Abstract
The importance of viruses in marine microbial ecology has been established over the past decade. Specifically, viruses influence bacterial abundance and community composition through lysis and alter bacterial genetic diversity through transduction and lysogenic conversion. By contrast, the abundance and distribution of viruses in soils are almost completely unknown. This study describes the abundance and diversity of autochthonous viruses in six Delaware soils: two agricultural soils, two coastal plain forest soils, and two piedmont forest soils. Viral abundance was measured using epifluorescence microscopy, while viral diversity was assessed from morphological data obtained through transmission electron microscopy. Extracted soil virus communities were dominated by bacteriophages that demonstrated a wide range of capsid diameters (20 nm to 160 nm) and morphologies, including filamentous forms and phages with elongated capsids. The reciprocal Simpson's index suggests that forest soils harbor more diverse assemblages of viruses, particularly in terms of morphological distribution. Repeated extractions of virus-like particles (VLPs) from soils indicated that the initial round of extraction removes approximately 70% of extractable viruses. Higher VLP abundances were observed in forest soils (1.31 × 109 to 4.17 × 109 g−1 dry weight) than in agricultural soils (8.7 × 108 to 1.1 × 109 g−1 dry weight). Soil VLP abundance was significantly correlated to moisture content (r = 0.988) but not to soil texture. Land use (agricultural or forested) was significantly correlated to both bacterial (r = 0.885) and viral (r = 0.812) abundances, as were soil organic matter and water content. Thus, land use is a significant factor influencing viral abundance and diversity in soils.
Historically, the central focus of research concerning viruses in soils has been the fate, transport, and detection of pathogenic viruses exogenous to soils. From this body of work, it is known that factors influencing viral adsorption encompass characteristics of the soil solution, including ionic strength and composition (36, 47, 66), pH (38), and presence of dissolved organic matter (46); characteristics of the virus, such as isoelectric point (16, 23) and hydrophobicity (9); and soil features, such as water content (33), clay and organic matter contents (40), and the presence of organic coatings (65). These findings have been extremely important in understanding the complex interactions between viruses and soil surfaces and in devising means of detecting viruses in soil. However, almost all of this information was gathered using viruses of enteric bacteria or phages otherwise exogenous to soils. Consequently, little is known regarding the behavior and ecology of autochthonous soil viruses.
By contrast, research indicates that viruses are abundant in the world's oceans (105 to 107 ml−1 [10, 27, 59]) and have significant impacts on biogeochemical cycles (8, 11, 25, 42) as well as on marine microbial communities (15, 42, 59). Viruses are even more abundant in marine sediments (109 to 1013 kg−1 [20, 29, 45]). Transmission electron microscopy (TEM) studies have demonstrated that viral communities of marine sediments have higher morphological diversity than those of the water column (41), and recent metagenomic analyses indicated that marine sediments harbored the most diverse assemblage of viruses yet known (12, 13). While viruses appear to control bacterial mortality in sediments (28), the extent and importance of viral activity in sediments are more poorly constrained. Compared to these marine paradigms, however, our understanding of the ecological significance of viruses in soils is extremely limited.
Thus far, studies of soil phage ecology have focused on population dynamics of cultivable soil phages and their hosts (6, 7, 44). While these model systems have provided valuable insights into phage-host interactions in soils, culture-independent detection and analyses will be critical in obtaining a more complete understanding of the ecological impacts of viruses in soils. As recently as 2003, the abundance of autochthonous viruses in soil was unknown (5, 57). Previous assessments of viral abundance in soils were based on determination of PFU by using susceptible indicator strains (64), microscopic enumeration of optically active viruses such as baculoviruses (53), or PCR amplification of viral nucleic acids by using specific primers (43). These approaches are unsatisfactory for determining the abundance of autochthonous soil viruses, since each method targets only a specific fraction of the total viral community.
In the present study, virus abundances in six different Delaware soils were estimated by direct counting of virus-like particles (VLPs) under epifluorescence microscopy (EFM). Viral diversity was determined based on morphological data gathered using TEM. Viral abundance and diversity were compared across soils in order to determine how these data correlate with soil biotic and abiotic factors.
MATERIALS AND METHODS
Sample collection.
Composite 1.5-kg samples were collected from the A horizons of six Delaware soils. The sample set consisted of two agricultural soils planted with corn, two forested Atlantic coastal plain soils, and two forested piedmont soils (Table 1).
TABLE 1.
Global properties of soils
| Soil | Land usea | pHb | OMb,c | % Wd | Sandb | Siltb | Clayb | VLPsf, 107 (SE) | Bacteriaf, 105 (SE) | VBRg | Capsid diamh | Morphological grouph |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Matapeake silt loam | A | 5.9 | 1.7 | 20 | 9 | 75 | 16 | 110 (8.1) | 3.96 (0.22) | 2,750 | 3.46 | 2.86 |
| Evesboro loamy sand | A | 6.1 | 0.1 | 12 | 85 | 12 | 3 | 87 (5.0) | 2.67 (0.43) | 3,346 | 5.70 | 2.44 |
| Elkton silt loam | F | 3.9 | 4.38 | 61 | 31 | 51 | 18 | 294 (13.1) | 2,603 (766) | 11 | 3.51 | 2.84 |
| Woodstown loamy sand | F | 5.4 | 3.91 | 21 | 79 | 14 | 7 | 131 (23) | 2,828 (707) | 4.6 | 2.89 | 3.59 |
| Piedmont wetlande | F | 5.9 | 3.0 | 60 | 59 | 36 | 5 | 417 (120) | 3,376 (1,309) | 12 | 4.75 | 4.12 |
| Piedmont uplande | F | 5.0 | 4.5 | 45 | 60 | 29 | 11 | 148 (19.2) | 1,301 (248) | 11.3 | 6.10 | 3.25 |
A, agricultural usage; F, forested.
Average for duplicate samples; all associated standard errors were within 5%.
Organic matter content on a percent dry weight basis.
Gravimetric water content (average for triplicate subsamples).
Soil is sandy loam by textural class.
Per gram (dry weight) of soil, based on single extractions.
Virus-to-bacterium ratio, based on grand means of VLPs and bacterial direct counts.
Diversity assessed by reciprocal Simpson's index (1/D).
(i) Agricultural soils.
Matapeake silt loam was obtained from the University of Delaware Agricultural Experimental Station, Newark, and Evesboro loamy sand was collected from the University of Delaware Research and Extension Center, Georgetown.
(ii) Coastal plain soils.
Elkton silt loam was collected from a wetland site and Woodstown loamy sand was obtained from an adjacent upland slope in Blackbird Forest, New Castle County.
(iii)Piedmont soils.
The two unclassified piedmont loamy sands were collected from a wetland site and from an adjacent upland slope in White Clay Creek Preserve, New Castle County.
All soils were homogenized and passed through a 4-mm sieve prior to analysis.
Extraction conditions.
Viruses were extracted from triplicate subsamples of each soil as described by Williamson et al. (57). Briefly, 5-g samples of field-moist soil were weighed into 25-ml Teflon-coated polyethylene centrifuge tubes, and 15 ml of one of the following eluants was added: 1% potassium citrate (containing, per liter, 10 g potassium citrate, 1.44 g Na2HPO4 · 7H2O, 0.24 g KH2PO4, pH 7), 10 mM sodium pyrophosphate (pH 7), or 250 mM glycine (pH 8). All tubes were vortexed, sonicated on ice for 3 min (with each minute interrupted by 30 s of manual shaking), and centrifuged at 10, 000 × g to sediment soil particles. Supernatants were passed through 0.20-μm syringe filters to remove bacteria and small soil particles. To assess potential differences in extractability of viruses across soils, sequential extractions were performed on triplicate subsamples of each soil, using 1% potassium citrate. After removing the supernatant from the initial extraction, soil pellets were resuspended in fresh eluant and the extraction procedure was repeated two more times.
Bacteria were extracted from triplicate subsamples of each soil by using methods adapted from those of van Elsas and Smalla (54). Ten grams of moist soil was transferred to a glass bottle containing 95 ml of 1% potassium citrate, representing a 10−1 dilution. Serial 10-fold dilutions were prepared in glass bottles containing sterile 1% potassium citrate. Bottles were manually shaken for 3 min and allowed to settle for 15 seconds prior to each transfer.
EFM. (i) VLPs.
VLPs were enumerated as described by Williamson et al. (57). Aliquots of the 0.2-μm-filtered soil extracts (100 μl) were suspended in 900 μl of sterile deionized water and vacuum filtered (∼50 mm Hg) through a stack of 25-mm filters consisting of a 0.02-μm Anodisc (Whatman International Ltd., Maidstone, England), which was supported by a 0.22-μm Supor (Pall Corporation, Ann Arbor, MI), and a glass fiber filter (Pall Corporation, Ann Arbor, MI). Anodisc filters containing captured virus particles were stained by adding 400 μl of 1× SYBR Gold (Molecular Probes, Eugene, Oregon) directly to the filter on the vacuum manifold. Filters were incubated for 15 min in the dark and analyzed by EFM using a Zeiss Axioskop2 microscope (Carl Zeiss Microimaging, Inc., Thornwood, NY) with a fluorescein isothiocyanate excitation filter. Ten fields per sample were digitally photographed at a magnification of ×1,000 using a Hamamatsu ORCA-ER camera (Hamamatsu Corporation, Bridgewood, NJ). Virus-like particles were counted using the Fovea Pro plug-in for Adobe Photoshop. VLPs were discriminated from bacteria or detritus based on pixel dimensions: the pixel area of the smallest known bacterium was established as a maximum cutoff; all objects smaller than this were counted as VLPs. The percentage of viruses eluted during a given extraction in a sequence is given by (VN/VT) × 100, where VN is the average number of VLPs counted from extraction N and VT = Σ(VN). Average VLP counts were calculated based on the grand mean for three replicate filters per soil.
(ii) Bacteria.
Ten-milliliter portions of 10−4 soil dilutions were filtered through a stack of 25-mm filters consisting of a 0.22-μm black polycarbonate filter (Pall Corporation, Ann Arbor, MI), which was supported by a 0.22-μm Supor (Pall Corporation, Ann Arbor, MI), and a glass fiber filter (Pall Corporation, Ann Arbor, MI). Filters were stained with SYBR Gold as described above, and bacteria were enumerated under EFM by manually counting cells in 40 fields per filter at a magnification of ×1,000. Average bacterial counts were calculated based on a grand mean of three replicate filters per soil.
TEM.
Six 50-g subsamples of each soil were extracted once with 1% potassium citrate as described above. Supernatants were pooled and purified via CsCl density gradient centrifugation (48). Aliquots (100 μl) of purified virus extracts were suspended in 900 μl of deionized water and spun down onto Formvar-coated 650 copper mesh grids as described by Wommack et al. (60). Grids were stained with 1% uranyl acetate for 1 min and examined at a magnification of ×85, 000 in a Zeiss CEM 902 transmission electron microscope (Carl Zeiss Microimaging, Inc., Thornwood, NY). Four hundred fifty individual virus particles were observed on triplicate grids from each soil and categorized by morphotype according to International Committee on Taxonomy of Viruses guidelines (14) (tailless [e.g., φX174], Podoviridae [e.g., T7], Siphoviridae [e.g., λ], Myoviridae [e.g., T4], and filamentous [e.g., M13]). An additional “elongate” category was created for head-tail (i.e., all but filamentous) phages with elongated capsids. All nonfilamentous phage particles were additionally categorized according to capsid diameter measurements in the width dimension.
Statistical analyses.
Diversity of soil virus communities was evaluated based on distribution of capsid diameters and morphological group, using the reciprocal Simpson's index (39). Analysis of variance (ANOVA) was performed using Prism 4 (GraphPad Software, Inc., San Diego, CA) to test for differences in bacterial and VLP counts across soils based on single extractions and for differences in VLP abundance among multiple virus extractions. Pearson's product-moment correlation was used to test for linear correlation between VLP abundance, viral diversity, bacterial abundance, and soil physical factors, using SPSS 11.0 (SPSS, Inc., Chicago, IL).
RESULTS
Multiple extractions of VLPs from soil.
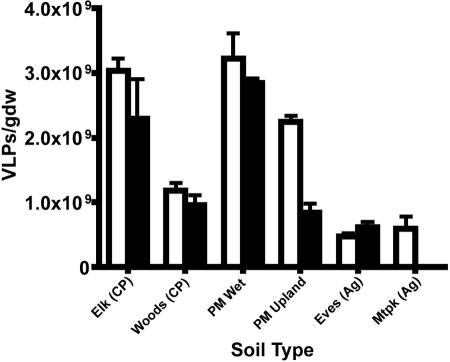
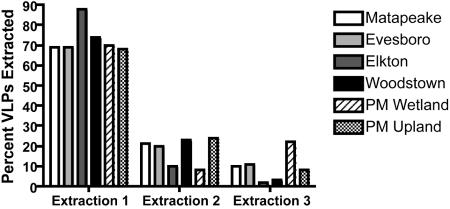
Previous experiments comparing EFM and TEM counts of VLPs extracted from soils, as well as the effects of DNase on EFM counts, confirmed that particles appearing under EFM are viruses or at least are virus-like (57). Glycine buffer proved incompatible with every soil in this sample set except the Evesboro loamy sand. EFM micrographs of glycine-extracted VLPs contained extremely high background fluorescence, which prohibited enumeration of VLPs. Potassium citrate (1%) yielded slightly higher numbers of VLPs (Fig. 1), as well as clearer micrographs than did 10 mM sodium pyrophosphate; thus, 1% potassium citrate was used for the remainder of the VLP extractions in this study. When soil samples were sequentially extracted three times, approximately 70% of the VLPs were eluted in the first extraction, 20% were eluted in the second extraction, and 10% were eluted in the third extraction (Fig. 2). This trend was consistent over all six soils used in this study. To confirm these observations, triplicate samples of piedmont wetland soil were sequentially extracted seven times. The first three extractions removed 64.7%, 23.4%, and 10.7% of total extractable VLPs, respectively. By the fourth extraction, fewer than 1% of the extractable VLPs remained in the soil pellet (data not shown). Results of sequential extractions indicate that measures of VLP abundance based on single extractions are representative of total extractable VLPs for the soils used in this study.
FIG. 1.
Abundance of virus-like particles per gram dry weight (gdw) in six Delaware soils based on single extractions with 1% potassium citrate (white bars) or 10 mM sodium pyrophosphate (black bars). Soils used were Elkton sandy loam (Elk), Woodstown loamy sand (Woods), piedmont wetland soil (PM Wet), piedmont upland soil (PM Upland), Evesboro loamy sand (Eves), and Matapeake silt loam (Mtpk). CP, coastal plain; Ag, agricultural. Bars represent standard errors.
FIG. 2.
Viral abundances in sequential soil extractions. Virus-like particles were extracted from six Delaware soils with 1% potassium citrate. The average percent VLPs extracted, based on triplicate subsamples, is shown; the sum of three extractions equals 100%.
VLP abundance in soils based on single extractions.
Based on single extractions with 1% potassium citrate, the abundance of VLPs was highest in the wetland forest soils (P < 0.01 for piedmont wetland soil and Elkton silt loam). Upland forest soils (piedmont upland and Woodstown loamy sand) contained more VLPs than the agricultural soils (P < 0.01 for Matapeake silt loam and Evesboro loamy sand) (Table 1; Fig. 1). Bivariate analysis of the data in Table 1 by Pearson product-moment correlation revealed that VLP abundance and soil water content were significantly correlated (Table 2). A strong, but not statistically significant, correlation was observed between bacterial abundance and VLP abundance (r = 0.782). Weaker correlations were identified between VLP abundance and soil organic matter content and between VLP abundance and soil pH. For this sample set, higher VLP abundances were observed in wetter, forested soils and lower VLP abundances were observed in drier, agricultural soils. No significant correlations were found between VLP abundance and soil texture (sand, silt, or clay content).
TABLE 2.
Two-tailed Pearson correlations between viral abundance, diversity, and soil physical factors
| Parameter | Correlationa with:
|
|||||
|---|---|---|---|---|---|---|
| Morphological group (1/D) | Capsid diam (1/D) | % Wb | % OMc | pH | Land used | |
| VLP abundance | 0.563 | 0.063 | 0.988** | 0.687 | −0.571 | 0.812* |
| % OM | 0.480 | −0.212 | 0.665 | 1.000 | −0.758 | 0.911* |
| Bacterial abundance | 0.790 | −0.326 | 0.699 | 0.686 | −0.393 | 0.885* |
*, significant at 95% confidence interval; **, significant at 99% confidence interval.
Gravimetric water content.
Organic matter content on a dry weight basis.
Land use was denoted as either agricultural or forested.
Virus-to-bacterium ratios (VBRs) were extremely high in the agricultural soils (ca. 3,000), while the forested soils had VBRs of about 10 (Table 1). All VBRs in this study were higher than those previously reported for soils or sediments (37).
Bacterial abundance in soils.
The bacterial abundance was significantly lower (P < 0.01) in the agricultural soils (Matapeake and Evesboro) than in the forest soils (Table 1). Bivariate analysis by Pearson product-moment correlation indicated that bacterial abundance was significantly correlated to land use (whether soil was agricultural or forested) but not to soil water content (Table 2). A moderate correlation was identified between bacterial abundance and soil organic matter content. Bacterial abundance and VLP abundance demonstrated similar trends with respect to soil water content, land use, and organic matter content, although soil water content showed a stronger correlation to VLP abundance than to bacterial abundance. As with VLP abundance, no significant correlation was found between bacterial abundance and soil texture.
Viral diversity.
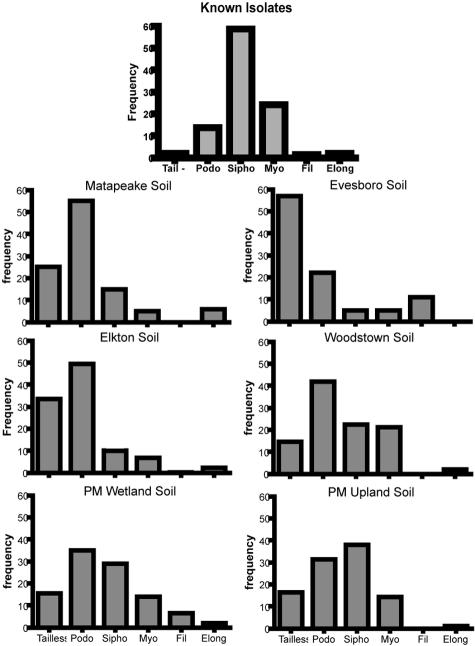
A cumulative diversity curve, which estimates the sample size needed to adequately represent population diversity, was constructed for viruses extracted from piedmont wetland soil (data not shown). The estimated minimum sample size was 400 individuals; thus, 450 individual virus particles were examined from each soil. Using the reciprocal Simpson's index (1/D), capsid size diversity was highest in the virus community sampled from piedmont upland soil and did not appear to follow any distinct trend across soils (Table 1). No significant Pearson product-moment correlations were found between capsid size diversity and VLP abundance or soil physical factors (Table 2). The distribution of virus morphotypes across soils indicates that the majority of soil viruses are bacteriophages belonging to the order Caudovirales (Fig. 3). Furthermore, the morphological distribution of soil virus communities is much different than that observed among some 4,950 known phage isolates (1). Filamentous viruses and viruses with elongated capsids were observed at a much higher frequency in autochthonous soil virus communities than among cultured isolates (Fig. 3). Morphological diversity was highest in the virus community sampled from piedmont wetland soil. For the soils in this study, viral morphological diversity was higher in forested soils than in agricultural soils (Table 1). A weak relationship was indicated by Pearson product-moment correlation between viral morphological diversity and VLP abundance (Table 2).
FIG. 3.
Frequency distributions of viral morphologies from six Delaware soils and known phage isolates. Categories include tailless (Tail−), Podoviridae (Podo), Siphoviridae (Sipho), Myoviridae (Myo), filamentous (Fil), and elongate (Elong). The elongate category is a subset of all icosahedral morphologies except filamentous. Histograms are based on measurements of 450 individual viruses per soil. Morphological data for known isolates are from reference 1.
DISCUSSION
Viral and bacterial abundances.
Sequential extractions were performed on triplicate samples of each soil to determine whether differences in measured VLP abundance across soils were due to true differences in abundance or merely to differences in VLP extraction efficiency from a particular soil. Approximately 70% of the total extractable VLPs were eluted during the first round of extraction, regardless of soil type (Fig. 2). Since the initial extraction appeared to be representative of true VLP abundance for the soils used in this study, differences across soils were evaluated based on single extractions.
The highest VLP abundances were observed in the wetland soil samples (piedmont wetland and Elkton silt loam) (Table 1). Since these soils also harbored the highest numbers of bacteria, it is tempting to conclude that a higher cell density resulted in a larger fraction of infected cells releasing viruses. However, it remains unclear whether more viruses were actively produced in these soils or whether viruses merely persist longer in these soils due to physical and/or biological factors.
Previous studies examining survival of enteric viruses in soil indicate that intact virions persist longer in wetter soils (32, 52, 55, 63). Increased virus survival times in wet soils are likely due to thicker water films and smaller interfacial areas (air-water interfaces or triple phase boundaries), since such interfaces are known to inactivate virions (26, 33, 66). The Pearson product-moment correlation between VLP abundance and soil water content was highly significant (Table 2); thus, soil water is an important factor contributing to measured VLP abundance in the soils in this study. Soil organic matter content was moderately correlated with VLP abundance, likely due to the water-holding capacity that organic matter imparts to soil (30), although organic matter may also play an independent role in virus survival in soils (26, 65). Another potential factor contributing to the high abundance of VLPs in wetland soils is their lowland topography. In addition to viruses being produced in situ, exogenous viruses may be transported in runoff from upland sites during rainfall events.
Higher VLP abundances may have been observed for forested soils in general because viruses persist longer at cooler temperatures (32, 52, 63). Canopy cover from trees and ground cover from leaf litter results in a consistently lower soil temperature than for bare soil or low crop cover encountered in agricultural fields. Forested upland soils contained fewer VLPs than the wetland soils but more VLPs than the agricultural soils (Table 1). Comparisons of the interplay between virus survival, soil water content, and soil temperature indicate that the intermediate VLP abundances of the upland forest soils could be due to the fact that these soils were cooler than the agricultural soils but drier than the wetland soils.
The observation of lower VLP abundance in managed agricultural soils mirrors previous reports correlating tillage practices and soil organic matter with microbial abundance (3, 4, 24). In the present study, bacterial abundance was significantly lower and the virus-to-bacterium ratio was significantly higher in the agricultural soils than in the forested soils (P < 0.01) (Table 1). Since the majority of these viruses are likely bacteriophages, it is not surprising that land use practices should affect soil viral and bacterial populations in similar ways. While tillage is not the only plausible explanation for lower virus abundance in the agricultural soils (i.e., the significant correlation between virus abundance and soil water content [Table 2]), it should be noted that levels of both viruses and bacteria were significantly higher (P < 0.05) in Woodstown soil (forested) than in Matapeake soil (agricultural), even though both soils had similar water contents (Table 1).
The VBR (37) has been used as a metric for determining the overall significance of free viruses within a particular ecosystem (18, 22, 62) and for interpreting the metabolic state of host bacteria (21). Typically, the VBR of marine waters is about 10 (59), while the VBRs of soil and marine sediments have been closer to 1 (20, 21, 37). However, it is unclear whether the lower VBRs of soil and sediments are due to the higher extraction efficiency of viruses relative to bacteria or to a habitat-dependent shift in VBR relative to aquatic systems. Measures of VBR in this study reveal striking differences between the agricultural and forested soils: the VBRs for both agricultural soils are extremely high, while the VBRs for the four forested soils are much lower (Table 1). This could mean that soil viral communities are less adversely affected by tillage than coexisting bacterial communities. However, the extremely high VBRs in agricultural soils owe more to low bacterial abundance than to high viral abundance; thus, it seems more likely that bacteria are simply more difficult to efficiently extract from agricultural soils, leading to highly variable VBR measurements.
In attempting to explain differences in VLP abundance across the six Delaware soils, it is difficult to clearly distinguish between biotic and abiotic factors affecting these measurements. Among biotic factors, the effects of bacterial community activity, abundance, and diversity on phage production and survival in soils are of particular interest. Conflicting results have been obtained in studies comparing survival rates of enteric viruses and bacteriophages in sterile and nonsterile soil microcosms. In some cases, the presence of bacteria decreased virus survival, likely due to production of proteolytic enzymes which degraded viral capsids (31, 63). However, in at least one other case, the presence of bacteria prolonged virus survival in soil relative to sterile controls (44). Currently, there are no estimates of viral production rates in soil. If the majority of autochthonous viruses in soil are bacteriophages, then bacterial abundance and activity should have a direct and proportional bearing on viral production. Since tillage is known to reduce soil bacterial abundance, bacterial diversity, and soil organic matter content (which also contributes significantly to soil water content), the collective data in this study implicate land management practices as an important underlying factor controlling viral abundance in soils.
Viral diversity.
Current methods for assessing genetic diversity of prokaryotes exploit sequence polymorphism in conserved genes, such as 16S rRNA genes. Thus, a census based on DNA sequences of even unculturable bacteria can be obtained from environmental samples. Unfortunately, such an approach is untenable in assessing global viral diversity in the environment, since virus genomes do not contain a single, universally conserved marker gene. However, some researchers have been able to use genes conserved across viral groups, such as those for DNA polymerase (17), RNA-dependent RNA polymerase (19), and the portal protein (56), to explore genetic diversity among members of a specific group. Even without knowledge of specific conserved sequences, random primers may be used to generate community DNA fingerprints (i.e., randomly amplified polymorphic DNA) that provide a measure of viral diversity for comparative studies (58). Outside the realm of PCR-based techniques, pulsed-field gel electrophoresis (PFGE) has been a useful method for fingerprinting viral communities and monitoring community-level changes in rumen (35) and in marine environments (50, 51, 61). Viral diversity has also been quantified through morphological measurements taken using TEM (2, 34, 49). The requirement of clean preparations of viral DNA for molecular techniques such as PCR or PFGE poses significant challenges to the study of viruses in soils. For the time being, such approaches demand further optimization and method development.
The distribution of morphotypes observed under TEM indicated distinct differences among the viral communities of the six soils (Fig. 3). More significantly, morphological distribution revealed that viral communities of all six soils were dominated by bacteriophages, based on the presence of tailed viruses (2). Phages with elongated capsids were observed in all soils except Evesboro loamy sand. In particular, approximately 10% of the viral community in Matapeake silt loam consisted of elongate-capsid phages. This morphotype has been rarely reported among environmental samples and comprises fewer than 2% of known phages (1). Filamentous viruses are another rarely reported morphotype; however, filamentous VLPs were seen in five of the six soils examined in this study (Fig. 3). Based on these observations, it is apparent that phages in culture collections probably do not reflect the true diversity of autochthonous soil phages.
Phage particles were additionally categorized on the basis of capsid diameter (data not shown). While the mean capsid diameter was the same for all six phage communities (ca. 50 nm), the distribution about this mean was characteristic of each individual phage community. Using the reciprocal Simpson's index to gauge viral diversity, the four forested soils had the greatest diversity in terms of morphology, while no consistent trend was observed among the six different soils in terms of capsid diameter (Table 1). No significant correlations were found between viral capsid size diversity and other soil biotic or abiotic factors. However, a moderate correlation was observed between bacterial abundance and viral morphological diversity (Table 2), lending additional support to the conclusion that most autochthonous soil viruses are bacteriophages.
In the analysis of phage morphology under TEM, one major concern is the potential miscategorization of phages due to tail breakage. The aggressive extraction procedure may have resulted in truncation or loss of phage tails, resulting in histograms which are not representative of the true morphological diversity. For example, the apparent dominance of tailless phages observed in Evesboro loamy sand might simply be an artifact resulting from a higher incidence of phage tail breakage during extraction from that particular soil (Fig. 3). Thus, future work will aim to quantify the extent of phage tail breakage in extracting and preparing soil viral extracts for TEM. In addition, research will focus on developing protocols for the application of PCR- and PFGE-based analyses to the study of soil viral ecology.
Acknowledgments
This research was funded by a U.S. EPA Science to Achieve Results (STAR) graduate fellowship.
We thank Kirk Czymmek and Debbie Powell, Delaware Biotechnology Institute Bio-Imaging Center, for their patient aid in electron microscopy and Bruce Vasilas, University of Delaware Department of Plant and Soil Sciences, for indispensable assistance in soil sampling and characterization.
REFERENCES
- 1.Ackermann, H. W. 2001. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 146:843-857. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann, H. W. 1998. Tailed bacteriophages: the order caudovirales. Adv. Virus Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aon, M. A., D. E. Sarena, J. L. Burgos, and S. Cortassa. 2001. Interaction between gas exchange rates, physical and microbiological properties in soils recently subjected to agriculture. Soil Tillage Research. 60:163-171. [Google Scholar]
- 4.Arias, R. S., F. A. Galizzi, M. A. Sagardoy, N. Peinemann, and A. Ares. 1998. Bacteria related to the nitrogen cycle in salt-affected soils of Argentina. J. Basic Microbiol. 38:159-171. [Google Scholar]
- 5.Ashelford, K. E., M. J. Day, and J. C. Fry. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashelford, K. E., J. C. Fry, M. J. Bailey, A. R. Jeffries, and M. J. Day. 1999. Characterization of six bacteriophages of Serratia liquefaciens CP6 isolated from the sugar beet phytosphere. Appl. Environ. Microbiol. 65:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashelford, K. E., S. J. Norris, J. C. Fry, M. J. Bailey, and M. J. Day. 2000. Seasonal population dynamics and interactions of competing bacteriophages and their host in the rhizosphere. Appl. Environ. Microbiol. 66:4193-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azam, F., T. Fenchel, J. G. Field, J. S. Gray, L. A. Meyer Reil, and F. Thingstad. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263. [Google Scholar]
- 9.Bales, R. C., S. M. Li, K. M. Maguire, M. T. Yahya, and C. P. Gerba. 1993. Ms-2 and poliovirus transport in porous media: hydrophobic effects and chemical perturbations. Water Resource Res. 29:957-963. [Google Scholar]
- 10.Borsheim, K. Y. 1993. Native marine bacteriophages. FEMS Microbiol. Ecol. 102:141-159. [Google Scholar]
- 11.Bratbak, G., F. Thingstad, and M. Heldal. 1994. Viruses and the microbial loop. Microb. Ecol. 28:209-221. [DOI] [PubMed] [Google Scholar]
- 12.Breitbart, M., B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. R. Soc. London Ser. B 271:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchen-Osmond, C. 2000, posting date. International Committee on Taxonomy of Viruses: list of criteria demarcating different virus taxa. [Online.] www.ncbi.nlm.nih.gov/ICTVdb/Ictv/index.htm.
- 15.Buckling, A., and P. B. Rainey. 2002. The role of parasites in sympatric and allopatric host diversification. Nature 420:496-499. [DOI] [PubMed] [Google Scholar]
- 16.Chattopadhyay, S., and R. W. Puls. 2000. Forces dictating colloidal interactions between viruses and soil. Chemosphere 41:1279-1286. [DOI] [PubMed] [Google Scholar]
- 17.Chen, F., C. A. Suttle, and S. M. Short. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ. Microbiol. 62:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corinaldesi, C., E. Crevatin, P. Del Negro, M. Marini, A. Russo, S. Fonda-Umani, and R. Danovaro. 2003. Large-scale spatial distribution of virioplankton in the Adriatic Sea: testing the trophic state control hypothesis. Appl. Environ. Microbiol. 69:2664-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Culley, A. I., A. S. Lang, and C. A. Suttle. 2003. High diversity of unknown picorna-like viruses in the sea. Nature 424:1054-1057. [DOI] [PubMed] [Google Scholar]
- 20.Danovaro, R., A. Dell'anno, A. Trucco, M. Serresi, and S. Vanucci. 2001. Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 67:1384-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danovaro, R., E. Manini, and A. Dell'Anno. 2002. Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl. Environ. Microbiol. 68:1468-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danovaro, R., and M. Serresi. 2000. Viral density and virus-to-bacterium ratio in deep-sea sediments of the Eastern Mediterranean. Appl. Environ. Microbiol. 66:1857-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowd, S. E., S. D. Pillai, S. Y. Wang, and M. Y. Corapcioglu. 1998. Delineating the specific influence of virus isoelectric point and size on virus adsorption and transport through sandy soils. Appl. Environ. Microbiol. 64:405-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey, S. D., E. T. Elliott, and K. Paustian. 1999. Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol. Biochem. 31:573-585. [Google Scholar]
- 25.Fuhrman, J. 1992. Bacterioplankton roles in cycling of organic matter: the microbial food web, p. 361-383. In P. G. Falkowski and A. D. Woodhead (ed.), Primary productivity and biogeochemical cycles in the sea. Plenum Press, New York, N.Y.
- 26.Gerba, C. P. 1984. Applied and theoretical aspects of virus adsorption to surfaces. Adv. Appl. Microbiol. 30:133-168. [DOI] [PubMed] [Google Scholar]
- 27.Heldal, M., and G. Bratbak. 1991. Production and decay of viruses in aquatic environments. Mar. Ecol. Prog. Ser. 72:205-212. [Google Scholar]
- 28.Hewson, I., and J. A. Fuhrman. 2003. Viriobenthos production and virioplankton sorptive scavenging by suspended sediment particles in coastal and pelagic waters. Microb. Ecol. 46:337-347. [DOI] [PubMed] [Google Scholar]
- 29.Hewson, I., J. M. O'Neil, J. A. Fuhrman, and W. C. Dennison. 2001. Virus-like particle distribution and abundance in sediments and overlying waters along eutrophication gradients in two subtropical estuaries. Limnol. Oceanogr. 46:1734-1746. [Google Scholar]
- 30.Hillel, D. 1998. Environmental soil physics. Harcourt Brace and Company, San Diego, Calif.
- 31.Hurst, C. J. 1988. Influence of aerobic microorganisms upon virus survival in soil. Can. J. Microbiol. 34:696-699. [DOI] [PubMed] [Google Scholar]
- 32.Hurst, C. J., C. P. Gerba, and I. Cech. 1980. Effects of environmental variables and soil characteristics on virus survival in soil. Appl. Environ. Microbiol. 40:1067-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin, Y., and M. Flury. 2002. Fate and transport of viruses in porous media. Adv. Agron. 77:39-51. [Google Scholar]
- 34.Klieve, A. V., and T. Bauchop. 1988. Morphological diversity of ruminal bacteriophages from sheep and cattle. Appl. Environ. Microbiol. 54:1637-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klieve, A. V., and R. A. Swain. 1993. Estimation of ruminal bacteriophage numbers by pulsed-field gel electrophoresis and laser densitometry. Appl. Environ. Microbiol. 59:2299-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lance, J. C., and C. P. Gerba. 1984. Effect of ionic composition of suspending solution on virus adsorption by a soil column. Appl. Environ. Microbiol. 47:484-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemke, M. J., C. E. Wickstrom, and L. G. Leff. 1997. A preliminary study on the distribution of viruses and bacteria in lotic habitats. Arch. Hydrobiol. 141:67-74. [Google Scholar]
- 38.Loveland, J., J. N. Ryan, G. L. Amy, and R. W. Harvey. 1996. The reversibility of virus atachment to mineral surfaces. Colloid Surface A 107:205-221. [Google Scholar]
- 39.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, N.J.
- 40.Meschke, J. S., and M. D. Sobsey. 2003. Comparative reduction of Norwalk virus, poliovirus type 1, F+ RNA coliphage MS2 and Escherichia coli in miniature soil columns. Water Sci. Technol. 47:85-90. [PubMed] [Google Scholar]
- 41.Middelboe, M., R. N. Glud, and K. Finster. 2003. Distribution of viruses and bacteria in relation to diagenetic activity in an estuarine sediment. Limnol. Oceanogr. 48:1447-1456. [Google Scholar]
- 42.Middelboe, M., N. O. G. Jorgensen, and N. Kroer. 1996. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl. Environ. Microbiol. 62:1991-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monpoeho, S., A. Maul, B. Mignotte-Cadiergues, L. Schwartzbrod, S. Billaudel, and V. Ferre. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pantastico-Caldas, M., K. E. Duncan, C. A. Istock, and J. A. Bell. 1992. Population dynamics of bacteriophage and Bacillus subtilis in soil. Ecology 73:1888-1902. [Google Scholar]
- 45.Paul, J. H., J. B. Rose, S. C. Jiang, C. A. Kellogg, and L. Dickson. 1993. Distribution of viral abundance in the reef environment of Key Largo, Florida. Appl. Environ. Microbiol. 59:718-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powelson, D. K., J. R. Simpson, and C. P. Gerba. 1991. Effects of organic matter on virus transport in unsaturated flow. Appl. Environ. Microbiol. 57:2192-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quignon, F., F. Thomas, C. Gantzer, A. Huyard, and L. Schwartzbrod. 1998. Virus adsorption in a complex system: an experimentally designed study. Water Res. 32:1222-1230. [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sharma, R. S., A. Mohmmed, and C. R. Babu. 2002. Diversity among rhizobiophages from rhizospheres of legumes inhabiting three ecogeographical regions of India. Soil Biol. Biochem. 34:965-973. [Google Scholar]
- 50.Steward, G. F., and F. Azam. 1998. Presented at the Eighth International Symposium on Microbial Ecology, Halifax, Canada.
- 51.Steward, G. F., J. L. Montiel, and F. Azam. 2000. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol. Oceanogr. 45:1697-1706. [Google Scholar]
- 52.Straub, T. M., I. L. Pepper, and C. P. Gerba. 1993. Virus survival in sewage-sludge amended desert soil. Water Sci. Technol. 27:421-424. [Google Scholar]
- 53.Taverner, M. P., and E. F. Connor. 1992. Optical enumeration technique for detection of baculoviruses in the environment. Environ. Entomol. 21:307-313. [Google Scholar]
- 54.van Elsas, J. D., and K. Smalla. 1997. Methods for sampling soil microbes, p. 383-390. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental micobiology. ASM Press, Washington, D.C.
- 55.Vaughn, J. M., and Landry, E. F. 1983. Viruses in soils and groundwaters., p. 163-210. In G. Berg (ed.), Viral pollution of the environment. CRC Press, Inc., Boca Raton, Fla.
- 56.Wang, K., and F. Chen. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microb. Ecol. 34:105-116. [Google Scholar]
- 57.Williamson, K. E., K. E. Wommack, and M. Radosevich. 2003. Sampling natural viral communities from soil for culture-independent analyses. Appl. Environ. Microbiol. 69:6628-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winget, D., and K. E. Wommack. 2003. Presented at the 103rd General Meeting of the American Society for Microbiology, Washington, D.C.
- 59.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wommack, K. E., R. T. Hill, M. Kessel, E. Russek-Cohen, and R. R. Colwell. 1992. Distribution of viruses in the Chesapeake Bay. Appl. Environ. Microbiol. 58:2965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wommack, K. E., J. Ravel, R. T. Hill, and R. R. Colwell. 1999. Population dynamics of Chesapeake Bay virioplankton: total community analysis using pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wommack, K. E., J. Ravel, R. T. Hill, and R. R. Colwell. 1997. Presented at the 97th General Meeting of the American Society for Microbiology 1997, Miami Beach, Fla.
- 63.Yates, M. V., L. D. Stetzenbach, C. P. Gerba, and N. A. Sinclair. 1990. The effect of indigenous bacteria on virus survival in ground-water. J. Environ. Sci. Health Part A 25:81-100. [Google Scholar]
- 64.Yin, X., L. R. Zeph, and G. Stotzky. 1997. A simple method for enumerating bacteriophages in soil. Can. J. Microbiol. 43:461-466. [DOI] [PubMed] [Google Scholar]
- 65.Zhuang, J., and Y. Jin. 2003. Virus retention and transport as influenced by different forms of soil organic matter. J. Environ. Qual. 32:816-823. [DOI] [PubMed] [Google Scholar]
- 66.Zhuang, J., and Y. Jin. 2003. Virus retention and transport through Al-oxide coated sand columns: effects of ionic strength and composition. J. Contam. Hydrol. 60:193-209. [DOI] [PubMed] [Google Scholar]