Abstract
Human flora-associated (HFA) mice have been considered a tool for studying the ecology and metabolism of intestinal bacteria in humans, although they have some limitations as a model. Shifts in dominant species of microbiota in HFA mice after the administration of human intestinal microbiota was revealed by 16S rRNA gene sequence and terminal restriction fragment length polymorphism (T-RFLP) analyses. Characteristic terminal restriction fragments (T-RFs) were quantified as the proportion of total peak area of all T-RFs. Only the proportion of the T-RF peak at bp 366, identified as the Gammmaproteobacteria group and the family Coriobacteriaceae, was reduced in this study. Increased T-RFs over time at bp 56, 184, and 196 were affiliated with the Clostridium group. However, most of the isolated bacteria with unique population shifts were phylotypes. The vertical transmission of the intestinal microbiota of the mouse offspring was also investigated by dendrogram analysis derived from the similarity of T-RFLP patterns among samples. As a result, the intestinal microbiota of HFA mice and their offspring reflected the composition of individual human intestinal bacteria with some modifications. Moreover, we revealed that human-derived lactobacilli (HDL), which have been considered difficult to colonize in the HFA mouse intestine in previous studies based on culture methods, could be detected in the HFA mouse intestine by using a lactic acid bacterium-specific primer and HDL-specific primers. Our results indicate that the intestinal microbiota of HFA mice represents a limited sample of bacteria from the human source and are selected by unknown interactions between the host and bacteria.
Intestinal microbiota is composed of many kinds of bacteria. These indigenous intestinal bacteria play an important role in the health and disease of hosts, including nutrition, host immunity, and carcinogenesis. Experimental animal studies are indispensable when formulating basic concepts of the microbial ecology in the intestinal tracts of humans. However, the intestinal microbiota in experimental animals is quite different in composition from that in humans (23, 31). A recent study revealed the existence of many unidentified microorganisms, including some bacteria specific to the mouse intestine, by molecular techniques (2, 17, 35). Therefore, data obtained in animal experiments cannot be directly applied to humans.
Human flora-associated (HFA) mice have been considered a tool for studying the ecology and metabolism of human intestinal bacteria (6, 7, 30), although they have some limitations as a model (12). These limitations include differences in enzyme activity, concentrations of putrefactive products, and immunological activation by the composition of fecal bacteria (13, 15). Previous studies reported that 60% to 80% of the observable bacteria in human intestines could not be cultivated (10, 37). These limitations may therefore be caused by composition differences in microbiota of intestinal contents between humans and HFA mice. However, there are few reports about the use of molecular techniques with HFA mice or rats to determine the composition of microbiota including unidentified bacteria (5, 7, 15).
The genera Lactobacillus and Bifidobacterium are well known as beneficial bacteria for probiotics. These bacteria derived from humans seem to hardly colonize in the mouse intestine (12, 28, 41). In particular, it has been reported that lactobacilli have strong host specificity (24, 19, 39). The possibility has been considered that this phenomenon is caused by a difference in physiological condition and balance of microbes between humans and mice, but this has not been clarified.
In this study, 16S rRNA gene sequence and terminal restriction fragment length polymorphism (T-RFLP) analyses were used to reveal the shifts in the dominant bacteria of the intestinal microbiota in HFA mice after the administration of human fecal specimens. T-RFLP analysis is a useful molecular approach for the rapid assessment and comparison of diverse complex bacterial communities, such as those in soil, feces, and oral microbiota (20, 32, 33, 34). The horizontal transmission of intestinal microbiota in their offspring was also investigated. Moreover, lactobacilli were characterized, and the human-derived lactobacilli (HDL) were screened and detected in the intestines of HFA mice by use of genus-specific and the HDL-specific primers. This report provides new information regarding the intestinal microbiota of HFA mice.
MATERIALS AND METHODS
Animals.
Germfree BALB/c mice, from the Department of Infectious Diseases, Tokai University School of Medicine, Kanagawa, Japan, were used in all examinations. They were housed in vinyl isolators with sterilized Clean tip (CLEA Japan, Inc., Tokyo) as bedding and given sterilized water and sterilized commercial CL-2 pellets (CLEA Japan, Inc.) ad libitum. The diet was sterilized with an autoclave (121°C, 30 min). Five male and five female mice were inoculated at 4 weeks of age into the stomach by a metal catheter with 0.5 ml of a 10−1 suspension of feces obtained from an apparently healthy human volunteer (male, aged 59 years). The mice were mated at 8 weeks of age, and their offspring were weaned at 3 weeks of age.
Sampling.
Fecal samples from mice were taken 1, 2, and 4 weeks after administration of the fecal suspension. Eight weeks after inoculation, the mice were sacrificed by use of diethyl ether, and the contents of the small intestine, cecum, and colon were removed as samples of the intestinal microbial community. Collected samples were stored immediately at −80°C until use. Intestinal content samples from the offspring were collected at 6 weeks of age by the same methods.
Cell lysis and DNA isolation from samples.
DNA extraction and purification were based on the methods described by Clement and Kitts (3), using an Ultra Clean Soil DNA isolation kit (Mo Bio Laboratories, Inc., Solana Beach, CA) with some modification, as described previously (17).
T-RFLP analysis.
A pair of universal primers, 5′ FAM (6-carboxyfluorescein)-labeled 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) (Applied Biosystems, Tokyo, Japan) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) (18), were used. PCR was performed as described previously (17). Purified PCR products were digested with 20 U of HhaI (Takara Bio Inc.) in a total volume of 10 μl at 37°C for 3 h. The lengths of the terminal restriction fragments (T-RFs) were determined using standard size markers GS500 ROX and GS 1000 ROX (Applied Biosystems) with an ABI PRISM 310 genetic analyzer (Applied Biosystems) and GeneScan analysis software (Applied Biosystems). Cluster analysis was performed using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) based on T-RFLP patterns. The Jaccard matching coefficient was used for objective interpretation of the difference in T-RF patterns. The distances of similarity among samples were represented graphically by constructing a dendrogram. The unweighted pair-group method with arithmetic mean (UPGMA) was used to establish the dendrogram type. T-RFs were quantified as the proportion of total peak area of all T-RFs. The significant differences between the samples at 1 week after administration and other time points were calculated using Student's t test (P < 0.05). Characteristic peaks were identified by direct cloning of T-RFs as previously described (22). Reproducibility of T-RFLP patterns was previously investigated in detail (25, 34).
16S rRNA gene sequences.
The human inoculum, mouse fecal samples 1 and 4 weeks after administration, and mouse colon samples 8 weeks after administration were used for cloning and sequencing to compare with T-RFLP analysis results. Sequencing of the 16S rRNA genes was performed based on a previous study (17). All sequences were compared with similar sequences of the reference organisms by BLAST (1) and FASTA (27) searches and checked for possible chimeric artefacts by the CHIMERA CHECK program of the Ribosomal Database Project-II (4). All sequences underwent fragment analysis with restriction enzymes by computer simulation and were compared by T-RFLP analysis. The term “phylotype” is used for a cluster of clone sequences that differs from the sequence of a known species by approximately 2%, and these clusters were at least 98% similar to numbers within a cluster of clone sequences (26).
PCR and sequencing for the detection of HDL.
Three primer sets were used for the detection of HDL. One primer and one primer set used in this study were designed in previous studies of the detection of lactic acid bacteria (LAB) or Lactobacillus spp. (11, 40). A reverse primer; S-G-Lab-0677-a-A-17 (Lab-0677r, 5′-CACCGCTACACATGGAG-3′), was designed by Heilig et al. (11). Two primers, Lac 1 (5′-AGCAGTAGGGAATCTTCCA-3′) and Lac 2 (5′-CATGTGTAGCGGTGRAAT-3′), were designed by Walter et al. (40). Lab-0677r was used with 5′ FAM-labeled 27f for LAB-specific T-RFLP analysis of the human inoculum and intestinal contents of HFA mice. LAB-specific T-RFLP analysis was performed with four restriction enzymes, HhaI, MspI, HaeIII, and AluI. The 27f-Lab-0677r primer set and Lac 1-Lac 2 primer set were also used in cloning to identify amplicons. The amplification program conformed to the description of each previous study (11, 40). After the identification of HDL, we designed the forward primer HDL-f (5′-AGGATAGAGGC-3′) to amplify the HDL. Nested PCR was performed to detect the HDL. Lac 1 and Lac 2 were used for first-round PCR. HDL-f and Lac 2 were used for second-round PCR. The program included 94°C for 3 min; 30 cycles consisting of 94°C for 30 s, 51°C for 30 s, and 72°C for 30 s; and a final extension period at 72°C for 3 min. Ten nanograms of DNA from the first-round PCR amplicon was subjected to a second PCR in a 50-μl reaction mixture. Amplified DNA was verified by 1.5% agarose gel electrophoresis and sequencing.
Nucleotide sequence accession numbers.
Sequences of the 16S rRNA genes of new phylotypes derived from humans in this study were deposited with the GenBank database under accession numbers AB191009 to AB191022. Representative sequences of the 16S rRNA genes confirmed in this study, i.e., of HDL derived from human inoculum, of an Aerococcus sp. derived from HFA mouse intestine, and of HDL derived from HFA mouse intestine, were deposited in GenBank under accession numbers AB191025 to AB191027, AB191028 to AB191030, and AB191023 to AB191024, respectively.
RESULTS
Dynamics of intestinal microbiota in HFA mice.
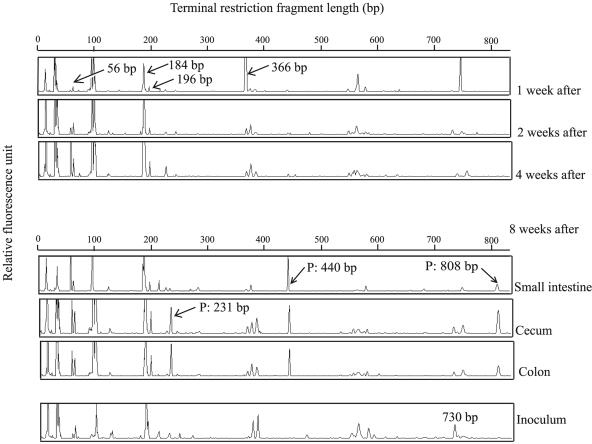
The shift of intestinal microbiota in HFA mice after the administration of human feces was determined by T-RFLP patterns. Unique changes over time were revealed by T-RFLP profiles of samples derived from HFA mice (Fig. 1). Common or characteristic T-RFs were detected in all samples. Characteristic T-RFs detected in all samples were confirmed numerically, and significance was calculated by Student's t test (P < 0.05) (Table 1). Increases in T-RFs over time were at bp 56, 184, and 196; in contrast, decreases in T-RFs with time were at bp 366. Although the T-RFs at bp 730 were detected at a high rate in human inoculum, they were not detected at such a high rate in HFA mouse samples. T-RFs at bp 231, 440, and 808 were detected in samples of intestinal contents but were not detected or were detected in small populations in fecal samples. We designated these T-RFs as “persisting.” 16S rRNA gene clone library analysis and direct cloning of T-RFs were used to identify these unique movements (Table 1). All sequences obtained from each sample had a fragment position confirmed by computer simulation. Almost all the T-RFs were presumed to represent species or phylotypes detected by 16S rRNA gene clone library analysis. The same identification was confirmed by direct cloning of T-RFs (data not shown).
FIG. 1.
Movement of intestinal microbiota in HFA mice by T-RFLP analysis. T-RFLP patterns of 16S rRNA genes from samples digested with HhaI. The minimum and maximum values of the ordinate are 0 to 500 fluorescence units. The unique T-RFs are shown by arrows. The peaks detected in only the intestinal contents of HFA mice are indicated as persisting (P).
TABLE 1.
Dynamics and persistence of characteristic T-RFs and identification of the T-RFs
| Position (bp) | Ratio of T-RF area at indicated time after administrationa
|
Trendb | Ratio of T-RF area in inoculum feces | Identification by 16S clone libraries | |||||
|---|---|---|---|---|---|---|---|---|---|
| Feces
|
Small intestine | Cecum | Colon | ||||||
| 1 wk | 2 wk | 4 wk | |||||||
| 56 | 0.17 ± 0.02 | 0.63 ± 0.08* | 2.72 ± 0.89* | 3.91 ± 1.09* | 1.50 ± 0.31* | 1.23 ± 0.34* | ↑ | 0.32 ± 0.02 | NDc |
| 184 | 0.41 ± 0.07 | 0.70 ± 0.06* | 3.87 ± 1.81* | 3.24 ± 0.64* | 1.82 ± 0.58* | 1.84 ± 0.98 | ↑ | 0.84 ± 0.10 | Uncultured Clostridium cluster XVIa |
| 196 | 0.60 ± 0.06 | 0.66 ± 0.06 | 0.91 ± 0.06* | 1.16 ± 0.17* | 0.89 ± 0.20 | 1.09 ± 0.27 | ↑ | ND | Uncultured Clostridium cluster XVIa |
| 231 | ND | ND | 0.20 | 14.3 ± 11.6 | 1.29 ± 0.36 | 1.79 ± 0.75 | P | ND | Uncultured Clostridium cluster VI |
| 366 | 20.7 ± 5.37 | 1.03 ± 0.08* | 1.07 ± 0.37* | 1.45 ± 0.60* | 1.08 ± 0.32* | 0.85 ± 0.16* | ↓ | 0.64 ± 0.13 | “Gammaproteobacteria”and Coriobacteriaceae |
| 440 | 0.31 ± 0.01 | 0.32 ± 0.05 | 0.24 ± 0.01* | 6.25 ± 1.17* | 2.62 ± 0.21* | 2.52 ± 0.54* | P | ND | Uncultured firmicutes |
| 730 | 0.33 ± 0.05 | 0.85 ± 0.17 | 0.52 ± 0.09 | ND | 0.91 ± 0.15* | 0.61 ± 0.17 | 1.37 ± 0.26 | Uncultured Clostridium clusters VI and IX | |
| 808 | ND | ND | ND | 2.88 ± 1.25 | 1.59 ± 0.30 | 1.24 ± 0.32 | P | ND | ND |
Values are ratios of a characteristic T-RF area to total area (mean ± standard error). Values for small intestine, cecum, and colon were determined at 8 weeks. *, Significantly different by paired t test from the value for the sample at 1 week.
Changes in T-RF area after administration of human inoculum over time, ↑, increase; ↓, decrease; P, persistent in intestine.
ND, not detected as T-RFs or clones.
16S rRNA gene sequencing of intestinal microbiota in the human inoculum and HFA mice.
The numbers of clones detected in samples of the inoculum, in a fecal sample 1 and 4 weeks after administration, and in a colon sample 8 weeks after administration are shown in Table 2. One hundred eighty-seven clones from the inoculum were analyzed. Sequences were classified into 12 phylogenetic groups. Although no exact 16S rRNA gene similarity limits exist to define specific taxa such as genus and species, species definition in general requires sequence similarities of greater than 98% (36). All clones were divided into 81 species, including phylotype. About 83% of clones detected in samples of the inoculum belonged to phylotypes. About 40 clones derived from fecal samples at 1- and 4-week time points were analyzed because the T-RFLP patterns at these two time points in HFA mice resembled each other. Additionally, about 40 clones derived from the colon sample at 8 weeks in HFA mice were analyzed. Clones identified as the Bacteroides group, the Clostridium cluster IV, or the Clostridium cluster XIVa were detected at a high rate in all samples, including the human inoculum. Clones belonging to the “Gammaproteobacteria” group, the Verrucomicrobium group, and the Coriobacteriaceae were detected only in samples from HFA mice.
TABLE 2.
Numbers of species and clones detected in 16S rRNA sequences
| Bacterial group | Human sequences
|
HFA mouse sequences
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 wka
|
4 wk
|
8 wk
|
||||||||||||||
| No.
|
Ratio (%)
|
No.
|
Ratio (%)
|
No.
|
Ratio (%)
|
No.
|
Ratio (%)
|
|||||||||
| s/pb | Clone | s/p | Clone | s/p | Clone | s/p | Clone | s/p | Clone | s/p | Clone | s/p | Clone | s/p | Clone | |
| Bacteroides | 11 | 19 | 13.6 | 10.4 | 6 | 7 | 37.5 | 25.9 | 4 | 4 | 50.0 | 30.8 | 5 | 16 | 23.8 | 38.1 |
| Clostridium cluster I | 2 | 3 | 2.5 | 1.6 | NDc | ND | ND | ND | ND | ND | ||||||
| Clostridium cluster IV | 23 | 54 | 28.4 | 29.5 | 3 | 4 | 18.8 | 14.8 | 1 | 4 | 12.5 | 30.8 | 3 | 3 | 14.3 | 7.1 |
| Clostridium cluster IX | 6 | 14 | 7.4 | 7.7 | ND | ND | ND | ND | ND | ND | ||||||
| Clostridium cluster XIVa | 29 | 53 | 35.8 | 29.0 | 6 | 8 | 37.5 | 29.6 | 2 | 2 | 25.0 | 15.4 | 7 | 7 | 33.3 | 16.7 |
| Clostridium cluster XIVb | 1 | 2 | 1.2 | 1.1 | ND | ND | ND | ND | ND | ND | ||||||
| Clostridium cluster XVI | 1 | 24 | 1.2 | 13.1 | ND | ND | ND | ND | ND | ND | ||||||
| Clostridium cluster XVIII | 1 | 1 | 1.2 | 0.5 | ND | ND | ND | ND | ND | ND | ||||||
| Unclass Clostridium | 3 | 7 | 3.7 | 3.8 | ND | ND | ND | ND | ND | ND | ||||||
| Streptococcus | 2 | 3 | 2.5 | 1.6 | ND | ND | ND | ND | ND | ND | ||||||
| Actinobacteria | 1 | 2 | 1.2 | 1.1 | ND | ND | ND | ND | ND | ND | ||||||
| “Deltaproteobacteria” | 1 | 1 | 1.2 | 0.5 | ND | ND | ND | ND | 1 | 1 | 4.8 | 2.4 | ||||
| “Gammaproteobacteria” | ND | ND | 1 | 8 | 6.3 | 29.6 | ND | ND | ND | ND | ||||||
| Verrucomicrobium | ND | ND | ND | ND | 1 | 3 | 12.5 | 23.1 | 4 | 14 | 19.0 | 33.3 | ||||
| Coriobacteriaceae | ND | ND | ND | ND | ND | ND | 1 | 1 | 4.8 | 2.4 | ||||||
| Total | 81 | 183 | 100 | 100 | 16 | 27 | 100 | 100 | 8 | 13 | 100 | 100 | 21 | 42 | 100 | 100 |
The time point after administration.
s/p, species or phylotype.
ND, not detected as clones.
Cluster analysis of intestinal microbiota in the offspring of HFA mice.
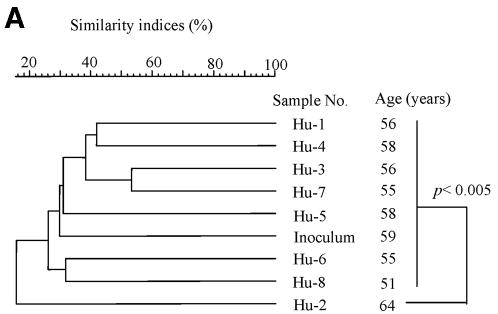
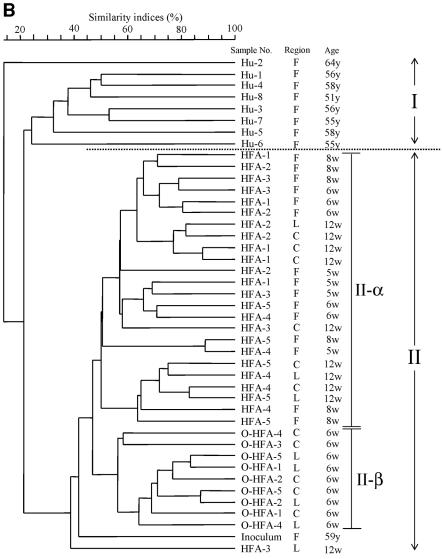
We performed dendrogram analysis based on Jaccard matching coefficients of T-RFLP profiles to assess the similarity of microdiversity in the intestine among the samples. UPGMA was used for the dendrogram (Fig. 2). The human inoculum was compared with other human fecal samples (Fig. 2A). The difference in similarity indices among the samples, except for the Hu-2 sample, was not significant by t test. In addition, T-RFLP patterns of samples from HFA mice and their offspring were determined in addition to those of human samples (Fig. 2B). The dendrogram was divided into two clusters (I and II). Cluster I was formed by only human samples, excluding the human inoculum. Cluster II was formed by samples from HFA mice and their offspring and the human inoculum. Moreover, for similarity indices of about 45%, cluster II was divided into two subclusters (II-α and II-β). The inoculum sample became an outgroup of cluster II. Subcluster II-α was composed only of samples from HFA mice, and subcluster II-β was composed only of samples from offspring.
FIG. 2.
Relationship of intestinal microbiota based on T-RFLP patterns among human samples, including the inoculum (A), and among samples from HFA mice and their offspring (B). The dotted line represents the boundary between the cluster of samples from other humans and samples from HFA mice, including the inoculum and their offspring. Similarity indices (Jaccard coefficients) are indicated at the scale bar of the tree (UPGMA). Hu, human samples; O-HFA, offspring of HFA mice; F, fecal sample; C, cecal sample; L, colon sample; y, years; w, weeks.
Detection of HDL by specific primers.
To confirm the existence of HDL in the intestine of HFA mice, three primer sets were used in this study. The first set, primer 27f and the Lab-0677r primer designed by Heilig et al. (11), was used for LAB-specific T-RFLP analysis with four restriction enzymes (HhaI, MspI, HaeIII, and AluI). T-RFs derived from HFA mouse samples by use of this primer set appeared at bp 588 to 589, 556 to 560, 263 to 264, and 683 to 684, digested by HhaI, MspI, HaeIII, and AluI, respectively.
From the computer simulation results, any bacteria belonging to the genus Lactobacillus were not assigned to these restriction sites. On the other hand, T-RFs derived from the human inoculum appeared at bp 691 (digested by HhaI), at bp 134 and 164 (digested by MspI), at bp 213 (digested by HaeIII), and at bp 68 and 246 (digested by AluI). The T-RFLP analysis results were different for the inoculum and HFA samples. However, these restriction sites were also not assigned. To identify the composition of bacteria in these T-RFs, amplicons from HFA mouse samples were confirmed by cloning and sequencing. All clones derived from HFA samples were more than 98% similar to the species Aerococcus viridans at Escherichia coli positions 27 to 677. The restriction fragment lengths of the Aerococcus viridans clones were 581, 555, 266, and 1,069 bp, digested by HhaI, MspI, HaeIII, and AluI, respectively. There was no restriction site for AluI at positions 27 to 677. This result was identical with LAB-specific T-RFLP results for HFA mouse samples. We found no lactobacilli in HFA samples by use of the 27f-Lab-0677r primer set. Therefore, another primer set, Lac 1-Lac 2, was used. This primer set was designed by Walter et al. (40) for the amplification of LAB from samples. These amplicons were confirmed by cloning and sequencing. Clones derived from the human inoculum were more than 98% similar to the species Lactobacillus delbrueckii at Escherichia coli positions 355 to 644. These clones were designated HDL. However, clones derived from HFA mice were not similar to HDL but were similar to Aerococcus spp. This result was identical to the result determined with another primer set (27f-Lab-0677r). Therefore, in the second PCR, a primer (HDL-f) was designed to discriminate between HDL and other LAB. HDL were detected in seven of all the samples from HFA mice (Table 3). All amplified products were confirmed by sequencing.
TABLE 3.
Detection of HDL by use of each primer set
| Sample no. | Time pointa(wk) | Tractb | Detection by primer setc:
|
|
|---|---|---|---|---|
| Lac 1-Lac 2 | HDL-f-Lac 2 | |||
| HFA-1 | 1 | F | + | ND |
| 2 | F | + | ND | |
| 4 | F | + | + | |
| 8 | S | + | + | |
| C | + | ND | ||
| L | + | ND | ||
| HFA-2 | 1 | F | + | ND |
| 2 | F | + | ND | |
| 4 | F | + | + | |
| 8 | S | + | + | |
| C | + | ND | ||
| L | + | ND | ||
| HFA-3 | 1 | F | + | ND |
| 2 | F | + | + | |
| 4 | F | + | ND | |
| 8 | S | + | ND | |
| C | + | ND | ||
| L | + | ND | ||
| HFA-4 | 1 | F | + | ND |
| 2 | F | + | ND | |
| 4 | F | + | ND | |
| 8 | S | + | ND | |
| C | + | + | ||
| L | + | ND | ||
| HFA-5 | 1 | F | + | ND |
| 2 | F | + | ND | |
| 4 | F | + | + | |
| 8 | S | + | ND | |
| C | + | ND | ||
| L | + | ND | ||
The time point after administration of human feces.
Intestinal tracts of samples derived from HFA mice. F, feces; S, small intestine; C, cecum; L, large intestine.
+, detected; ND, not detected by PCR.
DISCUSSION
Horizontal transmission of intestinal microbiota in HFA mice.
HFA mice have been exploited and used as a tool reflecting human intestinal microbial ecology (6, 7, 30). Although the development of intestinal microbiota in HFA mice has been revealed by culture methods (12), there are few reports of the use of molecular methods (5, 7, 15). Our work has revealed the movement and persistence of human intestinal microbiota in formerly germfree mice by T-RFLP analysis. Additionally, some bacteria in unique T-RFs were identified in a comparison with 16S rRNA gene sequences and direct cloning of T-RFs (Table 1). Only a proportion of the T-RF peak at bp 366, identified as the “Gammmaproteobacteria” group and/or Coriobacteriaceae, was reduced. We also confirmed that the increased T-RFs were affiliated with the Clostridium group. According to the culture method (12), the number of Enterobacteriaceae decreased rapidly after the third day after inoculation. Conversely, anaerobic bacteria were dominant in HFA mice. Our results are in agreement with previous studies (12) because the “Gammmaproteobacteria” group belonging to Enterobacteriaceae and bacteria of the Clostridium group are anaerobic bacteria. However, most of the bacteria isolated in this study belonged to phylotypes. Briefly, most of human intestinal bacteria with unique shifts in the intestines of HFA mice have not yet been cultured.
HFA mice have limitations as a model because some of their enzyme activities and products of intestinal microbiota are different from those of humans. This result indicates that the limitations of HFA mice are caused by the differential composition of bacteria belonging to phylotypes. Bacteria in the Clostridium cluster XIVa and the Clostridium cluster IV were dominant in specific-pathogen-free mice and in conventional mice (9, 16, 17, 29). In spite of different environments, Clostridium group bacteria can inhabit the mouse intestine. Therefore, the physiological conditions of the mouse intestine might be suitable for these bacteria.
The change of indigenous bacteria in the intestine.
The 16S rRNA gene clone sequences were used to identify microbiota of the human inoculum and of samples from HFA mice (Table 2). Clones belonging to the Bacteroides group, the Clostridium cluster IV, and the Clostridium cluster XIVa were detected in all samples at a high rate. However, clones belonging to the “Gammmaproteobacteria” group, the Verrucomicrobium group, the “Deltaproteobacteria” group, and the Coriobacteriaceae were detected only in samples from HFA mice. Godon et al. (8) reported the relationship between the number of sequences and the cumulative number of operational taxonomic units. In this study, despite the analysis of a small number of clones, many clones of these bacteria were detected only in HFA mice. This indicated the possibility that the population of these bacteria was changed between the intestines of humans and HFA mice.
Almost all T-RFs of bacteria in the Bacteroides group in the database, which were digested with HhaI, are located from bp 94 to 104. Moreover, the computer simulation confirmed that clones in the Verrucomicrobium group corresponded to T-RFs at bp 98 with HhaI digestion. Therefore, T-RFs of both groups overlapped at bp 98. Although the movement of the T-RFLP pattern at bp 98 was not confirmed, the dominant bacterial group in HFA mice might change from those present in inoculated human feces.
Movement of intestinal microbiota to the offspring of HFA mice.
By the use of culture methods, an earlier study revealed that HFA mice could be reproduced by breeding (14). In the present study, T-RFLP analysis was used to confirm the transfer of intestinal microbiota to the offspring of HFA mice. To compare the whole T-RFs in samples, we used dendrogram analysis derived from the similarity of T-RFLP patterns among samples. The human inoculum was compared with other human fecal samples (Fig. 2A). The inoculum sample was not specific among human fecal samples because it was not an outgroup of the cluster. This result was confirmed by a t test of similarity indices among T-RFLP patterns. T-RFLP patterns of samples from HFA mice and their offspring were determined in addition to those from human samples (Fig. 2B). The dendrogram was divided into two large clusters. Cluster I was composed only of human samples. Cluster II was composed of samples from HFA mice, their offspring, and the human inoculum. This result indicated that the intestinal microbiota of HFA mice and their offspring was more similar to that of the inoculum than to that of other human samples. Moreover, this result indicated that the composition of intestinal microbiota of HFA mice and their offspring reflected the individual differences of human intestinal microbiota. On the other hand, cluster II was divided into two subclusters. Subcluster II-α was composed of samples from HFA mice, and subcluster II-β was composed only of samples from the offspring. These results indicated that the microbiota in HFA mice was changed by host-specific modification from the bacterial composition of the inoculum in their intestines, although the intestinal microbiota of HFA mice and their offspring reflected the composition of the inoculum as in previous studies (14). Moreover, the intestinal microbiota of offspring showed greater modifications than did the microbiota of their parents.
Existence of the HDL in HFA mice.
The genera Lactobacillus and Bifidobacterium derived from humans seem to hardly colonize in the mouse intestine (13, 28, 41). In particular, although lactobacilli are the predominant bacteria in the mouse intestine, previous studies could not detect them in HFA mice (13, 28) by culture methods. We used three primer sets in this study (27f-Lab-0677r, Lac 1-Lac 2, and HDL-f-Lac 2). Two LAB-specific primer sets (27f-Lab-0677r and Lac 1-Lac 2) were used to detect LAB in the human inoculum and in the HFA mouse intestine. HDL was detected in the human inoculum by LAB-specific primer sets. However, clones detected from the HFA mouse intestine were identified not as HDL but as bacteria of the genus Aerococcus by both primer sets. The clone identified as the Aerococcus species was not detected in the human inoculum. From the results of the differential detection, our results suggested that the populations of HDL and Aerococcus bacteria in samples were different between the human inoculum and the HFA mouse intestine. Therefore, a specific primer for HDL was designed to detect HDL in the intestines of HFA mice, and we performed nested PCR for increased sensitivity of detection. Consequently, HDL were detected in seven samples from HFA mice (Table 3) and revealed that HDL could exist in HFA mice. Hirayama et al. (13) considered that the bacterial balance in the intestines of HFA mice might be controlled by physiological conditions of the mouse intestine and not by the balance of microbes in human feces. HDL in this study were most similar to L. delbrueckii. This bacterium can be established in the murine cecum (21, 38). Therefore, HDL may also be established in the intestines of humans and mice under different physiological conditions. Imaoka et al. (15) improved HFA mice for the evaluation of functional food, and they reported that the HFA mice with segmented filamentous bacteria were able to retain HDL for 14 days. In this study, the HDL were detected at random and regardless of time point or intestinal tract. Our results indicated that the intestines of HFA mice have difficulty in retaining HDL or that HDL are difficult to detect in intestinal samples from HFA mice.
This study showed the dynamics of human intestinal microbiota in formerly germfree mice, and the movement and persistence of many unidentified bacteria were also shown. These results indicated that although HFA mice reflect the composition of individual human intestinal bacteria, there are differences between dominant bacterial populations. Moreover, we revealed that HDL could be established in the HFA mouse intestine. This report showed the intestinal bacteria of HFA mice by the use of molecular techniques in regions and time points, and a new concept could therefore be introduced. The composition of intestinal microbiota of colonized HFA mice was selected from a limited sample of bacteria derived from human inoculum. This finding would be reflected by a host-bacterium interaction, physiological conditions, and diet differences between humans and mice. We need to further the establishment of a suitable model for study and to clarify the details of the interaction between the host and the bacteria based on this research.
Acknowledgments
This work was supported by a grant from the Junior Research Associate Program of RIKEN, Japan.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Apajalahti, J. H. A., H. Kettunen, A. Kettunen, W. E. Holben, P. H. Nurminen, N. Rautonen, and M. Mutanen. 2002. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl. Environ. Microbiol. 68:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clement, B. G., and C. L. Kitts. 2000. Isolating PCR-quality DNA from human feces with a soil DNA kit. BioTechniques 28:640-646. [DOI] [PubMed] [Google Scholar]
- 4.Cole, J. R., B. Chai, T. L. Marsh, et al. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards, C. A., C. Rumney, M. Davies, A. M. Parrett, J. Dore, F. Martin, J. Schmitt, B. Stahl, E. Norin, T. Midtvedt, I. R. Rowland, P. Heavey, H. Köhler, B. Stocks, and H. Schroten. 2003. A human flora-associated rat model of the breast-fed infant gut. J. Pediatr. Gastroenterol. Nutr. 37:168-177. [DOI] [PubMed] [Google Scholar]
- 6.Fujiwara, S., T. Hirota, H. Nakazato, T. Mizutani, and T. Mitsuoka. 1991. Effect of konjac mannan on intestinal microbial metabolism in mice bearing human flora and in conventional F344 rats. Food Chem. Toxicol. 29:601-606. [DOI] [PubMed] [Google Scholar]
- 7.Gérard, P., F. Béguet, P. Lepercq, L. Rigottier-Gois, V. Rochet, C. Andrieux, and C. Juste. 2004. Gnotobiotic rats harboring human intestinal microbiota as a model for studying cholesterol-to-coprostanol conversion. FEMS Microbiol. Ecol. 47:337-343. [DOI] [PubMed] [Google Scholar]
- 8.Godon, J. J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris, M. A., C. A. Reddy, and G. R. Carter. 1976. Anaerobic bacteria from the large intestine of mice. Appl. Environ. Microbiol. 31:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi, H., M. Sakamoto, M. Kitahara, and Y. Benno. 2003. Molecular analysis of fecal microbiota in elderly individuals using 16A rDNA library and T-RFLP. Microbiol. Immunol. 47:557-570. [DOI] [PubMed] [Google Scholar]
- 11.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirayama, K. 1999. Ex-germfree mice harboring intestine microbiota derived from other animal species as an experimental model for ecology and metabolism of intestinal bacteria. Exp. Anim. 48:219-227. [DOI] [PubMed] [Google Scholar]
- 13.Hirayama, K., K. Itoh, E. Takahashi, and T. Mitsuoka. 1995. Comparison of composition of faecal microbiota and metabolism of faecal bacteria among ‘human-flora-associated’ mice inoculated with faeces from six different human donors. Microb. Ecol. Health Dis. 8:199-211. [Google Scholar]
- 14.Hirayama, K., K. Miyaji, S. Kawamura, K. Itoh, E. Takahashi, and T. Mitsuoka. 1995. Development of intestinal flora of human-flora-associated (HFA) mice in the intestine of their offspring. Exp. Anim. 44:219-222. [DOI] [PubMed] [Google Scholar]
- 15.Imaoka, A., H. Satoyama, A. Takagi, S. Matsumoto, and Y. Umesaki. 2004. Improvement of human faecal flora-associated mouse model for evaluation of the functional foods. J. Appl. Mocrobiol. 96:656-663. [DOI] [PubMed] [Google Scholar]
- 16.Itoh, K., T. Mitsuoka, K. Sudo, and K. Suzuki. 1983. Comparison of fecal flora of mice based upon different strains and different housing conditions. Z. Versuchstierkd. 25:135-146. [PubMed] [Google Scholar]
- 17.Kibe, R., M. Sakamoto, H. Hayashi, H. Yokota, and Y. Benno. 2004. Maturation of the murine cecal microbiota as revealed by terminal restriction fragment length polymorphism and 16S rRNA gene clone libraries. FEMS Microbial. Lett. 235:139-146. [DOI] [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. R. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons Ltd., Chichester, United Kingdom.
- 19.Lin, J. H., and D. C. Savage. 1984. Host specificity of the colonization of murine gastric epithelium by lactobacilli. FEMS Microbiol. Lett. 24:67-71. [Google Scholar]
- 20.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McConnell, M. A., and G. W. Tannock. 1991. Lactobacilli and azoreductase activity in the murine cecum. Appl. Environ. Microbiol. 57:3664-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mengoni, A., E. Grassi, and M. Bazzicalupo. 2002. Cloning method for taxonomic interpretation of T-RFLP. BioTechniques 33:990-992. [DOI] [PubMed] [Google Scholar]
- 23.Mitsuoka, T. 1982. Recent trends in research on intestinal flora. Bifidobacteria Microflora 1:3-24. [Google Scholar]
- 24.Morishita, Y., T. Mitsuoka, C. Kaneuchi, S. Yamamoto, and M. Ogata. 1971. Specific establishment of Lactobacilli in the digestive tract of germ-free chickens. Jpn. J. Microbiol. 15:531-538. [DOI] [PubMed] [Google Scholar]
- 25.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 26.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raibaud, P., R. Ducluzeau, F. Dubos, S. Hudault, H. Bewa, and M. C. Muller. 1980. Implantation of bacteria from the digestive tract of man and various animals into gnotobiotic mice. Am. J. Clin. Nutr. 33:2440-2447. [DOI] [PubMed] [Google Scholar]
- 29.Roach, S., and G. W. Tannock. 1980. Anaerobic fusiform shaped bacteria isolated from the caecum of conventional mice. J. Appl. Bacteriol. 48:115-123. [DOI] [PubMed] [Google Scholar]
- 30.Rowland, I. R., and R. Tanaka. 1993. The effects of transgalactosylated oligosaccharides on gut flora metabolism in rats associated with a human faecal microflora. J. Appl. Bacteriol. 74:667-674. [DOI] [PubMed] [Google Scholar]
- 31.Rumney, C. J., and I. R. Rowland. 1992. In vivo and in vitro models of the human colonic flora. Crit. Rev. Food Sci. Nutr. 31:299-331. [DOI] [PubMed] [Google Scholar]
- 32.Sait, L., M. Galic, R. A. Strugnell, and P. H. Janssen. 2003. Secretory antibodies do not affect the composition of bacterial microbiota in the terminal ileum of 10-week-old mice. Appl. Environ. Microbiol. 69:2100-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakamoto, M., H. Hayashi, and Y. Benno. 2003. Terminal restriction fragment length polymorphism analysis for human fecal microbiota and its application for analysis of complex bifidobacterial communities. Microbiol. Immunol. 47:133-142. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto, M., Y. Takeuchi, M. Umeda, I. Ishikawa, and Y. Benno. 2003. Application of terminal RFLP analysis to characterize oral bacterial flora in saliva of healthy subjects and patients with periodontitis. J. Med. Microbiol. 52:79-89. [DOI] [PubMed] [Google Scholar]
- 35.Salzman, N. H., H. de Jong, Y. Paterson, H. J. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148:3651-3660. [DOI] [PubMed] [Google Scholar]
- 36.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 37.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tannock, G. W., M. P. Dashkevicz, and S. D. Feighner. 1989. Lactobacilli and bile salt hydrolase in the murine intestinal tract. Appl. Environ. Microbiol. 55:1848-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tannock, G. W., O. Szylit, Y. Duval, and P. Raibaud. 1982. Colonization of tissue surfaces in the gastrointestinal tract of gnotobiotic animals by lactobacillus strains. Can. J. Microbiol. 28:1196-1198. [DOI] [PubMed] [Google Scholar]
- 40.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong, W. C., D. J. Hentges, and S. H. Dougherty. 1996. Adequacy of the human faecal microbiota associated mouse as a model for studying the ecology of the human intestinal tract. Microb. Ecol. Health Dis. 9:187-198. [Google Scholar]