Abstract
The genome for the marine pseudotemperate member of the Siphoviridae φHSIC has been sequenced using a combination of linker amplification library construction, restriction digest library construction, and primer walking. φHSIC enters into a pseudolysogenic relationship with its host, Listonella pelagia, characterized by sigmoidal growth curves producing >109 cells/ml and >1011 phage/ml. The genome (37,966 bp; G+C content, 44%) contained 47 putative open reading frames (ORFs), 17 of which had significant BLASTP hits in GenBank, including a β subunit of DNA polymerase III, a helicase, a helicase-like subunit of a resolvasome complex, a terminase, a tail tape measure protein, several phage-like structural proteins, and 1 ORF that may assist in host pathogenicity (an ADP ribosyltransferase). The genome was circularly permuted, with no physical ends detected by sequencing or restriction enzyme digestion analysis, and lacked a cos site. This evidence is consistent with a headful packaging mechanism similar to that of Salmonella phage P22 and Shigella phage Sf6. Because none of the phage-like ORFs were closely related to any existing phage sequences in GenBank (i.e., none more than 62% identical and most <25% identical at the amino acid level), φHSIC is unique among phages that have been sequenced to date. These results further emphasize the need to sequence phages from the marine environment, perhaps the largest reservoir of untapped genetic information.
Phages are now recognized as the most abundant form of life on this planet, with their total numbers estimated at 1031 in the biosphere (43). Undoubtedly the largest reservoir of viruses is the oceans, where they range in concentration from 104 to over 107/ml in the water column, and in excess of 108/cm3 in the sediment (6, 40, 53). Although viruses can modulate the abundance and diversity of microbial populations in the oceans through their lytic activity, their greatest role may be in silent viral infections resulting from lysogeny (24, 25, 36, 49, 50, 51, 52, 53). Lysogeny has the potential to alter microbial phenotypes through conversion. Currently, lysogeny is detectable in oceanic populations only through prophage induction by the addition of DNA-damaging agents (38). Preliminary evidence suggests that environmental factors may control the lysogenic switch (i.e., the decision between lytic and lysogenic gene expression) in marine microbial populations (23, 32, 52).
By the definition of Ackermann and DuBow (1), pseudolysogeny occurs when there is simultaneous occurrence of high numbers of phages and hosts in the same culture. Ackermann and DuBow attributed this to a mixture of either sensitive and resistant host cells or virulent and temperate phage. In many ways, most marine environments behave like cultures of pseudolysogens in that they are combinations of sensitive and resistant hosts as well as temperate and virulent phages. However, other definitions of pseudolysogeny have been put forward, such as a condition of starved cells, whereby phage-infected cells lack sufficient energy to lysogenize or have a lytic infection (42). Moebus (33) described a phage host system which simultaneously produced high phage titers and had unimpeded host cell growth, resulting in sigmoidal growth curves, attributed to partially resistant host cells.
We have previously isolated φHSIC and its host, a Listonella pelagia strain, from ocean waters near Oahu, Hawaii (25). Members of this taxon, formerly known as Vibrio pelagius, are fish pathogens that have been shown to produce tetrodotoxin (45) and piscicidal extracellular proteases (16) and to cause massive fish mortality in aquaculture facilities (48). We have previously described the interaction of φHSIC with its host as a relatively unstable pseudolysogenic relationship, which yielded >1011 phage/ml while enabling host growth to >2 × 109 cells/ml in broth cultures (51). The pseudolysogen is not sensitive to φHSIC in plaque agar overlays, nor can additional prophages be induced by mitomycin C treatment (51). Probing of Southern transfers of lysogenized host DNA indicated that the phage genome appeared to be integrated into the host genome, with a portion of the hybridizing DNA in a plasmid-sized fraction comigrating with undigested viral DNA (51). Because there is a dearth of sequence information on marine phages and, to our knowledge, no sequence information for any marine pseudotemperate phage, we have sequenced and analyzed the φHSIC genome.
MATERIALS AND METHODS
Host and phage.
The isolation and cultivation conditions of phage φHSIC and its host, Listonella pelagia, have been described elsewhere (25, 51). For genomic sequencing, a single plaque from an agar overlay was selected and propagated on the host in liquid culture. Liquid culture was harvested at 10,000 × g, the supernatant was collected and 0.2-μm filtered, and viral DNA was purified by use of the Wizard Lambda Prep DNA purification kit (Promega, Madison, WI). The quality of the DNA was assessed by agarose gel electrophoresis, and DNA preparations were quantified by the Hoechst 33258 fluorometric method of Paul and Myers (39).
Library construction.
Restriction digestion and cloning using the pBluescript vector (Stratagene) were initially employed to generate a small number of clones for sequencing. A linker-amplified shotgun library with upwards of a million clones was constructed in the pSMART vector as described elsewhere (http://www.sci.sdsu.edu/PHAGE/LASL/) at Lucigen Corp. (Middleton, WI).
Sequencing.
Sequencing of the restriction library clones and primer-walking efforts were performed in an Applied Biosystems model 373 sequencer by the University of Florida core sequencing laboratory (University of Florida, Gainesville). Sequencing of inserts from the linker-amplified shotgun library (http://www.sci.sdsu.edu/PHAGE/LASL/) was performed using BigDye v.3.1 chemistry from ABI and analyzed on an ABI 3100 capillary sequencer at the San Diego State University Microchemical Core Facility (San Diego, CA).
Sequence analysis.
Sequencing reads were trimmed of vector using Sequencher 3.0 (Genecodes, Ann Arbor, MI) and further trimmed manually after chromatograms were inspected for ambiguities. Trimmed and edited sequences were assembled using Sequencher 3.0. Additional ambiguities were corrected by examining chromatograms, and a consensus for base calling was determined. Six rounds of primer walking were necessary to close the sequence. Closure was determined when one contig remained and further primer walking from one end of the contig yielded a sequence identical to the other end of the contig. The average coverage was 5.2-fold. Open reading frames (ORFs) were defined by use of KODON software (Applied Maths, Austin, TX) and ORF Finder software from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) using the bacterial code option. Putative ORFs were analyzed using BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi). Annotation was performed using the KODON software (Applied Maths, Austin, TX).
Protein analysis by mass spectroscopy.
Phage proteins were separated on a one-dimensional 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel run at 5 mA for 16 h essentially by the method of Laemmli (29). Gels were fixed and stained in methanol-acetic acid-water (50:10:40) using Coomassie brilliant blue R250 (Bio-Rad, Hercules, CA). Protein-containing bands were excised from the gel with a scalpel. The excised gel slices were subjected to in-gel trypsin digestion using the In-Gel Tryptic Digestion kit (Pierce) as per the manufacturer's instructions. The extracted peptides were cleaned up on ZipTip (Millipore) C18 microcolumns and spotted with α-cyano-4-hydroxycinnamic acid (matrix) on an Applied Biosystems 4700 Proteomics Analyzer MALDI plate (192-spot configuration). The spotted peptides were then analyzed by tandem mass spectrometry on the 4700 Proteomics Analyzer matrix-assisted laser desorption ionization—time-of-flight (MALDI-TOF-TOF) instrument. To identify the peptide sequences, the fragment ion patterns were analyzed using the DeNovo Explorer Software (Applied Biosystems) algorithms. Sequences with an identity score of 93% or better were used to screen the deduced proteome (a score of 93% means that 93% of the mass fragments produced agreed with the theoretical pattern of the deduced amino acid sequence). Proteins were initially identified by searching the proteome using the entire de novo sequence (12 to 15 amino acids [aa]), but a greater degree of success was obtained by using portions of the de novo sequence (4 to 6 aa). A match in the proteome was found if at least two tryptic fragments had appreciable identity and the molecular weight of the translated ORF was in agreement with that found on the gel.
PFGE.
For pulsed-field gel electrophoresis (PFGE) analysis, approximately 200 ng of HSIC DNA was digested with 5 U of the specified restriction enzymes. Digestions were performed with PvuI, SalI, ScaI, and double digests of PvuI plus SalI and PvuI plus ScaI (Promega). PFGE was performed in a 1% PFGE-certified agarose gel (Bio-Rad, Hercules, CA) and 1× Tris-acetate-EDTA. A CHEF-DR II PFGE system (Bio-Rad) was used with run parameters of 3.4 V/cm (113.3 V) for 23.5 h, with switch times ramped from 0.2 to 0.8 s. The tank buffer temperature was controlled at 14°C. The gel was stained with ethidium bromide after electrophoresis.
Phylogeny.
Individual ORFs were aligned with closely related sequences (obtained from BLASTP searches) using OMIGA 1.1 (Oxford Molecular Group, Oxford, United Kingdom) and a Clustal W pairwise weighted alignment method. Alignments were exported to Mega 2.0 beta (28), and trees were built by the neighbor-joining method using a gamma distribution (gamma parameter = 2.0) to correct for rate heterogeneity across sites. Trees were bootstrapped at 1,000 replicates, and an amino acid Poisson correction was employed.
Nucleotide sequence accession number.
The sequence of φHSIC has been deposited in GenBank under accession number AY772740.
RESULTS AND DISCUSSION
General characteristics of φHSIC.
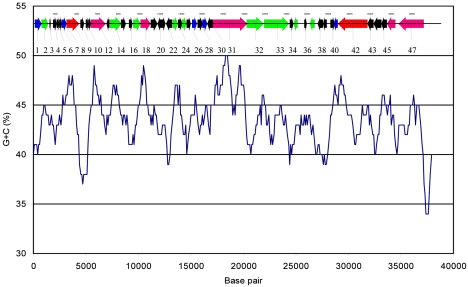
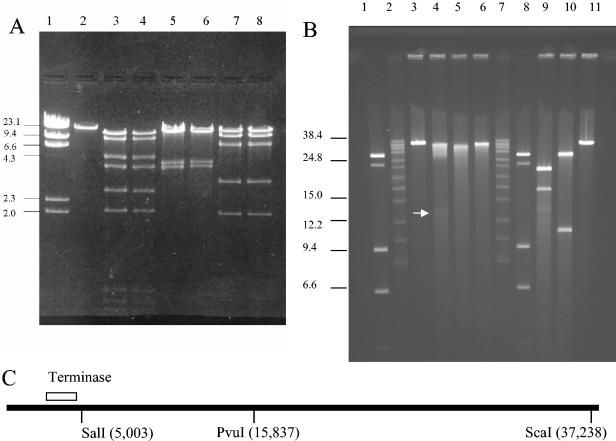
Transmission electron microscopy previously indicated that φHSIC has a morphology typical of the Siphoviridae (25), common among temperate phages (7, 9). The genome of φHSIC was determined to be 37,966 nucleotides in length with a G+C content of 44%. Figure 1 is a G+C plot along the genome length analyzed at 100-bp intervals. There is no evidence of variation in G+C content in large modules, although certain ORFs appear to have lower (ORFs 9 and 10) or higher (ORFs 30 and 31) G+C contents. No physical terminus of the genome was detected by multiple rounds of primer walking (the ends of the genome depicted in Fig. 2 and Table 1 are arbitrary). The genome was circularly permuted and assembled into a circle in silico. No cos site was found when restriction digests were followed by heating to 65°C and rapid cooling prior to electrophoresis (Fig. 3A). However, because ORFs similar to the large and small subunits of terminase were identified, we hypothesized that this phage may be packaged by a headful packaging mechanism. Undigested phage DNA ran as a single, sharp band on 1% agarose gels (Fig. 3A). Restriction analysis with enzymes that possess a unique restriction site in the genome (PvuI and ScaI) did not result in a difference in mobility on 1% agarose gels (data not shown), as would be expected for a covalently closed genome. This suggests that the genome is not supercoiled as occurs with plasmids or plasmid-like prophages (21). Restriction analysis of the purified phage with multiple cutting enzymes resulted in patterns identical to those predicted by restriction analysis in silico. For example, EcoRI digestion analysis of the φHSIC genome should produce fragments of 744, 824, 930, 2,055, 2,610, 3,796, 4,632, 9,088, and 13,287 bp, assuming a circularly permuted genome. Each of these nine bands is visible in Fig. 3. Similar results were obtained with EcoRV and XbaI, each of which yielded four bands upon digestion, the number predicted based on computational digestion. These observations further confirm that our assembly is correct. When unique cutters (PvuI, ScaI, and SalI) were used to digest the genome and the genome was run on a pulsed-field gel, the DNA ran similarly to the undigested DNA, with some smearing of the band; the degree of smearing depended on the enzyme employed (Fig. 3B). When unique cutters were combined in a double digestion (PvuI and ScaI), two bands of the expected sizes, 16.5 and 21 kb, were observed, as if the genome was in an apparent circle (Fig. 3B and C). Similar results were observed for codigestion with PvuI and SalI (Fig. 3B and C).
FIG. 1.
G+C content as a function of base pair. The scaled genome appears at the top of the figure. Colors of ORFs are explained in the legend to Fig. 2, and ORFs are numbered as in Table 1.
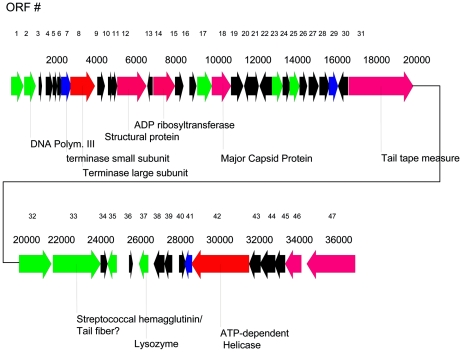
FIG. 2.
Putative ORFs of phage function on the φHSIC genome. Coloring of ORF arrows represents the color code of the BLASTP score used by the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST) (red, >200; magenta, 80 to 200; green, 50 to 80; blue, 40 to 50; black, 0 to 40). Numbers at the top are the ORF numbers; the second row of values indicates base pair locations.
TABLE 1.
Open reading frames of the φHSIC genome and best BLASTP hits
| ORF no./ orientationa | Nucleotide position (MALDI analysis) | Best BLASTP hit(s) (e value[s]) |
|---|---|---|
| 1/+ | 2-625 | Hypothetical protein, Streptococcus pyogenes, NP_268910 (3e-04); ORF188, Lactobacillus phage φAdh, NP_050127 (0.001) |
| 2/+ | 679-1254 | DNA polymerase III, beta subunit, Y. pestis, NP_407518 (7e-6) |
| 3/+ | 1393-1542 | Unknown |
| 4/+ | 1757-2101 | Unknown |
| 5/+ | 2095-2328 | Unknown |
| 6/+ | 2374-2568 | Unknown |
| 7/+ | 2586-3029 | Terminase small subunit (bacteriophage HK620), NP_112075 (0.009) |
| 8/+ | 3010-4236 | Phage terminase (Actinobacillus pleuropneumoniae serovar 1 strain 4074), ZP_00134759 (e-100); large terminase (Staphylococcus aureus phage phi 11), NP_803283 (8e-30) |
| 9/+ | 4354-4719 | Unknown |
| 10/+ | 4874-5107 | Unknown |
| 11/+ | 5104-5325 | Unknown |
| 12/+ | 5372-6799 (MALDI) | 60-kDa structural protein, Salmonella serovar Typhimurium phage MB78, CAB59889 (2e-20) |
| 13/− | 6829-7110 | Unknown |
| 14/+ | 7182-8252 | NAD-asparagine ribosyltransferase, Pseudomonas syringae, ZP_00127008 (7e-17); homolog of phage Mu protein gp30, Silicibacter sp. strain TM1040, ZP_00338818 (2e-13) |
| 15/+ | 8258-8662 | Unknown |
| 16/+ | 8984-9313 | Unknown |
| 17/+ | 9382-10,086 | P26, serovar Typhimurium phage MB78, CAA60565 (1e-5) |
| 18/+ | 10098-11045 (MALDI) | Hypothetical protein, Pseudomonas syringae, ZP_00127006 (e-110); putative phage protein, Y. pestis minor capsid, NP_405658 (e-103) |
| 19/+ | 11173-11669 | Unknown |
| 20/− | 11702-12130 | Unknown |
| 21/+ | 12164-12445 | Unknown |
| 22/− | 12474-13058 | Unknown |
| 23/+ | 13153-13650 | Hypothetical phage protein, Y. pestis, NP_405659 (5e-9) |
| 24/+ | 13654-14052 | Hypothetical phage protein, Y. pestis, AAM87563 (0.073) |
| 25/+ | 14027-14485 | Hypothetical phage protein, Y. pestis, AEM85762 (4e-7) |
| 26+ | 14485-14868 | Unknown |
| 27/+ | 14985-15461 (MALDI) | Unknown |
| 28/+ | 15530-16012 | Unknown |
| 29/+ | 16024-16428 | Unknown |
| 30/− | 16452-16928 | Unknown |
| 31/+ | 17022-20231 | Hypothetical protein, R. sphaeroides, ZP_00005117 (2e-32); putative phage tail tape measure protein, Acinetobacter sp. strain ADP1 YP_046779 (1e-16); ORF19 Pseudomonas phage D3, NP_061515 (5e-16); phagetail tape measure protein, lambda family, NP_743734 (3e-15) |
| 32/+ | 20231-21817 | Hypothetical protein, Novosphingobium, ZP_00305036 (6e-13) |
| 33/+ | 21870-24275 (MALDI) | Streptococcal hemagglutinin protein, Staphylococcus epidermidis AAO05891 (2e-6) |
| 34/+ | 24308-24643 | Unknown |
| 35/− | 24648-25079 | Cyanophage P60 protein, AAL73273 (7e-7) |
| 36/+ | 25708-25905 | Unknown |
| 37/− | 26214-26672 | gp5 baseplate hub subunit and tail lysozyme, phage 44RR2.8t, AAQ81457 (2e-11); lysozyme murein hydrolase, bacteriophage 44RR2.8t, AAQ81541 (1e-8) |
| 38/− | 26940-27488 | Unknown |
| 39/− | 27485-27829 | Unknown |
| 40/+ | 28235-28564 | Unknown |
| 41/− | 28476-28859 | Putative phage protein of enterobacteriophage P1, YP_006552 (0.001) |
| 42/− | 28856-31717 | Putative helicase, Erwinia carotovora, YP_50835 (4e-81); putative ATP-dependent helicase, E. coli, NP_416689 (4e-80); helicase precursor, Deinococcus radiodurans, AAN62835 (3e-20) |
| 43/− | 31723-32265 | Unknown |
| 44/− | 32262-33017 | Unknown |
| 45/− | 33017-33550 | Unknown |
| 46/− | 33557-34315 | Holliday junction resolvasome, helicase subunit, Magnetococcus, ZP_00289517, (3e-21); phage-associated protein, Streptococcus pyogenes, NP_268905 (3e-16) |
| 47/− | 34641-37088 | Predicted ATPase, Clostridium thermocellum, ZP_0311598 (9e-13); integrase, vibriophage VP2, YP_022428 (3e-10); primase, Bordetella phage BP-1, NP_996639 (3e-08); phage protein, S. pyogenes NP_268911 (3e-08) |
+, forward orientation; −, reverse orientation.
FIG. 3.
(A) Restriction digestion analysis of φHSIC genome. Lane 1, molecular weight markers (in thousands); lane 2, undigested φHSIC DNA; lanes 3 and 4, EcoRI-digested φHSIC DNA; lanes 5 and 6, EcoRV-digested φHSIC DNA; lanes 7 and 8, BstXI-digested φHSIC DNA. Lanes 4, 6, and 8 contain DNA that was digested, heated to 80°C, and then chilled on ice before electrophoresis. (B) Pulsed-field gel analysis of restriction enzyme-digested φHSIC genome. Lanes 1, 2, 7, and 8, molecular weight markers (in thousands); lanes 3 and 11, undigested φHSIC DNA; lanes 4 through 6, φHSIC DNA digested with PvuI, ScaI, and SalI, respectively; lanes 9 and 10, double digestions (PvuI plus ScaI and PvuI plus SalI, respectively). (C) Restriction map for unique cutters for φHSIC in relation to the terminase gene.
These results are similar to those found for headful packaging phages such as P22 and Sf6. Sf6 has been shown to be a headful packaging phage with no cos site or recognizable pac fragment and is also circularly permuted (10). In such phages, the site of initiation of a packaging series is so variable that it is ±1,000 bp around the packaging recognition site, not enabling a pac site-containing fragment to be recognized by staining. Because the physical ends of these chromosomes occur at different places on different molecules, restriction digests do not reveal the location of the physical ends. In such phages, restriction analysis behaves as for a circular genome (10). We further hypothesize that the preferred pac site is somewhere near the terminase, as found for Sf6 and similar phages. This is based on the degree of smearing observed in the single digestions (Fig. 3B). When the genome was digested with SalI, which has a digestion site adjacent to the terminase (Fig. 3C), little smearing occurred, suggesting that this site was close to the preferred site for initiation of packaging (the physical end of many copies of the genome). When the genome was digested with PvuI alone, which is ∼18 kb from the terminase gene (Fig. 3C), the most smearing occurred, as well as a faint 18-kb band (also observed in PvuI-plus-ScaI codigestions), which might be the fragment between the PvuI site and the preferred pac site. Upon single digestion with ScaI, the site for which is about 33 kb (or 5 kb in the circularly permuted genome) from the hypothetical pac site, an intermediate amount of smearing was observed. We propose that φHSIC can initiate packaging throughout its genome, with the preferred site being upstream of the terminase. The observation that φHSIC is almost certainly packaged by a headful packaging mechanism is consistent with the observation that it can function as a generalized transducing phage (22). Its status as a pseudotemperate phage would give it greater opportunities to interact with the host genome and be an agent of horizontal gene transfer in the marine environment.
ORF analysis.
ORF analysis indicated the presence of 47 ORFs. Table 1 shows the results of ORF analysis, and Fig. 2 is a map of the ORFs of putative phage function. In general, the “lefthand” side of the genome has most of the ORFs in the forward direction, while the ORFs on the “righthand” side (starting with ORF 35) are generally in the opposite orientation. There are 17 ORFs with expect (e) values lower than 0.001. These are divided between ORFs that are similar to genes of known function and those of unknown function (generally phage-related proteins). In Table 1 a few ORFs with higher e values from BLASTP searches are also listed if they are similar to phage-like proteins.
ORF 2: DNA polymerase III, β subunit.
ORF2 is probably a portion or remnant of a DNA polymerase gene, as evidenced by its length (195 aa compared to 366 to 367 aa for most microbial β subunits of DNA polymerase). A neighbor-joining tree was generated from an alignment of several DNA polymerase β subunits (data not shown). The putative φHSIC gene was not closely related to any of the existing sequences, as it resided on a fairly deeply rooted branch on this tree (the closest BLASTP hit was for Yersinia pestis DNA polymerase). The presence of a DNA polymerase III on a phage genome is not known to occur widely (31).
ORF 18: major capsid protein.
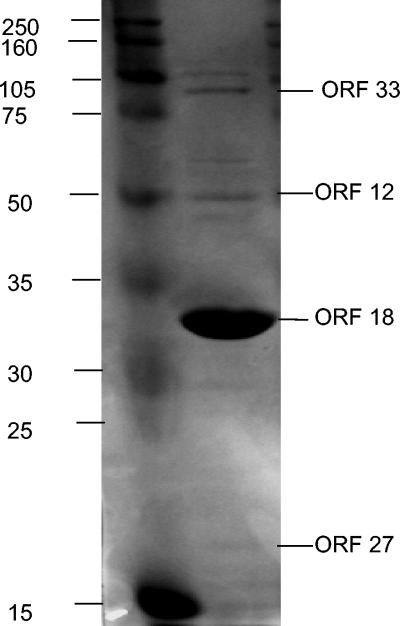
The best BLASTP hit for ORF 18 was a hypothetical protein of Pseudomonas syringae (62% identity), followed by a putative phage protein of Y. pestis that was similar to a minor capsid protein (58% identity). Protein sequence data obtained from trypsin protease fragments identified by MALDI-TOF analysis of the major protein band in one-dimensional PAGE gels (Fig. 4) of the phage yielded two de novo peptide sequences (CAADNLAVFNENSR and YGPYETTEEAFK). A close inspection of all translations yielded identical sequences in ORF 18 (DNLAVFNENSR and YGPYETTEEAFK, respectively), corresponding to nucleotide positions 10173 and 10371 (aa 27 and 92, respectively, in the translation of ORF 18). The major protein band had a molecular mass of ∼34.2 kDa, which agrees with the predicted molecular mass of 33,760 Da (Fig. 4). Thus, we concluded that ORF 18 is the major capsid protein.
FIG. 4.
SDS-PAGE of proteins associated with φHSIC particles (right lane). Left lane, molecular size standards (in kilodaltons). ORF designations on the right of the gel are based on MALDI-TOF-TOF de novo amino acid sequence analysis of trypsin fragments of the indicated band.
ORFs 7 and 8: terminase small and large subunits.
The terminase gene product is involved in the ATP-dependent packaging of the concatameric DNA into phage heads (5). In most phages the enzyme is a two-protein complex, with the small subunit possessing DNA recognition specificity and the large subunit possessing nucleolytic and ATPase activities (30). ORF 7 yielded a BLASTP hit similar to the small-subunit terminase for coliphage HK620 (e = 0.009; 28% identity). HK620 is a lambdoid, temperate phage classified in the Podoviridae group (12). Although the e value is above 0.001, we have included this designation in Table 1 because of the proximity of this gene to the putative terminase large subunit (ORF 8), and the location of terminase small subunits in other phages (9, 12, 30, 44).
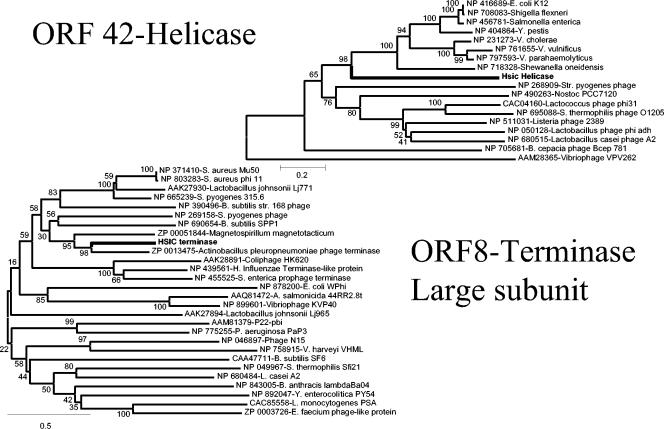
The best BLASTP hit for ORF 8 was a putative phage terminase from Actinobacillus pleuropneumoniae serovar 1 strain 4074, yielding an e value of e-100 with an amino acid identity of 47%. Neighbor-joining analysis indicated that the φHSIC terminase was also closely related to a terminase-like gene found in Magnetospirillum magnetotacticum (Fig. 5).
FIG. 5.
Neighbor-joining trees of ORF 42 (putative helicase) and ORF 8 (putative terminase large subunit), and closely related sequences from the results of BLASTP searches. Each branch has the GenBank accession number. The φHSIC proteins are boldfaced. The numbers at the internal nodes are the bootstrap value scores (0 to 100%) obtained from 1,000 bootstrap replicates.
ORF 14: NAD-asparagine ADP ribosyltransferase.
ORF 14 had a BLASTP hit most closely related to the NAD-asparagine ADP ribosyltransferase gene of Pseudomonas syringae (ZP_00127008) (3e-11). ADP ribosyltransferases are key components of bacterial pathogenic mechanisms and are parts of the toxin systems in diphtheria toxin, Pseudomonas exotoxin A, pertussis toxin, cholera toxin, and Escherichia coli heat-labile enterotoxins (reviewed in reference 19). In fact, the fish pathogen Aeromonas salmonicida subsp. salmonicida has an extracellular toxin, AexT, which is also an ADP ribosyltransferase that is secreted by a recently identified type III secretion system (8). The second-best hit was an uncharacterized bacterial protein that was a homolog of phage Mu protein GP30.
ORF 31: tail tape measure protein.
Tail tape measure proteins play a critical role in the assembly and determination of length of the phage tail (20, 27, 41). The best BLASTP hit for ORF 31 was a methyl-accepting chemotaxis protein of Rhodobacter sphaeroides. However, the closest phage-related proteins were a series of tail tape measure proteins, which encompassed aa 40 to 288. The remainder of the ORF hit other putative phage tail fiber proteins (aa 169 to 938).
ORF 33.
ORF 33 had a BLASTP similarity to a streptococcal hemagglutinin of Staphylococcus epidermidis (2e-6; 20% identity). However, no other hemagglutinin proteins from GenBank showed any similarity to ORF 33. MALDI-TOF-TOF analysis indicated that a ∼87-kDa band on protein gels (Fig. 4) was most likely this gene product (i.e., five of seven tryptic fragments from this band had appreciable sequence identity to de novo sequences), which had a predicted molecular mass of 85,569 Da. Because the proteins associated with the structural components of the phage would be expected to be found in phage particles and thus on the gel (while a prophage-encoded converting gene such as a hemagglutinin would not), we assume that this is a structural protein of some sort. Hemagglutinin proteins are inherently fibrous, and tail fibers are fibrous (as well as being diverse); thus, this ORF may have a tail fiber function (Matthew Sullivan, personal communication). However, the transcription of four toxin genes in staphylococcal phage φSa3ms increased upon mitomycin C induction. Virulence factor production in this phage was regulated by processes that governed lysogeny (47). Thus, a hemagglutinin that is produced during prophage induction could possibly appear in gels of phage particle proteins.
ORF 37: phage lysozyme.
ORF 37 was most closely related to the gp5 baseplate hub subunit and tail lysozyme of Aeromonas salmonicida bacteriophage 44RR2.8t (AAQ81457.1) (e value = 9e-8), followed by the murein lysozyme of bacteriophage Aeh1 of Aeromonas hydrophila (AAQ17989) (e value = 1e-4; 27% identical). Alignment of ORF 37 with these and the T4-like gp5 protein suggested that the HSIC protein was homologous to only a portion of the gp5-like proteins (approximately 150 of the 600 aa of the gp5-like proteins). However, alignment with general phage lysozymes (which are only ∼150 aa long), including that of Aeh1 and the phage-related lysozyme of Synechococcus elongatus 7942 (ZP_00163918), suggests that this protein (155 aa) is most likely a general lysozyme involved in general cell lysis, not just in the infection/penetration process like the gp5 proteins.
ORF 42: ATP-dependent helicase.
DNA helicases are molecular motor enzymes that unwind and transport DNA. Upon BLASTP searching with ORF 42, it was apparent that this ORF (954 aa) contained two putative proteins, a helicase (aa 375 to 889) and a helicase precursor (aa 54 to 373). The best BLASTP similarities for the helicase were those to Erwinia carotovora (38% identical) and E. coli K-12 (4e-80; 35% identical), while the helicase precursor was similar to that of Deinococcus radiodurans (accession no. AAN62835) (3e-20; 28% identical). The helicase precursor of D. radiodurans is primarily an intein that is believed to be spliced out during joining of the helicase protein and helicase-related protein exteins (46). It is not known if inteins occur in φHSIC. Figure 5 is a neighbor-joining tree generated from the aa-375-to-889 fragment of ORF 42 (i.e., the helicase portion). As with all ORFs in the φHSIC genome, the putative helicase is deeply branched, equidistant between the bacterial helicases and the phage sequences appearing in Fig. 5.
ORF 46: Holliday junction helicase.
ORF 46 had a best BLASTP hit to a helicase subunit of a Holliday junction resolvasome of Magnetococcus strain MC1 (e value = 3e-21; 30% identity). Holliday junction resolvasomes are protein complexes involved in recombination events, and in E. coli they are composed of the RuvABC proteins (14). The process involves branch migration (catalyzed by RuvB) that is targeted by the structure-specific RuvA protein and resolution catalyzed by the RuvC endonuclease (14). This enables resolution of the Holliday structure intermediate and ensures completion of recombination-dependent repair. Resolvase function in phage integration is also catalyzed by certain phage integrases (i.e., the serine integrases; see below). However, certain phages contain separate resolvases involved in recombination, including the T4 endonucleases VII (3). The second-best hit for this ORF was a phage-related protein from a Streptococcus pyogenes M1 GAS putative prophage.
ORF 47.
ORF 47 had a best BLASTP hit to an ATPase from Clostridium thermocellum, while the best phage-related hits were for a primase of Bordetella phage BIP-1 and integrases from vibriophages VP2 and VP5 of Vibrio cholerae (26% identical; 3e-8). The latter two ORFs in turn were similar to ORF64 of the Vibrio parahaemolyticus phage VP16T (44), and the annotation of this ORF as a putative integrase was also used to “label” the VP2 and VP5 ORFs. VP16T ORF64 was annotated in GenBank as a putative integrase, not on the basis of similarity to integrases, but rather on the basis of possessing the conserved active-site residues of tyrosine recombinases in the correct order and with roughly appropriate spacing (44). The tyrosine-type integrases are known to contain a catalytic tyrosine (Y342 in λ) and the conserved pentad RKHR(H/W) at positions R212, K235, H308, R311, and H333 (sometimes replaced by a tryptophan) (15, 18). In ORF 47, a potentially catalytic tyrosine was observed at Y417, and the pentad RKHRW was observed at positions R272, K289, H379, R382, and W402—generally in the same relative positions, but shifted by 50 to 60 aa downstream in the φHSIC sequence. However, PSI-BLAST analysis has shown that ORF 47 is distantly related to a diverse array of proteins, and an integrase function cannot be ascertained based on this information.
Phage-related proteins.
ORFs 1, 12, 17, 23 to 25, 35, and 41 had BLASTP hits that were most closely related to unidentified ORFs from microbial and/or phage genomic sequencing studies. Some of these were detected in SDS-PAGE gels by MALDI-TOF-TOF analysis, including ORF 12 (BLASTP hit, a 62-kDa structural protein of Salmonella enterica serovar Typhimurium phage MB78), and ORF 27 (a very weak BLASTP similarity to a single-stranded-DNA-specific exonuclease of Oceanobacillus iheyensis HTE831 [data not shown]). Some of these phages are temperate Siphoviridae (i.e., φadh_15 of Lactobacillus gasseri [2]; Pseudomonas aeruginosa D3). Several ORFs were similar to phage sequences found in the Streptococcus pyogenes genome (ORFs 1, 46, and 47) (4), the highly virulent phage MB78 of Salmonella serovar Typhimurium (ORFs 12 and 17), and putative prophages occurring in the genome of Yersinia pestis (ORFs 18 and 23 to 25). For ORFs 18 and 23, these genes were in a putative integrated temperate-phage genome of Y. pestis CO92 of about 47 kb that included a putative integrase, an excisionase, an antirepressor, and a phage terminase (37). For ORFs 24 and 25, these were found in a putative prophage of Y. pestis KIM of ∼41,000 bp which contained phage tail proteins, a tape measure protein, antirepressor proteins, a phage ninG-like protein, and other phage proteins (13). ORFS 1, 46, and 47 also share similarity to phage-like genes occurring in the genome of Streptococcus pyogenes M1 GAS (17). These genes were clustered over a 37,968-bp region of the S. pyogenes genome that contained an integrase, structural proteins, a holin, and CI and cro-like regulatory elements. These distant relationships with other phages or prophages support our hypothesis that φHSIC is not closely related to any currently existing phage for which genomic sequencing data exists.
Comparative phage genomics.
Twelve of the 20 ORFs with BLASTP hits possessing low expect values were similar to genes from temperate Siphoviridae or other temperate phages (ORFs 1, 7, 8, 31, and 41) or were identified as phage-related proteins from prophage-like regions in bacterial genome annotations (ORFs 14, 18, 23 to 25, 46, and 47). Thus, 60% of the ORFs with significant counterparts in GenBank were most likely derived from or related to temperate-phage genes. Of the 12 temperate-phage-like ORFs, only 5 could be assigned putative functions: the terminase small and large subunits (ORFs 7 and 8), the tail tape measure protein (ORF 31), the major capsid protein (ORF 18), and the helicase (ORF 42). This is most likely because of the lack of marine temperate-phage genomic information accumulated to date. This may also account for our inability to definitively recognize the components of a lysogeny module. Cells of the pseudolysogen were previously shown to contain chromosomally integrated and unintegrated copies of the prophage (51).
Only 4 of the 20 ORFs with low expect values (ORF 12, 17, 35, and 37) were closely related to lytic-phage genes. ORFs 12 and 17 are similar to proteins of Salmonella serovar Typhimurium phage MB78 (ORF 12 is a structural gene), ORF 35 is a putative gene of unknown function similar to that occurring in the marine podocyanophage P60 (11), and ORF 37 is the lysozyme of Aeromonas salmonicida phage RR442.8t (Nolan et al., unpublished). Thus, even though φHSIC is pseudotemperate, it shares more similarities with temperate than with lytic phages, based on the present BLAST analyses.
Temperate Siphoviridae have been shown to possess some conservation in genomic organization, at least in the gene cluster that encodes the virion assembly proteins (9). These are typically in the following order: terminase, portal, protease, scaffold, major head shell protein, head/tail-joining proteins, tail shaft protein, tape measure protein, tail tip/baseplate proteins, tail fiber (9). There are “missing pieces” in this gene organization in φHSIC, but this is most likely caused by our inability to recognize the genes, particularly the portal, scaffold, protease, and tail shaft genes.
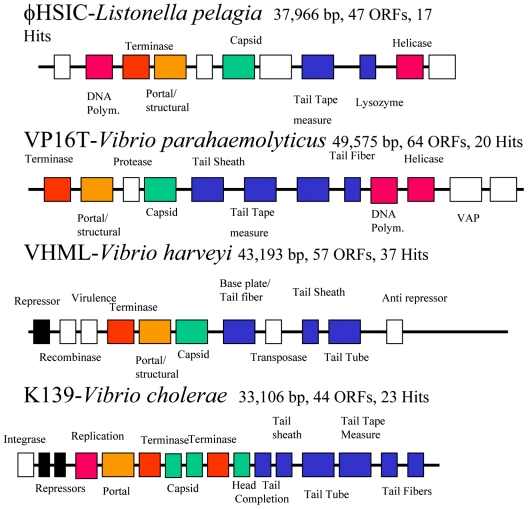
In order to compare genomic organization among marine vibriophages, we have shown the relative arrangement of functional genes in three vibriophages in comparison to that of φHSIC (Fig. 6). Vibriophage VP16T, infective for V. parahaemolyticus strain 16, has a 49,575-bp genome, a G+C content of 59%, and 64 putative ORFs (44). Although phage VP16T resembles a myovirus by electron microscopy, it possesses an organization close to that of the Siphoviridae. Vibriophage VHML infects Vibrio harveyi and is a member of the Myoviridae with 43,193 bp and 57 ORFs (34, 35), while K139 infects V. cholerae, contains 44 ORFs, and is 33,106 bp in length (26). All three are members of the Myoviridae, and VHML and K139 are clearly temperate while VP16T may be temperate. Even though few of the ORFs are closely related to one another between these phages, there is a certain degree of conservation of organization: terminase, portal/structural, capsid, and tail fiber, with replication genes preceding or following this grouping (or both). In K139, the terminase large and small subunits are separated by capsid protein genes (Fig. 6). Each of these phages contains genes that may play a role in host pathogenicity: for φHSIC, an ADP ribosyltransferase; for VP16T, a virulence-associated protein; and for K139 and VHML, N6-DAM (DNA adenine methlytransferase). For all four phages, many of the genes were more closely related to temperate Siphoviridae than to virulent Myoviridae. However, there is a fundamental difference in the relative similarity to existing phage sequences between the three vibriophages and φHSIC. Each of the former was more closely related to a single type or group of phages, whereas φHSIC was not closely related to any individual phage. VP16T was most closely related to a putative prophage of Ralstonia solanacearum (44). Both VHML and K139 were most closely related to the temperate P2 or its closest relatives. In VHML, 12 of 57 ORFs were P2-like, and in K139, 27 of 44 ORFs were similar to P2 (26, 35). For φHSIC, ORFs were related to a variety of phages, including unidentified phages of Streptococcus pyogenes and Y. pestis, Lactobacillus phage φAdh, an Actinobacillus pleuropneumoniae phage, Salmonella serovar Typhimurium phage MB78, Staphylococcus aureus phage phi 11, Pseudomonas phage D3, phages of Bordetella pertussis and Bordetella bronchiseptica, and cyanophage P60. Many of these bacteria are pathogens, and in many cases the phages play a role in the mechanisms of pathogenesis. It is tempting to conclude that φHSIC is related to these phages of pathogens, yet it is more likely that phages of pathogenic bacteria have been sequenced more frequently than marine phages. That is, φHSIC is probably more closely related to marine phages, the genomes of which have yet to be sequenced. It may also be that the other vibriophages, isolated primarily from coastal/estuarine waters, have greater similarity to the temperate Siphoviridae of terrestrial origin. This is in contrast to φHSIC and its host, Listonella pelagia, which were isolated from the waters surrounding Oahu (25). It may be for this reason that φHSIC was found to be unrelated to any phage for which sequence information resides in GenBank. These results further emphasize the tremendous opportunity afforded by sequencing of phages from the marine environment, perhaps the largest reservoir of untapped genetic information on the planet.
FIG. 6.
Schematic representation of the organization of four completed marine vibriophage genomes. Color code: red, DNA replicative genes; orange, terminases; yellow, portal/structural proteins; green, capsid genes; blue, tail-associated proteins. Polym., polymerase. Information for VP16T, VHML, and K139 is taken from references 44, 35, and 26, respectively.
Acknowledgments
This research was supported by an NSF Biocomplexity Award to J.H.P. and A.M.S. and by a Parrothead Fellowship to S.J.W.
We are indebted to the Proteomics Core Facility of the Moffitt Cancer Center for performing the MALDI-TOF-TOF analysis and to Jen Mobberly for assistance in primer walking. We gratefully acknowledge Sherwood Casjens for discussion of the terminase and for sharing results of work prior to its publication.
REFERENCES
- 1.Ackermann, H. W., and M. S. DuBow. 1987. Viruses of prokaryotes, vol. I. General properties of bacteriophages. CRC Press, Inc., Boca Raton, Fla.
- 2.Altermann, E., J. R. Klein, and B. Henrich. 1999. Primary structure and features of the genome of the Lactobacillus gasseri temperate bacteriophage φadh. Gene 236:333-346. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution, and evolutionary trajectories. Nucleic Acids Res. 28:3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, L. W. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267-292. [DOI] [PubMed] [Google Scholar]
- 6.Breitbart, M., B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. R. Soc. Lond. Ser. B 271:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Büchen-Osmond, C. 2002. ICTVdB: the universal virus database of the International Committee on Taxonomy of Viruses. Index of viruses. [Online.] http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/ICTVindex.htm.
- 8.Burr, S. E., K. Stuber, and J. Frey. 2003. The ADP-ribosylating toxin, AexT, from Aeromonas salmonicida subsp. salmonicida is translocated via a type III secretion pathway. J. Bacteriol. 185:6583-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 10.Casjens, S., D. A. Winn-Stapley, E. B. Gilcrease, R. Morona, C. Kulewein, J. E. H. Chua, P. A. Manning, W. Inwood, and A. J. Clark. 2004. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 339:379-394. [DOI] [PubMed] [Google Scholar]
- 11.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, A. J., W. Inwood, T. Cloutier, and T. S. Dhillon. 2001. Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J. Mol. Biol. 311:657-679. [DOI] [PubMed] [Google Scholar]
- 13.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plana, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickman, M. J., S. M. Ingleston, S. E. Sedelnikova, J. B. Rafferty, R. G. Lloyd, J. A. Grasby, and D. P. Hornby. 2002. The RuvABC resolvasome. Eur. J. Biochem. 269:5492-5501. [DOI] [PubMed] [Google Scholar]
- 15.Esposito, D., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farto, R., M. J. Perez, A. Fernandez-Briera, and T. P. Nieto. 2002. Purification and partial characterization of a fish lethal extracellular protease from Vibrio pelagius. Vet. Microbiol. 89:181-194. [DOI] [PubMed] [Google Scholar]
- 17.Ferretti, J. J., W. M. McShan, D. Ajdic, D. Savic, G. Savic, K. Lyon, C. Primeaux, S. S. Sezate, A. N. Surorov, S. Kenton, H. Lai, S. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. E. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groth, A. C., and M. P. Calos. 2004. Phage integrases: biology and applications. J. Mol. Biol. 335:667-678. [DOI] [PubMed] [Google Scholar]
- 19.Han, S., A. S. Arvai, S. B. Clancy, and J. A. Tainer. 2001. Crystal structure and novel recognition motif of rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. J. Mol. Biol. 305:95-107. [DOI] [PubMed] [Google Scholar]
- 20.Hendrix, R. W. 1988. Tail length determination in double-stranded DNA bacteriophages. Curr. Top. Microbiol. Immunol. 136:21-29. [DOI] [PubMed] [Google Scholar]
- 21.Inal, J. M., and K. V. Karunakaran. 1996. φ20, a temperate bacteriophage isolated from Bacillus anthracis, exists as a plasmidial prophage. Curr. Microbiol. 32:171-175. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, S. C., and J. H. Paul. 1998. Gene transfer by transduction in the marine environment. Appl. Environ. Microbiol. 64:2780-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, S. C., and J. H. Paul. 1996. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 142:27-38. [Google Scholar]
- 24.Jiang, S. C., and J. H. Paul. 1998. Significance of lysogeny in the marine environment: studies with isolates and a model for viral production. Microb. Ecol. 35:235-243. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, S. C., C. A. Kellogg, and J. H. Paul. 1998. Characterization of marine temperate phage-host systems isolated from Mamala Bay, Oahu, Hawaii. Appl. Environ. Microbiol. 64:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapfhammer, D., J. Blass, S. Evers, and J. Reidl. 2002. Vibrio cholerae phage K139: complete genome sequence and comparative phage genomics of related phages. J. Bacteriol. 184:6592-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katsura, I. 1987. Determination of bacteriophage λ tail length by a protein ruler. Nature 327:73-75. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: molecular evolutionary genetic analysis. The Pennsylvania State University, University Park, Pa.
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lin, H., V. B. Rao, and L. W. Black. 1999. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J. Mol. Biol. 289:249-260. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Jimenez, M. I., P. Mesa, and J. C. Alonso. 2002. Bacillus subtilis tau subunit of DNA polymerase III interacts with bacteriophage SPP1 replicative DNA helicase G40P. Nucleic Acids Res. 30:5056-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniel, L., L. A. Houchin, S. J. Williamson, and J. H. Paul. 2002. Lysogeny in marine Synechococcus populations. Nature 415:496. [DOI] [PubMed] [Google Scholar]
- 33.Moebus, K. 1997. Investigations of the marine lysogenic bacterium H24. II. Development of pseudolysogeny in nutrient broth rich culture. Mar. Ecol. Prog. Ser. 148:229-240. [Google Scholar]
- 34.Munro, J., J. Oakey, E. Bromage, and L. Owens. 2003. Experimental bacteriophage-mediated virulence in strains of Vibrio harveyi. Dis. Aquat. Organ. 54:187-194. [DOI] [PubMed] [Google Scholar]
- 35.Oakey, H. J., B. R. Cullen, and L. Owens. 2002. The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. J. Appl. Microbiol. 93:1089-1098. [DOI] [PubMed] [Google Scholar]
- 36.Ortmann, A. C., J. E. Lawrence, and C. A. Suttle. 2002. Lysogeny and lytic viral production during a bloom of the cyanobacterium Synechococcus spp. Microb. Ecol. 43:225-231. [DOI] [PubMed] [Google Scholar]
- 37.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, A. V. Karlyshev, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 38.Paul, J. H., and S. C. Jiang. 2001. Lysogeny and transduction. Methods Microbiol. 30:105-125. [Google Scholar]
- 39.Paul, J. H., and B. Myers. 1982. The fluorometric determination of DNA in aquatic microorganisms employing Hoechst 33258. Appl. Environ. Microbiol. 43:1393-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul, J. H., J. B. Rose, S. C. Jiang, C. A. Kellogg, and L. Dickson. 1993. Distribution of viral abundance in the reef environment of Key Largo, Florida. Appl. Environ. Microbiol. 59:718-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen, M., S. Ostergaard, J. Bresciani, and F. K. Vogensen. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276:315-328. [DOI] [PubMed] [Google Scholar]
- 42.Ripp, S., and R. V. Miller. 1998. Dynamics of the pseudolysogenic response in slowly growing Pseudomonas aeruginosa cells. Microbiology 144:2225-2232. [DOI] [PubMed] [Google Scholar]
- 43.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seguritan, V., I.-W. Feng, F. Rohwer, M. Swift, and A. N. Segall. 2003. Genome sequences of two closely related Vibrio parahaemolyticus phages, VP16T and VP16C. J. Bacteriol. 185:6434-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simidu, U., K. Kta-Tsukamoto, T. Yasumoto, and M. Yotsu. 1990. Taxonomy of four marine bacterial strains that produce tetrodotoxin. Int. J. Syst. Bacteriol. 40:331-336. [DOI] [PubMed] [Google Scholar]
- 46.Southworth, M. W., and F. B. Perler. 2002. Protein splicing of the Deinococcus radiodurans strain R1 Snf2 intein. J. Bacteriol. 184:6387-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sumby, P., and M. K. Waldor. 2003. Transcription of the toxin genes present within the staphylococcal phage φSa3ms is intimately linked with the phage's life cycle. J. Bacteriol. 185:6841-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villamil, L., A. Figueras, A. E. Toranzo, M. Planas, and B. Novoa. 2003. Isolation of a highly pathogenic Vibrio pelagius strain associated with mass mortalities of turbot, Scophthalmus maximus (L.), larvae. J. Fish Dis. 26:293-303. [DOI] [PubMed] [Google Scholar]
- 49.Weinbauer, M. G., and C. A. Suttle. 1996. Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the Gulf of Mexico. Appl. Environ. Microbiol. 62:4374-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinbauer, M. G., and C. A. Suttle. 1999. Lysogeny and prophage induction in coastal and offshore bacterial communities. Aquat. Microb. Ecol. 18:217-225. [Google Scholar]
- 51.Williamson, S., M. R. McLaughlin, and J. H. Paul. 2001. Interaction of the phage HSIC with its host. Lysogeny or pseudolysogeny? Appl. Environ. Microbiol. 67:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, W. H., S. Turner., and N. H. Mann. 1998. Population dynamics of phytoplankton and viruses in phosphate-limited mesocosm and their effect on DMSP and DMS production. Estuar. Coast. Shelf Sci. 46:49-59. [Google Scholar]
- 53.Wommack, K. E., and R. R. Colwell. 2000. Viroplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]