Abstract
Recently a facile method for genotyping single nucleotide polymorphisms (SNPs) using MALDI mass spectrometry, termed the GOOD assay, was developed. It does not require any purification and is performed with simple liquid handling, thermal incubation and cycling steps. Although this method is well suited to automation and high-throughput analysis of SNPs, it did not allow full flexibility due to lack of certain reagents. A complete set of β-cyanoethyl phosphoramidites is presented herein that give this SNP genotyping method full sequence and multiplex capabilities. Applications to SNP genotyping in the prion protein gene, the β-2-adrenergic receptor gene and the angiotensin converting enzyme gene using the GOOD assay are demonstrated. Because SNP genotyping technologies are generally very sensitive to varying DNA quality, the GOOD assay has been stabilised and optimised for low quality DNA. A template extraction method is introduced that allows genotyping from tissue that was taken while placing an ear tag on an animal. This dramatically facilitates the application of genotyping to animal agricultural applications, as it demonstrates that expensive and cumbersome DNA extraction procedures prior to genotyping can be avoided.
INTRODUCTION
Single nucleotide polymorphism (SNP) genotyping technologies are sought after for a variety of reasons. Pharmacogenomics may become a powerful tool in medicine, where it is postulated that depending on a particular genotype, a patient will get the most effective medication (1). Genotyping of SNPs is thought to be ideally suited to association studies (2). Additionally, SNP genotyping will have applications in quality control of food and for agricultural applications. In order to make high-throughput SNP genotyping feasible, facile and affordable procedures are required (3).
Matrix-assisted laser desorption/ionisation mass spectrometry (MALDI-MS) has revolutionised the instrumental analysis of biomolecules (4). Several protocols for SNP genotyping using MALDI-MS as a detection method have been established (5–7). The major drawback of all of these procedures is that they rely heavily on purification procedures prior to MALDI analysis. These purification procedures do not lend themselves to easy automation and often contribute a significant proportion of the cost of an analysis.
Based on the principle that the analysis of nucleic acids is strongly dependent on the charge state, a 100-fold increase in sensitivity can be achieved if the analysed DNA product carries a single positive or negative charge (8–10). We recently introduced a sample preparation method for genotyping SNPs by MALDI mass spectrometry called the ‘GOOD assay’, which does not require any purification (11). The GOOD assay fluently connects the reaction steps of PCR, shrimp alkaline phosphatase digestion of unreacted nucleotide triphosphates from the PCR, primer extension with positive or negative ‘charge tag’ carrying primers and phosphorothioate (di)deoxynucleotides, 5′-phosphodiesterase digestion to remove unmodified extension primer DNA and alkylation for charge neutralisation of the phosphorothioate backbone of the products. These products are then measured by MALDI-MS.
A limitation of the GOOD assay in positive ion mode detection was the commercial availability of only UNH2 for introduction of a charge tag near the 3′-end of the extension primer. This restricted positioning of the primer for the extension reaction. This limitation does not exist for procedures using negative ion mode, but the degree of freedom for multiplexing is much higher for the positive ion mode GOOD assay. This is of great importance for the design of efficient assays for SNP genotyping.
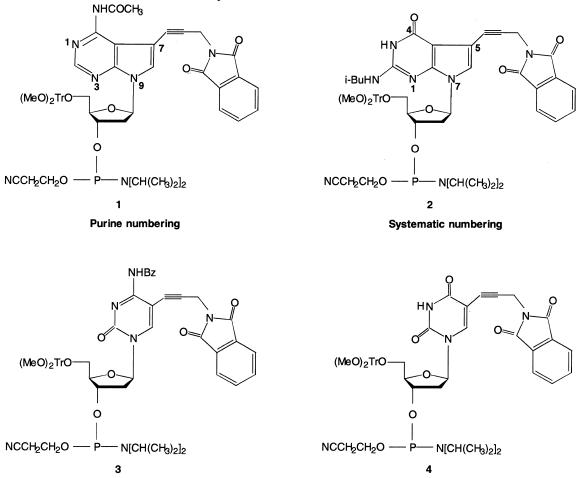
To increase the flexibility of the GOOD assay we have used four β-cyanoethyl phosphoramidites (compounds 1–4, Fig. 1) that can easily be converted into positively charged amides. A detailed description of the synthesis of these compounds will be published elsewhere.
Figure 1.
Scheme of ANH2, GNH2, CNH2 and UNH2 phosphoramidites.
Prion diseases are transmissible neurodegenerative disorders that affect a range of mammalian species such as sheep, cattle and humans (12,13). They can be transmitted between mammals by inoculation or dietary exposure to infected tissues. Bovine spongiform encephalopathy (BSE), for example, has caused a huge public upset because it may represent a threat to public health through the potential induction of Creutzfeldt–Jacob disease after the ingestion of BSE-infected beef. BSE is caused by ingestion of prion contaminated feed by cattle. The possibility of cross-species transfer of this disease is still contested, but there are indications that BSE prions can induce disease in humans.
To demonstrate SNP genotyping, we used the well-studied SNP 129 in the prion protein gene, a common polymorphism that is known to be involved in genetic susceptibility to human prion diseases. The presented assay was established to facilitate epidemiological studies and to identify high risk groups. Sequence homology between human and cattle in the prion protein gene is sufficient to allow the use of the same reagents to genotype position 129. In order to make bovine DNA amenable to genotyping, a template extraction method was introduced that allows genotyping from tissue that was taken while placing an ear tag on the animal. This dramatically facilitates the application of genotyping for animals, as it demonstrates that expensive and cumbersome DNA extraction procedures in combination with the GOOD assay can be avoided.
As other examples of the GOOD assay in positive ion mode, we present assays for SNPs –20 and 79 in the human β-2-adrenergic receptor gene and SNP 33730 in the angiotensin converting enzyme (ACE) gene. The human β-2-adrenergic receptor belongs to a family of seven-transmembrane receptors (14). SNPs in this receptor are associated with a genetic predisposition to hypertension and are found to be relevant for cardiovascular disease (15,16). ACE is a zinc metalloprotease widely distributed on the surface of endothelial and epithelial cells. It is one of the major genes of the renin–angiotensin and kallikrein–kinin systems and it is a candidate gene for hypertension and cardiovascular disease (17,18).
MATERIALS AND METHODS
Oligonucleotides for PCR were synthesised by an in-house synthesis service and were all HPLC purified. dNTPs, Taq DNA polymerase and 5′-phosphodiesterase (calf spleen) were purchased from Boehringer (Mannheim, Germany). Platinum™ Taq DNA polymerase high fidelity (THPF) was purchased from Gibco BRL (Karlsruhe, Germany). Proteinase K, α-S-dNTPs and α-S-ddNTPs were provided by Amersham (Little Chalfont, UK). Thermosequenase and shrimp alkaline phosphatase were purchased from Amersham Buchler (Braunschweig, Germany). Chemical reagents were purchased from Aldrich (Steinheim, Germany). The amino-modified phosphoramidites, charge tag reagents and matrix are available from Bruker Saxonia Analytik GmbH (Leipzig, Germany).
The β-cyanoethyl phosphoramidites 1–4 (Fig. 1) were employed in solid phase synthesis of oligonucleotides. The coupling yield of the modified DNA building block, as determined by monitoring the 4,4′-dimethoxytrityl cation at 498 nm, was always >96%. After deprotection with concentrated aqueous NH3 solution (8 h at 60°C), the 5′-(MeO)2Tr-protected oligomers were purified, detritylated, and desalted on oligonucleotide purification cartridges (Perkin Elmer, Applied Biosystems Division, Weiterstadt, Germany). Molecular masses of synthesised oligonucleotides were determined by MALDI-TOF mass spectrometry (BIFLEX III, equipped with a SCOUT MTP ion source; Bruker Saxonia Analytik GmbH).
The amino functionality of the synthesised primers were used for attaching a positive charge tag (6-trimethylammoniumhexyryl-N-hydroxy-succinimidyl ester) according to Gut et al. (9) and Bartlett-Jones et al. (19). UNH2 is converted to UCT, ANH2 to ACT, CNH2 to CCT and GNH2 to GCT. The primers containing the amino functionality were dissolved in 1% TE buffer to 500 pmol/µl. An aliquot of 30 µl of this solution was mixed with 1.5 µl of 2 M triethylammoniumhydrogencarbonate (pH 8.0) and 24 µl fresh 1% 6-trimethylammoniumhexyryl-N-hydroxy-succinimidyl ester. This reaction mixture was incubated at 0°C for 30 min. Afterwards the reaction mixture was lyophilised and resuspended in 15 µl of 300 mM ammonium acetate. Ethanol (60 µl) was added and precipitation of the oligonucleotide was completed by incubation at –20°C for 2 h. After microcentrifugation at 14 000 r.p.m. for 2 min the pellet was washed twice with 60 µl of 80% ethanol and resuspended in 30 µl of bidistilled water. The concentration was measured by UV spectroscopy and the quality of charge tagging was routinely verified by MALDI-MS. A saturated solution of hydroxypicolinic acid in 1:1 (v/v) acetonitrile and 0.1 M diammonium citrate was used as matrix. Aliquots of 0.4 µl of the matrix and 0.2 µl of the purified oligonucleotide were transferred to the MALDI target using the dried droplet sample preparation method. The charge-tagged oligonucleotides were measured in positive ion mode. Usually no residual starting material was detected, from which we conclude that >95% of the amino-modified oligonucleotides were converted into charge-tagged primers.
GOOD assay protocol
Preparation of bovine DNA. Cow ear tissue samples (5 mm3) were directly loaded straight into a sample tube with a Biopsytak (Biopsytec GmbH, Berlin, Germany). The Biopsytak pushes a projectile through the ear of the cow and lodges the projectile with some tissue in the sample tube (Biopsycap). In order to add reagents the projectile has to be punctured. Aliquots of 20 µl of buffer (50 mM Tris–HCl, 20 mM NaCl, 1 mM EDTA and 1% SDS, buffered to pH 8) and 1 µl of proteinase K (20 mg/ml) were added. The reaction was incubated for 30 min at 55°C. The reaction was then stopped by heat denaturation (100°C for 5 min), followed by addition of 200 µl water. An aliquot of 1 µl of this solution was used for the following PCR without further purification.
PCR. In the case of amplification of the prion protein gene, either 1 µl of the proteinase K-digested material or 20 ng human DNA purified as described in Sambrook et al. (20) was mixed with 5 pmol each of the forward, d(CAGCCCCATGGTGGTGGC), and reverse, d(GGTTGGGGTAACGGTGC), primers, 40 mM Tris base, 32 mM (NH4)2SO4, 50 mM KCl, 2 mM MgCl2, 200 µM dNTPs and 0.2 U Taq DNA polymerase in a 10 µl volume. The reaction was denatured for 1 min at 94°C, then thermocycled 35 times for 15 s at 94°C, 30 s at 56°C and 1 min at 68°C.
In the case of amplification of the gene for the β-2-adrenergic receptor, 3.5 pmol each of the forward, d(GACAAGCTGAGTGTGCAGGA), and reverse, d(AACTTGGCAATGGCTGTGAT), primers were used. The same reaction conditions as above were used. The reaction was denatured for 2 min, then thermocycled for 20 s at 95°C, 30 s at 68°C and 30 s at 72°C, repeating the cycle 35 times.
In the case of amplification of the ACE gene, 7.5 pmol each of the forward, d(GACAAGCTGAGTGTGCAGGA), and reverse, d(GCCAACATGATTAAACCCC), primers were mixed together with 10 ng genomic DNA, 1 µl of TPHF (10×) buffer, 1 µl of MgSO4 (50 mM), 200 µM dNTPs and 0.25 U TPHF DNA polymerase in a 10 µl volume. The reaction was denatured for 4 min at 94°C, then thermocycled for 30 s at 94°C, 45 s at 65°C and 30 s at 72°C, repeated 30 times.
Shrimp alkaline phosphatase digest. An aliquot of 0.5 µl (1 U/µl) of shrimp alkaline phosphatase was added to the PCR reaction and incubated for 1 h at 37°C, followed by denaturation at 90°C for 10 min.
Primer extension reaction. An aliquot of 25 pmol of the modified primer [in the case of the prion protein gene: d(GGGGCCTTGGTGGCTPTACTPTC); in the case of the β-2-adrenergic receptor gene d(CGCGCAGTCTGGCAGPTGCTPTT) (position –20) and d(CGGACCACGTCACGCPTACTPTG) (position 79); in the case of the ACE gene d(CATGTCCTCTAAATGGTTTPTCCTPTC)] was added. These primers contain two phosphorothioate bridges, indicated by PT. In the case of the human prion gene there is a mismatch at position 11. The primer contains a T while the human sequence contains a C. Nevertheless, this did not decrease the efficiency of the assay. To this reaction mixture 2 mM MgCl2, 0.2 mM MnCl2, 100 µM respective α-S-ddNTPs and 0.5 U Thermosequenase were added. The reaction volume was increased to 20 µl by the addition of water. An initial denaturing step of 2 min at 95°C was used, followed by 35 cycles of 10 s at 95°C, 30 s at 58°C and 15 s at 72°C.
Primer digestion, alkylation reaction and sample preparation for MALDI analysis. These steps were performed as previously described (11).
Mass spectrometric analysis. Spectra were recorded on a Bruker Reflex III time-of-flight mass spectrometer. This mass spectrometer is equipped with a Scout MTP™ ion source with delayed extraction. Spectra were recorded in positive ion linear time-of-flight mode. A typical acceleration potential was 18 kV. For delayed extraction, the extraction delay was 200 ns.
RESULTS
We recently described a procedure for the facile analysis of SNPs called the ‘GOOD assay’ (11). This protocol was previously only applicable to versatile SNP genotyping in negative ion MALDI-MS mode. In this regime instrument performance is slightly less favourable. In positive ion mode this assay was limited by the commercial availability of only UNH2 for attaching charge tags.
Here we have used a set of β-cyanoethyl phosphoramidites (Fig. 1) that can be easily charge tagged and used in the GOOD assay. To demonstrate the robust performance of this assay, low quality DNA (not extracted and non-purified) was used.
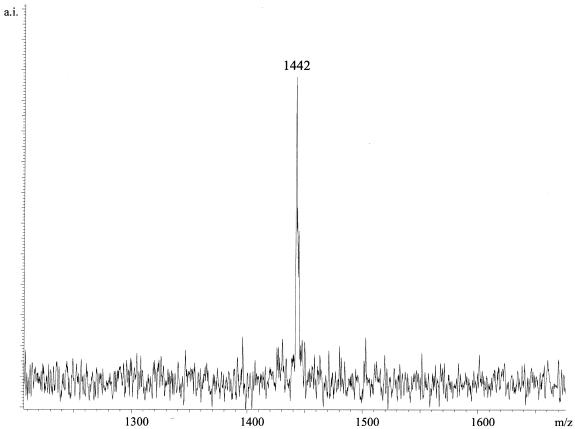
The GOOD assay was established for genotyping SNP 129 in the prion protein gene. In the case of PCR with bovine DNA, the procedure started with template obtained by proteinase K digestion of tissue taken from the animal’s ear without purification. An aliquot of 1 µl of this preparation was taken for the PCR reaction. Applying the same PCR conditions and materials, amplification of a stretch of the human and bovine prion protein gene containing SNP 129 was achieved. The PCR product was used for the primer extension reaction of the GOOD assay under standard conditions. Figure 2 shows analysis of heterozygous and homozygous human DNA at SNP 129 of the prion protein gene. Figure 3 shows the spectrum of a cattle homozygous for the A allele at SNP 129. Eight human and four cattle DNAs were used for this study. The sequences were confirmed by DNA sequencing.
Figure 2.
The top trace shows the MALDI-MS analysis of an individual homozygous for G at SNP 129 of the prion protein gene. The resulting product of the GOOD assay d(GptACTptGptdG) has a mass of 1458 Da. In the middle, the analysis of an individual homozygous for A with the resulting product d(GptACTptGptdA) and corresponding mass 1442 Da is shown. At the bottom, the analysis of heterozygous DNA is shown.
Figure 3.
The GOOD assay applied to SNP 129 of the prion protein gene on bovine DNA. The assay started with tissue samples that were only proteinase K digested and not purified.
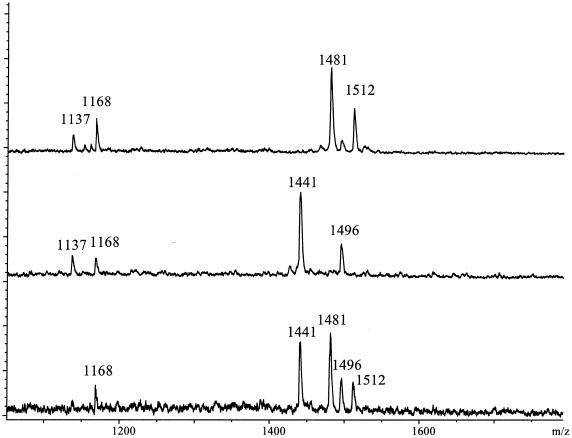
In Figure 4, analysis of two SNPs of the β-2-adrenergic receptor in a duplex GOOD assay is shown. The GOOD assay in this case was executed with primers for the primer extension reaction containing a GCT and an ACT. Twenty human DNAs were used for this study and controlled by DNA sequencing. In Figure 5 the analysis of a SNP in ACE is shown. The primer extension was done with a primer containing a CCT. Ninety-six DNAs were used in this study and the results were verified by DNA sequencing.
Figure 4.
MALDI-MS genotyping of SNPs –20 and 79 in the human β-2-adrenergic receptor gene. Product masses of the alleles are d(GptGCTptTptdA), 1496 Da and d(GptGCTptTptdG), 1512 Da for SNP –20 and d(CptACTptGptdG), 1481 Da and d(CptACTptGptdC), 1441 Da for SNP 79. At 1437 and 1168 Da residual primers of the primer extension reaction are observed. The three spectra show a homozygous individual for either allele and a heterozygous individual.
Figure 5.
MALDI-MS genotyping of SNP 33710 in ACE. Product masses of the alleles are d(TptCCTptCptdA), 1417 Da and d(TptCCTptCptdG), 1433 Da. At 1090 Da residual primer of the primer extension reaction is found. The three spectra show a homozygous individual for either allele and a heterozygous individual.
All spectra exhibited near baseline isotopic resolution and a more than sufficient signal-to-noise ratio for easy allele calling.
DISCUSSION
In this study the potential of the GOOD assay in positive ion mode detection was enhanced by several new developments.
Using a set of propargylamino-modified phosphoramidites, ANH2, GNH2, CNH2 and UNH2, introduced in primer synthesis, full flexibility of positioning of a primer for the primer extension reaction is now possible. Using charge tag reagents with different masses, such as 6-trimethylammoniumpentyryl-N-hydroxy-succinimidyl ester and similar compounds, results in a mass/charge tag system which allows much higher multiplexing than the ‘3-fold-plex’ factor that is achievable in the negative ion mode version of the GOOD assay (11). The achievable degree of multiplexing in positive ion mode is currently under investigation and will be reported at a later date. Furthermore, we have shown that it is possible to measure positive and negative charge-tagged products in the same sample preparation (10). Therefore, by switching the ion mode in the mass spectrometer, the potential for multiplexing is doubled.
The GOOD assay procedure was adapted to allow the use of low quality DNA without sacrificing robustness. This is interesting as it allows this method to be extended to streamlined genetic fingerprinting of cattle.
Establishing the GOOD assay using the same conditions for genotyping SNP 129 in the prion protein of cows and humans, we benefited from the high homology of the prion protein gene. The assay for this polymorphism in bovine DNA could be applied to the study of large numbers of cattle in order to determine genetic susceptibility linked with this polymorphism. Alternatively, the basis is provided for establishing genotyping of other SNPs of the prion protein gene that might have a bearing on the development of BSE in cattle.
The identification of genes involved in polygenic diseases, such as cardiovascular diseases, hypertension and obesity, are rather difficult to identify, in contrast to monogenic diseases (21). Their identification requires statistically relevant numbers of DNA samples from >1000 individuals for association studies. The GOOD assay for SNP genotyping is a solution for high-throughput genotyping, which is a prerequisite for efficient identification of genes involved in polygenic diseases. During the next few years the efforts of the SNP consortium will provide large numbers of SNP markers to choose from for SNP genotyping applications (22).
Currently, commercially available MALDI mass spectrometers are capable of recording 20 000 spectra per day. With a multiplex factor of 3, approximately 60 000 genotypes from up to 20 000 individuals can be generated per day in one mass spectrometer at only a fraction of the cost of other mass spectrometric technologies using expensive purification procedures.
Mass spectrometric methods for SNP genotyping, in contrast to gel-based techniques such as conventional DNA sequencing, have the advantage that each allele found gives an absolute, measured mass; a physical property. This fact contributes significantly to the potential of multiplexibility and avoids artifacts in allele determination that have been observed in gel-based detection systems (23).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Thomas Wenzel (Bruker Saxonia Analytik GmbH, Germany) for providing the β-cyanoethyl phosphoramidites and Roman Pawlik for synthesis of oligonucleotides. α-S-ddNTPs were kindly provided by Dr Alan Hamilton (Amersham, UK). This work was supported by Bruker-Daltonik GmbH (Bremen, Germany) in the project ‘Genwaage’ of the Bundesministerium für Bildung und Forschung (BMBF) and the Ministère de l’Education, Recherche et Technologie (MENRT).
REFERENCES
- 1.Evans W.E. and Relling,M.V. (1999) Science, 286, 487–491. [DOI] [PubMed] [Google Scholar]
- 2.Landegren U., Kaiser,R., Caskey,C.T. and Hood,L. (1988) Science, 242, 229–237. [DOI] [PubMed] [Google Scholar]
- 3.Schafer A.J. and Hawkins,J.R. (1998) Nature Biotechnol., 16, 33–39. [DOI] [PubMed] [Google Scholar]
- 4.Karas M. and Hillenkamp,F. (1988) Anal. Chem., 60, 2299–2303. [DOI] [PubMed] [Google Scholar]
- 5.Tang K., Fu,D.-J., Julien,D., Braun,A., Cantor,C.R. and Köster,H. (1999) Proc. Natl Acad. Sci. USA, 96, 10016–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin T.J., Hall,J.G., Prudent,J.R. and Smith,L.M. (1999) Proc. Natl Acad. Sci. USA, 96, 6301–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross P., Hall,L., Smirnov,I. and Haff,L. (1998) Nature Biotechnol., 16, 1347–1351. [DOI] [PubMed] [Google Scholar]
- 8.Gut I.G. and Beck,S. (1995) Nucleic Acids Res., 23, 1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gut I.G., Jeffery,W.A., Pappin,D.J.C. and Beck,S. (1997) Rapid Commun. Mass Spectrom., 11, 43–50. [Google Scholar]
- 10.Berlin K. and Gut,I.G. (1999) Rapid Commun. Mass Spectrom., 13, 1739–1743. [DOI] [PubMed] [Google Scholar]
- 11.Sauer S., Lechner,D., Berlin,K., Lehrach,H., Escary,J.L., Fox,N. and Gut,I.G. (2000) Nucleic Acids Res., 28, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter N. (1997) Trends Microbiol., 5, 331–334. [DOI] [PubMed] [Google Scholar]
- 13.Collinge J., Sidle,K.C.L., Meads,J., Ironside,J. and Hill,A.F. (1996) Nature, 383, 685–690. [DOI] [PubMed] [Google Scholar]
- 14.Johnson M. (1998) Am. J. Respir. Crit. Care Med., 158, 146–153. [DOI] [PubMed] [Google Scholar]
- 15.Timmermann B., Mo,R., Luft,F.C., Gerdts,E., Busjahn,A., Omvik,P., Li,G.H., Schuster,H., Wienker,T.F., Hoehe,M.R. and Lund-Johansen,P. (1998) Kidney Int., 53, 14514–14560. [DOI] [PubMed] [Google Scholar]
- 16.Busjahn A., Li,G.H., Faulhaber,H.D., Rosenthal,M., Becker,A., Jeschke,E., Schuster,H., Timmermann,B., Hoehe,M.R. and Luft,F.C. (2000) Hypertension, 35, 555–560. [DOI] [PubMed] [Google Scholar]
- 17.Soubrier F., Nadaud,S. and Williams,T.A. (1994) Eur. Heart J., 15, 24–29. [DOI] [PubMed] [Google Scholar]
- 18.Soubrier F. and Cambien,F. (1994) Curr. Opin. Nephrol. Hypertens., 3, 25–29. [DOI] [PubMed] [Google Scholar]
- 19.Bartlett-Jones M., Jeffery,W.A., Hansen,H.F. and Pappin,D.J.C. (1994) Rapid Commun. Mass Spectrom., 8, 737–742. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Weeks D.E. and Lathrop,G.M. (1995) Trends Genet., 11, 513–519. [DOI] [PubMed] [Google Scholar]
- 22.Marshall E. (1999) Science, 284, 406–407. [DOI] [PubMed] [Google Scholar]
- 23.Ronaghi M., Nygren,M., Lundeberg,J. and Nyrén,P. (1999) Anal. Biochem., 267, 65–71. [DOI] [PubMed] [Google Scholar]