Abstract
The gram-negative, purple nonsulfur, facultative photosynthetic bacterium Rhodobacter capsulatus is a widely used model organism and has well-developed molecular genetics. In particular, interposon mutagenesis using selectable gene cartridges is frequently employed for construction of a variety of chromosomal knockout mutants. However, as the gene cartridges are often derived from antibiotic resistance-conferring genes, their numbers are limited, which restricts the construction of multiple knockout mutants. In this report, sacB—5-fluoroorotic acid (5FOA)—pyrE-based bidirectional selection that facilitates construction of unmarked chromosomal knockout mutations is described. The R. capsulatus pyrE gene encoding orotate phosphoribosyl transferase, a key enzyme of the de novo pyrimidine nucleotide biosynthesis pathway, was used as an interposon in a genetic background that is auxotrophic for uracil (Ura−) and hence resistant to 5FOA (5FOAr). Although Ura+ selection readily yielded chromosomal allele replacements via homologous recombination, selection for 5FOAr to replace pyrE with unmarked alleles was inefficient. To improve the latter step, 5FOAr selection was combined with sucrose tolerance selection using a suicide plasmid carrying the Bacillus subtilis sacB gene encoding levansucrase that induces lethality upon exposure to 5% (wt/vol) sucrose in the growth medium. Sucrose-tolerant, 5FOAr colonies that were obtained carried chromosomal unmarked mutant alleles of the target gene via double crossovers between the resident pyrE-marked and incoming unmarked alleles. The effectiveness of this double selection was proven by seeking insertion and deletion alleles of helC involved in R. capsulatus cytochrome c biogenesis, which illustrated the usefulness of this system as a genetic means for facile construction of R. capsulatus unmarked chromosomal mutants.
The gram-negative facultative photosynthetic bacterium Rhodobacter capsulatus belongs to the group of α-proteobacteria and is evolutionarily closely related to mitochondria (43). This bacterium grows under a variety of conditions and has well-developed genetics (17, 34). It has been used as a suitable model organism for a multitude of studies, including studies of respiration and photosynthesis, biosynthesis, and metabolism, as well as regulation of gene expression in response to various environmental stimuli (9). In R. capsulatus, isolation of chromosomal knockout mutations can be carried out by using gene transfer agent (GTA)-mediated interposon mutagenesis (34), and a large number of genetic and biochemical studies have benefited from this technique. However, as antibiotic resistance gene cassettes (e.g., cassettes for resistance to kanamycin [Kmr], gentamicin [Gmr], spectinomycin [Spcr], or tetracycline [Tetr] in R. capsulatus) are used as selectable markers, the number of chromosomal knockout mutations that can be generated simultaneously in a given strain is limited. This could be a restrictive factor for generation of multiple-knockout mutations in a given strain. In addition, some interposons might be polar on the expression of the downstream genes, which might cause undesired complications. Thus, a method to alleviate these limitations would be a valuable addition to the arsenal of genetic tools available for R. capsulatus.
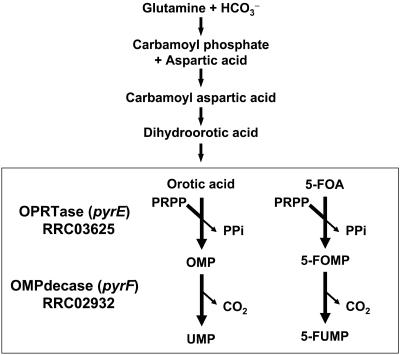
In various organisms, genes of the pyrimidine biosynthesis pathway provide an interesting opportunity for genetic studies due to the mode of action of 5-fluoroorotic acid (5FOA), which is a bactericidal pyrimidine analogue. The key enzymes of this pathway, orotate phosphoribosyl transferase (OPRTase) (encoded by pyrE) and orotidine 5′-monophosphate decarboxylase (OMPdecase) (encoded by pyrF), are at the basis of this toxicity. 5FOA is converted to 5-fluorouridine monophosphate and can be incorporated into RNA, as well as further metabolized to 5-fluoro-2′-dUMP to act as a potent inhibitor of thymidylate synthase, causing cessation of DNA synthesis (Fig. 1). Mutants that lack either of these enzymes are unable to metabolize 5FOA and hence are resistant to 5FOA (5FOAr), but they are uracil auxotrophs (Ura−) for growth. These properties can be used for selection in both directions (i.e., Ura+ and 5FOAr) and have been exploited successfully in Saccharomyces cerevisiae genetics (5). More recently, similar approaches have been extended to other organisms, including the thermophilic gram-negative bacterium Thermus thermophilus (39), the halophilic archaeon Haloferax volcanii (4), and the hyperthermophilic archaea Pyrococcus abyssi (20) and Thermococcus kodakaraensis (31).
FIG. 1.
Late steps of the de novo pyrimidine biosynthesis pathway of R. capsulatus. The last two reaction steps of the de novo pyrimidine biosynthesis pathway deduced from the R. capsulatus genome sequence that involve pyrE and 5FOA are enclosed in a box. The toxicity of 5FOA is considered to appear via its conversion to 5-fluoroorotidine monophosphate (5FOMP) and 5-fluorouridine monophosphate (5FUMP) by OPRTase and OMPdecase, respectively. OMP, orotidine 5′-monophosphate; PRPP, 5-phosphoribosylpyrophosphate; UMP, uridine monophosphate; PPi, pyrophosphate.
We thought that the development in R. capsulatus of a similar 5FOA-pyrE-based genetic selection system might be useful for facilitating construction of unmarked chromosomal knockout mutants. Therefore, we cloned the R. capsulatus pyrE gene and constructed a Ura− 5FOAr mutant carrying a chromosomal insertion-deletion [Δ(pyrE::kan)] allele. By using pyrE as an interposon, we then isolated a pyrE-marked chromosomal knockout mutation in helC [Δ(helC::pyrE)], a gene that is involved in cytochrome c biogenesis in R. capsulatus (1). To facilitate the isolation of a mutant with unmarked chromosomal knockout mutations in helC (helC or ΔhelC), we combined 5FOAr selection with sucrose lethality provided by the Bacillus subtilis sacB gene encoding levansucrase (7, 37). Selection for sucrose-tolerant and 5FOAr colonies in the presence of uracil readily yielded chromosomal unmarked helC or ΔhelC alleles, illustrating the efficiency of sacB-5FOA-pyrE selection as a useful genetic tool for facile isolation of unmarked chromosomal knockout mutants of R. capsulatus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used are listed in Table 1. Escherichia coli strains and their plasmid-harboring derivatives were grown in Luria broth (LB) containing appropriate antibiotics (ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; tetracycline, 12.5 μg/ml; streptomycin, 25 μg/ml; gentamicin, 10 μg/ml) at 37°C. R. capsulatus strains were grown at 35°C in Siström's minimal medium A (MedA) (36) or enriched medium (MPYE) (17) supplemented as needed with 40 μg/ml rifampin, 10 μg/ml kanamycin, 2.5 μg/ml tetracycline, or 1.25 μg/ml gentamicin, either chemoheterotrophically in the dark (Res conditions) or photoheterotrophically in the light (Ps conditions) using anaerobic jars and BBL GasPaks. When grown in MedA, pyrE mutants were supplemented with 0.1 mg/ml uracil and 0.1 mg/ml Casamino Acids.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Relevant phenotype | Reference or source |
|---|---|---|---|
| E. coli strains | |||
| HB101 | F−mcrB mrr hsdS20(rB− mB−) recA13 leu ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20(Smr) supE44 λ | 29 | |
| DH5α | hsdR17 (rK− mK+) deoR thi-1 phoA supE44 λ−gyrA96 relA1 | Invitrogen | |
| XL1-Blue | F′::Tn10(Tetr) proA+B+lacIq (lacZ)M15/recA1 endA1 gyrA96(NaIr) thi-1 hsdR17 (rK− mK+) glnV44 relA1 lac | Tetr | Stratagene |
| JM109(λpir) | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-proAB)F′ [traD36 proAB+laqIqlacZΔM15] λpirR6K | 22 | |
| S17-1 (λpir) | thi pro hsdR hsdM+recA::(RP4-2-Tc::Mu-Km::Tn7) λpir | Smr Tpr Str | 27 |
| R. capsulatus strains | |||
| MT1131 | crtD121 Rifr | Wild type | 35 |
| Y262 | GTA overproducer | 44 | |
| TJK11 | crtD121 Rifr Δ(pyrE::kan) | Ps+ Nadi+ PyrE− Kmr | This study |
| TJK11R | crtD121 Rifr Δ(pyrE::kan) | Ps+ Nadi+ PyrE− Kmr | This study |
| TCK1 | crtD121 Rifr Δ(pyrE::kan) helC::pyrE | Ps− Nadi−, all cytochrome c−, Kmr | This study |
| TCK2 | crtD121 Rifr Δ(pyrE::kan) helC::pyrE | Ps− Nadi−, all cytochrome c−, Kmr | This study |
| TCK4 | crtD121 Rifr Δ(pyrE::kan) helC | Ps− Nadi−, all cytochrome c−, Kmr | This study |
| TCK5 | crtD121 Rifr Δ(pyrE::kan) ΔhelC | Ps− Nadi−, all cytochrome c−, Kmr | This study |
| TRK1 | crtD121 Rifr Δ(pyrE::kan) helC::pyrE, pZJDhelABCDX integrated | Ps+ Nadi+ Kmr Gmr | This study |
| TRK2 | crtD121 Rifr Δ(pyrE::kan) helC::pyrE, pZJDΔhelC integrated | Ps− Nadi−, all cytochrome c−, Kmr Gmr | This study |
| Plasmids | |||
| pRK415 | Broad-host-range plasmid | Tetr | 8 |
| pCHB500 | pRK415 with a cycA promoter | Tetr | 3 |
| pRK2013 | Helper | Kmr | 8 |
| pUC4K | Kanamycin cassette | Ampr Kmr | Amersham |
| pCR-Blunt | pCR-Blunt II-TOPO | Kmr | Invitrogen |
| pBluescript | pBluescript II SK(+) | Ampr | Stratagene |
| pTZ18U | Ampr | BioRad Laboratories | |
| pZJD29a | Mobilizable suicide vector | Gmr | 38 |
| pCR-PyrE | pCR-Blunt with 782-bp PCR product of R. capsulatus pyrE including 60 bp of its upstream sequence | Kmr | This study |
| pCR-PyrESD | pCR-Blunt with 737-bp PCR product of R. capsulatus pyrE with Shine-Dalgarno sequence | Kmr | This study |
| pCR-PyrENde | pCR-Blunt with 718-bp PCR product of R. capsulatus pyrE without Shine-Dalgarno sequence | Kmr | This study |
| pCR-PyrC | pCR-Blunt with 726-bp KpnI-XhoI PCR product of R. capsulatus pyrC | Kmr | This study |
| pCR-DnaB | pCR-Blunt with 756-bp XhoI-XbaI PCR product of R. capsulatus dnaB | Kmr | This study |
| pPyrE01 | pBluescript with the ClaI-BamHI fragment of pCR-PyrE | Ampr | This study |
| pPyrE02 | pBluescript with the EcoRI fragment of pCR-PyrE | Ampr | This study |
| pRK-PyrE-PB | pRK415 with the PstI-BamHI fragment of pPyrE02 | Tetr | This study |
| pRK-PyrE-PH | pRK415 with the PstI-HindIII fragment of pPyrE02 | Tetr | This study |
| pRK-PyrENde | pRK415 with the BamHI-PstI fragment of pCR-PyrENde | Tetr | This study |
| pRK-PyrESD | pRK415 with the BamHI-PstI fragment of pCR-PyrESD | Tetr | This study |
| pΔPyrE::Km | pBluescript with the pyrC-Km-dnaB fragment | Ampr | This study |
| pRK-ΔPyrE::Km | pRK415 with the KpnI-XbaI fragment of pΔPyrE::Km | Tetr | This study |
| p2helABCDX | pUC118 with R. capsulatus helABCDX cluster | Ampr | R. Kranz |
| pCS1530 | pRK415 with 830-bp XbaI-KpnI PCR product containing R. capsulatus helC | Tetr | 30 |
| pCS1650#3 | pCS1530 with the pyrE cassette in the BamHI site of helC gene (polar orientation) | Tetr | This study |
| pCS1650#4 | pCS1530 with the pyrE cassette in the BamHI site of helC gene (nonpolar orientation) | Tetr | This study |
| pCS1652 | pCS1530 with a frameshift at the BamHI site of helC gene | Tetr | This study |
| pZJDhelABCDX | pZJD29a with the SacI-XbaI 3.5-kb fragment of pUChelABCDX | Gmr | This study |
| pZJDΔhelC | pZJD29a with the SacI-XbaI 830-bp fragment of pCS1652 | Gmr | This study |
| pZJDhelABΔCDX | pZJDhelABCDX with 250-bp in-frame deletion in helC | Gmr | This study |
Bacterial genetic techniques.
Plasmid pRK415 derivatives were conjugated from E. coli to R. capsulatus, and GTA-mediated crosses were performed as described previously (17). The transferable suicide plasmid pZJP29a, which carries a Gmr cassette and the B. subtilis sacB gene encoding levansucrase expressed from the R. capsulatus pucAB promoter, was a gift from C. Bauer (Indiana University). As pZJP29a is an R6K derivative, it cannot replicate in R. capsulatus and becomes toxic in the presence of 5% (wt/vol) sucrose in the growth medium. This toxicity is due to the expression of sacB (21, 38) and the production of branched-chain fructose-derived compounds (levans) (7, 37). All pZJP29a derivatives were maintained in E. coli JM109(λpir) and spot mated from E. coli S17-1(λpir) into R. capsulatus strains on nonselective MPYE plates, using a recipient-to-donor cell ratio of 10:1. Gmr transconjugants were subsequently selected on MPYE plates containing 1.25 μg/ml gentamicin and 40 μg/ml rifampin, which yielded presumably single-crossover chromosomal integrants. The colonies were then subcultured in nonselective MPYE at 35°C so that there was a second homologous recombination event to excise the plasmid DNA. The culture was serially diluted in MPYE and spread onto MPYE agar plates containing either 5% (wt/vol) sucrose or 5% (wt/vol) sucrose and 0.1 mg/ml 5FOA. Colonies that appeared on these plates were then characterized in detail.
Molecular cloning techniques.
All DNA manipulations were carried out according to standard protocols described by Sambrook et al. (29). Extraction of DNA fragments from agarose gels was performed according to the manufacturer's protocol using QIAGEN gel extraction kits. DNA sequences were determined by automated DNA sequencing with a Big-Dye terminator cycle sequencing kit (Applied Biosystems Inc., Foster City, CA) used according to the supplier's specifications by using the T7, M13 forward, and M13 reverse primers, as well as custom-designed primers, as needed.
Southern hybridization analyses of R. capsulatus mutants.
R. capsulatus chromosomal DNA was isolated from fully grown cultures by using a QIAGEN DNeasy kit and was digested with appropriate restriction enzymes (New England Biolabs Inc., Beverly, MA). DNA fragments were separated by electrophoresis on a 0.75% (wt/vol) agarose gel and blotted onto a MAGNA nylon membrane (MSI, Westborough, MA). DNA probes were purified from appropriate plasmids after restriction enzyme digestions and agarose gel electrophoresis and were labeled with digoxigenin (DIG)-dUTP with a DIG High Prime DNA labeling kit (Roche Inc., Indianapolis, IN). DNA hybridizations were performed at 55°C. The membranes were washed twice with 2× SSC buffer containing 0.1% sodium dodecyl sulfate (SDS) at room temperature for 10 min and twice with 0.5× SSC buffer containing 0.1% SDS at 65°C for 15 min (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Detection of hybridized probes was carried out by using anti-digoxigenin-alkaline phosphatase with a ready-to-use substrate (CPSD) according to the manufacturer's recommendations (Roche Inc., Indianapolis, IN).
PCR analysis of R. capsulatus mutants.
Chromosomal DNA isolated using a QIAGEN DNeasy kit from appropriate R. capsulatus mutants was used as a template, and the helC locus of the chromosome was amplified using primers helC2 (5′-CTT ATT CTA GAT TGA TGC GAG T-3′) and helC3 (5′-GAC GCA AGG TAC CTG ACG GCG TAT TTT C-3′) (the underlining indicates newly introduced XbaI and KpnI sites, respectively) and Platinum Taq DNA polymerase enzyme (Invitrogen Inc., Carlsbad, CA). The PCR mixture contained 1× reaction buffer (without MgCl2), each deoxynucleoside triphosphate at a concentration of 0.2 mM, 1.5 mM MgCl2, each PCR primer at a concentration of 1.0 μM, 0.8 ng/μl chromosomal DNA, 35% (vol/vol) glycerol, and 0.05 U/μl Taq DNA polymerase. The reaction conditions were as follows: denaturation at 98°C for 4 min, 30 cycles of amplification (98°C for 0.5 min, 55°C for 1 min, and 72°C for 1.5 min), and one final elongation step at 72°C for 4 min. PCR products were analyzed by 1.0% (wt/vol) agarose gel electrophoresis, extracted from gels, and cloned into the pCR 2.1-TOPO vector using a TOPO TA cloning kit (Invitrogen Inc., Carlsbad, CA).
Cloning and sequencing of the pyrE locus of R. capsulatus.
An approximately 2.0-kb DNA fragment encompassing R. capsulatus pyrE was PCR amplified using either Pfu Turbo DNA polymerase (Stratagene Inc., La Jolla, CA) or Platinum Pfx DNA polymerase (Invitrogen Inc., Carlsbad, CA) according to the manufacturer's protocols. As appropriate, primers PYR1FKPN (5′-GCG GTA CCC CGC CGC GCG ACG CCC GGG C-3′), PYR1RXHO (5′-GCC TCG AGG CAT CCG CGC GGT CAA CCG G-3′), PYR2FCLA (5′-CGA TCG ATG GCA CCA CGG TCG AAA AGA G-3′), PYR2RBAM (5′-GCG GAT CCC CAC AGG CGT ATC CCC GGG G-3′), PYR3FXHO (5′-GCC TCG AGG AGC GCC CCG CGC GCC TGG C-3′), and PYR3RXBA (5′-GCT CTA GAG CGC GCC CTC GGT CTT GAC G-3′) (the underlining indicates newly introduced KpnI, XhoI, ClaI, BamHI, XhoI, and XbaI sites, respectively) were employed as follows. Amplification using PYR1FKPN and PYR1RXHO yielded a 726-bp DNA fragment, which contained 560 bp of the 3′ end of pyrC, the 106-bp intergenic region between pyrC and pyrE, and the 54-bp 5′ end of pyrE. Using PYR2FCLA and PYR2RBAM, a 782-bp DNA fragment that contained the entire pyrE gene and its 60-bp upstream region was obtained. With PYR3FXHO and PYR3RXBA, a 756-bp DNA fragment which consisted of 43 bp of the pyrE 3′ end, the 245-bp intergenic region between pyrE and dnaB, and the 462-bp 5′ end of dnaB, was amplified. These three PCR products overlapped each other, yielding a total of 2,050 bp of DNA sequence surrounding pyrE. Appropriate PCR products were cloned into the pCR-Blunt II TOPO vector using a Zero Blunt TOPO PCR cloning kit (Invitrogen Inc., Carlsbad, CA) (Table 1), and their DNA sequences were determined and compared with that of the R. capsulatus genome to avoid any possible genome sequencing errors.
Construction of a Δ(pyrE::kan) knockout allele and various pyrE cassettes.
First, the 726-bp fragment of pCR-PyrC was restricted with KpnI and XhoI and ligated into the respective sites of pBluescript II, yielding pBluescript(pyrC), into which the 756-bp fragment of pCR-DnaB restricted with XhoI and XbaI was ligated at the corresponding sites to obtain pBluescript(pyrC-DnaB). A Kmr cassette was then digested from vector pUC4K as a SalI fragment and inserted at the compatible XhoI site of pBluescript(pyrC-DnaB). A plasmid containing the Kmr cassette in the same orientation as the pyrC, pyrE, and dnaB genes was selected and designated pΔPyrE::Km, from which a DNA fragment containing pyrC, pyrE::kan, and dnaB was obtained after digestion with KpnI and XbaI and transferred into the corresponding sites of pRK415 (Table 1) to obtain plasmid pRK-ΔPyrE::Km.
Aside from the pyrE cassette, which contains 60 bp of the 5′ upstream sequence of the pyrE start codon and hence encompasses its own expression region as described above, two additional variants, pyrE-SD and pyrE-Nde, were also constructed. pyrE-SD contained only 20 bp of the 5′ upstream sequence of the pyrE start codon, including a putative ribosome binding sequence, which was amplified by PCR using primers PYR2FSD (5′-GCA TCG ATA CAG CAT GAG GAC AAG CAC-3′) and PYR2RBAM (5′-GCG GAT CCC CAC AGG CGT ATC CCC GGG G-3′) (the underlining indicates newly introduced ClaI and BamHI sites, respectively). pyrE-Nde contained only the coding sequence of pyrE starting at the first ATG, which was amplified by PCR using primers PYR2FNDE (5′-GTC ATA TGA TCC CCT CCT CCT ATC-3′; the underlining indicates a newly introduced NdeI site) and PYR2RBAM. pyrE-SD and pyrE-Nde were both cloned into the pCR-Blunt II TOPO cloning vector, as shown in Table 1.
Construction of helC::pyrE and ΔhelC knockout alleles.
A pyrE-marked knockout allele of R. capsulatus helC was constructed by inserting the 790-bp pyrE cassette, retrieved from pPyrE02 by BamHI digestion, into the unique BamHI site in a 850-bp helC gene fragment of the pRK415 derivative pCS1530 (30) (Table 1). This yielded two marked knockout alleles of helC (helC::pyrE in pCS1650#3 and pCS1650#4), in which pyrE was inserted in different orientations (Table 1). The unmarked allele of helC was constructed by self-ligation of pCS1530 after its unique BamHI site was blunted with T4 DNA polymerase, yielding pCS1652, which contained a frameshift mutation in helC, as verified by DNA sequencing. Finally, the suicide plasmids pZJDhelABCDX and pZJDΔhelC were obtained by transferring the SacI and XbaI fragments of p2helABCDX (Table 1), containing the entire R. capsulatus helABCDX genes, and pCS1652, respectively, into the corresponding sites of pZJD29a.
Biochemical techniques.
Cytoplasmic membrane vesicles or chromatophores were prepared in 20 mM morpholinepropanesulfonic acid (MOPS)-KOH buffer (pH 7.0) containing 1 mM KCl, 5 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride using a French pressure cell as described previously (15). Protein concentrations were determined by the method of Bradford (6) using the Bio-Safe Coomassie blue stain from Bio-Rad Laboratories (Hercules, CA) or a BCA kit from Pierce Biotechnology Inc. (Rockford, IL) Membrane proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (16.5% T, 6% C) as described by Schägger and von Jagow (32), and the c-type cytochromes were revealed by their endogenous peroxidase activities using tetramethylbenzidine and H2O2 (41). The cytochrome c oxidase activity of the colonies was detected colorimetrically using the Nadi stain, as described by Keilin (16).
For determination of the OPRTase activity, cytoplasmic fractions prepared from R. capsulatus cells grown at 35°C to the late exponential phase in 5 ml of MedA were used. Cells were harvested in microtubes by centrifugation at 14,000 rpm for 15 min, washed once in 50 mM MOPS-KOH buffer (pH 7.0) containing 100 mM KCl and 1.0 mM phenylmethylsulfonyl fluoride, resuspended in 1 ml of the same buffer, and disrupted by ultrasonication with a Sonifier (Misonix, New York) in the 1-s pulse mode at 15% power output for a total of 1 min on ice. The treated suspensions were then ultracentrifuged in a 75Ti rotor at 184,000 × g for 60 min, and the cleared supernatant was used for enzyme assays. OPRTase activity was measured as described by Beckwith et al. (2) with a reaction mixture containing 20 mM Tris-HCl (pH 8.0), 2 mM MgCl2, 30 mM sodium orotidylic acid, 100 μM 5-phosphoribosylpyrophosphate, and 0.6 U/ml yeast OMPdecase (Sigma, St. Louis, MO). The reaction was initiated by adding cytoplasmic fractions prepared from appropriate R. capsulatus strains, and changes in absorbance at 295 nm were monitored spectrophotometrically at 25°C. The OPRTase activity was determined using the initial reaction rate and an extinction coefficient of 3.67 μM−1 cm−1. One unit of enzyme was defined as the amount that removed 1 μmol of orotate per h, and the specific activity was expressed in units of enzyme per mg of cytoplasmic proteins.
RESULTS
De novo pyrimidine biosynthesis pathway of R. capsulatus and pyrE.
Examination of the R. capsulatus genome (http://ergo.integratedgenomics.com) revealed that nine open reading frames have been annotated to encode the six essential enzymes of the de novo pyrimidine nucleotide biosynthesis pathway. These genes are not contiguous on the chromosome with the exception of pyrC (RRC03624), which encodes a second copy of dihydroorotic acid synthase, and pyrE (RRC03625), which encodes OPRTase (Fig. 2). Inactivation of pyrE or pyrF (RRC02932), which encodes orotic acid monophosphate decarboxylase and catalyzes the last step of the de novo pyrimidine biosynthesis pathway, is known to result in 5FOAr and uracil auxotrophy (Ura−) in some organisms (4, 5, 20, 31, 39) (Fig. 1). Whether this is also the case in R. capsulatus was tested by determining the inhibitory effect of 5FOA on aerobic growth of wild-type strain MT1131 in the dark on MedA plates containing 0, 0.1, 0.2, 0.3, 0.5, and 1.0 mg/ml 5FOA. In the presence of 0.2 mg/ml 5FOA or a higher concentration, growth of wild-type R. capsulatus was significantly inhibited for 3 to 4 days, after which 5FOAr colonies appeared, especially in the presence of 0.1 mg/ml of uracil and 0.1 mg/ml of Casamino Acids, at a frequency of ∼10−7. In any event, the sensitivity to 5FOA suggested that R. capsulatus contains a de novo pyrimidine biosynthesis pathway similar to that of E. coli (24, 25).
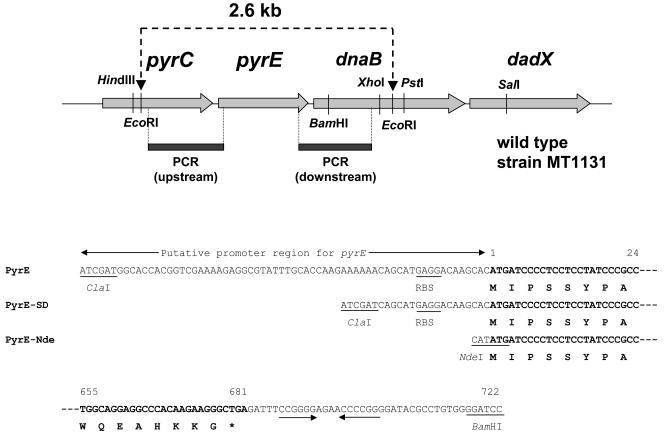
FIG. 2.
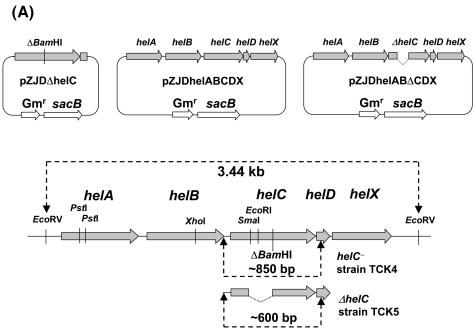
R. capsulatus pyrE locus and pyrE gene cassettes. (Top panel) Physical map of the pyrE locus of the wild-type R. capsulatus chromosome. pyrC (RRC03624) encoding a second copy of dihydroorotic acid synthase and dnaB (RRC03625) encoding a probable replicative DNA helicase plus dadX (RRC03627) encoding alanine racemase are present upstream and downstream of pyrE, respectively. Two flanking DNA segments encompassing pyrC and dnaB, as indicated by solid bars, were amplified by PCR and used to construct the knockout plasmid pRK-ΔPyrE::Km. Restriction enzyme recognition sites for BamHI, EcoRI, HindIII, PstI, SalI, and XhoI are indicated. Digestion of the wild-type chromosomal DNA with EcoRI generated a 2.6-kb fragment containing the 3′ half of pyrC, all of pyrE, and the 5′ half of dnaB, as indicated. (Bottom panel) Molecular structure of R. capsulatus pyrE gene cassettes. The 5′- and 3′-terminal DNA sequences of the three pyrE gene cassettes are shown. The coding region is indicated by boldface type, and the putative ribosome binding sequences (RBS) are underlined. The ClaI, NdeI, and BamHI restriction enzyme recognition sites are also indicated. The inverted arrows indicate a hairpin structure found immediately after the translation stop codon of pyrE, which might form a stem-loop structure with a predicted ΔGo of −14.9 kcal/mol.
Δ(pyrE::kan) knockout mutants of R. capsulatus are Ura− and 5FOAr.
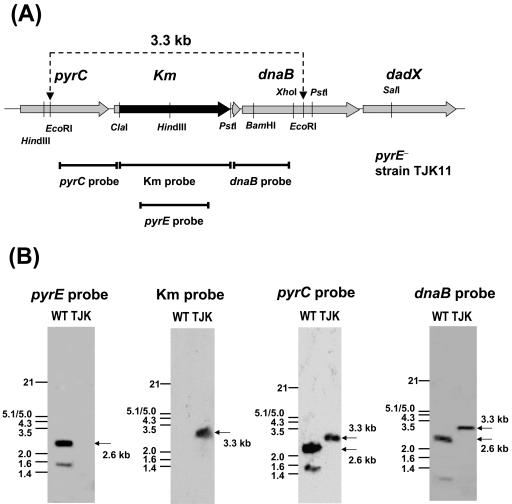
In order to examine whether an R. capsulatus pyrE knockout mutant is indeed Ura− and 5FOAr, two DNA fragments flanking pyrE located 103 bp downstream from pyrC and 250 bp upstream of dnaB (RRC03625), which has been annotated as the replicative DNA helicase gene (Fig. 2), were amplified by PCR using MT1131 chromosomal DNA as a template. The PCR products were assembled with a kanamycin gene cassette to construct a pyrE knockout plasmid, pRK-ΔpyrE::Km, carrying a Δ(pyrE::kan) allele, which was then integrated into the chromosome of MT1131 via GTA crosses with selection for Kmr colonies on MedA plates supplemented with 0.1 mg/ml uracil. One Kmr and Ura− colony, TJK11, was retained (Table 1) and examined by Southern hybridization analysis (Fig. 3). The data indicated that the 2.6-kb EcoRI DNA fragment carrying the wild-type pyrE allele in MT1131 was replaced in TJK11 with a 3.3-kb DNA fragment corresponding to the expected Δ(pyrE::kan) allele, whereas the upstream and downstream regions containing pyrC and dnaB, respectively, remained intact.
FIG. 3.
Southern hybridization analyses of TJK11 [Δ(pyrE::kan)]. (A) Physical map of the pyrE locus in TJK11. A 2.6-kb wild-type pyrE allele of R. capsulatus MT1131 was replaced in TJK11 by a Δ(pyrE::kan) allele via GTA crosses. Hybridization probes used for the Southern hybridization analyses are indicated by solid bars. In TJK11, EcoRI digestion generated a 3.3-kb DNA fragment containing the 3′ half of pyrC, the entire Kmr gene cassette, and the 5′ half of dnaB, which was 0.7 kb longer than its counterpart in the wild-type strain (see Fig. 2). (B) Southern hybridization analyses of R. capsulatus wild-type strain MT1131 (WT) and PyrE null mutant TJK11 (TJK). Genomic DNA was isolated from MT1131 and TJK11, digested with EcoRI, and analyzed by using DIG-labeled pyrE, kanamycin (Km), pyrC, and dnaB gene probes. The molecular sizes of the positive blots are indicated by arrows.
As expected, TJK11 was 5FOAr (up to a concentration of 1 mg/ml) and exhibited virtually no OPRTase activity. In addition to uracil, uridine (0.5 mg/ml) could also support the growth of TJK11, whereas 5′-UMP, adenosine, guanosine, cytosine, thymine, and 5′-GMP could not support growth (data not shown). In liquid MedA, the growth of TJK11 was significantly poorer than it was on agar plates supplemented with uracil alone. Addition of Casamino Acids (1.0 mg/ml) or yeast extract (1.0 mg/ml) improved the growth, which still depended on supplementation with uracil. The additional requirement of an R. capsulatus pyrE mutant for Casamino Acids or yeast extract is intriguing, but this issue was outside the scope of this work and was not pursued further.
Complementation of TJK11 [Δ(pyrE::kan)] to Ura+ and 5FOAs phenotypes by pyrE.
Three different constructs, designated PyrE, PyrE-SD, and PyrE-Nde, as described in Materials and Methods, were used to complement the Ura− and 5FOAr mutant TJK11. One of these, PyrE-Nde, contained only the coding sequence of pyrE from its putative initiating codon ATG at position 1, while PyrE-SD and PyrE carried an additional 18 bp (encompassing a putative ribosome binding site) and 60 bp (including a putative promoter sequence) 5′ upstream of this ATG, respectively (Fig. 2). Only plasmids containing PyrE but not PyrE-SD and PyrE-Nde were able to render TJK11 Ura+ and 5FOAs (Table 2), and only the strains harboring PyrE exhibited levels of OPRTase activity (0.26 to 0.31 μmol/h/mg of proteins) similar to those found in MT1131 (0.39 μmol/h/mg of proteins). Moreover, TJK11 was complemented with both plasmid pRK-PyrE-PB and plasmid pRK-PyrE-PH, which harbored pyrE in opposite orientations. Therefore, the data indicated that the Ura− and 5FOAr phenotypes of TJK11 were due to inactivation of pyrE and that the DNA fragment carrying the 60-bp 5′ upstream sequence was necessary and sufficient to mediate pyrE expression.
TABLE 2.
Phenotypes of various R. capsulatus pyrE and helC mutant strains and their derivatives
| Straina | Plasmida | MedAb
|
MedA + uracilb
|
MedA + uracil + 5FOA (Res)b | Nadid | ||
|---|---|---|---|---|---|---|---|
| Resc | Psc | Res | Ps | ||||
| MT1131 (wild type) | None | + | + | + | + | − | + |
| TJK11 [Δ(pyrE::kan)] | None | − | − | + | + | + | + |
| pRK-PyrE-PB | + | + | + | + | − | NDf | |
| pRK-PyrE-PH | + | + | + | + | − | ND | |
| pRK-PyrESD | − | − | + | + | + | ND | |
| pRK-PyrENde | − | − | + | + | + | ND | |
| pCS1650#3 | + | + | + | + | − | ND | |
| pCS1650#4 | + | + | + | + | − | ND | |
| TCK1 or TCK2 [Δ(pyrE::kan) helC::pyrE]e | None | + | − | + | − | − | − |
| pCS1650#3 | + | − | + | − | ND | − | |
| pCS1650#4 | + | − | + | − | ND | − | |
| pCS1530 | + | + | + | + | ND | + | |
| TCK4 [Δ(pyrE::kan) helC]e | None | − | − | + | − | + | − |
| pCS1650#3 | + | − | + | − | − | − | |
| pCS1650#4 | + | − | + | − | − | − | |
| pCS1530 | − | − | + | + | ND | + | |
| TCK5 [Δ(pyrE::kan) ΔhelC]e | None | − | − | + | − | + | − |
The strains and the plasmids that they carry are described in Table 1.
MedA was the minimal growth medium. MedA + uracil contained 0.1 mg/ml uracil. MedA + uracil + 5FOA contained 0.1 mg/ml uracil and 0.1 mg/ml 5FOA.
Res and Ps indicate respiratory and photosynthetic growth conditions, respectively. +, strain was able to grow; −, strain was not able to grow.
The Nadi reaction was examined for the strains grown on MedA containing uracil under Res conditions. +, Nadi reaction; −, no Nadi reaction.
Strains TCK1 and TCK2 are phenotypically identical, although they differ genotypically. The same is true for TCK4 and TCK5 (for details see Materials and Methods).
ND, not determined.
Isolation of helC::pyrE chromosomal knockout derivatives of TJK11.
We chose to isolate a chromosomal knockout allele of helC of R. capsulatus in order to assess the usefulness of pyrE as an interposon. The helC gene is located in the orf124-helABCDX cluster encoding several components required for cytochrome c biogenesis (1). These genes are tightly packed together, and recent work has indicated that their transcriptional organization is more complex than a simple operon structure (1, 30). The helABCD cluster encodes an ATP-binding cassette containing a transporter complex believed to export heme across the cytoplasmic membrane (12-14, 33), whereas helX encodes a periplasmic, membrane-anchored thioredoxin-like protein involved in the apocytochrome c thioreduction pathway (18, 23). The pyrE interposon was inserted at a unique BamHI site of helC in both orientations (pCS1650#3 and pCS1650#4) (Table 1) to obtain two helC::pyrE insertion alleles, which were then integrated into the chromosome of TJK11 via GTA crosses with selection for Ura+ colonies on kanamycin-containing MedA plates in the absence of uracil. Two such colonies, TCK1 with pCS1650#3 and TCK2 with pCS1650#4, were retained for further analyses.
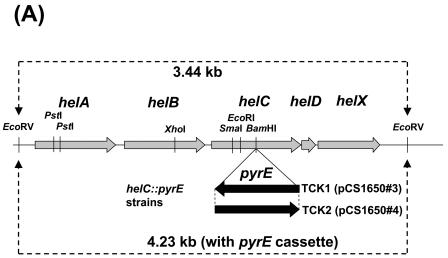
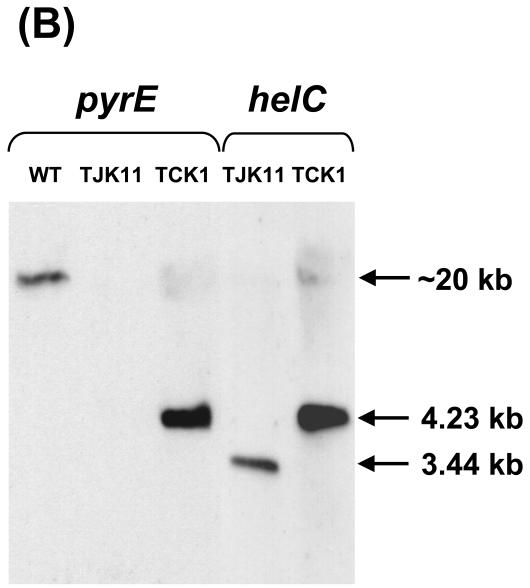
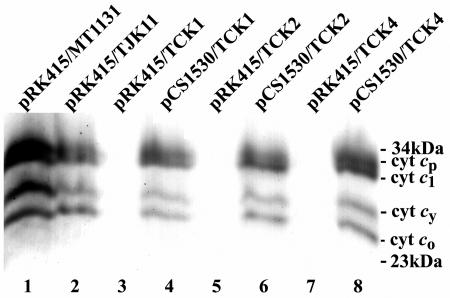
Both TCK1 and TCK2 were 5FOAs and grew slowly on enriched or appropriately supplemented minimal media under respiratory conditions; however, they were unable to grow under Ps conditions. Furthermore, they were Nadi− and excreted intermediates of heme biosynthesis into the growth media, unless they carried a wild-type copy of helC harbored by pCS1530 (Table 2). These phenotypes were consistent with both TCK1 and TCK2 being HelC− and unable to produce c-type cytochromes. Indeed, Southern hybridization analyses confirmed that in these mutants, a 4.23-kb EcoRV fragment carrying helC::pyrE, which was 0.79 kb longer than the wild-type helC (3.44 kb) of parental strain TJK11, replaced the wild-type chromosomal helC (Fig. 4). Similarly, TMBZ-SDS-PAGE data revealed that TCK1 and TCK2 derivatives carrying the cloning vector pRK415 as a control were unable to produce various c-type cytochromes (cytochromes cp, c1, co, and cy in the chromatophore membranes), but the derivatives that carried pCS1530 were able to produce these cytochromes (Fig. 5). Therefore, a pyrE cassette can be used as an interposon to knock out a desired gene, provided that the parental strain is auxotrophic for uracil, so that Ura+ selection could be applied.
FIG. 4.
Southern hybridization analyses of the helC::pyrE mutants TCK1 and TCK2. (A) Physical map of the hel gene cluster of the helC::pyrE constructs. Wild-type chromosomal DNA digested with EcoRV generated a 3.44-kb DNA fragment that contained the entire helABCDX gene cluster. In helC::pyrE mutants TCK1 and TCK2, the pyrE gene cassette was inserted at a unique BamHI site of helC in opposite orientations, as indicated by the solid arrows. In these mutant strains, EcoRV digestion generated a 4.23-kb DNA fragment containing the entire helABCDX gene cluster and the inserted pyrE gene cassette. The restriction enzyme recognition sites for BamHI, EcoRI, EcoRV, PstI, SmaI, and XhoI are indicated. (B) Southern hybridization analyses of R. capsulatus wild type (WT), the PyrE− mutant TJK11, and the HelC null mutant TCK1. Genomic DNA isolated from MT1131, TJK11, and TCK1 were digested with EcoRV, and the digests were analyzed by using DIG-labeled pyrE and helC gene probes. The molecular sizes of the positive blots are indicated. The high-molecular-weight band (∼20 kb) in the wild-type lane corresponds to the chromosomal pyrE locus.
FIG. 5.
TMBZ-SDS-PAGE analyses of various R. capsulatus strains. Chromatophore membranes prepared from R. capsulatus wild-type strain MT1131, the pyrE mutant TJK11, and the helC mutants TCK1, TCK2, and TCK4 (harboring either pRK415 or pCS1530 carrying an expressed helC gene) grown in MPYE under respiratory conditions were subjected to TMBZ-SDS-PAGE analyses as described in Materials and Methods. Approximately 75 μg of proteins was loaded in each lane, and the positions of the molecular mass markers (in kDa) are indicated on the right. The four c-type cytochromes detected in this gel system correspond to cytochrome c1 (cyt c1) of the cytochrome bc1 complex, the membrane-anchored cytochrome cy (cyt cy), and cytochromes cp (cyt cp) and co (cyt co) of the cbb3-type cytochrome c oxidase.
Isolation of a chromosomal unmarked ΔhelC knockout mutant.
The availability of a strain like TCK1 (helC::pyrE pyrE::kan), which is HelC− and 5FOAs, allowed us to probe whether homologous recombination between an unmarked ΔhelC allele and helC::pyrE could yield an unmarked HelC− chromosomal mutant simply by selection for 5FOAr. For this purpose, strong selection for the loss of pyrE was applied by combining 5FOAr with sucrose lethality conferred by the B. subtilis sacB gene. An 850-bp helC gene containing a frameshift mutation at its unique BamHI site (Table 1) was cloned into the suicide plasmid pZJD29a harboring sacB expressed via an R. capsulatus pucAB promoter. The sacB gene is known to be toxic to this species in the presence of 5% (wt/vol) sucrose in the growth medium (7, 21, 37, 38). As a control, pZJDhelABCDX that contained a 3.46-kb DNA fragment encompassing the entire wild-type helABCDX cluster was also constructed (Fig. 6A and Table 1). These nonreplicative plasmids were conjugated into the R. capsulatus HelC− mutant TCK1, as described in Materials and Methods, and Rifr and Gmr colonies were obtained on appropriate MPYE plates under Res growth conditions at a frequency of about 10−3. Transconjugants TRK1 and TRK2 with pZJDhelABCD and pZJDΔhelC, respectively, corresponding to integrants of the nonreplicative plasmids into the chromosome, were retained for further analysis.
FIG. 6.
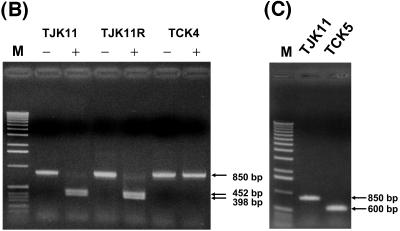
Genotype analyses of strains TJK11, TJK11R, TCK4, and TCK5. (A) Physical maps of suicide plasmids pZJDΔhelC, pZJDhelABCDX, and pZJDhelABΔCDX and of the hel gene cluster loci of TCK4 and TCK5. pZJDΔhelC contained an 850-bp fragment encompassing the helC gene, in which the unique BamHI site was deleted as described in Materials and Methods. The pZJDhelABCDX contained a 3.4-kb DNA fragment encompassing the wild-type hel gene cluster, and pZJDhelABΔCDX was the same as pZJDhelABCDX except that the helC gene contained a 250-bp in-frame deletion. These plasmids harbored the B. subtilis sacB gene that conferred lethality in the presence of 5% (wt/vol) sucrose in the growth medium. (B) The chromosomal helC region was PCR amplified by using the helC2 and helC3 primers and the chromosomal DNA isolated from the individual strains as a template. The PCR fragments were either digested with BamHI (+) or not treated (−) and were separated by agarose gel electrophoresis. The molecular sizes of the DNA fragments are indicated by arrows on the right. (C) The chromosomal helC region of TJK11 and TCK5 was amplified by PCR as described above for panel B, and the PCR products were analyzed by agarose gel electrophoresis without restriction enzyme treatment. For a description of the strains see Table 1. Lane M, DNA molecular weight marker.
TRK1 was Ps+ Nadi+ and did not excrete porphyrin, indicating that it became HelC+, whereas TRK2 was Ps− Nadi− and excreted porphyrin derivatives, remaining HelC− like its parent, TCK1 (data not shown). TRK1 and TRK2 were subcultured under Res growth conditions in MPYE in the absence of gentamicin and plated on enriched MPYE plates containing either 5% (wt/vol) sucrose or 5% (wt/vol) sucrose and 0.1 mg/ml 5FOA. In the case of TRK1, approximately 1,200 sucrose-tolerant and 350 sucrose-tolerant and 5FOAr colonies were obtained on the respective plates. Of the 46 sucrose-tolerant colonies tested, 39 were Gms and 5FOAr (∼85%), and 7 were Gmr and 5FOAs, and of the 52 sucrose-tolerant and 5FOAr colonies tested, 50 were Gms and 5FOAr (∼96%) and 2 were Gmr and 5FOAs. Therefore, the data indicated that single selection and double selection yielded similar results. One of the Gms and 5FOAr colonies from the double-selection plates, which exhibited wild-type phenotypes (HelC+, Ps+ Nadi+), was designated TJK11R and retained for further analysis. In the case of TRK2, 122 sucrose-tolerant colonies and only two sucrose-tolerant and 5FOAr colonies appeared on the respective plates. Of the 52 sucrose-tolerant colonies tested, only 1 was Gms and 5FOAr. Both of the colonies that appeared on sucrose- and 5FOA-containing plates were Gms and 5FOAr, suggesting that double selection was more efficient for identifying the clones that had evicted the integrated plasmid (i.e., Gms) and inactivated pyrE (i.e., 5FOAr). One of the three Gms and 5FOAr colonies, which were also Ps− Nadi− and excreted porphyrin derivatives (Table 2), was designated TCK4 and analyzed further.
The helC locus on the chromosome of TJK11R (HelC+) and TCK4 (HelC−) was compared to that of TJK11 (HelC+) by using PCR with the helC2 and helC3 primers (see Materials and Methods) to establish the molecular nature of the events leading to the sucrose-tolerant, Gms, and 5FOAr phenotypes. Although an approximately 850-bp DNA fragment encompassing helC was amplified by using chromosomal DNA of all three strains, only the fragments from TJK11 and TJK11R contained a unique BamHI site, like the wild-type helC (Fig. 6B). The absence of this enzyme recognition site was further confirmed by DNA sequencing of the PCR product, which was cloned into the pCR2.1-TOPO vector (pTOPO-helCΔBamHI) (data not shown). Strain TCK4 was also examined by Southern hybridization, which revealed that the 3.44-kb EcoRV DNA fragment containing helABCDX was devoid of pyrE (data not shown). Furthermore, TMBZ-SDS-PAGE data indicated that TCK4 was unable to produce c-type cytochromes, like TCK1 and TCK2, unless it was complemented with pCS1530, which carried a wild-type allele of helC (Fig. 5). Taken together, the overall data demonstrated that the helC::pyrE allele present in TCK1 was replaced in TCK4 with an unmarked ΔhelC allele devoid of the BamHI site. Therefore, the combined sacB-5FOA selection was a very efficient way to integrate unmarked alleles into the R. capsulatus chromosome.
DISCUSSION
De novo pyrimidine biosynthesis pathway of R. capsulatus and pyrE.
The R. capsulatus genome data indicated that the genes encoding the enzymes of the de novo pyrimidine biosynthetic pathway are scattered throughout the chromosome, except that pyrC and pyrE are adjacent to each other. Cloning and subsequent inactivation of RRC03625, which is annotated as pyrE, using a kan interposon (26, 40) yielded R. capsulatus strain TJK11, which is Ura−, 5FOAr, and devoid of OPRTase activity. As expected, TJK11 was fully complemented by an active copy of pyrE in trans, which conferred both the Ura+ and 5FOAs phenotypes. Therefore, RRC03625 is the only active copy of pyrE that is required for de novo pyrimidine biosynthesis in this species, and this metabolic pathway appears to be similar to that in E. coli (24, 25) and S. cerevisiae (19).
pyrE as a novel interposon in R. capsulatus.
The experiments reported here indicated that R. capsulatus pyrE is expressed autonomously and could itself be used as an interposon to inactivate any gene of interest, provided that the parental strain is Ura−. As interposon mutagenesis is a common tool used for construction of chromosomal knockout mutants, pyrE is a welcome addition to the antibiotic resistance gene cassettes that are available for R. capsulatus (17). Some of the antibiotic resistance-based interposons are available as transcriptionally polar or nonpolar forms (11, 28) and can be preferentially used depending on the expected outcomes of the gene knockout experiments. The pyrE cassette isolated here has a hairpin structure at its 3′ end, which might act as a rho-independent transcription termination signal (10, 42). Although we isolated helC::pyrE insertions in two opposite orientations, due to the complex operon structure of the helABCDX cluster with its multiple internal promoters (30), we could not deduce rigorously whether pyrE is a polar or nonpolar interposon with respect to transcription of the genes located downstream from its insertion site. Additional experiments and construction of variants of the pyrE interposon, which might be more desirable for various purposes, are required to settle this issue.
sacB-5FOA-pyrE as powerful bidirectional selection markers.
In knockout experiments with conventional antibiotic resistance-conferring interposons, such as kan and spe, removal of the inserted antibiotic marker from the chromosome is often a laborious process, if it is possible at all. A clear advantage of an interposon like pyrE is the ease with which one can excise it from the chromosome by selecting for 5FOAr derivatives. However, it should be kept in mind that this selection does not yield exclusively pyrE excisions via homologous recombination, as mutations inactivating pyrE also confer the 5FOAr phenotype. Although in the case of S. cerevisiae this does not appear to be a major drawback (5), in R. capsulatus, in which the frequency of the 5FOAr colonies is apparently high, it becomes a potential restricting factor when the loss of pyrE is selected for. However, combining 5FOAr and sucrose toxicity mediated by sacB readily overcomes this hurdle and yields chromosomal allele replacements even when the incoming unmarked allele is carried by a homologous flanking DNA fragment as short as ∼400 bp, as in the case of the ΔhelC construct used here. Conceivably, when larger DNA fragments with longer flanking homologous regions are used, even single 5FOAr might be sufficient to obtain the desired outcome without the need for sacB-mediated selection. Indeed, we observed that when a derivative of the suicide plasmid pZJD29a harboring the entire helABCDX gene cluster with a 250-bp in-frame deletion in helC (pZJDhelABΔCDX) was used, the desired mutants were obtained at comparable frequencies (approximately 500 colonies per plate) by selecting for either only 5FOAr colonies or 5FOAr and sucrose-tolerant colonies.
Another advantage of delivering a homologous DNA fragment via a nonreplicating plasmid instead of using GTA is the possibility that the double-crossover homologous recombination events can be divided into two parts. A first homologous recombination yields a chromosomal integrant selected using a marker carried by the nonreplicating plasmid. The chromosomal duplication generated in this way is then resolved via a second homologous recombination that can be selected for sucrose tolerance via the loss of sacB. Moreover, when this second step is further reinforced for the loss of pyrE by 5FOAr selection, then homologous replacement of a pyrE-marked allele of a chromosomal gene of interest by its unmarked allele becomes easy, as documented here by the construction of ΔhelC.
Finally, once the pyrE-marked allele of a chromosomal gene of interest is replaced by an unmarked allele, then the strain can be subjected to additional similar rounds of gene knockout experiments with the pyrE interposon as a selectable marker. The procedure described here could be used as many times as it is needed for introduction of various type of mutations (frameshifts, in-frame or out-of-frame deletions, site-specific mutations or tag insertions, or other mutations) into a chromosomal gene of interest. The sacB-5FOA-pyrE bidirectional selection system is, therefore, applicable to a variety of experiments, including structure-function studies of large multisubunit enzymes and elucidation of complex multicomponent pathways, and thus it is a useful addition to the well-developed arsenal of R. capsulatus genetic tools.
Acknowledgments
This work was supported by NIH grant GM30736 to T. Ohnishi (University of Pennsylvania) and by DOE grant ER9120053 to F.D.
We thank C. E. Bauer (Indiana University) for providing the suicide plasmid pZJD29a, and T.Y. thanks T. Ohnishi for her kind support.
REFERENCES
- 1.Beckman, D. L., D. R. Trawick, and R. G. Kranz. 1992. Bacterial cytochromes c biogenesis. Genes Dev. 6:268-283. [DOI] [PubMed] [Google Scholar]
- 2.Beckwith, J. R., A. B. Pardee, R. Austrian, and F. Jacob. 1962. Coordination of the synthesis of the enzymes in the pyrimidine pathway of E. coli. J. Mol. Biol. 5:618-634. [DOI] [PubMed] [Google Scholar]
- 3.Benning, C., and C. R. Somerville. 1992. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J. Bacteriol. 174:2352-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Dedonder, R. 1966. Levansucrase from Bacillus subtilis. Methods Enzymol. 8:500-505. [Google Scholar]
- 8.Ditta, G., T. Schmidhauser, E. Yakobson, P. Lu, X. W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 9.Elsen, S., L. R. Swem, D. L. Swem, and C. E. Bauer. 2004. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68:263-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farnham, P. J., and T. Platt. 1981. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 9:563-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 12.Goldman, B. S., D. L. Beck, E. M. Monika, and R. G. Kranz. 1998. Transmembrane heme delivery systems. Proc. Natl. Acad. Sci. USA 95:5003-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman, B. S., D. L. Beckman, A. Bali, E. M. Monika, K. K. Gabbert, and R. G. Kranz. 1997. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J. Mol. Biol. 268:724-738. [DOI] [PubMed] [Google Scholar]
- 14.Goldman, B. S., and R. G. Kranz. 2001. ABC transporters associated with cytochrome c biogenesis. Res. Microbiol. 152:323-329. [DOI] [PubMed] [Google Scholar]
- 15.Gray, K. A., P. L. Dutton, and F. Daldal. 1994. Requirement of histidine 217 for ubiquinone reductase activity (Qi site) in the cytochrome bc1 complex. Biochemistry 33:723-733. [DOI] [PubMed] [Google Scholar]
- 16.Keilin, D. 1966. The history of cell respiration and cytochrome. Cambridge University Press, Cambridge, United Kingdom.
- 17.Koch, H. G., H. Myllykallio, and F. Daldal. 1998. Using genetics to explore cytochrome function and structure in Rhodobacter. Methods Enzymol. 297:81-94. [Google Scholar]
- 18.Kranz, R., R. Lill, B. Goldman, G. Bonnard, and S. Merchant. 1998. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol. Microbiol. 29:383-396. [DOI] [PubMed] [Google Scholar]
- 19.Lacroute, F. 1993. The yeast pyrimidine pathway. Paths Pyrimidines 1:1-4. [Google Scholar]
- 20.Lucas, S., L. Toffin, Y. Zivanovic, D. Charlier, H. Moussard, P. Forterre, D. Prieur, and G. Erauso. 2002. Construction of a shuttle vector for, and spheroplast transformation of, the hyperthermophilic archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 68:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda, S., and C. E. Bauer. 2004. Null mutation of HvrA compensates for loss of an essential relA/spoT-like gene in Rhodobacter capsulatus. J. Bacteriol. 186:235-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monika, E. M., B. S. Goldman, D. L. Beckman, and R. G. Kranz. 1997. A thioreduction pathway tethered to the membrane for periplasmic cytochromes c biogenesis: in vitro and in vivo studies. J. Mol. Biol. 271:679-692. [DOI] [PubMed] [Google Scholar]
- 24.Neuhard, J., and R. A. Kellen. 1987. Biosynthesis and conversion of pyrimidines, p. 580-599. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 25.Neuhard, J., and P. Nygaard. 1987. Purines and pyrimidines, p. 445-473. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 26.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 27.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145-146. [DOI] [PubMed] [Google Scholar]
- 28.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 30.Sanders, C., M. Deshmukh, D. Astor, R. G. Kranz, and F. Daldal. Overproduction of CcmG and CcmFHRc fully suppresses the c-type cytochrome biogenesis defect of Rhodobacter capsulatus CcmI-null mutants. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 31.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 33.Schulz, H., H. Hennecke, and L. Thony-Meyer. 1998. Prototype of a heme chaperone essential for cytochrome c maturation. Science 281:1197-1200. [DOI] [PubMed] [Google Scholar]
- 34.Scolnik, P. A., and B. L. Marrs. 1987. Genetic research with photosynthetic bacteria. Annu. Rev. Microbiol. 41:703-726. [DOI] [PubMed] [Google Scholar]
- 35.Scolnik, P. A., M. A. Walker, and B. L. Marrs. 1980. Biosynthesis of carotenoids derived from neurosporene in Rhodopseudomonas capsulata. J. Biol. Chem. 255:2427-2432. [PubMed] [Google Scholar]
- 36.Siström, W. R. 1960. A requirement for sodium in the growth of Rhodopseudomonas spheroides. J. Gen. Microbiol. 22:778-785. [DOI] [PubMed] [Google Scholar]
- 37.Steinmetz, M., D. Le Coq, H. B. Djemia, and P. Gay. 1983. Genetic analysis of sacB, the structural gene of a secreted enzyme, levansucrase of Bacillus subtilis Marburg. Mol. Gen. Genet. 191:138-144. [DOI] [PubMed] [Google Scholar]
- 38.Swem, L. R., B. J. Kraft, D. L. Swem, A. T. Setterdahl, S. Masuda, D. B. Knaff, J. M. Zaleski, and C. E. Bauer. 2003. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 22:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamakoshi, M., T. Yaoi, T. Oshima, and A. Yamagishi. 1999. An efficient gene replacement and deletion system for an extreme thermophile, Thermus thermophilus. FEMS Microbiol. Lett. 173:431-437. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, L. A., and R. E. Rose. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 16:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, K. S., and P. H. von Hippel. 1995. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl. Acad. Sci. USA 92:8793-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yen, H. C., N. T. Hu, and B. L. Marrs. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 131:157-168. [DOI] [PubMed] [Google Scholar]