Abstract
Prokaryotes in marine sediments taken from two neighboring semienclosed bays (the Yamada and Kamaishi bays) at the Sanriku coast in Japan were investigated by the culture-independent molecular phylogenetic approach coupled with chemical and activity analyses. These two bays were chosen in terms of their similar hydrogeological and chemical characteristics but different usage modes; the Yamada bay has been used for intensive shellfish aquaculture, while the Kamaishi bay has a commercial port and is not used for aquaculture. Substantial differences were found in the phylogenetic composition of 16S rRNA gene clone libraries constructed for the Yamada and Kamaishi sediments. In the Yamada library, phylotypes affiliated with δ-Proteobacteria were the most abundant, and those affiliated with γ-Proteobacteria were the second-most abundant. In contrast, the Kamaishi library was occupied by phylotypes affiliated with Planctomycetes, γ-Proteobacteria, δ-Proteobacteria, and Crenarchaeota. In the γ-Proteobacteria, many Yamada phylotypes were related to free-living and symbiotic sulfur oxidizers, whereas the Kamaishi phylotype was related to the genus Pseudomonas. These results allowed us to hypothesize that sulfate-reducing and sulfur-oxidizing bacteria have become abundant in the Yamada sediment. This hypothesis was supported by quantitative competitive PCR (qcPCR) with group-specific primers. The qcPCR also suggested that organisms closely related to Desulfotalea in the Desulfobulbaceae were the major sulfate-reducing bacteria in these sediments. In addition, potential sulfate reduction and sulfur oxidation rates in the sediment samples were determined, indicating that the sulfur cycle has become active in the Yamada sediment beneath the areas of intensive shellfish aquaculture.
Prokaryotes in sediments play crucial roles in the decomposition and mineralization of organic matter in the coastal marine environment (16). A number of studies have examined prokaryotes in marine sediments, in which molecular ecological approaches based on PCR amplification and phylogenetic analysis of 16S rRNA gene fragments have been applied to investigate their diversity and community structures (4, 7, 17, 31, 42). In addition, group-specific 16S rRNA probes have been used for quantitative evaluation of microbial populations in marine sediments by fluorescence in situ hybridization (FISH) (3, 24, 32). These studies have given a fundamental view of the prokaryotic community in marine sediment, suggesting that δ-Proteobacteria, γ-Proteobacteria, Bacteroidetes, and Planctomycetes are abundant and ecologically important. Information obtained in these studies has been combined with results of activity measurements (19, 20, 34), suggesting that sulfate reduction is the most important electron-accepting process for organic matter decomposition in coastal and continental shelf sediments.
Even though much information is available that gives a general view of phylogenetic makeup and activity of prokaryotes in marine sediment, relatively little is known about the formation and transition of prokaryotic communities in response to changes in environmental conditions (27, 30, 33). Rosselló-Mora et al. have examined the response of the sediment microbial community to cyanobacterial biomass input under laboratory simulation conditions (33). A field study on sediments of eutrophic bays suggested that the sediment microbial community changes in response to eutrophication (30). Considering that prokaryotic populations rapidly respond to environmental perturbations and are reliably studied by application of molecular ecological tools, changes in sediment prokaryotes will be useful indices for assessing the state of coastal environments.
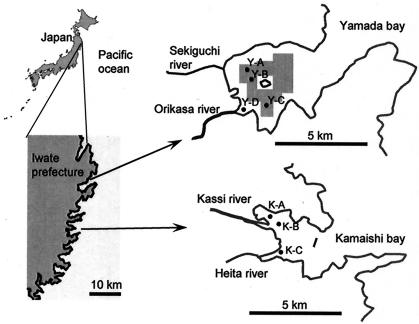
The Sanriku coast is situated in northeastern Japan (Fig. 1) and is famous for the complex Rias coastline. The Rias coastline is punctuated with numerous small, semienclosed bays, many of which have been used as fishing ports. In addition, some bays have also been utilized for intensive commercial shellfish aquaculture (mainly oysters and scallops) and seaweeds for over 30 years, which has become one of the important industries in the Sanriku region. Consequently, sustainable usage of the coastline has currently become an important issue for the Sanriku community, as is the case with other coastal communities.
FIG. 1.
Map of study sites.
This study was carried out to investigate possible influences of the aquaculture on sediment prokaryotes. Previously, Stenton-Dozey et al. investigated the macrobenthic community underneath mussel aquaculture in the Saldanha bay in South Africa, where sediment was affected by deposits of mussel feces and aquaculture-associated fouling organisms (40); unfortunately, they did not look at sediment prokaryotes. We considered that the Sanriku coast provides a good study site for such ecological investigation, since bays exist there which have similar hydrogeological and chemical characteristics but different usage modes. In this context, the present study chose the Yamada (intensive aquaculture zone) and Kamaishi (no-aquaculture zone) bays (Fig. 1). A culture-independent molecular phylogenetic approach was applied in combination with chemical and activity analyses.
MATERIALS AND METHODS
Sampling sites.
Coastal marine sediments were collected in the Yamada and Kamaishi bays (Fig. 1). The Yamada bay is situated approximately 20 km north of the Kamaishi bay and has been utilized for intensive shellfish aquaculture (oysters and scallops). In particular, oyster production in the Yamada bay is the largest among those in the Sanriku bays; approximately 5,000 tons of oysters have been produced annually. In Yamada aquaculture, no supplementation of nutrients is practiced.
Although the Yamada bay is larger in area (30 km2) than the Kamaishi bay (10 km2), they have similar enclosure indices (EI; 1.43 for the Yamada bay and 1.28 for the Kamaishi bay). An EI is estimated according to the equation 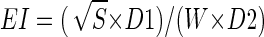 , where S is the bay area (in square kilometers), W is the width at the mouth of a bay (in kilometers), D1 is the depth at the center of a bay (in meters), and D2 is the depth at the mouth of a bay (in meters) (http://www.emecs.or.jp/closedsea-jp/closedsea-jp.htm). Since this index is correlated with the water exchange capacity, the index has been considered important for evaluating influences of biological activities in seawater to surrounding regions, including sediments. Water temperature in both bays ranged from 3°C (February and March) to 22°C (August and September). The degree of cleanliness of the seawater in these two bays is reported to be similar, as judged by chemical oxygen demand (COD) (<3 mg liter−1), total nitrogen (<0.4 mg liter−1) and total phosphorus (<0.04 mg liter−1) (http://www.pref.iwate.jp/∼hp0318/index.html). There has been no history of eutrophication in these bays.
, where S is the bay area (in square kilometers), W is the width at the mouth of a bay (in kilometers), D1 is the depth at the center of a bay (in meters), and D2 is the depth at the mouth of a bay (in meters) (http://www.emecs.or.jp/closedsea-jp/closedsea-jp.htm). Since this index is correlated with the water exchange capacity, the index has been considered important for evaluating influences of biological activities in seawater to surrounding regions, including sediments. Water temperature in both bays ranged from 3°C (February and March) to 22°C (August and September). The degree of cleanliness of the seawater in these two bays is reported to be similar, as judged by chemical oxygen demand (COD) (<3 mg liter−1), total nitrogen (<0.4 mg liter−1) and total phosphorus (<0.04 mg liter−1) (http://www.pref.iwate.jp/∼hp0318/index.html). There has been no history of eutrophication in these bays.
Figure 1 presents sampling points in these bays and the aquaculture area in the Yamada bay. As shown in this figure, sampling points Y-A, Y-B, and Y-C in the Yamada bay were situated beneath shellfish beds, while Y-D was situated at the mouth of the Orikasa river and out of the aquaculture zone. The water depths of the sampling sites were between 5 and 20 m (Table 1). Sediments in these two bays are composed of fine sand and silt.
TABLE 1.
Characteristics of marine sediment samples taken from the Yamada and Kamaishi baysa
| Sediment | Depth (m) | Temp (°C) | ORP (mV) | COD (mg g dry soil−1) | Sulfate (mg g dry soil−1) | TS (mg g dry soil−1) | TDC (108 cells g dry soil−1) |
|---|---|---|---|---|---|---|---|
| K-A | 8 | 18 ± 1.1 | −248 ± 47 | 5.9 ± 0.7 | 0.21 ± 0.07 | 0.09 ± 0.01 | 3.1 ± 1.9 |
| K-B | 15 | 18 ± 1.1 | −354 ± 16 | 5.8 ± 2.3 | 0.27 ± 0.07 | 0.81 ± 0.18 | 2.0 ± 0.9 |
| K-C | 12 | 18 ± 0.6 | −295 ± 28 | 3.7 ± 0.1 | 0.16 ± 0.09 | 0.73 ± 0.14 | 2.4 ± 1.7 |
| K-meanb | 11 ± 3.6 | 18 ± 1.0 | −299 ± 54 | 5.2 ± 1.6 | 0.21 ± 0.08 | 0.48 ± 0.38 | 2.5 ± 1.4 |
| Y-A | 10 | 17 ± 0.6 | −212 ± 7 | 14 ± 2.5 | 0.06 ± 0.03 | 0.40 ± 0.04 | 4.2 ± 1.2 |
| Y-B | 7 | 20 ± 0.4 | −293 ± 14 | 12 ± 0.4 | 0.10 ± 0.03 | 0.87 ± 0.39 | 4.5 ± 0.7 |
| Y-C | 10 | 20 ± 1.5 | −290 ± 46 | 6.9 ± 3.7 | 0.11 ± 0.02 | 0.22 ± 0.09 | 2.8 ± 1.8 |
| Y-meanc | 9 ± 1.7 | 19 ± 1.4 | −257 ± 45 | 11 ± 3.0 | 0.09 ± 0.03 | 0.50 ± 0.35 | 3.8 ± 1.4 |
| Y-D | 1 | 20 ± 1.3 | −177 ± 36 | 6.8 ± 2.2 | 0.18 ± 0.05 | 0.58 ± 0.27 | 1.9 ± 1.3 |
Values are means±SDs (n = 3 for each sediment sample and n = 9 for K-mean and Y-mean).
Means of values for the sediments taken at the K-A, K-B, and K-C sampling points.
Means of values for the sediments taken at the Y-A, Y-B, and Y-C sampling points (those beneath areas of aquaculture).
Sampling and characterization of sediment.
Sediment samples were collected from a fishing boat with a gravity core sampler. Sediments used for the clone library analysis were obtained at the K-B and Y-B points in March 2001 and August 2001, and those used for quantitative PCR and chemical and activity analyses were obtained at all sampling points in July 2004 and at some points in June 2003. At each point, sediment samples were obtained in triplicate. The surface sediment (approximately 0 to 5 cm from the surface) was collected, and the temperature and oxidation/reduction potential (ORP) were measured using a TOX-90i ORP meter (Toko Chemical Laboratories, Tokyo, Japan) immediately after sampling. It was observed that the sediment samples were anaerobic from at least 0.5 cm below the surface, and sulfate reduction was active in the top 5-cm core (unpublished data). This zone of a core (close to the surface) was considered to be influenced mainly by organic input from the aquaculture activity. Immediately after the temperature and ORP were measured, a sediment sample was put in a plastic bag to keep the sediment away from air and stored in an icebox.
The concentration of total sulfide (TS) in a sediment sample was measured using a Gastech detector (Gastech Corp., Kanagawa, Japan) according to the manufacturer's instructions. To determine the sulfate concentration in total sediment, a sediment sample was washed with 5 volumes of distilled water by being vigorously shaken. The washings were then pooled, and the sulfate concentration was determined by ion chromatography with an IA-100 ion analyzer (DKK Toa, Tokyo, Japan). For measurements of COD in sediment, approximately 30 mg of sediment was suspended in 2 ml of water and subjected to a COD analyzer (Hach, Loveland, CO). Total direct counts (TDCs) of microorganisms were determined by using fluorescence microscopy, after microorganisms were detached from sediment particles by gentle sonication (12) and stained with 4′,6′-diamidino-2-phenylindole (DAPI). These analyses were conducted within 6 h after sampling.
DNA extraction and purification.
For extraction of DNA, sediment samples were frozen within 2 h after sampling and stored in a freezer before use. DNA was extracted from 5 g of sediment according to a method described previously (48). The extraction procedure included proteinase K treatment, hot detergent treatment, three cycles of freezing/thawing, and phenol-chloroform extraction (37). The extracted nucleic acids were recovered by ethanol precipitation (37) and subjected to purification by being passed through a hydroxyl apatite column (39). The nucleic acids were finally treated with RNase (37). The quality and quantity of DNA were checked by measuring its UV absorption spectrum (37).
PCR, cloning, and sequencing of 16S rRNA gene.
PCR amplification of 16S rRNA genes was performed using B341f or U515f as a forward primer and U1492r as a reverse primer (Table 2). Amplification was performed with a Progene thermal cycler (Techne, Cambridge, England) by using a 50-μl mixture containing 1.25 U of Taq DNA polymerase (Amplitaq Gold; Applied Biosystems, Foster City, CA), 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% (wt vol) gelatin, each deoxynucleoside triphosphate at a concentration of 200 μM, 50 pmol of each primer, and 10 ng of DNA. The PCR conditions used were as follows: 10 min of activation of the polymerase at 94°C; 30 cycles, each consisting of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C; and finally 10 min of extension at 72°C.
TABLE 2.
PCR primers used in this study
| Primera | Sequence (5′ to 3′)b | Positionc | Specificity | Reference(s) |
|---|---|---|---|---|
| B341f | CCTACGGGIGGCIGCA | 341-357 | Bacteria | 46 |
| U515f | GTGYCAGCMGCCGCGGTAA | 515-533 | Universal | 46 |
| U533rd | TTACCGCGGCKGCTGRCAC | 515-533 | Universal | 46 |
| U1492r | GGYTACCTTGTTACGACTT | 1492-1510 | Universal | 11 |
| GAM660rd | AATTCCACTTCCCTCTAC | 660-674 | Sulfur-oxidizing bacteria and some other bacteria in Thiotrichales | 32 |
| SRB385mfd | CCTGACGCAGCRACGCC | 385-401 | Sulfate-reducing bacteria in δ-Proteobacteria and some other bacteria (e.g., Clostridium spp.) | 1, 29 |
| U926rd | CCGTCAATTCMTTTRAGTTT | 907-926 | Universal | 2 |
| Dv687rd | TCTACGGATTTCACTCCT | 687-704 | Desulfovibrio spp. | 10 |
| Sval428fd | GTAAAATCCTGTCAGATGG | 428-446 | Desulfotalea spp. and Desulfofustis spp. | 35 |
| DSS658rd | TCCACTTCCCTCTCCCAT | 658-685 | Desulfosarcina spp. and Desulfofaba spp. | 26 |
| T7W | TAATACGACTCACTATAGGGC | pGEM-T vector | 45 | |
| SP6W | ATTTAGGTGACACTATAGAATACTC | pGEM-T vector | 45 |
f, forward PCR primer; r, reverse PCR primer.
According to International Union of Biochemistry codes for bases. I, inosine.
Corresponding to the numbering in the sequence of the 16S rRNA gene of E. coli.
These primers were used for qcPCR.
Amplified fragments were purified by electrophoresis, ligated into the pGEM-T vector (Promega, Madison, WI), and cloned into Escherichia coli as described previously (45). Vector-harboring clones were selected on Luria-Bertani plates (37) supplemented with ampicillin (50 μg ml−1). PCR-amplified 16S rRNA gene fragments were recovered from each colony by PCR with primers T7W and SP6W (these primers target pGEM-T vector sequences flanking the insertion) (Table 2) as described previously (45). Clones containing appropriate insert sizes were selected by an electrophoretic analysis, and their nucleotide sequences were determined as described previously (45).
Nucleotide sequence analysis.
Database searches for related 16S rRNA gene sequences were conducted using the GenBank database. The profile alignment technique of ClustalW, version 1.7 (41), was used to align the sequences. Each alignment was refined by visual inspection, and secondary structures were considered for the refinement analysis (18). A phylogenetic tree was constructed by the neighbor-joining method (36) and the maximum-likelihood method (13). Nucleotide positions at which any sequence had a gap or an ambiguous base were not included in the phylogenetic calculations. Checks for chimeric sequences were conducted by the chimera check program in the Ribosomal Database Project database (25), and possible chimeras were excluded from further analyses.
Quantitative competitive PCR (qcPCR).
Primers used for qcPCR are listed in Tables 2, and qcPCR assays are summarized in Table 3. Competitor fragments were produced using the competitive DNA construction kit (Takara, Ohtsu, Japan). The composition of PCR was as described above, except for the competitor fragment being added at a known copy number. The PCR conditions were as described above except that 35 cycles of amplification were performed and the annealing temperatures were as described in Table 3. Two microliters of the PCR product was analyzed by electrophoresis through 1.5% (wt vol) agarose gels with TBE buffer (37), and the gels were photographed after being stained with SYBR Gold (FMC Bioproducts, Vallensbaek Strand, Denmark). The band intensities of the target and competitor fragments were quantified by using the Multianalyst program supplied with Gel Doc 2000 (Bio-Rad, Hercules, CA). At least three PCR assays with different concentrations of a competitor (generally, decimal dilutions) were conducted for estimating a copy number of a target fragment. A relative amplification efficiency of a target to a competitor was estimated by PCR with known numbers of target and competitor fragments. The copy number of the target was estimated by considering the band intensities, length of the fragment, relative amplification efficiency of the target to the competitor, and copy number of the competitor as described by Lee et al. (22).
TABLE 3.
Summary of qcPCR assays
| qcPCR | Primer | Annealing temp (°C) | Length of fragment (bp)
|
Target
|
||
|---|---|---|---|---|---|---|
| Target | Competitor | Phylotype | Phylogenetic group | |||
| BAC | B341f and U533r | 55 | 170-210 | 333 | Most of bacterial phylotypes | Bacteria |
| SOB | B341f and GAM660r | 58 | 319 | 254 | Y56, Y74, Y169, Y182, and Y222 | Thiotrichales |
| SRB | SRB385mf and U926r | 60 | 541 | 437 | δ-Proteobacterial phylotypes | δ-Proteobacteria |
| DSV | B341f and Dv687r | 55 | 346 | 394 | Y29 | Desulfovibrio spp. |
| SVA | Sva1428f and U926r | 55 | 498 | 439 | KY184 | Desulfotalea spp. and relatives |
| DSS | B341f and DSS658r | 58 | 317 | 394 | Y88 and Y183 | Desulfosarcina spp. and relatives |
Potential sulfate reduction rate (SRR).
Incubation for determining SRR was initiated within 3 h after sampling. A sediment sample (20 g [wet wt]) was suspended in an equal weight of deaerated natural seawater in a bottle (100-mlcapacity) under a nitrogen atmosphere, and the bottle was sealed with a butyl rubber septum and aluminum crimp cap. After being sealed, the bottle was supplemented with Na2S 9H2O (2 mM), resazurin (2 mg liter−1), and Na[35SO4] (74 kBq). After the bottle was gently agitated at 50 rpm at 20°C for 24 h, 5 ml of 20% zinc acetate was added to fix the sulfide, and the sediment and seawater were separated by centrifugation (5,000 × g for 5 min at room temperature [around 25°C]). The sediment was washed two times with natural seawater, and the three seawater fractions were mixed. A part (approximately 2 g) of sediment was subjected to a single-step chromium reduction method (14), and the total reduced inorganic sulfur was recovered in two zinc-acetate traps (5% solution; the two zinc-acetate solutions were later mixed). An appropriate amount of the seawater mixture or zinc-acetate solution was added to 10 ml of a ScintiVerse liquid scintillation cocktail (Fisher Scientific, Tokyo, Japan), and radioactivity was measured using a model 1900CA Tri-Carb liquid scintillation analyzer (Perkin-Elmer, Boston, MA). SRR was estimated from the radioactivities, sulfate concentration, sediment weight, incubation time (24 h), and isotope-fractionation ratio according to an equation described previously (14).
Potential sulfur oxidation rate (SOR).
Incubation for determining SOR was initiated within 3 h after sampling. The sediment sample (20 g) was suspended in an equal weight of sulfate-depleted artificial seawater composed per liter of NaCl, 28.5 g; MgCl2 · 6H2O, 5.4 g; CaCl2 · 2H2O, 1.47 g; KCl, 0.725 g; SrCl2 · 6H2O, 0.024 g; NaBr, 0.084 g; H3BO3, 0.027 g; NaF, 2.9 mg; and KI, 0.1 mg (pH 7.2). This slurry was transferred to a bottle (100 ml) under air and supplemented with 0.4 ml of a Na2S · 9H2O solution (0.2 M; pH 7.0) and 0.4 ml of a 1,8-dihydroxyanthraquinone solution (1 mM in acetone; an inhibitor for sulfate-reducing bacteria [SRB]) (8). The bottle was gently shaken on a rotary shaker (50 rpm) at 20°C for 2 days, and the sulfate concentration was periodically measured by ion chromatography. Autoclaved sediment was incubated under the same conditions, which served as the abiotic control for estimating rates for chemical oxidation. The SOR was estimated by subtracting the chemical oxidation rate from the total oxidation rate.
Statistics.
Data were statistically analyzed by the Student t test (P = 0.05 unless otherwise stated) using the Koneko program (Kodansha Scientific, Tokyo, Japan).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the DDBJ, EMBL, and National Center for Biotechnology Information nucleotide sequence databases under accession numbers AB116389 to AB116513.
RESULTS
Characteristics of sediment samples.
Table 1 summarizes characteristics of sediment samples taken from the Yamada and Kamaishi bays. Comparisons of the K-mean and Y-mean (K-mean and Y-mean are means of values for the sediments taken at the K-A, K-B, and K-C sampling points and the Y-A, Y-B, and Y-C sediments, respectively; Y-mean values represent those of sediment beneath aquaculture) values indicate that ORP, TS, and TDC values were not significantly different between the Yamada and Kamaishi sediment, while COD and sulfate concentrations were significantly different. The values for COD and sulfate concentrations of Y-mean weare also significantly different from the values for the Y-D sediment. Since sampling point Y-D was situated at the mouth of the Orikasa river and out of the aquaculture zone, it is likely that this sediment was mostly affected by organic loads from the river but not by the aquaculture activity.
16S rRNA gene clone library analysis.
Two sets of PCR primers (B341f/U1492r and U515f/U1492r) were used for amplification of prokaryotic 16S rRNA gene fragments, each of which successfully amplified a single fragment with the expected size from the sediment DNA. The16S rRNA gene clone libraries were constructed for the Kamaishi sediment obtained in August 2001 (the KM library) and Yamada sediments obtained in March 2001 (the Y3 library) and August 2001 (the Y8 library), each of which contained almost equal numbers of 16S rRNA gene clones derived from B341f/U1492r and U515f/U1492r-amplified fragments. Complete sequences of a total of 219 clones were determined (66 from the Y3 library, 79 from the Y8 library, and 76 from the K8 library), which produced 125 different phylotypes (a unique clone or group of clones with sequence similarity of >97%) (Table 4). Among them, 81 phylotypes (approximately 65%) comprised single clones, suggesting that the 16S rRNA gene clone libraries for the marine sediment prokaryotes were quite diverse.
TABLE 4.
Phylotypes obtained in this study
| Phylotype | No. of clones in a library
|
Phylogenetic affiliation | ||
|---|---|---|---|---|
| Y3 | Y8 | KM | ||
| Y5 | 1 | 2 | Rhodobacteraceae | |
| K11 | 1 | Hyphomicrobiaceae | ||
| KY25 | 1 | Methylocystaceae | ||
| KY40 | 1 | 1 | 1 | Hyphomicrobiaceae |
| Y42 | 2 | Bradyrhizobium | ||
| Y144 | 1 | Rhodobacteraceae | ||
| Y185 | 1 | α-Proteobacteria | ||
| K34 | 3 | Ralstonia | ||
| Y209 | 1 | Thiobacillus | ||
| Y2 | 1 | 3 | Cardiobacteriales | |
| Y17 | 1 | γ-Proteobacteria | ||
| Y56 | 1 | Thiotrichales | ||
| Y74 | 1 | Thiotrichales | ||
| Y94 | 1 | γ-Proteobacteria | ||
| Y95 | 1 | γ-Proteobacteria | ||
| Y121 | 1 | Thiotrichales | ||
| KY142 | 1 | 10 | Pseudomonas | |
| Y169 | 1 | Thiotrichales | ||
| Y182 | 5 | 3 | Thiotrichales | |
| Y189 | 1 | Thiotrichales | ||
| Y218 | 1 | Thiotrichales | ||
| Y222 | 2 | 2 | Thiotrichales | |
| Y223 | 1 | γ-Proteobacteria | ||
| K13 | 2 | Desulfuromonadales | ||
| Y29 | 2 | Desulfovibrionales | ||
| KY51 | 1 | 1 | Syntrophobacterales | |
| Y59 | 1 | Myxococcales | ||
| KY65 | 1 | 1 | δ-Proteobacteria | |
| Y88 | 4 | 2 | Desulfobacteraceae | |
| Y107 | 1 | Syntrophobacterales | ||
| Y117 | 1 | δ-Proteobacteria | ||
| Y137 | 2 | Desulfoarculaceae | ||
| Y148 | 1 | δ-Proteobacteria | ||
| Y150 | 1 | δ-Proteobacteria | ||
| Y171 | 1 | Syntrophobacterales | ||
| Y183 | 2 | 1 | Desulfobulbaceae | |
| KY184 | 5 | 3 | 2 | Desulfobulbaceae |
| Y188 | 1 | Desulfobacterales | ||
| Y196 | 1 | Syntrophobacterales | ||
| Y219 | 2 | Myxococcales | ||
| KY221 | 1 | 2 | δ-Proteobacteria | |
| Y149 | 1 | Sulfurospirillum | ||
| Y7 | 1 | Actinobacteria | ||
| Y8 | 1 | Actinobacteria | ||
| K33 | 1 | Actinobacteria | ||
| Y41 | 1 | Actinobacteria | ||
| KY50 | 1 | 1 | Rubrobacteridae | |
| Y57 | 3 | Actinobacteria | ||
| K106 | 1 | Actinobacteria | ||
| Y108 | 1 | Actinobacteria | ||
| Y193 | 1 | 1 | Rubrobacteridae | |
| Y194 | 1 | 1 | Actinobacteria | |
| KY204 | 1 | 2 | Actinomycetales | |
| KY206 | 2 | 1 | Actinobacteria | |
| K10 | 3 | Clostridium | ||
| Y110 | 1 | Firmicutes | ||
| Y181 | 1 | Bacillus | ||
| Y4 | 1 | Bacteroidetes | ||
| Y19 | 1 | Bacteroidetes | ||
| Y46 | 1 | 1 | Bacteroidetes | |
| Y49 | 1 | Flavobacteria | ||
| Y60 | 1 | 1 | Bacteroidetes | |
| Y61 | 1 | Flavobacteria | ||
| Y89 | 2 | Flavobacteria | ||
| Y93 | 1 | Bacteroidetes | ||
| Y122 | 1 | Flavobacteria | ||
| K200 | 1 | Sphingobacteriales | ||
| Y220 | 2 | Bacteriodetes | ||
| Y224 | 1 | Bacteriodetes | ||
| Y85 | 1 | Chlorobi | ||
| Y30 | 1 | Pirellula | ||
| K31 | 4 | Pirellula | ||
| K37 | 1 | Pirellula | ||
| Y70 | 1 | 1 | Pirellula | |
| K77 | 1 | Pirellula | ||
| Y98 | 1 | Pirellula | ||
| Y112 | 1 | 1 | Pirellula | |
| Y115 | 1 | Pirellula | ||
| Y119 | 1 | Planctomycetales | ||
| KY128 | 2 | 1 | 1 | Planctomycetales |
| KY130 | 1 | 1 | Pirellula | |
| K156 | 1 | Planctomycetales | ||
| Y164 | 1 | Isosphaera | ||
| K175 | 1 | Planctomycetales | ||
| K179 | 3 | Pirellula | ||
| Y225 | 1 | Planctomyces | ||
| K233 | 1 | Planctomyces | ||
| Y99 | 1 | Verrocomicrobia | ||
| Y123 | 1 | Verrocomicrobia | ||
| Y23 | 1 | 1 | Spirochaetes | |
| K35 | 1 | Chlamydia | ||
| Y166 | 1 | Chlamydia | ||
| K54 | 1 | Chloroflexi | ||
| Y66 | 1 | Chloroflexi | ||
| K101 | 1 | Chloroflexi | ||
| K102 | 1 | Chloroflexi | ||
| K103 | 1 | Chloroflexi | ||
| Y187 | 1 | Chloroflexi | ||
| Y217 | 1 | Chloroflexi | ||
| Y72 | 1 | Acidobacteria | ||
| Y90 | 1 | Acidobacteria | ||
| Y190 | 1 | Acidobacteria | ||
| Y192 | 1 | Acidobacteria | ||
| Y195 | 1 | Acidobacteria | ||
| KY12 | 1 | 2 | Candidate division OP8 | |
| K53 | 3 | Candidate division OP8 | ||
| Y139 | 1 | Candidate division OP8 | ||
| K134 | 4 | Candidate division OP9 | ||
| K199 | 1 | Candidate division OP9 | ||
| Y39 | 1 | Candidate division OP11 | ||
| K78 | 1 | Candidate division OP11 | ||
| Y116 | 1 | Candidate division OP11 | ||
| Y118 | 1 | Candidate division OP11 | ||
| Y161 | 1 | Candidate division OP11 | ||
| Y97 | 1 | Candidate division WS3 | ||
| K36 | 1 | Candidate division TM6 | ||
| Y27 | 1 | Unclassified bacterium | ||
| Y160 | 1 | Unclassified bacterium | ||
| KY177 | 1 | 2 | Unclassified bacterium | |
| Y67 | 1 | Crenarchaeota | ||
| K82 | 9 | Crenarchaeota | ||
| K131 | 1 | Crenarchaeota | ||
| Y162 | 1 | Crenarchaeota | ||
| Y165 | 1 | Crenarchaeota | ||
| Y170 | 1 | Crenarchaeota | ||
| Total | 66 | 79 | 76 | |
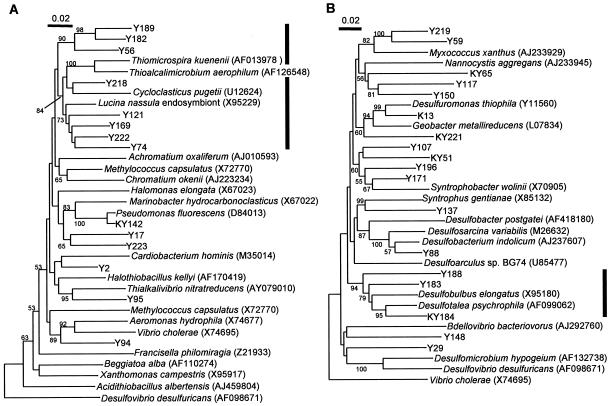
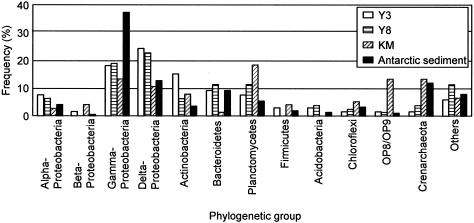
Table 4 also shows the phylogenetic affiliation of each phylotype, which is presented at the possible lowest level of rank determined by the BLAST database search and phylogenetic comparison with reference sequences retrieved from the databases. Figure 2 presents neighbor-joining trees showing the phylogenetic relationships among β-proteobacterial and δ-proteobacterial phylotypes and reference sequences. The phylogenetic relations appearing in Fig. 2 were confirmed by the maximum-likelihood algorithm (data not shown). Phylogenetic distributions of 16S rRNA gene clones in each of the Y3, Y8, and KM libraries at the phylum or subphylum level are presented in Fig. 3, in which clone distribution in the Antarctic library (4) is also shown for comparison. Although many phylotypes were present only in one library (Table 4), grouping analyses as shown in Fig. 3 revealed similarity in clone distribution between the Y3 and Y8 libraries, which was different from the KM library. In the Y3 and Y8 libraries, phylotypes affiliated with δ-Proteobacteria were the most abundant, and those with γ-Proteobacteria were the second. In contrast, the Kamaishi library was occupied evenly by phylotypes affiliated with Planctomycetes, γ-Proteobacteria, δ-Proteobacteria, and Crenarchaeota. Among them, those affiliated with Planctomycetes were the most abundant (18% in the KM library). In addition, the Kamaishi library contained many phylotypes affiliated with candidate divisions OP8 and OP9.
FIG. 2.
Neighbor-joining trees showing phylogenetic relationships among phylotypes obtained in this study and reference 16S rRNA gene sequences retrieved from the nucleotide sequence databases for γ-Proteobacteria (A) and δ-Proteobacteria (B). The bold vertical bar to the right of the tree in panel A represents the range of Thiotrichales, while the one to the right of the tree in panel B represents Desulfobulbaceae. The numbers at the branch nodes are bootstrap values per 100 trials; only values of >50 are shown. Accession numbers for the reference sequences are shown in parentheses.
FIG. 3.
Phylogenetic distribution of clones in each library. Y3, the Yamada March library; Y8, the Yamada August library; KM, the Kamaishi library (in August); Antarctic sediment, a 16S clone library for Antarctic continental shelf sediment (4).
As shown in Table 4 and Fig. 2A, many Yamada phylotypes in γ-Proteobacteria were affiliated with Thiotrichales (15) and related to free-living and symbiotic sulfur-oxidizing bacteria (SOB), whereas the Kamaishi phylotype (KY142) was related to the genus Pseudomonas. In δ-Proteobacteria (Table 4 and Fig. 2B), phylotype KY184 was the most abundant and included 10 clones that are distributed in all the three libraries; phylogenetic analysis indicated that this phylotype was closely related to the genus Desulofotalea (21), which includes psychrophilic SRB. Another phylotype found abundantly in the libraries was K82 (containing nine clones in the KM library); this phylotype and three other phylotypes (Y67, Y170, and Y162) were affiliated with Crenarchaeota and related to 16S rRNA gene clones in marine benthic group C (i.e., CRA9-27cm) (43). In addition, a significant number of phylotypes were related to the genus Pirellula in the Planctomycetes (38) (Table 4).
Quantification of rRNA gene copies by qcPCR.
The 16S rRNA gene clone library analysis detected differences in clone composition in libraries for the Kamaishi sediment (KM) and the Yamada sediment (Y3 and Y8) libraries; particularly, phylotypes related to potential sulfur oxidizers and those affiliated with δ-Proteobacteria (potential sulfate reducers) were detected in abundance in the Y3 and Y8 libraries (Table 4; Fig. 2 and 3). To further evaluate these differences, qcPCR assays with group-specific primers were developed and are summarized in Table 2. The utility and limitation of qcPCR assays for quantifying microbial populations in natural ecosystems have been described previously (22, 44). To check the specificities of these PCR primers and optimize PCR conditions, we carried out PCR experiments using Escherichia coli cells harboring sediment rRNA gene clones (i.e., 125 phylotypes). The results confirmed that these PCR assays could amplify fragments with the expected sizes only from those clones harboring the target phylotypes as listed in Table 3. Primers B341f and U533r were used in the BAC qcPCR (Table 3) for quantifying the total bacterial population, and ratios in copy number (relative abundances) of the target phylotypes to the total bacterial population (the bacterial artificial chromosome copy number) were estimated. This method can prevent a possible incorrect estimate of the copy number resulting from variation in the DNA extraction efficiency.
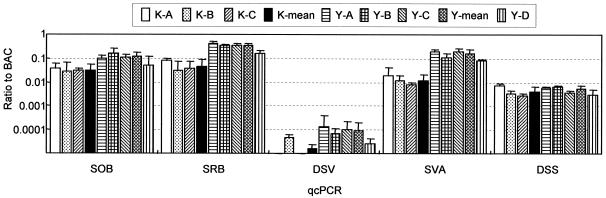
Results of qcPCR for sediments obtained in July 2004 are presented in Fig. 4. The variations in relative abundance of SOB, SRB, and SVA (Table 3) among the Kamaishi sediment samples (K-A, K-B, and K-C) were small, as were the variations among the Yamada sediment samples in the aquaculture zone (Y-A, Y-B, and Y-C). However, the in-site variations in the DSV and DSS data were large. Comparison of the K-mean with the Y-mean in Fig. 4 shows that the SOB, SRB, and SVA values of the Y-mean were significantly higher than those of the K-mean. Similar trends in qcPCR were observed for sediments obtained in 2003 (data not shown).
FIG. 4.
qcPCR assays for quantifying rRNA gene copies of phylotypes related to SOB and SRB. Ratios of specific rRNA gene copies to the total bacterial rRNA gene copies are shown. The names of qcPCR assays refer to Table 3. Data are means (n = 3 samples for each sediment sample and n = 9 samples for K-mean and Y-mean), and error bars represent standard deviations.
SOR and SRR.
The above results indicated that SOB and SRB were more abundant in the Yamada sediment in the aquaculture zone than those in the Kamaishi sediment and outside the aquaculture zone (Y-D). To further assess this observation, we also measured potential SOR and SRR in the Yamada and Kamaishi sediments obtained in July 2004 (Fig. 5). We found that in-site variations (variations in the K-A, K-B, and K-C samples and those in the Y-A, Y-B, and Y-C samples) were relatively small and not significant. Comparisons of the K-mean and Y-mean values indicated that SOR and SRR of the Yamada sediment in the aquaculture zone were significantly higher than SOR and SRR of the Kamaishi sediment. In particular, the difference in SRR between Y-mean and K-mean (also between Y-mean and Y-D) was great (P < 0.0001), showing that sulfate reduction was more intense in the Yamada sediment beneath the shellfish bed than in the Kamaishi sediment. Similar trends in SOR and SRR were observed for sediments obtained in 2003 (data not shown).
FIG. 5.
SOR and SRR of marine sediment samples. Data are means (n = 3 samples for each sediment sample and n = 9 samples for K-mean and Y-mean), and error bars represent standard deviations.
DISCUSSION
This study investigated prokaryotes in coastal marine sediments in the Sanriku region. Although the Yamada and Kamaishi bays are close to each other and have similar hydrogeological characteristics and water qualities, the bacterial community structure in the sediment of these two bays was substantially different. One prominent finding was that bacterial populations related to potential SOB in γ-Proteobacteria and those related to potential SRB in δ-Proteobacteria were more abundant in the Yamada sediment than those in the Kamaishi sediment. This finding was supported by several different lines of evidence, including the rRNA gene clone library analysis (Table 4; Fig. 2 and 3), the qcPCR assays (Fig. 4), and the activity measurements (Fig. 5). In addition, these differences were also observed between within (Y-A, Y-B, and Y-C) and outside (Y-D) the aquaculture zone. These results indicate that the sulfur cycle has become active in the Yamada sediment beneath the shellfish bed.
The 16S rRNA gene clone library analysis (Table 4) suggests that sediment prokaryotes in the Yamada and Kamaishi bays are quite diverse (125 phylotypes from 219 clones sequenced). Similar levels of prokaryotic diversity in the marine sediment have been reported previously (4, 31). Ravenschlag et al. analyzed bacterial populations in permanently cold marine sediment in the Arctic Ocean, showing that 140 different ARDRA (amplified rRNA gene restriction analysis) patterns were obtained from 353 clones in a 16S rRNA gene library (31). Bowman and McCuaig found that 496 phylotypes were present in a subsample of clones (1,046 clones) in a 16S rRNA gene library constructed from Antarctic marine sediments (4). A rarefraction analysis conducted for the Arctic sediment library (31) has, however, suggested that the prokaryotic diversity in the marine sediment can only be partially assessed at this scale of analysis (with several hundred clones). Nevertheless, when the results of clone library analyses are compared with those of quantitative community analyses, e.g., FISH (32), rRNA slot blot hybridization (3, 32), and qcPCR (this study), it can be seen that the clone libraries contained 16S rRNA gene types representing numerically major groups of microbial populations. It is therefore possible to obtain a fundamental insight into the major prokaryotic populations by clone-library analysis on a similar scale.
In our clone libraries, KY184 was the most abundant γ-proteobacterial phylotype (Table 4). This phylotype was closely related to the genus Desulfotalea that had been isolated from permanently cold Arctic marine sediments off the coast of Svalbard (21) and has been identified as psychrophilic SRB capable of incomplete oxidization of fatty acids (21). Related sequences have also been cloned from the Svalbard sediments (31), Antarctic shelf sediment (4), deep-sea sediments (23), and coastal sediments off Japan (42), indicating that this group of SRB is widely distributed in marine sediment. Our qcPCR assay using the SVA system (using a primer with the nucleotide sequence of probe Sval428) (35) estimated the relative abundance of this group (the SVA group) in the total bacterial population in the Sanriku sediments to be 1 to 19%. In addition, its relative abundance in the total SRB population was estimated to be 18 to 61%, indicating that it is the major sulfate-reducing population. A previous study has used the rRNA blot analysis for estimating this group of SRB in the Svalbard sediments (35), showing that they made up from 1.4 to 20.9% of the total eubacterial population. In the present study, it was also shown that the relative abundance of the SVA group (Fig. 4) corresponded to the SRR determined for the Yamada and Kamaishi sediments (Fig. 5). Based on these data, we concluded that SVA group SRB are the important sulfate-reducing population in the Sanriku coast sediment and suggest that their importance is not restricted to permanently cold marine sediments.
The qcPCR (Fig. 4) found that potential SOB made up from 9 to 16% of the total bacterial population in the Yamada bay sediments, while they only made up approximately 3 to 4% in the Kamaishi sediment. These values are compatible to estimates for SOB in the arctic Svalbard sediments (approximately 2% as determined by FISH using probe GAM660 and approximately 13% as determined by the rRNA blot analysis using probe GAM660) (32), while they are lower than the estimates for SOB in Antarctic shelf sediments (22 to 27% as determined by rRNA blot analysis with GAM660) (3). These data show that sulfur-oxidizing γ-Proteobacteria detectable with probe GAM660 are important members of the prokaryotic community in the marine sediment, while the variation in their abundance can be ascribed to the use of different methods for the detection and estimation (47) and/or actual differences in the community structure. We should also consider a possibility that some SOB in marine sediments escaped detection by the SOB qcPCR assay. For instance, 16S rRNA gene sequences of Thiomicrospira spp. have one to several mismatches to GAM660; this genus represents aerobic lithotrophic SOB inhabiting marine sediments (5, 6). In addition, phylotypes Y121, Y189, and Y218 that have one or two mismatches when compared with GAM660 formed a coherent cluster together with the GAM660 match phylotypes (Y56, Y74, Y169, Y182, andY222) and SOB-related reference sequences (Fig. 2A). Further studies are thus needed to understand the total population structure of SOB in marine sediments.
A previous study showed that mussel aquaculture affected the macrobenthic community (40), demonstrating that dominant species were shifted from suspension feeders to deposit feeders. Such a difference in macrobenthos populations has not been observed in the Kamaishi and Yamada bays; the major benthos in these bays have been reported to be lugworms affiliated with Polychaeta (http://www.sui.pref.iwate.jp/∼sui/frame2.htm). Other studies analyzed changes in compositions of biological molecules, e.g., phospholipids (28) and carbohydrates (9), in marine sediments in response to eutrophication. The results suggested that these biological signatures can be used as indices for the trophic states of the coastal environment. In the present study, we detected differences in the number and activity of sulfate reducers and sulfur oxidizers in marine sediments taken from two different bays, suggesting that these bacteria can serve as indices for assessing the organic load to the marine sediment. To further support this idea, comparative ecological surveys should be done, in which sediment samples will be taken from several different Sanriku bays with different levels of aquaculture and analyzed by the methods employed in the present study. In addition, it will also be necessary to examine contribution of other types of terminal electron-accepting processes (e.g., metal reduction) to organic matter decomposition. If this idea is fully supported by these analyses, sediment bacteria involved in the sulfur cycle (e.g., SVA PCR-detectable organisms) can be regarded as important indicators for assessing effects of aquaculture on the coastal environment in the Sanriku region.
Acknowledgments
We thank Ikuko Hiramatsu for technical assistance.
This work was supported by the New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chrisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., J. M. Stromley, R. Devereux, R. Key, and D. A. Stahl. 1992. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl. Environ. Microbiol. 58:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman, J. P., S. A. McCammon, J. A. E. Gibson, L. Robertson, and P. D. Nichols. 2003. Prokaryotic metabolic activity and community structure in Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2448-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, J. P., and R. D. McCuaig. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2463-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinkhoff, T., C. M. Santegoeds, K. Sahm, J. Kuever, and G. Muyzer. 1998. A polyphasic approach to study the diversity and vertical distribution of sulfur-oxidizing Thiomicrospira species in coastal sediments of the German Wadden Sea. Appl. Environ. Microbiol. 64:4650-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinkhoff, T., G. Muyzer, C. O. Wirsen, and J. Kuever. 1999. Characterization of Thiomicrospira kuenenii sp. nov. and Thiomicrospira frisia sp. nov., two mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacteria isolated from an intertidal mud flat. Int. J. Syst. Bacteriol. 49:385-392. [DOI] [PubMed] [Google Scholar]
- 7.Cifuentes, A., J. Anton, S. Benlloch, A. Donnelly, R. A. Herbert, and F. Rodriguez-Valera. 2000. Prokaryotic diversity in Zostera noltii-colonized marine sediments. Appl. Environ. Microbiol. 66:1715-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooling, F. B., III, C. L. Maloney, E. Nagel, J. Tabinowski, and J. M. Odom. 1996. Inhibition of sulfate respiration by 1,8-dihydroxyanthraquinone and other anthraquinone derivatives. Appl. Environ. Microbiol. 62:2999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dell'Anno, A., M. L. Mei, A. Pusceddu, and R. Danovaro. 2002. Assessing the trophic state and eutrophication of coastal marine systems: a new approach based on the biochemical composition of sediment organic matter. Mar. Pollut. Bull. 44:611-622. [DOI] [PubMed] [Google Scholar]
- 10.Devereux, R., M. D. Kane, J. Winfrey, and D. A. Stahl. 1992. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst. Appl. Microbiol. 15:601-609. [Google Scholar]
- 11.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epstein, S. S., and J. Rossel. 1995. Enumeration of sandy sediment bacteria: search for the optimal protocol. Mar. Ecol. Prog. Ser. 117:289-298. [Google Scholar]
- 13.Felsenstein, J. 1989. PHYLIP, phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 14.Fossing, H., and B. B. Jørgensen. 1989. Measurement of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry 8:205-222. [Google Scholar]
- 15.Garrity, G. M., and J. G. Holt. 2001. Taxonomic outline of the Archaea and Bacteria, p. 155-166. .In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y. [Google Scholar]
- 16.Gooday, A. J., and C. M. Turley. 1990. Responses by benthic organisms to inputs of organic material to the ocean floor: a review. Phil. Trans. R. Soc. Lond. B Biol. Sci. 331:119-138. [Google Scholar]
- 17.Gray, J. P., and R. P. Herwig. 1996. Phylogenetic analysis of the bacterial communities in marine sediments. Appl. Environ. Microbiol. 62:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutell, R. R. 1994. Collection of small subunit (16S and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 22:3502-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jørgensen, B. B. 1977. The sulfur cycle of a coastal marine sediment (Limfjorden, Denmark). Limnol. Oceanogr. 22:814-832. [Google Scholar]
- 20.Jørgensen, B. B. 1982. Mineralization of organic matter in the sea bed -the role of sulphate reduction. Nature 296:643-645. [Google Scholar]
- 21.Knoblauch, C., K. Sahm, and B. B. Jørgensen. 1999. Psychrophilic sulfate-reducing bacteria isolated from permanently cold Arctic marine sediments: description of Desulfofrigus oceanense gen. nov., sp. nov., Desulfofrigus fragile sp. nov., Desulfofaba gelida gen. nov., sp. nov., Desulfotalea psychrophila gen. nov., sp. nov., and Desulfotalea arctica sp. nov. Int. J. Syst. Bacteriol. 49:1631-1643. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. Y., J. Bollinger, D. Bezdicek, and A. Ogram. 1996. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl. Environ. Microbiol. 62:3787-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 24.Lobet-Brossa, E., R. Rosselló-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manz, W., M. Eisenbrecher, T. R. Neu, and U. Szewzyk. 1998. Abundance and spatial organization of gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25:43-61. [Google Scholar]
- 27.McCaig, A. E., C. J. Phillips, J. R. Stephen, G. A. Kowalchuk, S. M. Harvey, R. A. Herbert, T. M. Embley, and J. I. Prosser. 1999. Nitrogen cycling and community structure of proteobacterial β-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl. Environ. Microbiol. 65:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinturier-Geiss, L., L.Méjanelle, B. Dale, and D. A. Karlsen. 2002. Lipids as indicators of eutrophication in marine coastal sediments. J. Microbiol. Methods 48:239-257. [DOI] [PubMed] [Google Scholar]
- 29.Rabus, R., M. Fukui, H. Wilkes, and F. Widdel. 1996. Degradative capacities and 16S rRNA-targeted whole cell hybridization of sulfate-reducing bacteria in an anaerobic environment culture utilizing alkylbenzenes from crude oil. Appl. Environ. Microbiol. 62:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajendran, N., O. Matsuda, R. Rajendran, and Y. Urushigawa. 1997. Comparative description of microbial community structure in surface sediments of eutrophic bays. Mar. Pollut. Bull. 34:26-34. [Google Scholar]
- 31.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosselló-Mora, R., B. Thamdrup, H. Schafer, R. Weller, and R. Amann. 1999. The response of the microbial community of marine sediments to organic carbon input under anaerobic conditions. Syst. Appl. Microbiol. 22:237-248. [DOI] [PubMed] [Google Scholar]
- 34.Sagemann, J., B. B. J.ørgensen, and O. Greeff. 1998. Temperature dependence and rates of sulfate reduction in cold sediments of Svalbard, Arctic ocean. Geomicrobiol. J. 15:85-100. [Google Scholar]
- 35.Sahm, K., C. Knoblauch, and R. Amann. 1999. Phylogenetic affiliation and quantification of psychrophilic sulfate-reducing isolates in marine arctic sediments. Appl. Environ. Microbiol. 65:3976-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory PRess, Cold Spring Harbor, N.Y.
- 38.Staley, J. T., J. A. Fuerst, S. Giovannoni, and H. Schlesner. 1991. The order Planctomycetales and the genera Planctomyces, Pirellula, Gemmata, and Isosphaera, p. 554-582. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 4. Springer-Verlag, New York, N.Y. [Google Scholar]
- 39.Steffan, R. J., J. Goksoyr, A. K. Bej, and R. M. Atlas. 1988. Recovery of DNA from soils and sediments. Appl. Environ. Microbiol. 54:2908-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stenton-Dozey, J. M. E., L. F. Jackson, and A. J. Busby. 1999. Impact of mussel culture on macrobenthic community structure in Saldanha bay, South Africa. Mar. Pollut. Bull. 39:357-366. [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urakawa, H., K. Kita-Tsukamoto, and K. Ohwada. 1999. Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology 145:3305-3315. [DOI] [PubMed] [Google Scholar]
- 43.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe, K., M. Teramoto, and S. Harayama. 1999. An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl. Environ. Microbiol. 65:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe, K., K. Watanabe, Y. Kodama, K. Syutsubo, and S. Harayama. 2000. Molecular characterization of bacterial populations in petroleum-contaminated groundwater discharged from underground crude-oil-storage cavities. Appl. Environ. Microbiol. 66:4803-4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, K., Y. Kodama, and S. Harayama. 2001. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J. Microbiol. Methods 44:253-262. [DOI] [PubMed] [Google Scholar]
- 47.Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]