Abstract
The pathway oxoaverantin (OAVN) → averufin (AVR) → hydroxyversicolorone (HVN) → versiconal hemiacetal acetate (VHA) is involved in aflatoxin biosynthesis, and the cypX and moxY genes, which are present in the aflatoxin gene cluster, have been previously suggested to be involved in this pathway. To clarify the function of these two genes in more detail, we disrupted the genes in aflatoxigenic Aspergillus parasiticus NRRL 2999. The cypX-deleted mutant lost aflatoxin productivity and accumulated AVR in the mycelia. Although this mutant converted HVN, versicolorone (VONE), VHA, and versiconol acetate (VOAc) to aflatoxins in feeding experiments, it could not produce aflatoxins from either OAVN or AVR. The moxY-deleted mutant also lost aflatoxin productivity, whereas it newly accumulated HVN and VONE. In feeding experiments, this mutant converted either VHA or VOAc to aflatoxins but did not convert OAVN, AVR, HVN, or VONE to aflatoxins. These results demonstrated that cypX encodes AVR monooxygenase, catalyzing the reaction from AVR to HVN, and moxY encodes HVN monooxygenase, catalyzing a Baeyer-Villiger reaction from HVN to VHA as well as from VONE to VOAc. In this work, we devised a simple and rapid method to extract DNA from many fungi for PCR analyses in which cell disruption with a shaker and phenol extraction were combined.
Aflatoxins (AF) are a group of polyketide-derived secondary metabolites produced mainly by certain strains of the common molds Aspergillus flavus and Aspergillus parasiticus (16). Some other strains of Aspergillus nomius (13), Aspergillus pseudotamarii (8), Aspergillus bombycis (17), and Aspergillus ochraceoroseus (12) have also been reported to produce aflatoxins. These toxins are highly toxic and carcinogenic in animals and humans, leading to hepatotoxicity, teratogenicity, immunotoxicity, and even death (3). Among the naturally occurring aflatoxins, the four major ones are aflatoxin B1 (AFB1), AFB2, AFG1, and AFG2, of which AFB1 is the most toxic and carcinogenic compound. Contamination of food and feed crops, such as wheat, corn, cotton, peanuts, and tree nuts, with AFB1 presents not only a very serious health hazard but also an economic problem all over the world (9).
A. parasiticus generally produces both B-group (AFB1 and AFB2) and G-group (AFG1 and AFG2) aflatoxins, whereas A. flavus produces only B-group aflatoxins. The biosynthetic pathway of aflatoxins has been extensively studied, and most of the enzymes and corresponding genes involved in aflatoxin biosynthesis have been identified (14, 22, 27, 29). More than 18 enzymatic reactions are required for the conversion of acetyl coenzyme A to the final products, AFB1, AFB2, AFG1, and AFG2. It had been demonstrated that 25 genes are clustered within a 70-kb DNA region in the chromosomes of A. parasiticus and A. flavus and that the expression of most of these genes is regulated by the positive regulatory gene, aflR, and another regulatory gene, aflJ (31, 32). Sterigmatocystin (ST), produced by Aspergillus nidulans and many other species, is the penultimate precursor of aflatoxins. The ST pathway genes form a gene cluster of about 60 kb in A. nidulans, and the functions of the genes encoding enzymes in the ST cluster are similar to those of the genes in A. parasiticus and A. flavus (1, 10). However, the positions of individual genes are different from those of the homologous genes in the aflatoxin gene cluster. Although the AF/ST gene clusters have already been sequenced, the functions of several genes have not yet been clarified.
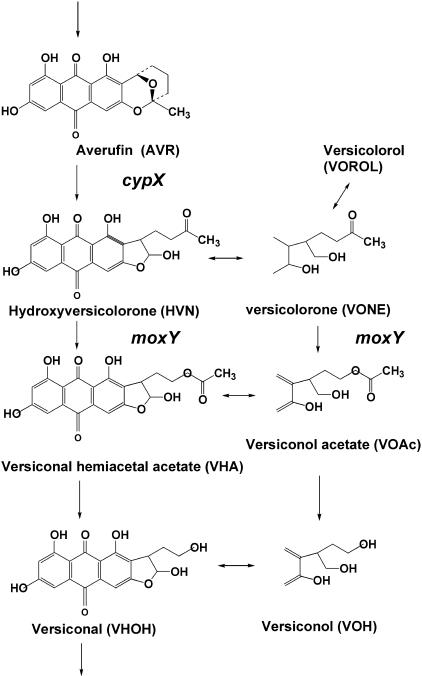
Several oxidative steps are required for AF/ST biosynthesis (11). In the early step, the conversion from averufin (AVR) to versiconal hemiacetal acetate (VHA) involves at least two monooxygenase reactions. Recently, we demonstrated that the conversion from AVR to hydroxyversicolorone (HVN) is catalyzed by a microsome monooxygenase which requires NADPH for its activity (26) (Fig. 1). This enzyme shows strict stereospecificity for the configuration of (1′S, 5′S)-AVR because (1′R, 5′R)-AVR does not serve as a substrate (26). The resulting HVN is converted to VHA by a cytosol monooxygenase which also requires NADPH as a cofactor (26). Furthermore, HVN and VHA commonly function as substrates for the VHA reductase enzyme to yield versicolorone (VONE) and versiconol acetate (VOAc), respectively. Finally, VONE, VOAc,HVN, and VHA comprise a metabolic grid in aflatoxin biosynthesis. VONE produced from HVN can be converted to versicolorol (VOROL) in the presence of NADPH (26).
FIG. 1.
Pathways and metabolites discussed in this work. The functions of the cypX and moxY genes determined in this work are also indicated.
Yu et al. cloned a gene, avfA, from A. parasiticus, A. flavus, and Aspergillus sojae (33). Gene complementation experiments using an AVR-accumulating mutant of A. parasiticus suggested that the avfA gene encodes an enzyme that is necessary for the conversion of AVR to VHA (18, 33). On the other hand, in the sterigmatocystin gene cluster of A. nidulans, it was suggested that the genes stcB and stcW encode a putative P450 monooxygenase and a putative flavin adenine dinucleotide-requiring monooxygenase, respectively (11). Disruption of either stcB or stcW led to the accumulation of AVR, suggesting that these genes are involved in the reaction from AVR to VHA. Also, the mutant A42355-OC-1 of A. nidulans, which had a translocation in the stcW gene and was blocked in ST biosynthesis, converted either VHA or versicolorin A to sterigmatocystin (6). In the aflatoxin gene cluster of A. parasiticus, the genes cypX and moxY were isolated as homologs of stcB and stcW, respectively, by Yu et al. (30). In spite of these extensive studies, the function of each gene has not been clarified.
In this work, we disrupted either cypX or moxY and then characterized both disruptants in order to clarify their functions in the aflatoxin biosynthetic pathway. We finally demonstrated that the cypX gene is involved in the reaction from AVR to HVN and that the moxY gene is involved in the reaction from HVN to VHA as well as that from VONE to VOAc.
MATERIALS AND METHODS
Microorganisms.
A. parasiticus SYS-4 (NRRL-2999), the wild-type aflatoxin-producing strain, was used as a recipient strain for gene disruption experiments. A. parasiticus NIAH-26, a mutant from SYS-4, was used in feeding experiments. NIAH-26 induced all enzymes during the conversion of norsolorinic acid to aflatoxins in an aflatoxin-inducing medium, although it produced neither aflatoxins nor anthraquinone or xanthone precursors (28).
Standard samples.
HVN and VONE were isolated from mycelia of A. parasiticus mutant WE-47 (hvn-1) (20). VHA and VOAc were purified from mycelia of the mutant A. parasiticus strain NIAH-9, which had been cultured in YES liquid medium (2% yeast extract, 20% sucrose) supplemented with dichlorvos (24). AVR was prepared from mycelia of A. versicolor (Vuillemin) Tiraboschi (5). Oxoaverantin (OAVN) was purified from hydroxyaverantin (HAVN) (19). The concentrations of these metabolites were determined based on the UV absorption spectra in methanol with the following absorption coefficients (λmax): for OAVN, 8,500 M−1 cm−1 (466 nm); AVR, 10,500 M−1 cm−1 (454 nm); HVN, 5,300 M−1 cm−1 (476 nm); VONE, 6,000 M−1 cm−1 (468 nm); VHA, 7,250 M−1 cm−1 (480 nm); and VOAc, 8,500 M−1 cm−1 (453 nm).
Construction of the cypX double-crossover gene disruption vector.
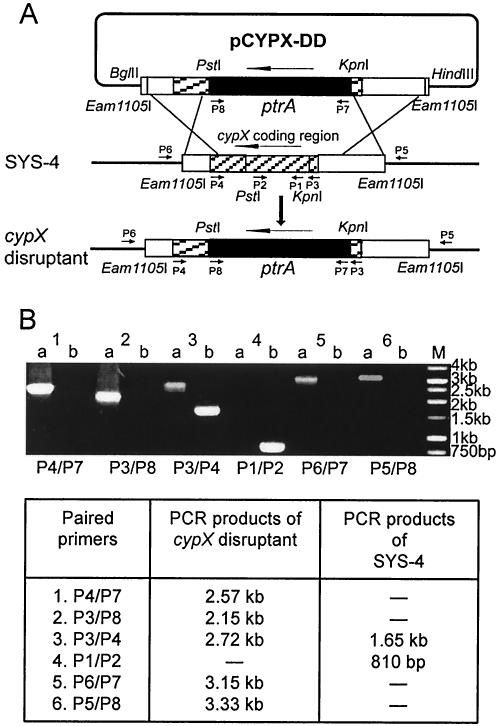
A 3-kb fragment containing a 1.65-kb cypX coding region and 5′- and 3′-flanking regions was amplified with KOD-plus-enzyme (Toyobo Co., Osaka, Japan) using cypX-HindIII-F (no. 377, CCCCAAGCTTCTCACTGCCGCCAACG) and cypX-BglII-R (no. 376, GAAGATCTGCGTTGCATACCCTTCCC) as primers and using genomic DNA of the strain SYS-4, prepared with Nucleon PhytoPure (Amersham Life Science), as the template. The resultant HindIII/BglII PCR fragment was then ligated into the corresponding sites in the pSP72 vector (Promega). The inside 920-bp PstI/KpnI fragment of the cypX gene was excised from the resultant vector and then replaced with the 2-kb pyrithiamine-resistant gene ptrA, which had been amplified by PCR with a KOD enzyme and primers PTRI-KpnI-F (no. 311, GGGGTACCGGGCAATTGATTACGGGATCCCA) and PTRI-PstI-R2 (no. 322, AAAACTGCAGTGACGATGAGCCGCTCTTGC) from the vector pPTRI (Takara). The resulting cypX double-crossover disruption vector, pCYPX-DD, was linearized by Eam1105I (Fig. 2A). The final linear replacement construct, which contained the 2-kb selectable marker ptrA flanked by a 910-bp fragment containing the cypX 3′-flanking region and a 570-bp 3′-coding region and a 1.14-kb fragment containing the cypX 5′-flanking region and a 150-bp 5′-coding region, was then used to transform A. parasiticus SYS-4.
FIG. 2.
Disruption of cypX via double-crossover recombination. (A) Strategy for the disruption of the cypX gene. The gene disruption vector pCYPX-DD was constructed as described in the text. The vector was linearized and then transformed into wild-type strain SYS-4. The double-crossover recombination events resulted in the replacement of most of the internal section of the target gene cypX with the selectable marker ptrA gene. Long arrows, gene direction; short arrows, positions of primers used for confirmation of gene disruptions; vertical arrow, gene replacement. (B) PCR analysis using different combinations of primers was done to confirm that the cypX gene was deleted in the cypX disruptant CYPX-DD-55. Lanes: a, cypX disruptant CYPX-DD-55; b, the recipient strain SYS-4; M, 1-kb molecular marker. The expected lengths of the PCR products are shown at the bottom of panel B.
Construction of the moxY disruption vector pMOXY-DD.
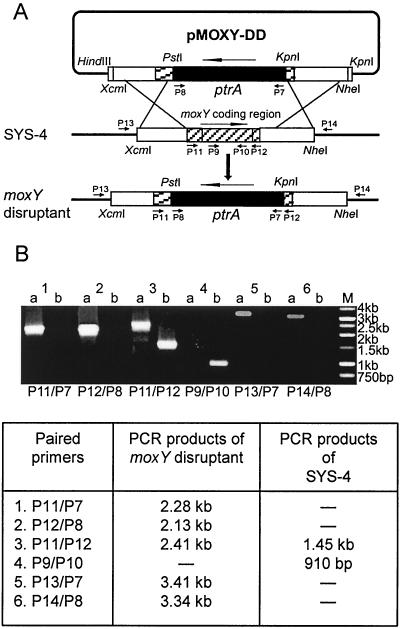
The moxY disruption vector was constructed by a three-step procedure. First, the 2-kb PstI/KpnI PCR fragment of selectable marker gene ptrA was inserted into the corresponding sites in the pSP72 vector. Second, the 1.24-kb KpnI fragment containing a moxY 3′-flanking region and a 132-bp moxY 3′-coding region, which had been amplified with a KOD enzyme and primers moxYR-KpnI-F (no. 380, GGGGTACCCCGTAGTTTCGTGCCGC) and moxYR-KpnI-R (no. 382, GGGGTACCGCCACACTGTACCATCGC) from the genomic DNA of SYS-4, was inserted into the KpnI site of the resultant vector. The desired insertion direction of the moxY 3′-region in the vector was then selected by either restriction enzyme digestion or PCR analysis. Finally, the resultant vector was ligated with a 1.26-kb HindIII/PstI fragment containing a moxY 5′-flanking region and a 278-bp moxY 5′-coding region, which had been amplified with KOD-plus-enzyme using moxYL-HindIII-F (no. 378, CCCCAAGCTTCCACGGGATCGGCAATG) and moxYL-PstI-R (no. 379, AAAACTGCAGGCATACCCGGTCGAGG) as primers and the genomic DNA of SYS-4 as the template, to obtain the moxY disruption vector pMOXY-DD (Fig. 4A). The linear replacement construct containing the 2-kb ptrA gene flanked by a 1.2-kb segment containing the moxY 5′-region and a 1.13-kb segment containing the moxY 3′-region was excised from pMOXY-DD using XcmI and NheI and then used to transform A. parasiticus SYS-4.
FIG. 4.
Disruption of the moxY gene via double-crossover recombination. (A) To disrupt the moxY gene, gene disruption vector pMOXY-DD was constructed as described in the text. The vector was linearized and then transformed into the wild-type strain SYS-4. The double-crossover recombination resulted in the replacement of most of the internal section of the target gene moxY with the ptrA gene. (B) PCR analysis of the moxY disruptant was conducted with genomic DNA of MOXY-DD-69 and the recipient strain SYS-4 by using different combinations of primers. Lanes: a, the moxY disruptant MOXY-DD-69; b, the recipient strain SYS-4; M, 1-kb molecular marker. The expected lengths of the PCR products are shown at the bottom of panel B.
Fungal transformation.
The transformation of fungal protoplasts with plasmid DNAs was performed as described by Gomi et al. (4) and Horng et al. (7) with some modifications. For the preparation of the protoplast, approximately 108 conidia of SYS-4 were inoculated into 100 ml Czapek-Dox (CD) liquid medium and incubated for 18 to 20 h at 28°C in a stationary culture. Then, 0.6-cm glass beads were added to a shaking flask and incubated for another 24 h at 30°C with shaking at 130 rpm. Mycelia were harvested, washed two to three times with 0.8 M NaCl, resuspended in 10 ml freshly prepared protoplast medium (15 mg/ml Yatalase [Takara], 5 mg/ml cellulase Onozuka R-10 [Yakult Honsha], 0.8 M NaCl, 10 mM phosphate buffer [pH 6.0], and 1 mM dithiothreitol), and then incubated for 3 to 4 h at 30°C with gentle shaking at 130 rpm. After incubation, the residual mycelia were removed by filtration through Miracloth (Calbiochem). Protoplasts were harvested by centrifugation at 4°C and 2,500 × g for 5 min and gently washed in ice-cold solution 1 (0.8 M NaCl, 10 mM CaCl2, and 10 mM Tris-HCl [pH 8.0]). The pellet was resuspended in solution 1 at a concentration of 2 × 108/ml; then, 0.2 volume of solution 2 (40% polyethylene glycol 4000, 50 mM CaCl2, 10 mM Tris-HCl [pH 8.0]) was added with gentle mixing. For transformation, 0.1 ml protoplast suspension was transferred to a new 15-ml polypropylene tube. DNA was added in a volume of less than 15 μl (approximately 4 to 7 μg), and each mixture was placed on ice for 30 min. Then, 0.5 ml of solution 2 was added with gentle mixing, followed by incubation for 20 min at room temperature. The suspensions were diluted with 5 ml solution 1 and centrifuged at 4°C and 2,500 × g for 5 min. The pellet was resuspended in 0.1 ml solution 1, plated on CD regeneration medium containing 0.8 M NaCl and 2% agar, and then overlaid with CD soft agar (0.5% low-melting-point agar, 30°C). The plates were incubated at 28°C for 4 to 7 days. Pyrithiamine-resistant transformants were selected on CD medium plates containing 0.1 mg/liter pyrithiamine.
Rapid and simple DNA extraction for PCR analysis.
Conidia of transformants were inoculated into 300 μl YES medium in a 2-ml tube (USA/Scientific Plastics, Milton Keynes, England) with a sterile toothpick. After 2 days of incubation at 28°C, the medium was removed and discarded, and then 150 μl Tris-EDTA buffer, 150 μl Tris-EDTA-saturated phenol, and zirconium beads (diameter, 0.5 mm; Nikkato) were added to the remaining mycelia in the tube. The mycelia with/without spores in the tube were completely disrupted with FastPrep FP100A (Q-BIO 101) at a speed of 6.5 m s−1 for 45 s. After cooling on ice, the whole tube was centrifuged at 15,000 × g for 15 min at 4°C. The aqueous upper layer containing DNA was transferred to a new microcentrifuge tube, followed by a fourfold dilution with sterile water. The resulting DNA solution was used for PCR analysis.
Screening for cypX or moxY gene disruptants by PCR analyses and detection of pigments in their mycelia.
PCRs were performed using DNA from the resulting pyrithiamine-resistant transformants with moxY primers P9 (no. 432, moxY-F1, GAAGACCGCGGAGAATGG) and P10 (no. 433, moxY-R1, GGCCCAATGACACTGCC) or with cypX primers P1 (no. 434, cypX-F1, CGCAAGATTCCTGGTCCC) and P2 (no. 435, cypX-R1, CCAGCTAGGAGCAACGC). The transformants showing a faint band or no band corresponding to the expected product from the recipient strain SYS-4 were selected as candidates for gene-deleted mutants. At the same time, the conidia of each pyrithiamine-resistant transformant were inoculated onto aflatoxin-inducible GY agar medium (2% glucose, 0.5% yeast extract, and 2% agar) to check for pigment production. After 2 to 4 days of incubation at 28°C, mutants accumulating pigments in the mycelia were selected as candidate disruptants. After comparison with the results of PCR analyses, the possible mutants were then purified three times by single-colony isolation on GY medium, and the spores of the resulting colonies were stored at −80°C. The pigments that accumulated in the mycelia of the mutants were extracted and analyzed by thin-layer chromatography (TLC) as described below.
Confirmation of gene disruption by PCR analyses.
Gene disruption events were confirmed by PCR analyses using different combinations of primers, depending on the different gene constructs in the genome between the mutants and the recipient strain SYS-4. In most assays, a 0.5-μl diluted DNA sample in an 8-μl PCR volume resulted in good amplification even if the PCR products were larger than 3 kb. The primers used for PCR analyses were as follows: P1 (no. 434, cypX-F1), CGCAAGATTCCTGGTCCC; P2 (no. 435, cypX-R1), CCAGCTAGGAGCAACGC; P3 (no. 212, cypX-XhoI-F), CCGCTCGAGATGACCAACACTGCGCCAAG; P4 (no. 257, cypX-HindIII-R), CCCCAAGCTTCTACAGCTGAATGGCACAAC; P5 (no. 456, cypX-F2), CTTGGTAGTCGTCGGGC; P6 (no. 457, cypX-R2), CAGGTCGCATCAGGAGC; P7 (no. 311, PTRI-KpnI-F), GGGGTACCGGGCAATTGATTACGGGATCCCA; P8 (no. 322, PTRI-PstI-R2), AAAACTGCAGTGACGATGAGCCGCTCTTGC; P9 (no. 432, moxY-F1), GAAGACCGCGGAGAATGG; P10 (no. 433, moxY-R1), GGCCCAATGACACTGCC; P11 (no. 213, moxY-XhoI-F), CCGCTCGAGATGGACCCAGCCAACCGC; P12 (no. 256, moxY-HindIII-R), CCCCAAGCTTCAAGTTAGACGTGGCCGTC; P13 (no. 454, moxYL-F), CGTACAGCTTGCGTCGG; and P14 (no. 455, moxYR-R), CGCTGGAGGATGTCTCG. An 8-μl reaction mixture consisted of 0.5 μl of the diluted fungal DNA extract, 4 μl of 2× PCR Master Mix (Promega), 2.5 μl of nuclease-free water, and 0.5 μl of each primer (each 12.5 pmol/μl). PCR cycling depended on the size of the target fragment. PCR conditions for amplifying fragments shorter than 2.5 kb were (i) 94°C for 5 min; (ii) 35 cycles of 94°C for 40 s, 56°C for 40 s, 72°C for n min (determined by the size of the target fragment); and (iii) 72°C for 10 min. For fragments larger than 2.5 kb, PCR conditions were (i) 94°C for 5 min; (ii) 40 cycles of 94°C for 1 min, 56°C for 1 min, 72°C for n min; and (iii) 72°C for 10 min. PCR products were visualized by performing electrophoresis on a 1% agarose gel.
Characterization of the accumulating pigments in the mycelia of cypX or moxY disruptants.
A spore suspension (about 2 × 106 spores) of the disruptants or SYS-4 was inoculated into 100 ml YES medium in a bottle (10 cm in length, 4.5 cm in width, and 15 cm in depth), and the bottle was laid on its side to maximize the surface area of the medium. After a 3-day stationary culture at 28°C, the medium was removed and partially saved, and the remaining wet mycelia were extracted with 30 ml acetone. The acetone extract was collected, and the pigments in the extract were analyzed by TLC. The medium or resultant acetone extract (10 μl each) was then spotted onto silica gel TLC plates (Kieselgel 60, no. 5721; Merck & Co., Rahway, N.J.), and then each plate was developed with benzene:ethyl acetate (7:3, vol/vol) equilibrated with 10% aqueous acetic acid. The pigments were detected as visible yellow spots or fluorescent spots under UV light (365 nm). The fluorescence pictures were taken using a Fluor-S MAX MultiImager (Bio-Rad Laboratories, CA). To recover the pigments from each spot, the acetone extract was spotted onto the TLC plate in a line. After development with the same solution, the part corresponding to each spot was scraped, and pigments on the silica gel were extracted with acetone. The acetone extract was supplemented with 0.05 volume of water and then kept at −20°C until use. To identify the pigments, the resulting acetone extract was injected into a high-performance liquid chromatography (HPLC) apparatus (Shimadzu HPLC LC-6A system) equipped with an octyldecyl silane column (150 by 4.6 mm, S type; STR ODS-II; Sinwa Chemical Industries Ltd.). The flow solution was acetonitrile:tetrahydrofuran:water:acetic acid (25:25:50:1, vol/vol/vol/vol). The flow rate and column temperature were 1 ml min−1 and 35°C, respectively. The retention times of AVR, HVN, and VONE were compared with those of standard samples. Typical retention times were as follows: VONE, 3.4 min; HVN, 3.9 min; and AVR, 20.2 min.
Feeding experiments.
Each of the mutants—cypX-deleted CYPX-DD-55, moxY-deleted MOXY-DD-90, and NIAH-26—was cultured in 200 μl YES medium supplemented with 10 μM OAVN, 2.8 μM AVR, 10 μM HVN, 20 μM VONE, 10 μM VHA, or 19 μM VOAc or with the same volume of methanol as a solvent at 28°C for 4 days using the tip culture method (23, 28). Aflatoxin formation was measured by HPLC analysis with a silica gel HPLC column following the extraction of the medium with chloroform as described previously (25).
RESULTS
Isolation of the cypX disruptants.
After the transformation of SYS-4 with the linear replacement construct of pCYPX-DD in three separate experiments, 136 pyrithiamine-resistant transformants were obtained. Since other researchers have previously suggested that the disruption of the cypX gene did not show obvious changes of the phenotype (32), we did a PCR analysis using DNA extract from 115 transformants and the cypX internal primers P1 and P2 to select for the deleted cypX gene as a first screening. One transformant, transformant 55, showed a drastic decrease of the amount of the PCR product, suggesting that it was the desired mutant. We also inoculated the spores of each transformant on GY plates and cultured them to detect the accumulation of pigments in the mycelia. After culture for more than 2 days, only transformant 55 made an orange colony. The other 21 of the 136 transformants were similarly investigated, and another mutant, transformant 127, was also isolated as a candidate.
To confirm cypX deletion in these two mutants, we performed PCR analysis using a variety of primers (Fig. 2A). When combinations of primers P4/P7 and P3/P8 were used for PCR analysis of the purified mutant 55, a 2.57-kb band and a 2.15-kb band appeared, respectively, whereas the same bands were not detected when the genomic DNA of the wild-type strain SYS-4 was used as the template (Fig. 2B). In contrast, when primer pair P1/P2 located within the deletion region of the cypX gene was used, only SYS-4 produced the 810-bp PCR fragment as predicted. On the other hand, the mutant 55 produced a 2.72-kb band with primers P3/P4, whereas SYS-4 produced a 1.65-kb band with the same primers. Also, only mutant 55 generated a 3.15-kb band and a 3.33-kb band with primers P6/P7 and P5/P8, respectively, indicating that the cypX replacement construct was inserted into the desired position in the chromosome of the mutant. These results confirmed that transformant 55 is a cypX double-crossover gene disruptant in which the cypX gene is mostly deleted and replaced with the selectable marker ptrA by double-crossover recombination. The same results were obtained when transformant 127 was used (data not shown). We named mutants 55 and 127 CYPX-DD-55 and CYPX-DD-127, respectively.
Characterization of the cypX disruptants.
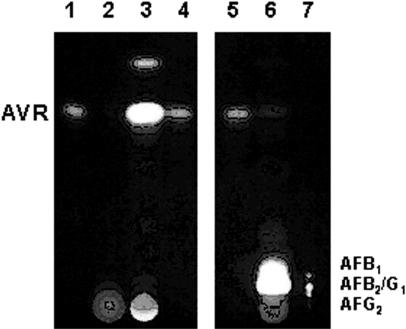
The culture medium and acetone extract of the mycelia of the CYPX-DD-55 mutant were analyzed by TLC (Fig. 3, lanes 2 and 3). The cypX-deleted mutant CYPX-DD-55 did not form aflatoxin, in contrast to results with the recipient strain SYS-4 (lane 6). Instead, it accumulated three kinds of pigments in the mycelia, the primary pigment of which has an Rf corresponding to that of AVR (Fig. 3, lane 3). Another pigment corresponding to the higher spot visible in Fig. 3 did not convert to aflatoxins in the preliminary feeding experiment, suggesting that this substance may not be related to aflatoxin production (data not shown). Its identity is still unknown. The third pigment was present very close to the origin on the TLC plate, indicating that it is a very polar substance. The Rf of this substance was different from that of OAVN and HAVN. When this pigment was analyzed again by TLC or HPLC following the extraction from the spot, AVR newly appeared. Although it may be related to AVR, the function of this substance still remains unclear. The accumulation of AVR in the mutant CYPX-DD-127 was also confirmed by TLC and HPLC analyses after the extraction of the major substance (data not shown).
FIG. 3.
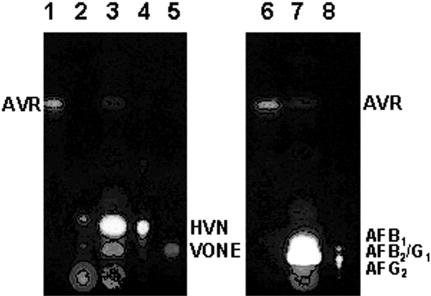
Accumulation of AVR in the cypX disruptant. Metabolites produced by the cypX-deleted mutant or the recipient strain were analyzed by TLC. Lanes 1, 4, and 5, standard for AVR; lane 2, culture medium of the cypX-deleted mutant CYPX-DD-55; lane 3, acetone extract of the same mutant; lane 6, acetone extract of the recipient strain SYS-4; lane 7, standards for aflatoxins as indicated in the figure. Aflatoxins showed blue or green fluorescence (lanes 6 and 7), and other substances showed orange or yellow fluorescence under UV light.
Isolation of the moxY disruptants.
The transformation of A. parasiticus SYS-4 with the linear replacement construct of pMOXY-DD (Fig. 4A) in two separate experiments generated 128 pyrithiamine-resistant transformants. We checked the deletion of moxY in the genome by using PCR with primers P9 and P10 and found that 12 transformants lost the 910-bp PCR band. When all 128 transformants were separately inoculated onto GY plates, two transformants, transformants 69 and 90, which were included in the 12 transformants mentioned above, accumulated bright yellow pigments in the mycelia.
When the purified mutant 69 was also analyzed by PCR using primers P11/P7, P12/P8, and P11/P12, PCR products of 2.28, 2.13, and 2.41 kb, respectively, which were specific for the replacement of the moxY gene with the ptrA gene, were formed (Fig. 4B). When primer pair P9/P10 was used, only SYS-4 produced the 910-bp PCR product. All PCR results indicated that the moxY replacement construct was inserted into the desired position in the chromosome of the mutant. Transformant 90 showed the same results as those of mutant 69 (data not shown). These results demonstrated that these two mutants were the moxY disruptants; we named them MOXY-DD-69 and MOXY-DD-90.
Characterization of the moxY disruptants.
When MOXY-DD-90 was cultured in YES medium, two kinds of pigments accumulated mainly in the mycelia (Fig. 5, lane 3), corresponding to HVN (lane 4) and VONE (lane 5). These results were further confirmed using HPLC analyses (data not shown). The medium as well as the acetone extract of this mutant did not contain any aflatoxins, indicating that a step catalyzed by the moxY gene product was completely blocked. Small amounts of HVN and VONE were observed in the medium (Fig. 5, lane 2), indicating that these substances were partially excreted from the mycelia due to their high polarities. The same results were obtained with MOXY-DD-69 (data not shown).
FIG. 5.
Accumulation of HVN and VONE in the moxY disruptant. Metabolites produced by the moxY disruptant or the recipient strain were analyzed by TLC. Lanes 1 and 6, standard samples of AVR; lane 2, culture medium of the moxY disruptant MOXY-DD-90; lane 3, acetone extract of the same mutant; lane 4, standard of HVN; lane 5, standard for VONE; lane 7, acetone extract of the recipient strain, SYS-4; lane 8, standards for aflatoxins as indicated in the figure. Aflatoxins (lanes 7 and 8) showed blue or green fluorescence, and other substances showed orange or yellow fluorescence under UV light.
Feeding experiments of the deleted mutants.
We performed feeding experiments using precursors related to the pathway reaction from OAVN to VHA in aflatoxin biosynthesis. CYPX-DD-55 could not convert either OAVN or AVR to aflatoxins. In contrast, it converted HVN, VONE, VHA, and VOAc to aflatoxins (Table 1), indicating that cypX is involved in the reaction from AVR to HVN. In contrast, MOXY-DD-90 could not convert OAVN, AVR, HVN, or VONE to aflatoxins, whereas it converted either VHA or VOAc to aflatoxins. These results indicated that the moxY gene is involved in the reaction from HVN to VHA or from VONE to VOAc.
TABLE 1.
Aflatoxin formation of mutants in feeding experiments
| Strain | Total concn of aflatoxin formed (μg/250 μl culture medium)a
|
|||||
|---|---|---|---|---|---|---|
| OAVN | AVR | HVN | VONE | VHA | VOAc | |
| CYPX-DD-55 | NDb | ND | 17.2 ± 0.9 | 0.9 ± 0.2 | 6.9 ± 1.0 | 4.3 ± 0.4 |
| MOXY-DD-90 | ND | ND | ND | ND | 8.4 ± 1.1 | 3.7 ± 0.6 |
| NIAH-26 | 53.5 ± 0.7 | 14.1 ± 1.3 | 21.2 ± 0.3 | 3.5 ± 0.2 | 43.9 ± 6.4 | 6.1 ± 3.1 |
Values are means ± standard deviations.
ND, not detected.
DISCUSSION
This work showed that the cypX gene encodes AVR monooxygenase, which catalyzes the reaction from AVR to HVN (Fig. 1). Since the cypX gene is a homolog of stcB in the sterigmatocystin gene cluster of A. nidulans (11), StcB may catalyze the same reaction in sterigmatocystin biosynthesis. Based on amino acid homology, the cypX gene codes for a cytochrome P450 monooxygenase (30). We previously reported that the reaction from AVR to HVN is catalyzed by a microsomal AVR monooxygenase enzyme (26). The accumulation of AVR in the mycelia of the cypX-deleted mutant matches these biochemical data well (Fig. 3). Characterization of the other two unknown substances that accumulated along with AVR in the cypX disruptant is now in progress in our laboratory.
We also demonstrated that the moxY gene encodes the HVN monooxygenase catalyzing the reaction from HVN to VHA as well as from VONE to VOAc (Fig. 1). The moxY gene is a homolog of stcW in the sterigmatocystin gene cluster of A. nidulans (11), indicating that the enzyme encoded by the stcW gene may catalyze the same reactions in sterigmatocystin biosynthesis. moxY is commonly involved in the two reactions, from HVN to VHA and from VONE to VOAc, because the moxY-deleted mutant accumulated both HVN and VONE (Fig. 5). Both reactions are Baeyer-Villiger reactions, in which an oxygen atom is inserted into a C—C bond adjacent to the carbonyl group of an aliphatic or alicyclic ketone. Microbial enzymes carrying Baeyer-Villiger reactions are known to contain flavin adenine dinucleotide in their molecules (15). Therefore, moxY as well as stcW encodes the HVN monooxygenase in aflatoxin/sterigmatocystin biosynthesis. Although VOROL can be produced from VONE in the aflatoxin biosynthetic pathway (26), the accumulation of VOROL was not detected, which may be due to the short (3-day) culture time.
The pathway following VHA or VOAc was intact in the moxY-deleted mutant, because VHA as well as VOAc could be converted to aflatoxins in the feeding experiments (Table 1). Keller et al. reported that the disruption of stcW caused the accumulation of AVR and that a stcW-inactivated mutant did not change 14C-labeled norsolorinic acid to HVN. However, if stcW is a homolog of moxY, the stcW-inactivated mutant should have changed 14C-labeled norsolorinic acid to HVN because only the next step after HVN should have been blocked. The discrepancies among these results should be studied in more detail.
The avfA gene was suggested above to be involved in the step from AVR to VHA by a complementation experiment with the AVR-accumulating mutant. When the AVR-accumulating mutant was transformed by cosmid clones containing the avfA gene and different lengths of its flanking regions, its aflatoxin-producing ability was restored (33). However, avfA does not have a sequence similar to either cypX or moxY. Furthermore, involvement of avfA in aflatoxin biosynthesis has not been directly determined by gene disruption or other methods. The relationship among these genes still remains to be studied.
The production of multinucleate conidia by A. parasiticus makes screening the mutant by PCR somewhat difficult. The transformants generated just after transformation are generally heterokaryotic, and untransformed or intact nuclei can coexist with transformed or mutant nuclei even in a conidium, which results in the generation of undesired PCR products even from desired mutants. Therefore, the selected mutant candidates with slight or no undesired PCR bands should be purified at least three times by single-colony isolation and then be analyzed by PCR again to confirm the mutation events.
Gene disruption followed by the characterization of the mutant usually provides useful information in determining the function of the gene. If we can anticipate the possible function of the target gene, the screening of the desired disruptant is relatively easy. However, if no such projection can be made, the selection of the desired transformants from many transformants is always difficult. Such was the case in this work because other groups have failed in the isolation of the cypX- or moxY-deleted mutants (32). Therefore, we decided to check for the deletion of the target gene in all obtained transformants by using PCR. For this examination, a simple, effective, and stable method for extracting genomic DNA from many (at least 100) fungi at the same time was required. Although some methods have already been reported, such as heating of the mycelia (2) and the protoplast formation method (21), we needed a much simpler and more stable method applicable for long as well as short PCR. We thus devised a simple method to extract genomic DNA from fungal mycelia, in which cell disruption with a shaker and phenol extraction were combined. This method was useful for even a slight amount of mycelia collected from a fungal colony on an agar plate. Any type of shaker was useful if it could effectively disrupt the mycelia as well as the conidia. A Vortex mixer did not seem to be enough for this purpose. We ultimately obtained genomic DNA from more than 100 transformants at the same time within 2 h. The resulting DNA could be used to detect PCR products of at least 4 kb. In this study, we found that 2 of 136 transformants, transformants 55 and 127, deleted the cypX gene; we also found that only these two mutants among all of the 136 transformants accumulated pigments in their mycelia. Furthermore, we found that two of 128 transformants, transformants 69 and 90, deleted the moxY gene; only these two mutants among all of the 128 transformants accumulated pigments in their mycelia. These results showed that PCR analysis data of the desired mutants were consistent with their phenotype changes. Therefore, this extraction method makes it possible to do so-called colony PCR of fungi, and it will certainly be useful for gene analyses of a number of organisms containing rigid cell walls like fungi and plants.
Acknowledgments
We thank Hiromitsu Nakajima for his critical reading of the manuscript and Ken-ichi Kusumoto for his advice on fungal transformation. The DNA search of the GenBank database was performed with the assistance of the Computer Center of Agriculture, Forestry, and Fisheries Research, MAFF, Japan.
This work was supported in part by grant-in-aid BDP-04-VI-1-2 (biodesign program) from the Ministry of Agriculture, Forestry, and Fisheries, Japan.
REFERENCES
- 1.Brown, D. W., J. H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiou, C.-H., M. Miller, D. L. Wilson, F. Trail, and J. E. Linz. 2002. Chromosomal location plays a role in regulation of aflatoxin gene expression in Aspergillus parasiticus. Appl. Environ. Microbiol. 68:306-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eaton, D. L., and J. D. Groopman (ed.). 1994. The toxicology of aflatoxins: human health, veterinary, and agricultural significance. Academic Press, San Diego, Calif.
- 4.Gomi, K., Y. Iimura, and S. Hara. 1987. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric. Biol. Chem. 51:2549-2555. [Google Scholar]
- 5.Hamasaki, T., Y. Hatsuda, N. Terashima, and M. Renbutsu. 1965. The structure of a new metabolite of Aspergillus versicolor. Agric. Biol. Chem. 29:696-697. [Google Scholar]
- 6.Hodges, R. L., H. S. Kelkar, X. Xuei, P. L. Skatrud, N. P. Keller, T. H. Adams, R. E. Kaiser, V. A. Vinci, and D. McGilvray. 2000. Characterization of an echinocandin B-producing strain blocked for sterigmatocystin biosynthesis reveals a translocation in the stcW gene of the aflatoxin biosynthetic pathway. J. Ind. Microbiol. Biotechnol. 25:333-341. [DOI] [PubMed] [Google Scholar]
- 7.Horng, J. S., P. K. Chang, J. J. Pestka, and J. E. Linz. 1990. Development of a homologous transformation system for Aspergillus parasiticus with the gene encoding nitrate reductase. Mol. Gen. Genet. 224:294-296. [DOI] [PubMed] [Google Scholar]
- 8.Ito, Y., S. W. Peterson, D. T. Wicklow, and T. Goto. 2001. Aspergillus pseudotamarii, a new aflatoxin producing species in Aspergillus section Flavi. Mycol. Res. 105:233-239. [Google Scholar]
- 9.Jelinek, C. F., A. E. Pohland, and G. E. Wood. 1989. Worldwide occurrence of mycotoxins in foods and feeds—an update. J. Assoc. Off. Anal. Chem. 72:223-230. [PubMed] [Google Scholar]
- 10.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 11.Keller, N. P., C. M. H. Watanabe, H. S. Kelkar, T. H. Adams, and C. A. Townsend. 2000. Requirement of monooxygenase-mediated steps for sterigmatocystin biosynthesis by Aspergillus nidulans. Appl. Environ. Microbiol. 66:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klich, M. A., E. J. Mullaney, C. B. Daly, and J. W. Cary. 2000. Molecular and physiological aspects of aflatoxin and sterigmatocystin biosynthesis by Aspergillus tamarii and A. ochraceoroseus. Appl. Microbiol. Biotechnol. 53:605-609. [DOI] [PubMed] [Google Scholar]
- 13.Kurtzman, C. P., B. W. Horn, and C. W. Hesseltine. 1987. Aspergillus nomius, a new aflatoxin-producing species related to Aspergillus flavus and Aspergillus tamarii. Antonie Leeuwenhoek 53:147-158. [DOI] [PubMed] [Google Scholar]
- 14.Minto, R. E., and C. A. Townsend. 1997. Enzymology and molecular biology of aflatoxin biosynthesis. Chem. Rev. 97:2537-2556. [DOI] [PubMed] [Google Scholar]
- 15.Miyamoto, M., J. Matsumoto, T. Iwaya, and E. Itagaki. 1995. Bacterial steroid monooxygenase catalyzing the Baeyer-Villiger oxidation of C21-ketosteroids from Rhodococcus rhodochrous: the isolation and characterization. Biochim. Biophys. Acta 1251:115-124. [DOI] [PubMed] [Google Scholar]
- 16.Payne, G. A., and M. P. Brown. 1998. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 36:329-362. [DOI] [PubMed] [Google Scholar]
- 17.Peterson, S. W., Y. Ito, B. W. Horn, and T. Goto. 2001. Aspergillus bombycis, a new aflatoxigenic species and genetic variation in its sibling species, A. nomius. Mycologia 93:689-703. [Google Scholar]
- 18.Prieto, R., G. L. Yousibova, and C. P. Woloshuk. 1996. Identification of aflatoxin biosynthesis genes by genetic complementation in an Aspergillus flavus mutant lacking the aflatoxin gene cluster. Appl. Environ. Microbiol. 62:3567-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakuno, E., K. Yabe, and H. Nakajima. 2003. Involvement of two cytosolic enzymes and a novel intermediate, 5′-oxoaverantin, in the pathway from 5′-hydroxyaverantin to averufin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 69:6418-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Townsend, C. A., K. A. Plavcan, K. Pal, S. W. Brobst, M. S. Irish, E. W. Ely, and J. W. Bennett. 1988. Hydroxyversicolorone: isolation and characterization of a potential intermediate in aflatoxin biosynthesis. J. Org. Chem. 53:2472-2477. [Google Scholar]
- 21.Van Zenji, C. M. J., E. H. M. van de Kamp, P. J. Punt, G. C. M. Selten, B. Hauer, R. F. M. van Gorcom, and C. A. M. J. J. van den Hondel. 1997. An improved colony-PCR method for filamentous fungi for amplification of PCR-fragments of several kilobases. J. Biotechnol. 59:221-224. [DOI] [PubMed] [Google Scholar]
- 22.Yabe, K. 2002. Pathway and genes of aflatoxin biosynthesis, p. 227-251. In F. Fierro and J. Francisco (ed.), Microbial secondary metabolites: biosynthesis, genetics and regulation. Research Signpost, Kerala, India.
- 23.Yabe, K., Y. Ando, and T. Hamasaki. 1988. Biosynthetic relationship among aflatoxins B1, B2, G1, and G2. Appl. Environ. Microbiol. 54:2101-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yabe, K., Y. Ando, and T. Hamasaki. 1991. A metabolic grid among versiconal hemiacetal acetate, versiconol acetate, versiconol and versiconal during aflatoxin biosynthesis. J. Gen. Microbiol. 137:2469-2475. [DOI] [PubMed] [Google Scholar]
- 25.Yabe, K., Y. Ando, and T. Hamasaki. 1991. Desaturase activity in the branching step between aflatoxins B1 and G1 and aflatoxins B2 and G2. Agric. Biol. Chem. 55:1907-1911. [Google Scholar]
- 26.Yabe, K., N. Chihaya, S. Hamamatsu, E. Sakuno, T. Hamasaki, H. Nakajima, and J. W. Bennett. 2003. Enzymatic conversion of averufin to hydroxyversicolorone and elucidation of a novel metabolic grid involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 69:66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yabe, K., and H. Nakajima. 2004. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 64:745-755. [DOI] [PubMed] [Google Scholar]
- 28.Yabe, K., H. Nakamura, Y. Ando, N. Terakado, H. Nakajima, and T. Hamasaki. 1988. Isolation and characterization of Aspergillus parasiticus mutants with impaired aflatoxin production by a novel tip culture method. Appl. Environ. Microbiol. 54:2096-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu, J., D. Bhatnagar, and K. C. Ehrlich. 2002. Aflatoxin biosynthesis. Rev. Iberonam. Micol. 19:191-200. [PubMed] [Google Scholar]
- 30.Yu, J., P.-K. Chang, D. Bhatnagar, and T. E. Cleveland. 2000. Genes encoding cytochrome P450 and monooxygenase enzymes define one end of the aflatoxin pathway gene cluster in Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 53:583-590. [DOI] [PubMed] [Google Scholar]
- 31.Yu, J., P.-K. Chang, J. W. Cary, M. Wright, D. Bhatnagar, T. E. Cleveland, G. A. Payne, and J. E. Linz. 1995. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl. Environ. Microbiol. 61:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu, J., P.-K. Chang, K. C. Ehrlich, J. W. Cary, D. Bhatnagar, T. E. Cleveland, G. A. Payne, J. E. Linz, C. P. Woloshuk, and J. W. Bennett. 2004. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70:1253-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, J., C. P. Woloshuk, D. Bhatnagar, and T. E. Cleveland. 2000. Cloning and characterization of avfA and omtB genes involved in aflatoxin biosynthesis in three Aspergillus species. Gene 248:157-167. [DOI] [PubMed] [Google Scholar]