Abstract
In the aflatoxin biosynthetic pathway, 5′-oxoaverantin (OAVN) cyclase, the cytosolic enzyme, catalyzes the reaction from OAVN to (2′S,5′S)-averufin (AVR) (E. Sakuno, K. Yabe, and H. Nakajima, Appl. Environ. Microbiol. 69:6418-6426, 2003). Interestingly, the N-terminal 25-amino-acid sequence of OAVN cyclase completely matched an internal sequence of the versiconal (VHOH) cyclase that was deduced from its gene (vbs). The purified OAVN cyclase also catalyzed the reaction from VHOH to versicolorin B (VB). In a competition experiment using the cytosol fraction of Aspergillus parasiticus, a high concentration of VHOH inhibited the enzyme reaction from OAVN to AVR, and instead VB was newly formed. The recombinant Vbs protein, which was expressed in Pichia pastoris, showed OAVN cyclase activity, as well as VHOH cyclase activity. A mutant of A. parasiticus SYS-4 (= NRRL 2999) with vbs deleted accumulated large amounts of OAVN, 5′-hydroxyaverantin, averantin, AVR, and averufanin in the mycelium. These results indicated that the cyclase encoded by the vbs gene is also involved in the reaction from OAVN to AVR in aflatoxin biosynthesis. Small amounts of VHOH, VB, and aflatoxins also accumulated in the same mutant, and this accumulation may have been due to an unknown enzyme(s) not involved in aflatoxin biosynthesis. This is the first report of one enzyme catalyzing two different reactions in a pathway of secondary metabolism.
Aflatoxins are toxic, carcinogenic, and mutagenic secondary metabolites mainly produced by certain strains of Aspergillus flavus and Aspergillus parasiticus. Contamination by aflatoxins in food and feed is a serious problem in many areas of the world. The biosynthetic pathway of aflatoxin has been studied extensively, and most of the steps have been clarified (reviewed in references 12, 16, 18, 19, and 29). Many enzymes and the genes encoding the enzymes have been isolated, and most of the enzyme genes were found to comprise a huge cluster over 70 kb long in the fungal genome (16, 25, 27, 29).
We recently reported that two enzymes are involved in the pathway from 5′-hydroxyaverantin (HAVN) to averufin (AVR); HAVN dehydrogenase catalyzes the conversion of HAVN to 5′-oxoaverantin (OAVN), and OAVN cyclase catalyzes the next reaction from OAVN to AVR (13). These enzymes have been purified and characterized. The identity of HAVN dehydrogenase with the gene product of adhA (3) was confirmed by tryptic digestion and matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis (13). The molecular masses of natural OAVN cyclase and denatured OAVN cyclase are 158 kDa and 79 kDa, respectively.
In this study, we determined the amino acid sequence of the enzyme to find the gene encoding the OAVN cyclase. Surprisingly, the N-terminal sequence of OAVN cyclase was the same as a stretch of the versiconal (VHOH) cyclase sequence that was deduced from the reported vbs gene (15). VHOH cyclase has been purified independently by Lin and Anderson (10) and by McGuire et al. (11). VHOH cyclase catalyzes the conversion of VHOH to versicolorin B (VB) and is also called VB synthase (11, 14, 15). This enzyme shows strict stereospecificity to the (1′R,2′S) configuration of VHOH, and it determines the configurations of the subsequent intermediates, as well as the final products, aflatoxins (11, 22). To establish the identity of OAVN cyclase and VHOH cyclase, we conducted the following three experiments: (i) competition between OAVN and VHOH for the enzyme in the cytosol fraction of A. parasiticus, (ii) transformation of Pichia pastoris with vbs and conversion of OAVN to AVR by the expressed VHOH cyclase, and (iii) disruption of the vbs gene in A. parasiticus, followed by characterization of the disruptant. This work is the first report of an enzyme that is involved in two distinct and disconnected reactions in aflatoxin biosynthesis.
MATERIALS AND METHODS
Microorganisms.
Aflatoxigenic strain A. parasiticus SYS-4 (= NRRL 2999) was used as a recipient strain for vbs gene disruption. A. parasiticus NIAH-26, a UV-irradiated mutant of A. parasiticus SYS-4 (20), was used as a source of the enzyme. In YES medium (2% yeast extract, 20% sucrose), A. parasiticus NIAH-26 produces all enzymes in the aflatoxin biosynthetic pathway from norsolorinic acid to aflatoxins, although it does not produce aflatoxins and anthraquinone and xanthone precursors (20-24).
Standard samples.
HAVN and averufanin (AVF) were isolated from cultures of Emericella heterothallica IFO30842 (24). OAVN was prepared by incubation of HAVN with the partially purified HAVN dehydrogenase obtained from the cytosol of A. parasiticus NIAH-26 (13). Versiconal hemiacetal acetate (VHA) was purified from mycelia of the mutant A. parasiticus NIAH-9 cultured in YES medium supplemented with dichlorvos (23). VB (6) and AVR (24) were prepared from mycelia of A. versicolor (Vuill.) Tiraboschi. VHOH was prepared by incubation of VHA with porcine liver esterase followed by extraction with ethyl acetate (1). Averantin (AVN) was prepared from an AVN-accumulating mutant (20). The concentrations of these metabolites were determined based on the UV absorption spectra in methanol by using the following molar absorption coefficients (λmax): for AVN, 6,700 M−1 cm−1 (453 nm); for HAVN, 7,100 M−1 cm−1 (466 nm); for AVF, 7,600 M−1 cm−1 (468 nm); for OAVN, 8,500 M−1 cm−1 (457 nm); for AVR, 10,500 M−1 cm−1 (454 nm); for VHA, 7,300 M−1 cm−1 (480 nm); and for VB, 8,700 M−1 cm−1 (450 nm).
Determination of N-terminal amino acid sequence of OAVN cyclase.
OAVN cyclase purified from the cytosol of A. parasiticus NIAH-26 as previously described (13) was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a 10% polyacrylamide gel (8, 13), and the proteins on the gel were blotted onto a polyvinylidene difluoride membrane (Immobilon P; Millipore) with a semidry blotting system (model AE 6675; Atto, Tokyo, Japan). The part of the membrane corresponding to the OAVN cyclase protein (79 kDa) was cut out and applied to an automated Edman degradation gas phase sequencer (model HP G1005A; Hewlett-Packard, Palo Alto, Calif.).
Competition experiment.
A cytosol fraction was prepared from the mycelia of A. parasiticus NIAH-26, which had been cultured in YES medium (5% yeast extract, 20% sucrose) at 28°C for 4 days (24). VHA (0.26 mM) was incubated with porcine esterase (0.24 mg of protein ml−1; Sigma) in solution A (60 mM potassium phosphate buffer, pH 7.5, containing 10% glycerol) at 30°C for 1 h, and the VHOH formed was then extracted with ethyl acetate. The ethyl acetate extract containing VHOH was transferred to a new tube and then dried by keeping the tube open to the atmosphere in the dark. This mild evaporation was needed to reduce the production of versicolorin C from VHOH by spontaneous dehydration (22). The VHOH residue was dissolved in solution A supplemented with 40 μM OAVN and incubated with the cytosol fraction of A. parasiticus NIAH-26 (final concentration, 0.18 mg of protein ml−1) at 30°C for 10 min. The reaction products were extracted with chloroform, dried, dissolved in a small volume of methanol, and then analyzed with a high-performance liquid chromatography (HPLC) apparatus (model CL-6A; Shimadzu Co., Kyoto, Japan) equipped with a Cosmosil-5Ph column (0.46 by 15 cm; Nacalai Tesuque, Kyoto, Japan). Seventy percent methanol in a 1% aqueous acetic acid solution was used as the solvent at a flow rate of 0.5 ml min−1. The column was kept at 30°C, and the absorbance at 290 nm was monitored. The retention times of standard samples were as follows: OAVN, 8.0 min; AVR, 19.7 min; VHOH, 6.2 min; and VB, 12.3 min.
Expression of recombinant VHOH cyclase in P. pastoris.
A Pichia expression kit (Invitrogen, Groningen, The Netherlands) was used to express VHOH cyclase. The coding region in the VHOH cyclase gene, vbs, was amplified by PCR using the cDNA of A. parasiticus NIAH-26 as a template and primers vbs-EcoRI-F (GGAATTCACCATGGGAGGAAACTGGTTCCCA) and vbs-EcoRI-R (GGAATTCTACTGCCCAGCCATCATTTCA), which had been designed based on the 5′- and 3′-terminal nucleotide sequences of the vbs gene (15). These primers each contained an additional EcoRI restriction site. The PCR product was ligated to the pHIL-D2 vector, and the resulting plasmid was transformed into the Pichia GS115 strain. Screening of the transformant, expression of the gene, and preparation of the cell extract were done according to the manufacturer's instructions accompanying the Pichia expression kit (Invitrogen). After the transformant was screened on MD medium (1.34% yeast nitrogen base with ammonium sulfate and without amino acids [YNB], 4 × 10−5% biotin, and 1% dextrose), the recombinant enzyme was induced by incubating the recombinant in BMMY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate [pH 6.0], 1.34% YNB, 4 × 10−5% biotin, 0.5% methanol) after growth in BMGY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate [pH 6.0], 1.34% YNB, 4 × 10−5% biotin, 1% glycerol). The intracellular fraction of the vbs recombinant was prepared by disrupting the yeast with glass beads. To confirm VHOH cyclase activity, we incubated 50 μM VHA with the cell extract (45 μg ml−1) of vbs-transformed cells and 0.2 mg ml−1 porcine esterase in solution A (total volume, 25 μl) at 30°C for 60 min. To check OAVN cyclase activity, we incubated 50 μM OAVN with the same cell extract. pHIL-D2 vector-transformed cell extract was used as a control. The reaction products were extracted with chloroform, and the chloroform solution was spotted onto a silica gel thin-layer chromatography (TLC) plate (Silica Gel 60; Merck, Rahway, N.J.), which was developed with benzene-ethyl acetate (1:1, vol/vol). Fluorescence of the reaction products was observed under 365-nm UV light.
Construction of the vbs disruption vector and transformation.
The replacement vector for the vbs gene was constructed by the following three-step procedure. First, the 4.3-kb fragment containing the vbs coding region and both the 5′ and 3′ flanking regions was amplified with vbs-HindIII-F (CCCCAAGCTTGGCGACATTTGAGCACCG) and vbs-EcoRI-R2 (GGAATTCCTCCTAGCTGGATCGGAC) as the primers and genomic DNA of strain SYS-4 (= NRRL 2999) as the template. Second, the resultant HindIII/EcoRI PCR fragment was ligated into the corresponding sites in the pUC19 vector. Third, the NheI/SmaI fragment containing the vbs gene was excised from the resultant vector and replaced with the pyrithiamine-resistant gene ptrA, which had been amplified by PCR with the KOD enzyme and primers PTRI-NheI-F (CCTAGCTAGCGGGCAATTGATTACGGGATCCCA) and PTRI-SmaI-R (TCCCCCGGGTGACGATGAGCCGCTCTTGC) from the vector pPTR I (Takara Biochem, Japan). The resultant double-crossover disruption vector, pVBS-DD1, was linealized with BglII and PstI and then transformed into A. paraciticus SYS-4 by using the method of Gomi's group (5). Conidia were collected from pyrithiamine-resistant transformants after at least 4 days of culturing and then inoculated onto GY agar (2% glucose, 0.5% yeast extract, 2% agar) to check the pigment production. The transformants accumulating yellow pigment on the GY agar plate were selected as suspected vbs disruptants. The mutants were then purified by three passages on GY medium, and the spores were stored at −80°C.
Confirmation of the gene disruptions using PCR.
The vbs gene disruption was confirmed by PCR analyses. Genomic DNA of suspected vbs disruptants and recipient strains were prepared as templates by using Nucleon PhytoPure (Amersham Life Science). The primer pairs utilized depended on the different vbs gene constructs in the chromosomes of disruptants and recipient strains. The primers used for checking the vbs double-crossover disruptants were primers P1 (vbs-5′in-F; GGCGGTGGTCCCCGTG), P2 (vbs-3′in-R; GCGACACCGGCGGAAGG), P3 (vbs-F3; CGCGCGAGGAGCTCG), P4 (vbs-R3; GCCGAGGGAGACCGG), P5 (vbsin-KpnI-F; GGGGTACCGGATGGCCTGGGCAGT), P6 (vbsin-KpnI-R; GGGGTACCGGCATCGATATCGGCG), P7 (vbspro-KpnI-F; GGGTACCTGTGTAGAAATGCTGCACAG), P8 (vbs-dis-R; GGGGAAGGGTATGCAACGC), P9 (ptrA-F1; CCACTGTGGCCGCTACC), and P10 (ptrA-R1; CACCGAAGGTAGGGCCC). The PCR conditions were (i) 94°C for 5 min, (ii) 94°C for 40 s, 56°C for 40 s, and 72°C for 3 min for 35 cycles, and (iii) 72°C for 10 min. PCR products were observed by electrophoresis on a 1% agarose gel.
Enzyme assay.
Cytosol fractions were prepared from the mycelia of the disruptants and the recipient strains, which had been cultured in YES medium at 28°C for 4 days. To remove contaminating pigments, we further purified the cytosol fraction through a Sephadex G-25 M column (PD-10; Pharmacia LKB Biotechnology, Uppsala, Sweden), which was equilibrated and then eluted with a solution containing 20 mM Tris-HCl (pH 7.5), 10% (vol/vol) glycerol, 10 mM MgCl2, 0.4 mM EDTA, and 1 mM mercaptoethanol. The cytosol fraction (0.15 mg of protein per ml) was inoculated with 6 μM VHOH or 15 μM OAVN in a reaction mixture containing 60 mM potassium phosphate buffer (pH 7.5) and 10% (vol/vol) glycerol. The total volume of the reaction mixture was 50 μl in a 0.5-ml microtube. After incubation at 30°C for 10 min or 30 min, the reaction was terminated by adding 70 μl of ethyl acetate and mixing the resulting solution with a Vortex mixer. After centrifugation, an aliquot of the ethyl acetate layer was injected into the HPLC apparatus. The O-methyltransferase II reaction catalyzing the reaction from sterigmatocystin (ST) to O-methylsterigamatocystin (OMST) was examined as the non-vbs-related enzyme reaction (26). The cytosol fraction was incubated with 60 μM ST and 0.3 mM S-adenosylmethionine in the same reaction mixture at 30°C for 10 or 30 min. After extraction with ethyl acetate, the reaction products were measured using a Shimadzu HPLC apparatus (LC-6A) equipped with an octadecyl silane column (0.46 by 15 cm; Inertsil ODS-2; GL Sciences Inc.). The column was kept at 40°C, and the flow rate was 1 ml min−1. Absorption at 290 nm was monitored, and the solvent system was acetonitrile-tetrahydrofuran-water (20:20:60, vol/vol/vol). The retention times of standard metabolites were as follows: OMST, 3.8 min; ST, 7.3 min; OAVN, 7.5 min; AVR, 35.5 min; VHOH, 4.3 min; and VB, 11.7 min.
Characterization of the accumulating pigments in the mycelia.
A spore suspension (about 2 × 106 spores) of disruptant DD1-16, -23, or -27 or the recipient strain SYS-4 was inoculated into 100 ml YES liquid medium (2% yeast extract, 20% sucrose) in a bottle (10 cm by 4.5 cm; depth, 15 cm), and the bottle was laid on one side to maximize the surface area of the medium. After 3 days of stationary culturing at 28°C, wet mycelia were extracted with 30 ml acetone. The acetone extract was collected, and 2 or 3 μl of the extract was then spotted onto a silica gel TLC plate (Kieselgel 60; no. 5721; Merck & Co., Rahway, N.J.) together with various standard precursors. The TLC plate was developed with benzene-ethyl acetate (7:3, vol/vol) equilibrated with 10% acetic acid. The accumulating pigments were detected as visible yellow spots or fluorescent spots under UV light (365 nm). Fluorescence photographs of red or orange anthraquinone pigments were taken using a Contax 167 camera with Kenko SL-39 UV and SO 56.2 (YA-3) filters on Kodak Tmax 400 film. Fluorescence photographs of blue or green aflatoxins were taken with the Contax 167 camera with Shott KV 450 and BG 12 filters. For preparation of the pigments from each spot, the acetone extract was spotted onto a TLC plate in line, and after development with the same solution, the part corresponding to each spot was scraped off and pigments on the silica gel were extracted with acetone. The acetone extract was supplemented with more than 5% (vol/vol) water and then kept at −20°C until it was used. To identify the pigments in the resultant extract, we injected the extract into an HPLC apparatus (Shimadzu LC-6A HPLC system) equipped with an octadecyl silane column (0.46 by 15 cm; STR-ODS-II; Shinwa Chemical Industries, Ltd.). The flow solution was acetonitorile-tetrahydrofuran-water-acetic acid (25:25:50:1, vol/vol/vol/vol). The flow rate and column temperature were 1 ml min−1 and 35°C, respectively. The retention times of metabolites were compared with those of standard samples. Typical retention times were as follows: VHOH, 3.1 min; HAVN, 4.3 min; OAVN, 4.7 min; VB, 7.1 min; AVR, 18.5 min; AVN, 21.0 min; and AVF, 23.1 min.
Tip culture
A spore suspension (5 μl) of SYS-4 or one of the mutants with vbs deleted was inoculated into 250 μl of YES medium. After 4 days of culturing, the aflatoxins excreted into the medium were measured by HPLC (20).
RESULTS
N-terminal amino acid sequence of the OAVN cyclase.
The OAVN cyclase was purified from the cytosol of A. parasiticus NIAH-26 as previously described (13). The enzyme was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the protein (79 kDa) on the gel was applied to an automated sequencer. Twenty-five amino acid residues at the N terminus of OAVN cyclase were sequenced (Fig. 1). BLAST analysis unexpectedly revealed that the resultant sequence was the same as that of a stretch (amino acids 42 to 66) of VHOH cyclase (VBS) that was deduced from the vbs gene of A. parasiticus SU-1 (accession no. U51327 and U51328) (14, 15). N-terminal amino acid residues 1 to 41 in the VHOH cyclase of SU-1 were not found in the OAVN cyclase of NIAH-26. We also confirmed that the purified OAVN cyclase catalyzed the production of VB from VHOH, when VHOH was used as the substrate instead of OAVN in a preliminary experiment (data not shown). During all steps through purification of the OAVN cyclase, we did not recognize OAVN cyclase activity other than the main activity (13), suggesting that the purified enzyme is the sole or at least a major enzyme involved in the reaction from OAVN to AVR or from VHOH to VB. These results suggested that the same enzyme catalyzes the two different reactions, from OAVN to AVR and from VHOH to VB, in aflatoxin biosynthesis.
FIG. 1.
Alignment of the N-terminal amino acid sequence of purified OAVN cyclase. The N-terminal sequence corresponded to part of VHOH cyclase (same as VB synthase [12, 13]) which was deduced from the vbs gene (GenBank accession no. U51328). Only the N-terminal sequence of VHOH cyclase is shown.
Competition between OAVN and VHOH for the same enzyme.
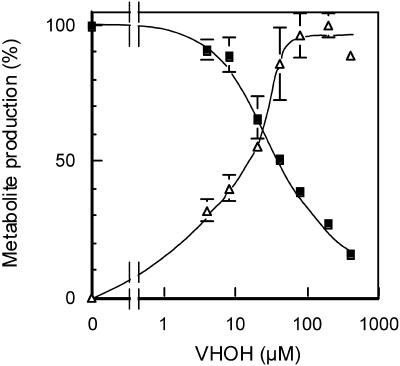
To determine the relationship between the enzyme activities of OAVN cyclase and VHOH cyclase in the cells, we performed a competition experiment between OAVN and VHOH using the cytosol fraction of A. parasiticus NIAH-26. AVR was exclusively produced from OAVN by the cytosol fraction. In contrast, when VHOH was added to the same reaction mixture, VB was also formed, and an increase in the concentration of VHOH resulted in an increase in the VB concentration, whereas the formation of AVR obviously decreased (Fig. 2). At a VHOH concentration of 400 μM, the amount of AVR was reduced to approximately 10%. These findings indicated that OAVN and VHOH compete for the same substrate binding site on the same enzyme in the cytosol.
FIG. 2.
Competition between OAVN and VHOH for OAVN cyclase. Cytosol was incubated in reaction mixtures containing 40 μM OAVN and various concentrations of VHOH, and the amounts of AVR (▪) or VB (▵) produced were then determined by HPLC. The amount of AVR produced (143 pmol/μl) in the reaction mixture without VHOH was defined as 100%. The amount of VB produced (28.5 pmol/μl) in the reaction mixture with 200 μM VHOH was defined as 100%, which was almost same as the amount in the absence of OAVN (data not shown).
Expression of recombinant VHOH cyclase.
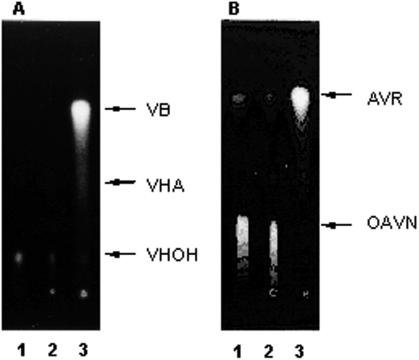
The expression vector pHIL-D2 containing the vbs (VHOH cyclase gene) coding region was transformed into P. pastoris. Cell extracts were prepared from the transformants, and the enzyme activities of both VHOH cyclase and OAVN cyclase were then determined. When VHA was incubated with the cell extract of P. pastoris containing the vbs gene, VB was formed in the presence of porcine esterase (Fig. 3A), by which VHOH was produced from VHA. In contrast, the cell extract from the control yeast strain containing only the vector pHIL-D2 did not produce any VB. These results confirmed that the vbs gene was successfully expressed in the yeast expression system. When OAVN instead of VHOH was incubated with the same cell extract of P. pastoris containing the vbs gene, AVR was newly formed (Fig. 3B). This reaction did not occur when the cell extract from yeast containing only the vector was used, although a small amount of AVR was present in all experiments because AVR was spontaneously produced from OAVN. Therefore, the enzyme encoded by vbs was confirmed to catalyze reactions of both OAVN cyclase and VHOH cyclase. We used the whole sequence of vbs containing the region encoding N-terminal amino acid residues 1 to 41 to express the gene, although we found that the N-terminal 41-amino-acid region of the purified OAVN cyclase was truncated. Therefore, truncation of the N-terminal 41-amino-acid region in the OAVN cyclase may not affect the substrate specificity for OAVN and VHOH.
FIG. 3.
Conversion of VHOH to VB and conversion of OAVN to AVR by the recombinant VHOH cyclase expressed in P. pastoris. VHA (A) or OAVN (B) was incubated with cell extract of P. pastoris. In panel A, porcine esterase was also added in order to convert VHA to VHOH for determination of the VHOH enzyme activity. The reaction products were extracted with chloroform and analyzed by TLC. Lane 1, no cell extract; lane 2, cell extract of P. pastoris transformed with only the vector; lane 3, cell extract of P. pastoris transformed with the vector containing the vbs gene. The small amount of VHOH in panel A was due to the low extraction efficiency of the high-polarity substance VHOH with chloroform.
Disruption of the vbs gene.
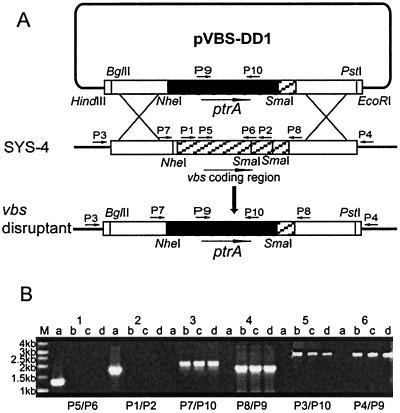
We disrupted the vbs gene of A. parasiticus SYS-4 (= NRRL 2999) using the double-crossover disruption vector pVBS-DD1, which had been constructed by a three-step procedure (Fig. 4A). About 70 pyrithiamine-resistant transformants were obtained by using 4 μg of the disruption vector. Among them, three mutants accumulated bright yellow pigments in the mycelia on a GY agar plate, and we named these mutants VBS-DD1-16, VBS-DD1-23, and VBS-DD1-27. The double-crossover recombination event in these mutants was also confirmed by PCR analysis (Fig. 4B). When we used primer pairs P7-P10 and P8-P9, only the three mutants generated the expected 1.8-kb and 1.7-kb PCR products, respectively, which are specific for replacement of most of the vbs gene with the pyrithiamine-resistant gene ptrA. The mutants also produced 2.83-kb and 2.8-kb bands with primer pairs P3-P10 and P4-P9, respectively, indicating that the vbs replacement construct was inserted into the desired position in the chromosome of the mutants. In contrast, when primer pairs P1-P2 and P5-P6 located within the deletion region of the vbs were used, only wild-type strain SYS-4 produced 1.6-kb and 1.2-kb PCR fragments, as predicted. These results confirmed that all three mutants were vbs double-crossover gene disruptants, in which most of the vbs gene containing the start codon was deleted and replaced with ptrA.
FIG. 4.
Disruption of the vbs gene. To disrupt the vbs gene, a double-crossover disruption vector, pVBS-DD1, was constructed (A). The vector was linearized and then transformed into A. parasiticus SYS-4 (= NRRL 2999). Double-crossover recombination led to replacement of most of the vbs gene containing the start codon with the selectable marker ptrA. The long arrows show gene directions. The short arrows indicate the positions of primers used for confirmation of gene disruption. The transformants accumulating yellow pigment on a GY agar plate were obtained as the vbs disruptants. The vbs gene disruptions were confirmed by PCR analyses (B) (lanes a, recipient train SYS-4; lanes b, c, and d, vbs disruptants VBS-DD1-16, VBS-DD1-23, and VBS-DD1-27, respectively; lane M, 1-kb molecular marker) with primer pairs P5-P6 (lanes 1), P1-P2 (lanes 2), P7-P10 (lanes 3), P8- P9 (lanes 4), P3-P10 (lanes 5), and P4-P9 (lanes 6).
Enzyme activities of the disruptants and the recipient strain.
To confirm that the vbs gene is involved in the reactions of both the OAVN cyclase and VHOH cyclase, we measured the enzyme activities (Table 1). Although the recipient strain showed enzyme activities in both reactions, cytosol from neither disruptant VBS-DD1-16 nor disruptant VBS-DD1-23 showed significant enzyme activity in the case of VHOH cyclase, indicating that the vbs disruption occurred correctly. Furthermore, the disruptants did not show any enzyme activity catalyzing the reaction from OAVN to AVR. In contrast, the activities of O-methyltransferase II catalyzing conversion of ST to OMST were not changed by the vbs disruption. These results demonstrated that the vbs gene is involved in both reactions, from OAVN to AVR and from VHOH to VB.
TABLE 1.
Enzyme activities of cytosol fractions from the recipient strain and mutants with vbs deleted
| Strain | Enzyme activity (nmol 50-μl reaction mixture−1 30 min−1)a
|
||
|---|---|---|---|
| VB from VHOH | AVR from OAVN | OMST from ST | |
| SYS-4 (= NRRL 2999) | 0.15 ± 0.003 | 1.82 ± 0.09 | 0.21 ± 0.01 |
| VBS-DD1-16 | NDb | ND | 0.22 ± 0.01 |
| VBS-DD1-23 | ND | ND | 0.25 ± 0.03 |
The values are means ± standard deviations for more than two measurements.
ND, Not detected.
Characterization of the pigments accumulated in mycelia of the disruptants.
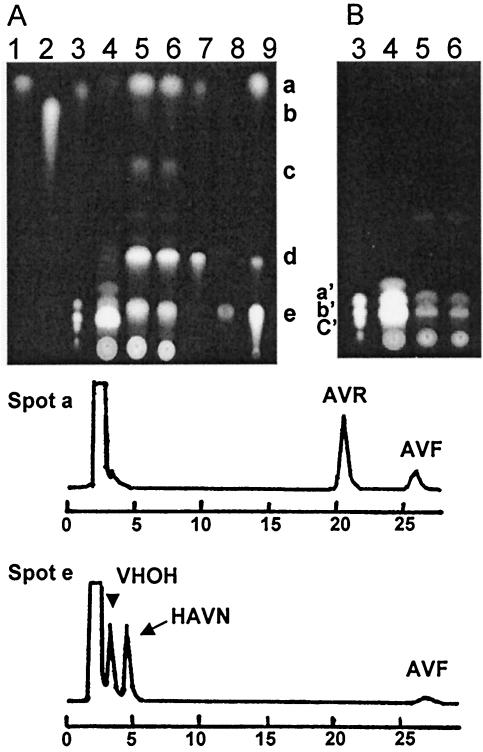
The vbs gene is a sole gene encoding a cyclase (dehydrase) in the aflatoxin gene cluster. If the same enzyme is involved in the OAVN cyclase and VHOH cyclase reactions, disruption of the vbs gene in the latter reaction may primarily cause inhibition of the former reaction, causing accumulation of the precursor of the early enzyme, that is, OAVN cyclase. The vbs disruptants, as well as the recipient strain SYS-4, were cultured in YES medium for 3 days. Pigments extracted with acetone were developed on TLC plates and observed under UV light (Fig. 5A and B). The disruptants accumulated five kinds of orange pigments (spots a to e), which corresponded to AVR, VB, AVN, OAVN, and HAVN when each Rf value was compared with the those of standard samples, but AVR overlapped with AVF in spot a and HAVN overlapped with VHOH in spot e. Therefore, pigments in each spot were analyzed further by HPLC (Fig. 5, chromatograms). Spot a contained more AVR than AVF. The AVF probably came from spontaneous dehydration of HAVN during TLC analysis. The faint spot b was VB. Spot c was AVN, and the brightest spot, spot d, was OAVN. Spot e contained VHOH, HAVN, and a small amount of AVF as determined by HPLC analysis, and the AVF seemed to have come from HAVN through extraction from the TLC plate. Among these pigments, AVR, OAVN, and HAVN were the pigments with major accumulations, and AVR and AVF were spontaneously produced from OAVN and HAVN, respectively, since both OAVN and HAVN are unstable substances. AVN, HAVN, and OAVN are precursors of AVR in the aflatoxin biosynthetic pathway. These results demonstrated that disruption of the vbs gene caused almost complete inhibition of the OAVN cyclase.
FIG. 5.
Characterization of the pigments in the vbs disruptant VBS-DD1-16. Acetone extracts of the mycelia of A. parasiticus SYS-4 (lane 4, 3 μl) and VBS-DD1-16 (lane 5, 3 μl; lane 6, 2 μl) were analyzed by TLC together with precursors (lane 1, AVR; lane 2, VB; lane 3, VA and aflatoxins B1, B2, G1, and G2; lane 7, OAVN; lane 8, VHOH; lane 9, HAVN). Two kinds of conditions for taking fluorescence photographs were used for the same TLC plate to detect red or orange fluorescence of the precursors (A) and blue or green fluorescence of aflatoxins (B). Each spot from the acetone extract of VBS-DD1-16 (A) was extracted and analyzed by HPLC. HPLC chromatograms of spots a and e are shown below the TLC plates. The first peak of the HPLC chromatogram corresponded to the solvent. The spots in panel A corresponded to the following compounds: spot a, AVR and AVF; spot b, VB; spot c, AVN; spot d, OAVN; and spot e, HAVN and VHOH. The standards in panel B were aflatoxin B1 (spot a′), aflatoxins G1 and B2 (spot b′), and aflatoxin G2 (spot c′)
We did detect a small amount of VHOH, as well as VB, indicating that the vbs deletion did not cause complete inhibition of OAVN cyclase. Since VHOH is also unstable, the VB probably came from spontaneous dehydration of VHOH during the TLC analysis. Furthermore, small amounts of aflatoxins were detected in the acetone extract of VBS-DD1-16 (Fig. 5B). Also, when this mutant was cultured in the tip culture, a small amount of aflatoxins still formed; the wild-type strain produced 9.90 ± 0.17 μg total aflatoxins per 250 μl of culture medium, and the mutants with vbs deleted produced 0.20 ± 0.05 μg aflatoxins per 250 μl of medium, which corresponded to 2.0% ± 0.2% of the amount produced by the recipient strain.
DISCUSSION
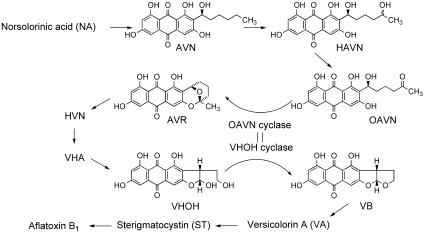
We recently purified OAVN cyclase that catalyzes the reaction from OAVN to AVR in A. parasiticus NIAH-26 (13). In this work we found that OAVN cyclase catalyzes two reactions, the reaction from OAVN to AVR and the reaction from VHOH to VB, in aflatoxin biosynthesis (Fig. 6). The molecular mass of the OAVN cyclase, a homodimer of 79-kDa subunits (13), was almost the same as that of the VHOH cyclase, a homodimer of 72-kDa subunits (10) or 78-kDa subunits (11). The molecular mass of VHOH cyclase deduced from the cDNA sequence was 70.27 kDa, and that of the matured enzyme after glycosylation is 78 kDa (11, 14, 15). Silva et al. suggested that the N terminus of the enzyme is posttranslationally modified because they could not sequence the N terminus of the purified VHOH cyclase in A. parasiticus SU-1 (15). In contrast, in the present study we succeeded in sequencing the N terminus of the OAVN cyclase and showed that the N terminus of the OAVN cyclase was Phe-42 of the deduced sequence reported for VHOH cyclase in A. parasiticus SYS-4 (15). The purified OAVN cyclase also showed enzyme activity of the VHOH cyclase. Processing of the enzyme seems to depend on the strain used. For transformation of P. pastoris with the vbs gene we used the whole sequence of the gene containing amino acid residues 1 to 41 and found that the recombinant VHOH cyclase showed enzyme activities of both OAVN cyclase and VHOH cyclase. Therefore, the processing at the N-terminal part does not affect the enzyme activities.
FIG. 6.
Involvement of the same cyclase in two independent reactions, the reaction from OAVN to AVR and the reaction from VHOH to VB.
One enzyme for one step is common in many metabolic pathways. Thus, even when we found that the N terminus of the OAVN cyclase was Phe-42 of VHOH cyclase, we still thought that two kinds of cyclase must be present in the cells. But for the following reasons we concluded that one enzyme catalyzes two different steps. First, no enzyme other than the enzyme purified was observed during purification of OAVN cyclase, even though the purified enzyme catalyzed both the conversion of VHOH to VB and the conversion of OAVN to AVR. Second, in the purification of VHOH cyclase by Lin and Anderson the enzyme activities were not separated (10). Third, there is only one cyclase gene (vbs) in the aflatoxin gene cluster, and the genes in the gene cluster are generally accepted to be sufficient to make aflatoxins. These three points indicate that the gene product of vbs catalyzes two different steps in aflatoxin biosynthesis. To confirm this, we conducted deletion experiments for the vbs gene. Disruption of the vbs gene caused the accumulation of OAVN and other precursors preceding the formation of OAVN, indicating that the enzyme encoded by the vbs gene is involved in the step from OAVN to AVR. However, the disruptant still produced small amounts of VHOH, as well as VB (Fig. 5A) and aflatoxins (Fig. 5B). Tip culture experiments also showed that the aflatoxin productivity of the vbs disruptant was 2.0% of that of the recipient strain. Therefore, another enzyme might attend to aflatoxin biosynthesis, but the activity of the enzyme seems to be too low to be detected in the cell-free system of the disruptant (Table 1), while the soluble enzymes involved in aflatoxin biosynthesis are generally active enough to be detected easily in cell-free experiments. Thus, the remaining activity in the disruptant should not be due to a specific enzyme for aflatoxin biosynthesis but to an unknown enzyme also present in the cells that has wide group specificity.
In similar gene disruption experiments, disruption of a monooxygenase gene such as the ordA (28) and verB genes (7, 29) caused complete inhibition of aflatoxin production. Monooxygenase usually shows rigid specificity for its substrate. However, remaining activity after deletion of a certain gene has also been reported. Deletion of the oxidoreductase nor1 gene (17) or the esterase estA gene (4) from the gene cluster did not cause complete inhibition of aflatoxin or precursor production. Most notably, about 10% of the esterase activity involved in the reaction from VHA to VHOH was carried out by an unknown cytosol enzyme(s) (4). Generally, oxidase, reductase, and esterase are common enzymes in cells, and many enzymes in these groups are present in cells. Also, they usually show wide group specificity for many substrates. In fungal cells, they may participate to a slight degree in aflatoxin biosynthesis due to its wide group specificity. In the case of the cyclase reaction from OAVN to AVR or from VHOH to VB, another unknown enzyme(s) may also nonspecifically and partially participate in the cyclization of OAVN or VHOH.
Furthermore, another possibility is that nonenzymatic dehydration of OAVN, as well as VHOH, might occur in cells. Both OAVN and VHOH are so unstable in the hydrophobic condition that they are easily converted to AVR and VB, respectively. A relatively hydrophobic microenvironment, such as a membrane surface, might provide the appropriate conditions for spontaneous cyclization. However, it is difficult to imagine this occurring in an aqueous environment in cells.
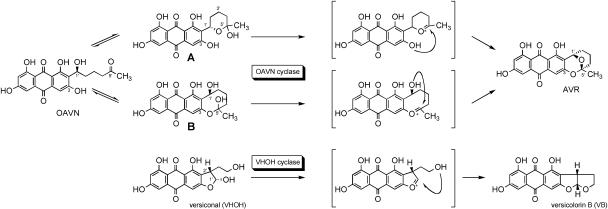
Although the same enzyme commonly catalyzes dehydration of OAVN and VHOH, these substrates appear to be quite different in their structures. The Km values of the enzyme for OAVN and VHOH were 20 μM (13) and 2.4 μM (11), respectively. As for the mechanism of the cyclase activity from OVAN to AVR, two possible pathways can be proposed (Fig. 7). OAVN is converted to AVR after two cyclizations. Compounds A and B are the intermediate candidates. Compound A is a hemiacetal of OAVN in which C-5′ is linked to C-1′ through an oxygen atom to form a six-member ether ring. Compound B is a hemiacetal of OAVN in which C-5′ is linked to C-3 via an oxygen atom to form an eight-member ether ring. It is likely that OAVN and compound A or OAVN and compound B are in equilibrium. It is, however, still unclear whether the OAVN cyclase works in hemiacetal formation. Considering the structural similarity to VHOH, this cyclase mainly catalyzes the step from the hemiacetal intermediate (compound A or B) to AVR. The cyclization of VHOH proceeds through the following steps. The hemiacetal OH leaves the molecule by dehydration to form a double bond between C-1′ and the oxygen atom adjacent to C-1′. The oxygen of the remaining 4′-OH group attaches to C-1′, and then C-4′ is linked to C-1′ via an oxygen atom. The involvement of two alcoholic hydroxyls in compound B in the ether ring formation is the same as that of VHOH, suggesting that compound B is a real substrate of the cyclase. VHOH cyclase shows strict stereospecificity to the (1′R,2′S) configuration of VHOH (22). Taking the stereochemistry of VB and AVR into account, 4′-OH must attach to C-1′ from the α-side in VHOH. On the other hand, 1′-OH must attach to C-5′ from the β-side in compound B. If this mechanism is true, the enzyme cannot recognize this stereochemical difference. More experiments are, however, needed to clarify the details of the reaction mechanisms.
FIG. 7.
Proposed mechanism for the reaction from OAVN to AVR and the reaction from VHOH to VB. Compounds A and B are hemiacetal forms of OAVN. OAVN may be converted to AVR through either intermediate compound A or intermediate compound B.
Involvement of the same enzyme in two different steps in aflatoxin biosynthesis is of interest from an evolutionary standpoint. Some part of the aflatoxin gene cluster region has been reported to be duplicated in some strains (2, 9), although the vbs region is not duplicated. This indicates that the gene cluster has changed after a long evolutionary time. Two genes encoding OAVN cyclase and VHOH cyclase might have been present in the genome in ancient fungi. During evolution, one of them was probably deleted, but this did not affect aflatoxin production because the product of the remaining vbs gene could catalyze both reactions. Although a beneficial function for aflatoxin biosynthesis in fungi has not been clarified, aflatoxin production catalyzed by the product of the remaining vbs gene might be necessary for fungus survival. The function and evolution of secondary metabolism remain to be studied.
Acknowledgments
We thank Y. Ando, National Institute of Animal Health, for taking fluorescence pictures and K. Gomi and K. Kusumoto for advice on fungal transformation. The BLAST search of the GenBank database was performed with the assistance of the Computer Center of Agriculture, Forestry and Fisheries Research, MAFF, Japan.
This work was supported in part by grant-in-aid BDP-04-VI-1-2 (Bio Design Project) from the Ministry of Agriculture, Forestry and Fisheries.
REFERENCES
- 1.Anderson, J. A., and C. H. Chung. 1990. Conversion of versiconal acetate to versiconal and versicolorin C in extracts from Aspergillus parasiticus. Mycopathologia 110:31-35. [DOI] [PubMed] [Google Scholar]
- 2.Chang, P.-K., and J. Yu. 2002. Characterization of a partial duplication of the aflatoxin gene cluster in Aspergillus parasiticus ATCC 56775. Appl. Microbiol. Biotechnol. 58:632-636. [DOI] [PubMed] [Google Scholar]
- 3.Chang, P.-K., J. Yu, K. C. Ehrlich, S. M. Boue, B. G. Montalbano, D. Bhatnagar, and T. E. Cleveland. 2000. adhA in Aspergillus parasiticus is involved in conversion of 5′-hydroxyaverantin to averufin. Appl. Environ. Microbiol. 66:4715-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, P.-K., K. Yabe, and J. Yu. 2004. The Aspergillus parasiticus estA-encoded esterase converts versiconal hemiacetal acetate to versiconal and versiconol acetate to versiconol in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70:3593-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomi, K., Y. Iimura, and S. Hara. 1987. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric. Biol. Chem. 51:2549-2555. [Google Scholar]
- 6.Hamasaki, T., Y. Hatsuda, N. Terashima, and M. Renbutsu. 1967. Studies on the metabolites of Aspergillus versicolor (Vuillemin) Tiraboschi. Part V. Isolation and structures of three new metabolites, versicolorin A, B and C. Agric. Biol. Chem. 31:11-17. [Google Scholar]
- 7.Kelkar, H. S., T. W. Skloss, J. F. Haw, N. P. Keller, and T. H. Adams. 1997. Aspergillus nidulans stcL encodes a putative cytochrome P-450 monooxygenase required for bisfuran desaturation during aflatoxin/sterigmatocystin biosynthesis. J. Biol. Chem. 272:1589-1594. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 9.Liang, S.-H., C. D. Skory, and J. E. Linz. 1996. Characterization of function of the ver-1A and ver-1B genes, involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 62:4568-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, B.-K., and J. A. Anderson. 1992. Purification and properties of versiconal cyclase from Aspergillus parasiticus. Arch. Biochem. Biophys. 293:67-70. [DOI] [PubMed] [Google Scholar]
- 11.McGuire, S. M., J. C. Silva, E. G. Casillas, and C. A. Townsend. 1996. Purification and characterization of versicolorin B synthase from Aspergillus parasiticus. Catalysis of the stereodifferentiating cyclization in aflatoxin biosynthesis essential to DNA interaction. Biochemistry 35:11470-11486. [DOI] [PubMed] [Google Scholar]
- 12.Minto, R. E., and C. A. Townsend. 1997. Enzymology and molecular biology of aflatoxin biosynthesis. Chem. Rev. 97:2537-2555. [DOI] [PubMed] [Google Scholar]
- 13.Sakuno, E., K. Yabe, and H. Nakajima. 2003. Involvement of two cytosolic enzymes and a novel intermediate, 5′-oxoaverantin, in the pathway from 5′-hydroxyaverantin to averufin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 69:6418-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva, J. C., and C. A. Townsend. 1997. Heterologous expression, isolation, and characterization of versicolorin B synthase from Aspergillus parasiticus. J. Biol. Chem. 272:804-813. [DOI] [PubMed] [Google Scholar]
- 15.Silva, J. C., R. E. Minto, C. E. Barry III, K. A. Holland, and C. A. Townsend. 1996. Isolation and characterization of the versicolorin B synthase gene from Aspergillus parasiticus. J. Biol. Chem. 271:13600-13608. [DOI] [PubMed] [Google Scholar]
- 16.Trail, F., N. Mahanti, and J. Linz. 1995. Molecular biology of aflatoxin biosynthesis. Microbiology 141:755-765. [DOI] [PubMed] [Google Scholar]
- 17.Trail, F., P. K. Chang, J. Cary, and J. E. Linz. 1994. Structural and functional analysis of the nor1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl. Environ. Microbiol. 60:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woloshuk, C. P., and R. Prieto. 1998. Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol. Lett. 160:169-176. [DOI] [PubMed] [Google Scholar]
- 19.Yabe, K., and H. Nakajima. 2004. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 64:745-755. [DOI] [PubMed] [Google Scholar]
- 20.Yabe, K., H. Nakamura, Y. Ando, N. Terakado, H. Nakajima, and T. Hamasaki. 1988. Isolation and characterization of Aspergillus parasiticus mutants with impaired aflatoxin production by a novel tip culture method. Appl. Environ. Microbiol. 54:2096-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yabe, K., Y. Matsuyama, Y. Ando, H. Nakajima, and T. Hamasaki. 1993. Stereochemistry during aflatoxin biosynthesis: conversion of norsolorinic acid to averufin. Appl. Environ. Microbiol. 59:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yabe, K., and T. Hamasaki. 1993. Stereochemistry during aflatoxin biosynthesis: cyclase reaction in the conversion of versiconal to versicolorin B and racemization of versiconal hemiacetal acetate. Appl. Environ. Microbiol. 59:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yabe, K., Y. Ando, and T. Hamasaki. 1991. A metabolic grid among versiconal hemiacetal acetate, versiconol acetate, versiconol and versiconal during aflatoxin biosynthesis. J. Gen. Microbiol. 137:2469-2475. [DOI] [PubMed] [Google Scholar]
- 24.Yabe, K., Y. Nakamura, H. Nakajima, Y. Ando, and T. Hamasaki. 1991. Enzymatic conversion of norsolorinic acid to averufin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 57:1340-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu, J., D. Bhatnagar, and T. E. Cleveland. 2004. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 564:126-130. [DOI] [PubMed] [Google Scholar]
- 26.Yu, J., J. W. Cary, D. Bhatnagar, T. E. Cleveland, N. P. Keller, and F. S. Chu. 1993. Cloning and characterization of a cDNA from Aspergillus parasiticus encoding an O-methyltransferase involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:3564-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, J., P.-K. Chang, J. W. Cary, M. Wright, D. Bhatnagar, T. E. Cleveland, G. A. Payne, and J. E. Linz. 1995. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl. Environ. Microbiol. 61:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, J., P.-K. Chang, K. C. Ehrlich, J. W. Cary, B. Montalbano, J. M. Dyer, D. Bhatnagar, and T. E. Cleveland. 1998. Characterization of the critical amino acids of an Aspergillus parasiticus cytochrome P-450 monooxygenase encoded by ordA that is involved in the biosynthesis of aflatoxins B1, G1, B2, and G2. Appl. Environ. Microbiol. 64:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu, J., P. K. Chang, K. C. Ehrlich, J. W. Cary, D. Bhatnagar, T. E. Cleveland, G. A. Payne, J. E. Linz, C. P. Woloshuk, and J. W. Bennett. 2004. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70:1253-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]