACGAACCGATTTGGGCTA | CCGCCGTATTGGATAATCAC |
| pykA | GTACGGATGCGGTAATGCTT | ACCTTCCATACGGTGACGAG |
| groEL | CGCAAATCAAAGCACGAGTA | ACCGGCTAATTTTGCAACAC |
| luxS | CGTGTTGCAAAAACGATGAC | GCATAAAGCCGGCAAATAAG |
| tbpA | ATTCAGCCTGCTCGCTATGT | GGCAAAACCGACTTGTTCAT |
| exbB | CCGTTCATTGGGTTATTTGG | AAATAGTCCCATTGCCGTTG |
| fhuD | TTTTATACGGAAGGCGATGC | ACAAGCGGTCGAATTTCATC |
Primer specificity.
A Rotor-Gene 3000 melting curve analysis was performed, which for all primer sets resulted in single product-specific melting curves. Gel electrophoresis and a melting curve analysis of PCR products showed that very little or no primer-dimers were generated during the runs.
Relative quantification.
The relative expression ratios were calculated by a mathematical model, which included an efficiency correction for real-time PCR efficiency of the individual transcripts (18), as follows:
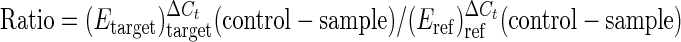 |
The relative expression ratio of a target gene was computed based on its real-time PCR efficiencies (E) and the crossing point difference (ΔCt) for an unknown sample versus a control. For each gene, cDNA dilution curves were generated and used to calculate the individual real-time PCR efficiencies (E = 10[−1/slope]). The geometric mean of the five internal reference genes was used to correct the raw values for the genes of interest.
Statistical analysis.
The stability of the mRNA expression of the reference genes was evaluated by using the freely distributed MS Excel application geNorm (27). This approach is based on the assumption that the expression ratio of two ideal control genes is the same for all samples, regardless of the experimental conditions, and that increasing variation in this ratio corresponds to decreasing expression stability. For each reference gene, the program generates the expression stability measure M, which represents the average pairwise variation for a particular reference gene with all other reference genes tested (27). This measure is used to select the most stable genes in a particular experimental situation. In general, the lower the M value, the more stable the housekeeping genes expressed and the more precise the quantification of the experiment. Stepwise exclusion of genes with the highest M values allows ranking of the genes tested according to their expression stabilities. A reliable normalization factor, averaging individual housekeeping gene-specific differences, is obtained by using the geometric mean of multiple relatively stable control genes (27). The geometric mean of the best-scoring reference genes was used as a normalization factor in the analysis.
The total expression ratio of the five genes of interest over 1 h was tested for significance by a randomization test implemented in the relative expression software tool (REST) (www.gene-quantification.info), which is an Excel-based application for groupwise comparison and statistical analysis of relative expression results in qRT-PCR (19).
RESULTS
Validation of reference genes.
To compare the expression levels of each reference gene, bacterial samples were taken at six different times, 0, 10, 20, 30, 40, and 60 min after addition of the iron chelator to the media. qRT-PCR analysis was performed twice for different RNA extracts from the same growth experiment. The geNorm program was used to evaluate the expression stability of each reference gene at the different times during growth. The results of the two qRT-PCR runs are ranked and shown in Table 2. Genes with the lowest M values have the most stable expression values. The stability of the genes as assessed by geNorm analysis was between 0.17 and 0.55 in the first run and between 0.35 and 0.87 in the second run; recF and glyA were the most stably expressed genes in the first run, and pykA and glyA were the most stably expressed genes in the second run.
TABLE 2.
geNorm stability resultsa
| Gene | geNorm stability measure (M)
|
|
|---|---|---|
| Run 1 | Run 2 | |
| rhoAP | 0.554 (3) | 0.868 (4) |
| pykA | 0.615 (4) | 0.346 (1) |
| tpiA | 0.401 (2) | 0.821 (3) |
| recF | 0.170 (1) | 0.508 (2) |
| glyA | 0.170 (1) | 0.346 (1) |
geNorm stability values were determined for the five housekeeping genes included in this study. Genes with the lowest M values have the most stable expression values. M is the standard deviation of the logarithmically transformed expression ratios with all other control genes. The ranks of the genes are indicated in parentheses.
Test reproducibility for RNA isolation and qRT-PCR.
Test precision (intra-assay variation) and test variability (interassay variation) were evaluated by the Ct variation from the mean Ct value. RNA isolation was performed with samples containing approximately 107 CFU. Repeated RNA extractions of the same bacterial sample resulted in an interassay variability between 0.04 and 13.9% and an average variability of 4.10%. Normalization by using the geometric mean of the five reference genes reduced the standard deviation of the total interassay variations for all genes of interest between one-third and one-fifth. The intra-assay precision was determined for three repeats within one Rotor-Gene run and was found to be between 0 and 6.3%, with an average variability of 1.14%.
Validation of reverse transcriptase inhibition and dynamic range.
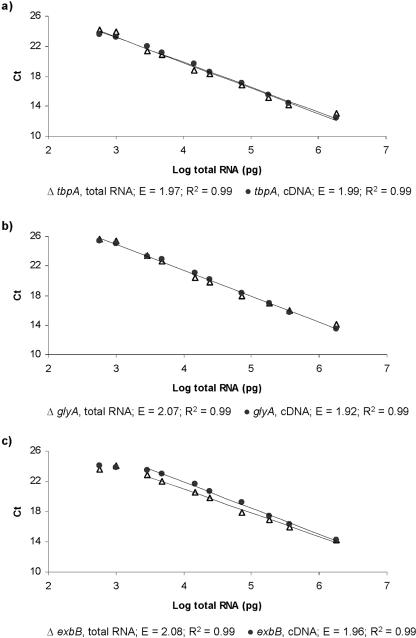
For three of the genes, tbpA, exbB, and glyA, standard curve analyses were performed for both cDNA and total RNA dilution series. The standard curves were all highly linear (R2 > 0.98) over a wide dilution range, with slopes greater than −3.1 and less than −3.6, indicating a PCR efficiency of 1.92 to 2.08
Figure 1 shows the Ct values for the two standard curves for the tbpA, exbB, and glyA genes. The input amount for the real-time PCR ranged between 1.8 μg and 0.6 ng of reverse-transcribed total RNA per reaction mixture. No significant difference in the efficiency levels between the cDNA and total RNA dilution series were observed for any of the genes. All the dilutions used are within the dynamic range of the glyA and tbpA genes. For the exbB gene, the dynamic range is 1.8 μg to 3.0 ng of reverse-transcribed total RNA.
FIG. 1.
Standard curves for total RNA and cDNA of tbpA (a), glyA (b), and exbB (c) generated by plotting the log of the reverse-transcribed total RNA concentration versus the threshold cycle (Ct). Linear regression analysis was used to determine the slope which corresponded to the amplification efficiency, E = 10(−1/slope) (18).
Expression of genes of interest after iron deprivation.
The total expression of the five genes of interest during 1 h after the addition of the iron chelator 2,2′-dipyridyl was analyzed by using the relative expression software tool (REST) (19), and the geometric means of the Ct values for each time for the five housekeeping genes were used to normalize the data. The results for the two runs of the experiment based on separate RNA extractions are shown in Table 3. In both runs, a significant increase (P < 0.05) in the expression of the tbpA, exbB, and fhuD genes was observed under iron-deprived conditions compared to growth in iron-replete media, and the mean levels of fhuD, tbpA, and exbB were up-regulated approximately two-, four-, and eightfold, respectively. The remaining two genes, luxS and groEL, showed no significant regulation (Table 3). For none of the genes did normalization alter the relative expression ratios significantly.
TABLE 3.
Data from a pairwise fixed reallocation randomization test (2,000 randomizations) of the relative expression of the five genes of interest, before and after normalization
| Run | Parameter | groEL | luxS | tbpA | exbB | fhuD |
|---|---|---|---|---|---|---|
| 1 | Expression ratio | 0.84 | 0.87 | 3.67 | 8.30 | 1.82 |
| P value | 0.501 | 0.223 | 0.016 | 0.002 | 0.016 | |
| Expression ratios (not normalized) | 0.82 | 0.85 | 3.57 | 8.10 | 1.78 | |
| P value (not normalized) | 0.560 | 0.314 | 0.009 | 0.002 | 0.004 | |
| 2 | Expression ratio | 1.04 | 0.94 | 4.00 | 8.11 | 1.94 |
| P value | 0.911 | 0.624 | 0.014 | 0.006 | 0.047 | |
| Expression ratio (not normalized) | 0.89 | 0.80 | 3.40 | 6.94 | 1.66 | |
| P value (not normalized) | 0.740 | 0.280 | 0.008 | 0.006 | 0.041 |
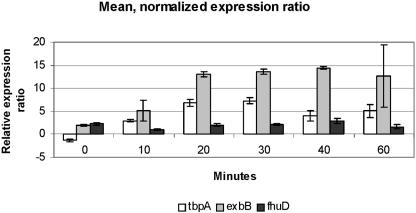
Figure 2 shows the mean normalized expression ratios of the three significantly up-regulated genes 0, 10, 20, 30, 40, and 60 min after addition of the iron chelator. The expression of the tbpA and exbB genes increased rapidly during the first 20 min, after which exbB expression remained relatively stable at the same level, while the expression of tbpA seemed to peak after 30 min of growth under iron-deprived conditions, after which it slowly started to level off. Besides a small drop after 10 min of growth under iron deprivation conditions, the level of expression of fhuD remained slightly up-regulated during the whole observation period.
FIG. 2.
Mean relative expression levels for the three significantly regulated genes at each time after addition of the iron chelator 2,2′-dipyridyl, normalized by the geometric mean for the five housekeeping genes. The data are the means ± standard deviations (n = 2).
DISCUSSION
The aims of the present investigation were to develop and test a sensitive and reproducible method for expression analysis of A. pleuropneumoniae and, as it has been shown that housekeeping gene expression can vary considerably (3, 24, 26), to validate that the reference genes used were minimally regulated in response to experimental treatment. In this context, qRT-PCR was used to monitor the expression of 10 selected genes from A. pleuropneumoniae cultivated under iron-restricted conditions. Previous studies of A. pleuropneumoniae expression by other methods (5, 25) have shown that the exbB and tpbA genes are expressed only under iron-limiting conditions. Consequently, these two genes were chosen with the aim of verifying previous results. Two other genes, fhuD and luxS, which might be expected to be up-regulated in response to diminished levels of iron, were included as genes of interest along with a chaperone gene, groEL, involved in protein folding and assembly.
In order to avoid coregulation, the five housekeeping genes in this study were selected from different functional groups. The tpiA gene codes for a protein involved in gluconeogenesis, while the pykA gene product is a key enzyme in glycolysis. The recF gene is involved in the minor recombination pathway, the rhoAP gene encodes a protein factor involved in the termination of transcription, and finally, the glyA gene product is a catabolic protein involved in converting serine to glycine.
A reliable normalization factor, which is the average of the individual specific expression differences of the genes, is obtained by using the geometric mean of multiple relatively stable housekeeping genes (27). As the levels of expression of the five candidate reference genes in the present study varied less than onefold from each other, they were all included in the calculation of a gene expression normalization factor. However, except for a reduction in the interassay variation, normalizing the genes of interest does not alter the results of the expression analysis significantly in this experimental setup.
As expected from previous investigations (5, 25), the exbB and tbpA genes are significantly up-regulated in response to diminished levels of iron, both before and after normalization. Also, the observed up-regulation of the two genes within 15 to 20 min after establishment of iron starvation (Fig. 2) is in accord with previous observations made by Deneer and Potter (5), who detected a similar rapid induction of iron-repressible outer membrane proteins. Although the exbB and tbpA genes are most likely transcribed on a single polycistronic mRNA (described for serotype 7 by Tonpitak et al. [25]), the observed mean expression of tbpA increased approximately fourfold, while that of the exbB gene increased eightfold in this study. Such a discrepancy has been observed in a number of prokaryotes (9, 10, 16) and might be explained by differential expression of cotranscribed genes due to variation in posttranscriptional mRNA degradation, which seems to be an important step in regulation of prokaryotic gene expression (21).
A previous study found that regulation of the ferrichrome receptor, encoded by fhuA, is independent of the iron supply (15). In this investigation, the mean expression of the fhuD gene, which belongs to the same operon as fhuA, was found to be up-regulated twofold in response to low iron levels. Again, this might be explained by differences in the rate of mRNA degradation, or the observation could be due to the higher sensitivity of the real-time qRT-PCR assay, which might have been able to detect changes in gene expression smaller than changes detected by the traditional RT-PCR method used by Mikael et al. (15). However, to clarify this, further investigation by real-time qRT-PCR, including all genes of the fhu operon, is probably needed. The levels of expression of the groEL and luxS genes did not increase significantly in response to low-iron conditions. Although luxS expression is apparently not stimulated by a reduction in the iron concentration, further investigations, involving construction of a luxS null mutant, are needed to evaluate the role of this gene in A. pleuropneumoniae growth under iron-restricted conditions.
Reverse transcriptase is known to inhibit PCR at low concentrations of template (4). In order to assess the effects of reverse transcription over the dynamic range of potential mRNA expression levels, comparative dilution series of total RNA and cDNA, ranging from approximately 1.8 μg to 0.6 ng of reverse-transcribed total RNA per reaction mixture, were amplified by one control gene, glyA, and two genes of interest, tbpA and exbB.
In this study no inhibitory effect of reverse transcriptase on the PCR were observed at any template concentration. Assuming that mRNAs rarely make up more than 5% of the total RNA, it was possible to detect and amplify at least down to 30 pg of mRNA and likely much less mRNA, considering that with no iron deprivation of the bacteria, two of the genes tested, tbpA and exbB, are expressed at very low levels in vitro (5, 25).
In conclusion, as previously stated (20, 26) ideal and universal housekeeping genes do not exist. Therefore, one should search for stably expressed genes specific to each experimental system. Although the geometric mean of the five housekeeping genes did not alter the results significantly in the present investigation, the genes were all found to be minimally regulated in this study. With this in mind, the present investigation demonstrated that qRT-PCR is an easy, applicable, and reproducible tool with which to study A. pleuropneumoniae gene expression under various biological conditions which this bacterium may encounter when it invades a host organism and that it is an independent method for verification of microarray expression data.
Acknowledgments
We thank Oystein Angen for supplying the A. pleuropneumoniae strain and Joanna Amenuvor for technical assistance.
Kirstine K. Nielsen is supported by a grant from the Danish Agricultural and Veterinary Research Council (project 23-02-0137).
REFERENCES
- 1.Baltes, N., I. Hennig-Pauka, and G. F. Gerlach. 2002. Both transferrin binding proteins are virulence factors in Actinobacillus pleuropneumoniae serotype 7 infection. FEMS Microbiol. Lett. 209:283-287. [DOI] [PubMed] [Google Scholar]
- 2.Baltes, N., W. Tonpitak, G. F. Gerlach, I. Hennig-Pauka, A. Hoffmann-Moujahid, M. Ganter, and H. J. Rothkotter. 2001. Actinobacillus pleuropneumoniae iron transport and urease activity: effects on bacterial virulence and host immune response. Infect. Immun. 69:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 4.Chandler, D. P., C. A. Wagnon, and H. Bolton. 1998. Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Appl. Environ. Microbiol. 64:669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deneer, H. G., and A. A. Potter. 1989. Effect of iron restriction on the outer membrane proteins of Actinobacillus (Haemophilus) pleuropneumoniae. Infect. Immun. 57:798-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong, K. P., L. Gao, and D. R. Demuth. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlach, G. F., S. Klashinsky, C. Anderson, A. A. Potter, and P. J. Willson. 1992. Characterization of two genes encoding distinct transferrin-binding proteins in different Actinobacillus pleuropneumoniae isolates. Infect. Immun. 60:3253-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haesebrouck, F., K. Chiers, O. Van, and R. Ducatelle. 1997. Actinobacillus pleuropneumoniae infections in pigs: the role of virulence factors in pathogenesis and protection. Vet. Microbiol. 58:239-249. [DOI] [PubMed] [Google Scholar]
- 9.Heck, C., H. Evguenieva, A. Balzer, and G. Klug. 1999. RNase E enzymes from Rhodobacter capsulatus and Escherichia coli differ in context- and sequence-dependent in vivo cleavage within the polycistronic puf mRNA. J. Bacteriol. 181:7621-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homuth, G., A. Mogk, and W. Schumann. 1999. Post-transcriptional regulation of the Bacillus subtilis dnaK operon. Mol. Microbiol. 32:1183-1197. [DOI] [PubMed] [Google Scholar]
- 11.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loynds, B. M., P. R. Langford, and J. S. Kroll. 1992. recF in Actinobacillus pleuropneumoniae. Nucleic Acids Res. 20:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikael, L. G., P. D. Pawelek, J. Labrie, M. Sirois, J. W. Coulton, and M. Jacques. 2002. Molecular cloning and characterization of the ferric hydroxamate uptake (fhu) operon in Actinobacillus pleuropneumoniae. Microbiology 148:2869-2882. [DOI] [PubMed] [Google Scholar]
- 15.Mikael, L. G., R. Srikumar, J. W. Coulton, and M. Jacques. 2003. fhuA of Actinobacillus pleuropneumoniae encodes a ferrichrome receptor but is not regulated by iron. Infect. Immun. 71:2911-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newbury, S. F., N. H. Smith, and C. F. Higgins. 1987. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 51:1131-1143. [DOI] [PubMed] [Google Scholar]
- 17.Oswald, W., D. V. Konine, J. Rohde, and G. F. Gerlach. 1999. First chromosomal restriction map of Actinobacillus pleuropneumoniae and localization of putative virulence-associated genes. J. Bacteriol. 181:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radonić, A., S. Thulke, I. M. Mackay, O. Landt, W. Siegert, and A. Nitsche. 2004. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313:856-862. [DOI] [PubMed] [Google Scholar]
- 21.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23:353-370. [DOI] [PubMed] [Google Scholar]
- 22.Savli, H., A. Karadenizli, F. Kolayli, S. Gundes, U. Ozbek, and H. Vahaboglu. 2003. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52:403-408. [DOI] [PubMed] [Google Scholar]
- 23.Taylor, D. J. 1999. Actinobacillus pleuropneumoniae, p. 343-354. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine. Iowa State University Press, Ames.
- 24.Thellin, O., W. Zorzi, B. Lakaye, B. De, B. Coumans, G. Hennen, T. Grisar, A. Igout, and E. Heinen. 1999. Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75:291-295. [DOI] [PubMed] [Google Scholar]
- 25.Tonpitak, W., S. Thiede, W. Oswald, N. Baltes, and G. F. Gerlach. 2000. Actinobacillus pleuropneumoniae iron transport: a set of exbBD genes is transcriptionally linked to the tbpB gene and required for utilization of transferrin-bound iron. Infect. Immun. 68:1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, and E. Van. 2001. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J. Bacteriol. 183:7094-7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandesompele, J., P. De, F. Pattyn, B. Poppe, R. Van, P. De, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1-0034.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vezina, G., M. Sirois, N. Clairoux, and M. Boissinot. 1997. Cloning and characterization of the groE locus from Actinobacillus pleuropneumoniae. FEMS Microbiol. Lett. 147:11-16. [DOI] [PubMed] [Google Scholar]