Abstract
Endogenous proteins secreted from Kluyveromyces lactis were screened for their ability to bind to or to hydrolyze chitin. This analysis resulted in identification of a nucleus-encoded extracellular chitinase (KlCts1p) with a chitinolytic activity distinct from that of the plasmid-encoded killer toxin α-subunit. Sequence analysis of cloned KlCTS1 indicated that it encodes a 551-amino-acid chitinase having a secretion signal peptide, an amino-terminal family 18 chitinase catalytic domain, a serine-threonine-rich domain, and a carboxy-terminal type 2 chitin-binding domain. The association of purified KlCts1p with chitin is stable in the presence of high salt concentrations and pH 3 to 10 buffers; however, complete dissociation and release of fully active KlCts1p occur in 20 mM NaOH. Similarly, secreted human serum albumin harboring a carboxy-terminal fusion with the chitin-binding domain derived from KlCts1p also dissociates from chitin in 20 mM NaOH, demonstrating the domain's potential utility as an affinity tag for reversible chitin immobilization or purification of alkaliphilic or alkali-tolerant recombinant fusion proteins. Finally, haploid K. lactis cells harboring a cts1 null mutation are viable but exhibit a cell separation defect, suggesting that KlCts1p is required for normal cytokinesis, probably by facilitating the degradation of septum-localized chitin.
Chitin, a β-1,4-linked unbranched polymer of N-acetylglucosamine (GlcNAc), is the second most abundant polymer on earth after cellulose. It is a major component of insect exoskeletons (21), the shells of invertebrate crustaceans, and fungal cell walls (23). Chitinases are enzymes that hydrolyze the β-1,4-glycosidic bond of chitin and are encoded in the genomes of many prokaryotic, eukaryotic, and viral organisms. In the yeast Saccharomyces cerevisiae, chitinase plays a morphological role in promoting efficient cell separation (19). Additionally, plants express chitinases as a defense against chitin-containing pathogens. In fact, the heterologous expression of chitinase genes in transgenic plants has been shown to increase their resistance to certain plant pathogens (7, 16, 18).
Chitinases belong to either family 18 or family 19 of the glycosylhydrolases based on their amino acid sequence similarities (13). Familial differences in chitinase catalytic domain sequences reflect the different mechanisms of chitin hydrolysis that result in either retention (family 18) or inversion (family 19) of the anomeric configuration of the product (24). Most chitinases have a modular domain organization with distinct catalytic and noncatalytic domains that function independent of each other. An O-glycosylated Ser-Thr-rich region often separates the two domains and may serve to prevent proteolysis or aid in secretion of the chitinase (1). Noncatalytic chitin-binding domains (ChBDs) belong to one of three structural classes (type 1, 2, or 3) based on protein sequence similarities (12). Depending on the individual chitinase, the presence of a ChBD can either enhance (19) or inhibit (11) chitin hydrolysis by the catalytic domain. The ability of a ChBD to function independent of the catalytic domain and to selectively bind chitin has led to the use of ChBDs as affinity tags to facilitate attachment of recombinant proteins to chitin-coated surfaces (3, 9).
Kluyveromyces lactis is an industrially important yeast due to its ability to grow to a very high cell density and abundantly secrete heterologous proteins (10, 25, 30, 31). We are interested in using K. lactis to secrete commercially important recombinant proteins carrying ChBD affinity tags to facilitate their capture on chitin beads. Thus, in the present study, we conducted a survey of endogenous secreted K. lactis proteins having chitinolytic or chitin-binding properties that would compromise such an application.
To date, the only described K. lactis protein having chitin-binding or chitinolytic activity is the secreted killer toxin (28), a heterotrimeric glycoprotein composed of α, β, and γ subunits encoded by the killer plasmid k1. Killer toxin is thought to aid K. lactis by inhibiting the growth of competitive yeasts (17, 26). Toxicity is associated with the γ subunit but is dependent upon the exochitinase activity of the α subunit because chitin deficiency (29) and the chitinase inhibitor allosamidin (6) render competitive yeasts resistant to killer toxin.
In this paper, we report identification and characterization of a second chitinase (KlCts1p) secreted from K. lactis cells. We demonstrate that KlCts1p is a nucleus-encoded endochitinase containing a family 18 catalytic domain and a type 2 ChBD. Genetic disruption of K. lactis CTS1 (KlCTS1) indicated that KlCts1p is required for efficient cell separation, although it is not required for cell viability. Additionally, K. lactis Δcts1 cells produce no detectable chitinase activity, secrete no chitin-binding proteins, and are still able grow to a high density, making this strain background suitable for production of recombinant ChBD-tagged proteins. Finally, we demonstrate the use of the ChBD derived from KlCts1p as an affinity tag for purification of recombinant proteins expressed in K. lactis.
MATERIALS AND METHODS
Yeast strains and culture conditions.
Strains of K. lactis and S. cerevisiae (Table 1) were routinely cultured in YPD medium (1% yeast extract, 2% peptone, and 2% glucose) or YPGal medium (1% yeast extract, 2% peptone, and 2% galactose) at 30°C. Transformation of K. lactis and S. cerevisiae was achieved using standard methods. Transformants of K. lactis were selected by growth on YPD agar containing 200 μg G418 ml−1, whereas S. cerevisiae transformants were obtained by growth on SD medium (0.67% yeast nitrogen base, 2% glucose) or SGal medium (0.67% yeast nitrogen base, 2% galactose) containing the appropriate supplements needed to complement strain auxotrophies.
TABLE 1.
Yeast strains used in this study
| Organism | Strain | Genotype | Source |
|---|---|---|---|
| K. lactis | GG799 | MATα [pGKI1+] | This study |
| K. lactis | PCKl1 | MATα cts1::KanR [pGKI1+] | This study |
| K. lactis | PCKl2 | MATα LAC4::PLAC4HSA-BcChBD [pGKI1+] | This study |
| K. lactis | PCKl3 | MATα LAC4::PLAC4HSA-KIChBD [pGKI1+] | This study |
| K. lactis | CBS 2359 | MATa [pGKI1+ pGKI2+] | ATCC 8585 |
| K. lactis | CBS 683 | MATα [pGKI1+ pGKI2+] | ATCC 56498 |
| S. cerevisiae | BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| S. cerevisiae | RG6947 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ 0cts1::kanMX4 | Research Genetics |
| S. cerevisiae | PCSc1 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ 0cts1::kanMX4 [pMW20-KICTS1] | This study |
Detection and isolation of secreted K. lactis chitin-binding proteins.
Western blotting was used to detect secreted K. lactis proteins that cross-reacted with a polyclonal anti-chitin-binding domain antibody (α-ChBD) raised against the chitin-binding domain derived from Bacillus circulans chitinase A1 (New England Biolabs, Beverly, MA). Spent culture medium was isolated from K. lactis GG799 cultures following 48 to 96 h of growth by centrifugation at 4,000 × g for 10 min. Proteins in the spent medium were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on a 4 to 20% Tris-glycine polyacrylamide gel (Daiichi Pure Chemicals Co., Tokyo, Japan) and were transferred to a Protran nitrocellulose membrane (Schleicher & Schuell, Keene, NH). The membrane was blocked overnight in phosphate-buffered saline containing 0.05% Tween 20 (PBS-T) supplemented with 5% (wt/vol) nonfat milk at 4°C and probed with α-ChBD polyclonal antibodies (1:2,000 in PBS-T containing 5% nonfat milk), followed by a horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:2,000 in PBS-T containing 5% nonfat milk; KPL, Gaithersburg, Md.). Protein-antibody complexes were visualized using LumiGlo detection reagents (Cell Signaling Technology, Beverly, MA).
To isolate secreted proteins that bind chitin, K. lactis GG799 cells were grown in 20 ml YPD medium for 96 h. Cells were removed from the culture by centrifugation, and the spent medium was transferred to a fresh tube containing 1 ml of water-washed chitin beads (New England Biolabs) and incubated at room temperature with gentle rotation for 1 h. The chitin beads were harvested by centrifugation, washed with 10 ml of water, and resuspended in 1 ml of water. Approximately 50 μl of protein-bound chitin beads was removed and boiled in SDS sample buffer (New England Biolabs) for 2 min to elute bound proteins, after which the mixture was microcentrifuged for 2 min to remove the chitin beads. The supernatant, containing eluted proteins, was separated by SDS-PAGE and subjected either to Western analysis with α-ChBD polyclonal antibodies or to amino-terminal protein sequencing.
Analysis of chitin-binding properties of KlCts1p ChBD.
KlCts1p from 20 ml of K. lactis GG799 spent culture medium was bound to 1 ml of chitin beads as described above. The KlCts1p-containing beads were washed with 10 ml water and resuspended in 1.5 ml of water. Minicolumns were prepared by dispensing 100-μl aliquots of the KlCts1p-containing beads into individual disposable columns (Bio-Rad, Hercules, CA). One milliliter (∼10 bed volumes) of each of the following buffers was passed over separate minicolumns: 50 mM sodium citrate (pH 3.0), 50 mM sodium citrate (pH 5.0), 100 mM glycine-NaOH (pH 10.0), unbuffered 20 mM NaOH (pH 12.3), 5 M NaCl, and 8 M urea. Each column was then washed with 2 ml water, after which the beads were resuspended in 200 μl of water, transferred to microcentrifuge tubes, and collected by brief centrifugation. The protein that remained bound to the chitin was eluted by boiling the beads in 50 μl of 3× SDS sample buffer (New England Biolabs) for 5 min. Eluted proteins were separated by SDS-PAGE and detected by Western analysis as described above.
A KlCts1p elution profile with various concentrations of NaOH (0 to 40 mM) was obtained in a similar manner. Aliquots (100 μl) of chitin-bound KlCts1p were prepared as described above and were distributed into 0.45-μm nylon Spin-X microcentrifuge spin columns (Corning Life Sciences, Corning, NY). The flowthrough was collected by microcentrifugation at 15,800 × g for 1 min and discarded. The beads were then resuspended in 100 μl of NaOH at each desired concentration (0 to 40 mM), and the eluates were collected by centrifugation at 15,800 × g for 1 min. The chitinase activity in each eluate was measured as described below.
Expression of ChBD-tagged fusion proteins.
To create a fusion between the ChBD of KlCts1p and human serum albumin (HSA), primers 5′-GGAAGATCTGACTCCTGGGCTGTTACAAGA-3′ (BglII site underlined) and 5′-ATAAGAATGCGGCCGCCTAGAAGACGACGTCGGGTTTCAAATA-3′ (NotI site underlined) were used to amplify a DNA fragment encoding the C-terminal 81 amino acids of KlCts1p with Deep Vent DNA polymerase (New England Biolabs). The KlCts1p ChBD fragment was cloned into the BglII-NotI sites of the K. lactis integrative expression plasmid pGBN2 (New England Biolabs) to produce pGBN2-KlChBD. HSA was amplified with primers 5′-CCGCTCGAGAAAAGAGATGCACACAAGAGTGAGGTTGCT-3′ (XhoI site underlined) and 5′-CGCGGATCCTAAGCCTAAGGCAGCTTGACTTGC-3′ (BamHI site underlined) and cloned into the XhoI-BglII sites of pGBN2-KlChBD. When integrated into the K. lactis genome, the resulting expression construct produced a single polypeptide consisting of the S. cerevisiae α-mating factor pre-pro secretion leader (present in pGBN2), HSA, and KlChBD.
A control construct that produced HSA containing a carboxy-terminal type 3 ChBD derived from B. circulans chitinase A1 (BcChBD) was assembled in a similar manner. Primers 5′-GGAAGATCTACGACAAATCCTGGTGTATCCGCT-3′ (BglII site underlined) and 5′ATAAGAATGCGGCCGCTTATTGAAGCTGCCACAAGGCAGGAAC-3′ (NotI site underlined) were used to PCR amplify BcChBD from pTYB1 (New England Biolabs). The amplified product was cloned into the BglII-NotI sites of pGBN2 to create pGBN2-BcChBD. HSA was amplified and cloned into the XhoI-BglII sites of pGBN2-BcChBD as described above.
Disruption of KlCTS1.
A PCR-based method was used to construct a linear DNA disruption fragment consisting of an ADH2 promoter-G418 resistance gene cassette having 80 to 82 bp of KlCTS1 DNA on either end. When integrated at the KlCTS1 locus, this fragment caused replacement of DNA encoding the first 168 amino acids of KlCts1p with the G418 resistance cassette. Primers containing DNA that hybridized to the ADH-G418 sequence (not underlined) and having tails consisting of the KlCTS1 DNA sequence (underlined), 5′-CCAGTAATGCAACTATCAATCATTGTGTTAAACTGGTCACCAGAAATACAAGATATCAAAAATTACTAATACTACCATAAGCCATCATCATATCGAAG-3′ and 5′-CCAAACTAGCGTATCCGGTTGGATTATTGTTTTCGATATCGAAATCGAAACCATCGACGACAGCAGTGTCGAATGGTCTTTCCCCGGGGTGGGCGAAGAACTCC-3′, were used to amplify the disrupting DNA fragment from the ADH2-G418-containing vector pGBN2 using Taq DNA polymerase. The amplified product was used to transform K. lactis GG799 cells, and colonies were selected on YPD agar containing 200 μg G418 ml−1. Whole-cell PCR using a KlCTS1-specific forward primer (5′-GGGCACAACAATGGCAGG-3′; designed upstream of the integration site) and a G418-specific reverse primer (5′-GCCTCTCCACCCAAGCGGC-3′) was used to amplify an ∼600-bp diagnostic DNA fragment from cells that had correctly integrated the disrupting DNA fragment at the KlCTS1 locus. Of 20 transformants tested in this manner, 2 Δcts1 K. lactis strains were identified and characterized further.
Heterologous expression of KlCTS1 in S. cerevisiae.
To express KlCTS1 in S. cerevisiae, the gene was PCR amplified with primers 5′-GGCGGATCCGCCACCATGTTTCACCCTCGTTTACTT-3′ (BamHI site underlined) and 5′-ACATGCATGCCTAGAAGACGACGTCGGGTTTCAA-3′ (SphI site underlined) and cloned into the BamHI-SphI sites of pMW20 (32) to place expression of KlCTS1 under the control of the galactose-inducible, glucose-repressible S. cerevisiae GAL10 promoter. This expression construct was introduced into S. cerevisiae Δcts1 (RG6947) cells to produce strain PCSc1. To induce production of KlCtsp1, 2-ml starter cultures were grown in SD medium containing 20 μg uracil ml−1 overnight at 30°C, after which 1 ml of each culture was used to inoculate 20-ml YPD medium and 20-ml YPGal medium cultures. Each culture was grown overnight with shaking at 30°C prior to analysis of the spent culture medium for chitinase production and cell morphology by microscopy.
Chitinase activity measurements.
The following chitin oligosaccharides containing one to four GlcNAc residues and derivatized with 4-methyl umbelliferone (4-MU) were used as substrates: 4-methylumbelliferyl NI-acetyl-β-d-glucosaminide (4MU-GlcNAc), 4-methylumbelliferyl NI,NII-diacetyl-β-d-chitobioside (4MU-GlcNAc2), 4-methylumbelliferyl NI,NII,NIII-triacetyl-β-d-chitotrioside (4MU-GlcNAc3), and 4-methylumbelliferyl NI,NII,NIII,NIV-tetraacetyl-β-d-chitotetraoside (4MU-GlcNAc4) (Sigma, St. Louis, MO, and EMD Biosciences, San Diego, CA). Chitinase activity was determined by measuring the release of 4-MU using a Genios fluorescent microtiter plate reader (Tecan, Maennedorf, Switzerland), a 340-nm excitation filter, and a 465-nm emission filter at 37°C. The 100-μl reaction mixture in each well of 96-well black microtiter plates contained 50 μM substrate, 1× McIlvaine's buffer (the pH ranged from 3 to 7 in different experiments), and 5 to 10 μl of sample. The initial rates of release were recorded, and enzyme activity was expressed as pmol of 4-MU released min−1. Standard curves for 4-MU (Sigma) were prepared under conditions used for the reactions for conversion from fluorescent units.
Microscopy.
Approximately 1 to 2 optical density units (600 nm) of cells was harvested and fixed in 1 ml of 2.5% (vol/vol) glutaraldehyde on ice for 1 h. Cells were washed twice with water and resuspended in approximately 100 μl mounting medium (20 mM Tris-HCl, pH 8.0, 0.5% N-propylgallate, 80% glycerol). In septum-staining experiments, calcofluor white (fluorescent brightener 28; Sigma) was added to the mounting medium to a final concentration of 100 μg ml−1. Cells were viewed with a Zeiss Axiovert 200 M microscope using light-phase Normaski imaging or fluorescent 4′,6′-diamidino-2-phenylindole (DAPI) filter settings.
Cellular chitin measurements.
Cells were extracted with KOH, and the chitin in the alkali-insoluble material was hydrolyzed to GlcNAc with chitinase for quantification by a micro Morgan-Elson assay as previously described (5).
RESULTS
Identification of KlCts1p.
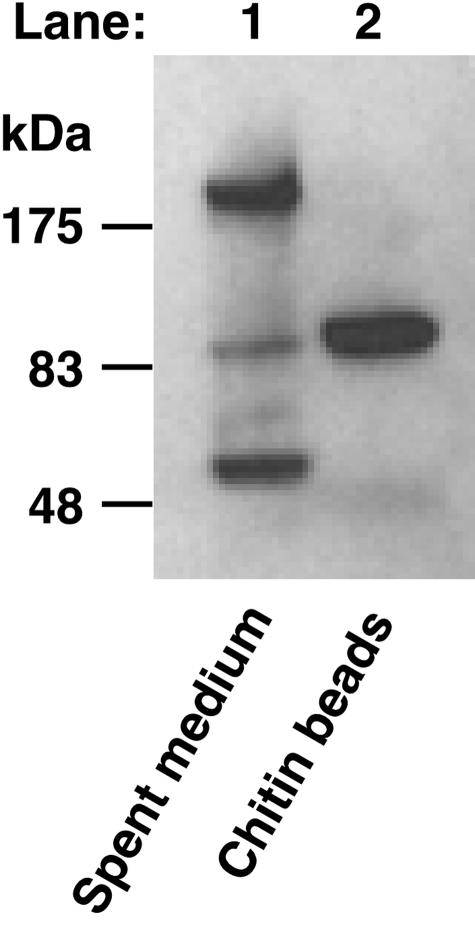
A polyclonal antibody raised against the B. circulans Chi1A chitin-binding domain was used in a Western blotting analysis of K. lactis GG799 spent culture medium in an effort to identify native secreted K. lactis proteins that contain a cross-reacting chitin-binding domain. Lane 1 in Fig. 1 shows that three secreted proteins with molecular masses of approximately >200, 85, and 50 kDa cross-reacted with the α-ChBD antibody. To test if any of these proteins was able to bind chitin, spent culture medium was mixed with chitin beads for 1 h at room temperature. Proteins that bound to the chitin beads were eluted by boiling the beads in SDS-PAGE sample buffer. Western blotting indicated that only the 85-kDa α-ChBD cross-reacting protein was able to bind chitin (Fig. 1, lane 2). Protein purified directly from chitin beads in this manner was subjected to amino-terminal protein sequencing, which resulted in identification of the first 20 amino acids of the mature protein (FDINAKDNVAVYWGQASAAT). A tBLASTn search using this amino acid sequence as the query to probe sequence databases identified a partially sequenced K. lactis gene having a translation sequence that exactly matched the query sequence and that had significant homology to the S. cerevisiae extracellular chitinase Cts1p (ScCts1p).
FIG. 1.
Identification of a secreted 85-kDa K. lactis chitin-binding protein. Secreted proteins in 10 μl of K. lactis GG799 spent culture medium (after 96 h of growth) were separated by SDS-PAGE and screened for the presence of a chitin-binding domain by Western blotting with α-ChBD antibodies (lane 1). Secreted proteins were incubated with chitin beads, after which chitin-bound proteins were eluted by boiling, separated by SDS-PAGE, and detected by α-ChBD Western blotting (lane 2).
Cloning KlCTS1 and sequence analysis.
At the onset of this work, the complete sequence of the K. lactis genome had not been reported yet. Therefore, we used a combination of Southern hybridization and anchored PCR (data not shown) to clone the remainder of a partial KlCTS1 sequence originally identified by database searches with the tBLASTn algorithm (see above). Our KlCts1p sequence is identical to the translated product of K. lactis open reading frame KLLA0C04730g in the recently reported K. lactis genome sequence (8).
KlCTS1 encodes a protein that is 53% identical and 82% similar to the S. cerevisiae Cts1p chitinase and has a similar modular domain organization consisting of a signal peptide, a catalytic domain, a Ser-Thr-rich domain, and a chitin-binding domain (Fig. 2). Signal P software (22) predicted the presence of a signal peptide that is cleaved after A19, in agreement with the amino-terminal protein sequence determined from purified secreted KlCts1p that begins at F20 (Fig. 2). The KlCts1p catalytic domain showed significant homology to family 18 chitinases (12, 13), including hevamine from the rubber tree and S. cerevisiae Cts1p (Fig. 2). Additionally, SMART domain prediction software (20, 27) indicated the presence of a C-terminal type 2 chitin-binding domain containing six conserved cysteine residues, as indicated in Fig. 2.
FIG. 2.
Domain organization and alignment of KlCts1p with family 18 chitinases. The KlCts1p sequence was aligned with family 18 chitinases from S. cerevisiae (Cts1p; GenBank accession no. A41035) and the rubber tree, Hevea brasiliensis (hevamine; GenBank accession no. AJ007701). The amino acids enclosed in boxes are identical in at least two of the three sequences. KlCts1p consists of a signal peptide that is cleaved after A19 (arrow), a family 18 chitinase catalytic domain (single line), a serine-threonine-rich domain (double line), and a type 2 chitin-binding domain (gray box). Conserved catalytic amino acids predicted from the hevamine crystal structure (4) are indicated by asterisks. Six cysteines conserved in type 2 ChBDs are indicated by solid dots. Hevamine (311 amino acids) does not contain a Ser-Thr-rich domain or a ChBD. Therefore, seven amino acids were truncated from its carboxy terminus to allow alignment of the Ser-Thr-rich domains of KlCts1p and ScCts1p.
KlCTS1 encodes a 551-amino-acid protein having a predicted molecular mass of 56 kDa, yet its apparent molecular mass after separation by SDS-PAGE is 85 kDa. In S. cerevisiae, the discrepancy between the predicted (60 kDa) and actual (130 kDa) molecular masses of ScCts1p has been attributed to extensive O-mannosylation of a Ser-Thr-rich domain (19). Because KlCts1p contains a similar Ser-Thr-rich domain, we speculate that O-mannosylation may be present in KlCts1p, but this was not tested experimentally. Additionally, the KlCts1p protein sequence contains one predicted Asn-linked glycosylation site; however, peptide N-glycosidase F (PNGase F) treatment did not alter the mobility of KlCts1p on an SDS-PAGE gel, indicating that it likely does not contain an Asn-linked glycan (data not shown).
Biochemical characterization of KlCts1p.
To define the types of chitinolytic activities produced by K. lactis cells, we used four 4-methylumbelliferyl-containing substrates that contained from one to four GlcNAc resides (4MU-GlcNAc[1-4]) in enzyme assays of spent culture medium. The hydrolysis of each substrate was measured as a real-time increase in fluorescence upon enzymatic release of 4-MU. These substrates allowed us to distinguish the activity of KlCts1p from the secreted exochitinase activity of the K. lactis secreted killer toxin and to determine if K. lactis cells produce a β-N-acetylhexosaminidase activity.
Vegetatively growing S. cerevisiae cells express and secrete only one chitinolytic activity (chitinase ScCts1p) and do not produce a β-N-acetyl-hexosaminidase activity. Therefore, we compared the extracellular chitinolytic activity of S. cerevisiae strain BY4741 with the activities of K. lactis strains GG799, CBS2359, and CBS683. No activity was detected using 4MU-GlcNAc1 as the substrate with K. lactis samples of either spent medium or cell homogenates at pH 4.5 or 6.0 (data not shown), indicating that K. lactis cells do not produce a detectable β-N-acetylhexosaminidase activity. Additionally, no killer toxin exochitinase activity was detectable using 4MU-GlcNAc2 at pH 6.0 for any of the three K. lactis strains. While 4MU-GlcNAc2 is the only substrate utilized by the killer toxin exochitinase, as reported by Butler et al. (6), the assays of these workers were performed with microgram amounts of purified killer toxin. Therefore, the K. lactis strains that we examined likely secrete insufficient amounts of killer toxin to be assayed directly from unconcentrated medium. Conversely, chitinolytic activity was readily detectable in all three K. lactis strains at pH 6.0 using both 4MU-GlcNAc3 and 4MU-GlcNAc4 as substrates. Furthermore, this activity was completely absent from the spent culture medium of K. lactis GG799 cells harboring a Δcts1 null mutation (strain PCKl1), indicating that it was entirely attributable to the KlCts1p chitinase.
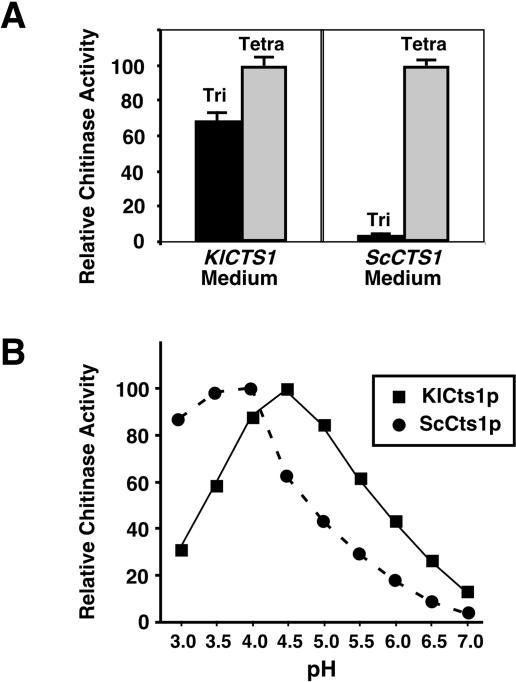
Given the similarity in protein sequence between KlCts1p and ScCts1p, we compared the catalytic properties of the two proteins. Because S. cerevisiae Cts1p has an acidic pH optimum (19), pH 4.5 was initially chosen for defining the substrate preferences of KlCts1p. As shown in Fig. 3A, chitinases from both yeasts hydrolyzed both 4MU-GlcNAc3 and 4MU-GlcNAc4. However, the K. lactis chitinase differed from ScCts1p in the extent to which it preferred 4MU-GlcNAc4 to 4MU-GlcNAc3. Results identical to those shown for K. lactis strain GG799 were observed for chitinase secreted from strains CBS2359 and CBS683 (data not shown). Additionally, KlCts1p showed maximum activity at pH 4.5, which was approximately 0.5 pH unit more alkaline than the pH for maximum ScCts1p activity (Fig. 3B).
FIG. 3.
Enzymatic properties of secreted KlCts1p. (A) Substrate preferences of KlCts1p and ScCts1p. Spent culture media from cultures of K. lactis GG799 and S. cerevisiae BY4741 cells were incubated with 50 μM 4MU-GlcNAc3 (Tri) (black bars) and with 50 μM 4MU-GlcNAc4 (Tetra) (gray bars) at pH 4.5 and 37°C. The relative rates of 4-MU release for each reaction were measured. (B) pH optima of KlCts1p and ScCts1p. The relative rates of 4-MU release were measured at 37°C for KlCts1p using 50 μM 4MU-GlcNAc3 and for ScCts1p using 50 μM 4MU-GlcNAc4 in McIlvaine's buffers with pHs ranging from 3.0 to 7.0.
KlCts1p ChBD-chitin affinity analysis.
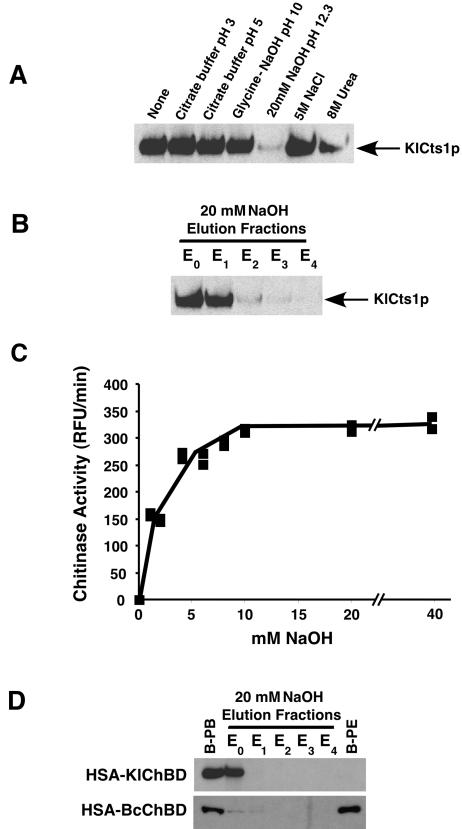
The association of KlCts1p and chitin beads was examined under a range of conditions. Figure 4A shows that the chitin association of KlCts1p was stable in 5 M NaCl and in buffers at pH 3, pH 5, and pH 10. In 8 M urea, which are conditions that normally denature proteins, only about 60% of chitin-bound KlCts1p dissociated from the chitin beads. However, complete dissociation was observed in 20 mM NaOH at pH 12.3. An elution profile of chitin-bound KlCts1p revealed that an equal amount of the protein eluted in the first two fractions (including the void volume), suggesting that destabilization of KlCts1p-chitin association occurs immediately in 20 mM NaOH (Fig. 4B). Surprisingly, KlCts1p eluted in this manner retained chitinolytic activity (Fig. 4C). In fact, measurement of chitinase activity before chitin binding and after elution with 20 mM NaOH showed that nearly 100% of the chitinase activity was recovered (data not shown).
FIG. 4.
KlCts1p dissociates from chitin at high pH. (A) KlCts1p was bound to chitin beads in minicolumns as described in Materials and Methods. The solutions were passed over separate minicolumns, after which KlCts1p that was still chitin bound was eluted by boiling, separated by SDS-PAGE, and detected by α-ChBD Western blotting. (B) KlCts1p elution from chitin with 20 mM NaOH. Chitin-bound KlCts1p was eluted in five successive 1-ml fractions of 20 mM NaOH (E0 to E4, where E0 represents the column void volume), separated by SDS-PAGE, and detected by α-ChBD Western blotting. (C) Base-eluted KlCts1p retains chitinolytic activity. Chitin-bound KlCts1p was eluted from chitin with various concentrations of NaOH, and the eluates were assayed for chitinase activity with 4MU-GlcNAc3 at pH 4.5 and 37°C. RFU, relative fluorescence units. (D) KlCts1p-ChBD can function as an elutable affinity tag. HSA fusions to ChBDs derived from KlCts1p or B. circulans ChiA1 were secreted from K. lactis, bound to chitin beads, and eluted with five 1-ml 20 mM NaOH fractions (E0 to E4). Proteins that were bound to chitin beads prior to elution (B-PB) or that were still bound after elution with 20 mM NaOH (B-PE) were eluted by boiling. All samples were separated by SDS-PAGE and examined by α-ChBD Western blotting.
To determine if the KlCts1p chitin-binding domain could function (i) independent of the KlCts1p catalytic domain and (ii) as an affinity tag on heterologously expressed proteins, HSA containing a C-terminal fusion to the ChBD derived from amino acids 470 to 551 of KlCts1p (KlChBD) was secreted from K. lactis strain PCKl3. For comparison, HSA was also fused to the B. circulans chitinase A1 type 3 ChBD (BcChBD) and secreted from K. lactis strain PCKl2 in the same manner. ChBD fusion proteins were bound to chitin beads as described in Materials and Methods, and their chitin affinities in the presence of 20 mM NaOH were determined. Figure 4D shows that the HSA-KlChBD fusion protein fully dissociated from chitin beads in 20 mM NaOH, whereas the HSA-BcChBD fusion protein remained bound to chitin even after extensive washing with 20 mM NaOH. These results indicate that KlChBD functions independent of the KlCts1p catalytic domain and that its dissociation from chitin in 20 mM NaOH is an intrinsic property. Furthermore, these data raise the possibility that KlChBD could be used as an elutable affinity tag for purification or reversible chitin immobilization of alkaliphilic or alkali-tolerant proteins.
Disruption of KlCTS1.
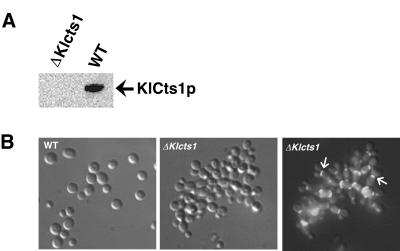
To examine the in vivo function of KlCts1p in K. lactis, the KlCTS1 allele was disrupted in haploid cells. A PCR-based method was used to assemble a DNA disruption fragment containing a kanamycin-selectable marker cassette as described in Materials and Methods. This fragment was used to transform K. lactis cells to G418 resistance. Transformants were screened by whole-cell PCR to obtain transformants that had the disrupting DNA fragment integrated at the KlCTS1 locus. Of 20 colonies tested, 2 had the disrupting DNA fragment correctly integrated (data not shown), indicating that KlCTS1 is not essential for the viability of K. lactis. Additionally, K. lactis Δcts1 cells (strain PCKl1) did not secrete chitinase, as demonstrated by the absence of KlCts1p (Fig. 5A) and chitinolytic activity in spent culture medium.
FIG. 5.
(A) K. lactis Δcts1 cells do not secrete KlCts1p. Proteins in spent culture medium from wild-type GG799 (WT) and Δcts1 K. lactis cells were incubated with chitin beads. Bound proteins were eluted by boiling and were detected by α-ChBD Western blotting. (B) KlCTS1 is required for efficient cell separation. Wild-type and Δcts1 K. lactis cells were grown in YPD medium and fixed in 2.5% glutaraldehyde. Septum-localized chitin was stained with calcofluor white and detected by fluorescence microscopy using a DAPI filter. The middle and right panels show the same cells visualized by phase-contrast and fluorescence microscopy, respectively. The arrows in the right panel indicate the locations of septa in certain cells.
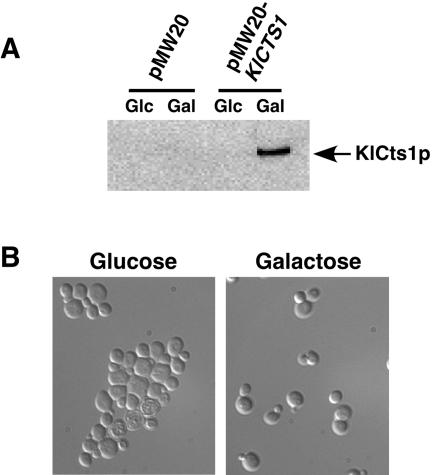
We also examined the growth and cell morphology of K. lactis wild-type strain GG799 and Δcts1 cells. In YPD medium, the Δcts1 strain grew as small clusters of loosely clumped cells that were easily dispersed into single cells by brief sonication. Fluorescence microscopy of cells stained with the chitin-binding dye calcofluor white showed that Δcts1 cells were joined via their septa (Fig. 5B, right panel), suggesting that these cells were unable to degrade septal chitin during cytokinesis. A similar phenotype has been observed for S. cerevisiae Δcts1 cells (19). Therefore, we tested the ability of KlCTS1 to restore normal cell separation to S. cerevisiae Δcts1 cells. KlCTS1 was placed under the control of the GAL10 galactose-inducible promoter in an S. cerevisiae expression vector. S. cerevisiae Δcts1 cells expressing KlCTS1 (strain PCSc1) secreted KlCts1p in galactose-containing medium (Fig. 6A) and did not form cell aggregates (Fig. 6B). Considered together, these data suggest that KlCTS1 and ScCTS1 encode functionally equivalent proteins that participate in cell separation, presumably by facilitating the degradation of septal chitin.
FIG. 6.
(A) S. cerevisiae Δcts1 cells expressing KlCTS1 secrete KlCts1p. S. cerevisiae Δcts1 cells harboring either an empty vector (pMW20) or a vector containing KlCTS1 under the control of a galactose-inducible glucose-repressible promoter (pMW20-KlCTS1) were grown in medium containing glucose or galactose overnight at 30°C. Spent culture medium was tested for the presence of KlCts1p by passage over chitin beads, elution of chitin-bound proteins by boiling, separation by SDS-PAGE, and α-ChBD Western blotting. (B) Expression of KlCTS1 corrects the aggregation phenotype of S. cerevisiae Δcts1 cells. S. cerevisiae Δcts1 cells harboring pMW20-KlCTS1 were grown in medium containing galactose or glucose. Cells were fixed in 2.5% glutaraldehyde and visualized by phase-contrast microscopy. Cells harboring an empty vector maintained an aggregation phenotype regardless of the carbon source (data not shown).
Because KlCts1p is abundantly secreted, we tested the accessibility of secreted KlCts1p to the septum by examining if exogenously added KlCts1p could abolish the cell aggregation phenotype. Purified KlCts1p was added to YPD medium prior to inoculation of K. lactis Δcts1 cells. After 48 h of growth, the cells were examined by microscopy. The aggregation of cells grown in the presence of exogenously added KlCts1p was alleviated but not completely ablated (data not shown). Thus, while exogenously supplied KlCts1p appeared to be able to access and cleave septal chitin, it did so less efficiently than KlCts1p that passed through the cell wall during secretion.
Finally, we examined the ability of Δcts1 cells to grow to a high culture density. The aggregation phenotype of K. lactis Δcts1 cells distorted measurements of cell density based on light absorbance at 600 nm so that they were less than 65% of those of wild-type cells. However, cultures of wild-type and Δcts1 cells grown for 48 h produced nearly identical dry weights of cells (data not shown). Additionally, the amounts of total cellular chitin did not differ significantly for the two strains. Strain GG799 yielded 21.8 ± 1.9 nmol GlcNAc per mg (dry weight) of cells, whereas the Δcts1 strain yielded 20.8 ± 1.0 nmol GlcNAc per mg (dry weight) of cells upon KOH extraction of cellular chitin and hydrolysis with chitinase. Therefore, despite their cell aggregation phenotype, Δcts1 cells remained capable of achieving the same cell densities in culture as wild-type cells, suggesting that this strain background would be suitable for commercial production of ChBD-tagged proteins.
DISCUSSION
In this study, we conducted a survey of native secreted K. lactis proteins that are capable of binding to chitin or that have chitinolytic activity. We discovered an abundantly expressed and previously undescribed extracellular protein (KlCts1p) that cross-reacted with an anti-ChBD polyclonal antibody, bound chitin with high affinity, and exhibited a chitinase activity distinct from that of the secreted killer toxin. We cloned and sequenced KlCTS1 and showed that it encodes a family 18 chitinase with a type 2 chitin-binding domain. We demonstrated that the ChBD derived from KlCts1p functions independent of the catalytic domain and readily dissociates from chitin in an alkaline environment. Finally, we showed that K. lactis cells require KlCts1p for proper cytokinesis, suggesting that cells use this protein to degrade septal chitin.
The small size (∼5 to 7 kDa), the substrate binding specificity, and the high avidity of ChBDs for chitin have led to utilization of these domains as affinity tags for immobilization of proteins to chitin surfaces. For example, the B. circulans chitinase A1 type 3 ChBD has been used to immobilize fusion proteins on chitin beads (9) and on chitin-coated microtiter dishes (3). However, a drawback of using the B. circulans ChBD as an affinity tag is that its binding to chitin is generally irreversible, which limits its utility for many applications, such as protein purification. In this study, we showed that the ChBD derived from KlCts1p can (i) bind to chitin in the absence of the catalytic domain, (ii) function as an affinity tag on a heterologously expressed protein in K. lactis, and (iii) dissociate from chitin in 20 mM NaOH, whereas BcChBD cannot do this. The latter observation likely reflects intrinsic structural differences between type 2 and type 3 ChBDs. For example, type 3 ChBDs (including BcChBD) contain conserved hydrophobic and aromatic amino acids that likely mediate their interaction with chitin (14), whereas type 2 ChBDs (including KlChBD) contain six conserved cysteine residues that mediate the tertiary structure, most likely through the formation of disulfide bridges (2). This observation also suggests that KlChBD could be used as an affinity tag for reversible immobilization or purification of alkaliphilic or alkali-tolerant proteins. Such proteins are widely used in the laundry detergent industry, where enzymes that function at a high pH (e.g., cellulases and proteases) are utilized to degrade common stains (15). Alternatively, it is possible to generate KlChBD mutants that dissociate from chitin at neutral pH. The plausibility of using mutagenesis to alter the chitin-binding characteristics of a ChBD is supported by a recent study in which a mutant type 3 ChBD that requires 2 M NaCl to bind chitin was created (9).
The ideal yeast strain background for secretion of recombinant ChBD-tagged proteins would (i) produce no abundant chitin-binding proteins or chitinolytic activity that would copurify during downstream applications, (ii) be capable of achieving a high cell density in culture, and (iii) efficiently secrete recombinant proteins. Our data indicate that despite a mild growth phenotype, K. lactis Δcts1 GG799 cells are capable of achieving the same cell density as wild-type cells in culture and produce no proteins with detectable chitin-binding or chitinolytic activities. Additionally, we have recently found that Δcts1 cells retain the ability to abundantly secrete recombinant maltose-binding protein (B. Taron, P. Colussi, and C. Taron, unpublished data). Thus, the K. lactis GG799 Δcts1 strain is well suited as a host background for production of recombinant ChBD-tagged proteins.
Our data suggest that KlCts1p is required for proper cytokinesis of K. lactis cells. Disruption of CTS1 in a haploid K. lactis strain produced viable cells that had a separation defect. During growth of this strain, cells remained joined together via their septa, which stained brightly for the presence of chitin. Thus, it is probable that KlCts1p is required for removal of septal chitin during cytokinesis. A similar phenotype has been described for S. cerevisiae Δcts1 cells (19). Therefore, we showed that KlCTS1 could restore normal morphogenesis to S. cerevisiae Δcts1 cells, indicating that KlCts1p and ScCts1p perform identical functions and that the two organisms likely have similar mechanisms for removing septum-localized chitin during cytokinesis.
Only a small fraction of total KlCts1p activity produced by K. lactis cells may be required for cytokinesis. Exogenous addition of purified KlCts1p to the culture medium prior to growth of K. lactis Δcts1 cells only partially alleviates the cell separation defect. Thus, exogenous KlCts1p may have a limited ability to access and cleave chitin in the septum. It is therefore possible that during secretion, some KlCts1p is retained in the cell wall, where it may act upon cell wall chitin during cell division. In support of this notion, we found that 99% of KlCts1p activity localizes to the culture medium, whereas ∼1% remains associated with a cell wall fraction of lysed cells (data not shown). Thus, it is likely that efficient cytokinesis requires only limited hydrolysis of cell wall chitin by KlCts1p. However, the abundance of medium-localized KlCts1p may reflect a second purpose for the protein. It is possible that secreted KlCts1p acts as an antimicrobial agent by digesting exposed chitin on competing fungi. Such a function was suggested by experiments in which transgenic tobacco plants expressing the S. cerevisiae Cts1 chitinase could inhibit spore germination and hyphal growth of the fungal pathogen Botrytis cinerea (7).
Acknowledgments
C.H.T. is grateful to Donald Comb for support. We thank J. Read for technical assistance, M. Cushing and J. Benner for amino-terminal protein sequencing, and L. Mazzola, J. Ware, and B. Slatko for nucleotide sequencing. Additionally, we thank B. Jack, P. Riggs, L. McReynolds, B. Taron, N. Bachman, and F. Mersha for comments on the manuscript.
REFERENCES
- 1.Arakane, Y., Q. Zhu, M. Matsumiya, S. Muthukrishnan, and K. J. Kramer. 2003. Properties of catalytic, linker and chitin-binding domains of insect chitinase. Insect Biochem. Mol. Biol. 33:631-648. [DOI] [PubMed] [Google Scholar]
- 2.Asensio, J. L., F. J.Canada, H. C. Siebert, J. Laynez, A. Poveda, P. M. Nieto, U. M. Soedjanaamadja, H. J. Gabius, and J. Jimenez-Barbero. 2000. Structural basis for chitin recognition by defense proteins: GlcNAc residues are bound in a multivalent fashion by extended binding sites in hevein domains. Chem. Biol. 7:529-543. [DOI] [PubMed] [Google Scholar]
- 3.Bernard, M. P., D. Cao, R. V. Myers, and W. R. Moyle. 2004. Tight attachment of chitin-binding-domain-tagged proteins to surfaces coated with acetylated chitosan. Anal. Biochem. 327:278-283. [DOI] [PubMed] [Google Scholar]
- 4.Bokma, E., H. J. Rozeboom, M. Sibbald, B. W. Dijkstra, and J. J. Beintema. 2002. Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis. Eur. J. Biochem. 269:893-901. [DOI] [PubMed] [Google Scholar]
- 5.Bulik, D. A., M. Olczak, H. A. Lucero, B. C. Osmond, P. W. Robbins, and C. A. Specht. 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2:886-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, A. R., R. W. O'Donnell, V. J. Martin, G. W. Gooday, and M. J. R. Stark. 1991. Kluyveromyces lactis toxin has an essential chitinase activity. Eur. J. Biochem. 199:483-488. [DOI] [PubMed] [Google Scholar]
- 7.Carstens, M., M. A. Vivier, and I. S. Pretorius. 2003. The Saccharomyces cerevisiae chitinase, encoded by the CTS1-2 gene, confers antifungal activity against Botrytis cinerea to transgenic tobacco. Trans. Res. 12:497-508. [DOI] [PubMed] [Google Scholar]
- 8.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44.15229592 [Google Scholar]
- 9.Ferrandon, S., T. Sterzenbach, F. B. Mersha, and M.-Q. Xu. 2003. A single surface tryptophan in the chitin-binding domain from Bacillus circulans chitinase A1 plays a pivotal role in binding chitin and can be modified to create an elutable affinity tag. Biochim. Biophys. Acta 1621:31-40. [DOI] [PubMed] [Google Scholar]
- 10.Fleer, R., X. J. Chen, N. Amellal, P. Yeh, A. Fournier, F. Guinet, N. Gault, D. Faucher, F. Folliard, H. Fukuhara, and J. Mayaux. 1991. High-level secretion of correctly processed recombinant human interleukin-1b in Kluyveromyces lactis. Gene 107:285-295. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto, M., T. Ikegami, S. Seino, N. Ohuchi, H. Fukada, M. Sugiyama, M. Shirakawa, and T. Watanabe. 2000. Expression and characterization of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12. J. Bacteriol. 182:3045-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henrissat, B. 1999. Classification of chitinases modules, p. 137-156. In P. Jolles and R. A. A. Muzzarelli (ed.), Chitin and chitinases. Birkhauser Verlag, Basel, Switzerland. [DOI] [PubMed]
- 13.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosylhydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikegami, T., T. Okada, M. Hashimoto, S. Seino, T. Watanabe, and M. Shirakawa. 2000. Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1. J. Biol. Chem. 275:13654-13661. [DOI] [PubMed] [Google Scholar]
- 15.Ito, S., T. Kobayashi, K. Ara, K. Ozaki, S. Kawai, and Y. Hatada. 1998. Alkaline detergent enzymes from alkaliphiles: enzymatic properties, genetics, and structures. Extremophiles 2:185-190. [DOI] [PubMed] [Google Scholar]
- 16.Itoh, Y., K. Takahashi, H. Takizawa, N. Nikaidou, H. Tanaka, H. Nishihashi, T. Watanabe, and Y. Nishizawa. 2003. Family 19 chitinase of Streptomyces griseus HUT6037 increases plant resistance to the fungal disease. Biosci. Biotechnol. Biochem. 67:847-855. [DOI] [PubMed] [Google Scholar]
- 17.Jablonowski, D., L. Fichtner, V. J. Martin, R. Klassen, F. Meinhardt, M. J. R. Stark, and R. Schaffrath. 2001. Saccharomyces cerevisiae cell wall chitin, the Kluyveromyces lactis zymocin receptor. Yeast 18:1285-1299. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J.-K., I.-C. Jang, R. Wu, W.-N. Zuo, R. S. Boston, Y.-H. Lee, I.-P. Ahn, and B. Hie Nahm. 2003. Co-expression of modified maize ribosome-inactivating protein and a rice basic chitinase gene in transgenic rice plants confers enhanced resistance to sheath blight. Trans. Res. 12:475-484. [DOI] [PubMed] [Google Scholar]
- 19.Kuranda, M., and P. W. Robbins. 1991. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 266:19758-19767. [PubMed] [Google Scholar]
- 20.Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz, R. Mott, F. Ciccarelli, R. R. Copley, C. P. Ponting, and P. Bork. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30:242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merzendorfer, H., and L. Zimoch. 2003. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 206:4393-4412. [DOI] [PubMed] [Google Scholar]
- 22.Nielson, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 23.Riccardo, A., and A. Muzzarelli. 1999. Native, industrial and fossil chitins, p. 1-6. In P. Jolles and R. A. A. Muzzarelli (ed.), Chitin and chitinases. Birkhauser Verlag, Basel, Switzerland. [DOI] [PubMed]
- 24.Robertus, J. D., and A. F. Monzingo. 1999. The structure and action of chitinases, p. 125-135. In P. Jolles and R. A. A. Muzzarelli (ed.), Chitin and chitinases. Birkhauser Verlag, Basel, Switzerland. [DOI] [PubMed]
- 25.Rocha, T., G. Paterson, K. Crimmins, A. Boyd, L. Sawyer, and L. A. Fothergill-Gilmore. 1996. Expression and secretion of recombinant ovine β-lactoglobulin in Saccharomyces cerevisiae and Kluyveromyces lactis. Biochem. J. 313:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffrath, R., and K. D. Breunig. 2000. Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet. Biol. 30:173-190. [DOI] [PubMed] [Google Scholar]
- 27.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark, M. J. R., and A. Boyd. 1996. The killer toxin of Kluyveromyces lactis: characterization of the toxin subunits and identification of the genes that encode them. EMBO J. 3:1995-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takita, M., and B. Castilho-Valavicius. 1993. Absence of cell wall chitin in Saccharomyces cerevisiae leads to resistance to Kluyveromyces lactis killer toxin. Yeast 9:589-598. [DOI] [PubMed] [Google Scholar]
- 30.van den Berg, J. A., K. J. van der Laken, A. J. J. van Ooyen, T. C. H. M. Renniers, K. Rietveld, A. Schaap, A. J. Brake, R. J. Bishop, K. Schultz, D. Moyer, M. Richman, and J. R. Shuster. 1990. Kluyveromyces as a host for heterologous gene expression: expression and secretion of prochymosin. Bio/Technology 8:135-139. [DOI] [PubMed] [Google Scholar]
- 31.Walsh, D. J., and P. L. Bergquist. 1997. Expression and secretion of a thermostable bacterial xylanase in Kluyveromyces lactis. Appl. Environ. Microbiol. 63:3297-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zieler, H. A., M. Walberg, and P. Berg. 1995. Suppression of mutations in two Saccharomyces cerevisiae genes by the adenovirus E1A protein. Mol. Cell. Biol. 15:3227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]