Abstract
A lacto-N-biose phosphorylase (LNBP) was purified from the cell extract of Bifidobacterium bifidum. Its N-terminal and internal amino acid sequences were homologous with those of the hypothetical protein of Bifidobacterium longum NCC2705 encoded by the BL1641 gene. The homologous gene of the type strain B. longum JCM1217, lnpA, was expressed in Escherichia coli to confirm that it encoded LNBP. No significant identity was found with any proteins with known function, indicating that LNBP should be classified in a new family. The lnpA gene is located in a novel putative operon for galactose metabolism that does not contain a galactokinase gene. The operon seems to be involved in intestinal colonization by bifidobacteria mediated by metabolism of mucin sugars. In addition, it may also resolve the question of the nature of the bifidus factor in human milk as the lacto-N-biose structure found in milk oligosaccharides.
The terms “probiotics” and “prebiotics” have become important in the area of food technology (9, 11, 32). A probiotic is defined as a live microbial food ingredient that has beneficial effects on health beyond basic nutrition, while prebiotics are food ingredients that function to increase probiotic intestinal bacteria in vivo. The gram-positive anaerobes bifidobacteria are the main targets of probiotics and prebiotics, along with the lactobacilli.
Intestinal colonization by bifidobacteria is important for human health, especially in pediatrics, because colonization seems to prevent infection by some pathogenic bacteria that cause diarrhea or other illnesses (4). An intestinal flora with a predominance of bifidobacteria is formed in breastfed infants within 1 week after birth, and they account for 95 to 99.9% of the intestinal flora (2, 30). In contrast, bottle-fed infants do not show such rapid colonization by bifidobacteria, and before the early 20th century, they often became infected by pathogenic bacteria (4). To improve the propagation of intestinal bifidobacteria, milk formulae are usually supplemented with oligosaccharides, such as lactulose (29), with the result that bottle-fed infants are now much healthier than before. However, the intestinal flora of bottle-fed infants, consisting of 90% bifidobacteria and 10% enterobacteria, is still different from that of breastfed infants (3).
There have been a number of studies of the growth factors for bifidobacteria in human milk. Initially, the growth of bifidobacteria was suggested to be dependent on nitrogen-containing sugar, but these results were later found to be due to the use of a specific Bifidobacterium bifidum strain, B. bifidum var. pennsylvanicus, that requires N-acetylglucosamine (GlcNAc) for its growth (17, 34). Further studies indicated that oligosaccharides in human milk (milk oligosaccharides) are candidates for the bifidus factor, which is responsible for increasing the bifidobacteria population (4). Cow's milk contains lactose as the sole saccharide, whereas human milk contains lactose, as well as various oligosaccharides (24, 28). However, it is yet to be determined which oligosaccharides or structures within them are responsible for the bifidus factor.
Intestinal colonization by bifidobacteria in the adult intestine is mediated by the recognition and metabolism of mucin carbohydrates (20, 25). Bifidobacteria produce factors that can hydrolyze mucin sugars. Derensy-Dron et al. reported the presence of an enzyme that reversibly phosphorolyzes β-d-galactopyranosyl-(1→3)-N-acetyl-d-glucosamine (lacto-N-biose I [LNB]) to α-d-galactopyranose 1-phosphate (Gal1-P) and N-acetyl-d-glucosamine in cell extracts of B. bifidum (13). The enzyme also phosphorolyzes β-d-galactopyranosyl-(1→3)-N-acetyl-d-galactosamine (galacto-N-biose [GNB]) to Gal1-P and N-acetyl-d-galactosamine (GalNAc), and it was named β-1,3-galactosyl-N-acetylhexosamine phosphorylase (EC 2.4.1.211) (13). Here, we propose the short name lacto-N-biose phosphorylase (LNBP) for this enzyme.
LNBP was found to be a useful tool to synthesize various derivatives of LNB and GNB (15). As LNB and GNB are the core structures of mucin sugar, the intracellular enzyme LNBP was considered to be related to intestinal colonization (13). There have, however, been no further reports regarding this enzyme. Therefore, we have purified and cloned the gene encoding the enzyme.
In the present study, we purified LNBP from B. bifidum JCM1254 and successfully cloned the gene from a strain of Bifidobacterium longum JCM1217. The enzyme was not homologous with any other known protein and should be classified in a new family. Here, we also discuss the role of LNBP, the gene for which is part of a putative operon in B. longum.
MATERIALS AND METHODS
Materials.
LNB and Gal1-P were purchased from Sigma (St. Louis, MO). The vector pET28a was obtained from Novagen (Madison, WI). Two strains of bifidobacteria, B. bifidum JCM1254 (DSM 20082) and B. longum JCM1217, were obtained from the Japan Collection of Microorganisms, The Institute of Physical and Chemical Research (Wako, Japan). Escherichia coli JM109 (Takara-Bio, Otsu, Japan) and BL21(DE3) (Novagen) were used as hosts for cloning and expression, respectively.
Measurement of LNBP activity.
LNBP activity was measured using routine methods by determining the increase in phosphate in reaction mixtures containing 10 mM Gal1-P and 10 mM GlcNAc in 50 mM MOPS (morpholinepropanesulfonic acid) buffer (pH 7.0) at 30°C by the method of Lowry and Lopez (27). One unit of activity was defined as the amount of enzyme that liberated 1 μmol of phosphate per minute under the above conditions. Protein concentrations were calculated from the theoretical A280 (1.823 liter · g−1 cm−1) based on the amino acid sequence of LNBP from B. longum JCM1217 (16). The value was used for both crude and purified enzymes.
Purification of LNBP.
B. bifidum JCM1254 was cultivated for 48 h at 37°C under anaerobic conditions (N2/CO2, 4:1) in a medium containing 16 g/liter nutrient broth, 5 g/liter yeast extract, 10 g/liter glucose, 3 g/liter K2HPO4 1 g/liter Tween 80, 10 g/liter sodium ascorbate, and 500 mg/liter cysteine-HCl. Cells were harvested from 4 liter of the culture, washed, and resuspended in 10 ml of 10 mM MOPS buffer (pH 7.0). To extract the enzyme, the cells were disrupted by sonication using a sonifier (Branson Ultrasonic Corporation, Danbury, CT), and the resulting mixture was centrifuged (17,000 × g for 30 min) to remove the cell debris. The supernatant of an ammonium sulfate precipitation (30% saturation) was applied to a Butyl-Toyopearl 650 M (Tosoh, Tokyo, Japan) column (8 ml) preequilibrated with 10 mM MOPS buffer containing 30% saturated ammonium sulfate, and the column was washed with 50 ml of the same buffer. The enzyme was then eluted with 100 ml of a linear gradient of ammonium sulfate from 30% saturation to 0%. The fractions exhibiting LNBP activity were collected, and ammonium sulfate was added to the solution to 60% saturation. The precipitate was resuspended in 10 mM MOPS buffer (pH 7.0), and the enzyme was purified by chromatography on a DEAE-Toyopearl 650 M (Tosoh) column (1 ml) with a linear gradient of NaCl from 150 mM to 350 mM (20 ml). The enzyme was purified further on a MonoQ column (1 ml; Amersham Biosciences, Picataway, NJ) under the same conditions described for the DEAE-Toyopearl procedure. The active fractions were collected and dialyzed against 10 mM MOPS buffer (pH 7.0), and the purity was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Amino acid sequence.
The N-terminal amino acid sequence of the purified LNBP was determined using an automated peptide sequencer (model G1001A; Hewlett-Packard, Corvallis, CA). To obtain internal amino acid sequences, the purified LNBP was digested in SDS-polyacrylamide gels by Staphylococcus V8 protease (Wako Pure Chemicals, Osaka, Japan) (10). After completion of SDS-PAGE, the peptide fragments were blotted onto polyvinylidene difluoride membranes (26). Several bands were cut out from the membranes, and their N-terminal amino acid sequences were determined using the automated peptide sequencer.
Cloning of LNBP gene.
The DNA fragment containing the whole LNBP gene, lnpA, was amplified from B. longum JCM1217 genomic DNA by PCR (27 cycles of 98°C for 10 s, 60°C for 10 s, and 74°C for 2 min) with the primers Fwd (TGC TGT TCC TCG GCC TCC AGC GCT ACT GGC) and Rev (ATG CCG AAT AAA ACT TCA TTG CTT TCG GTC), designed according to the sequences surrounding BL1641. The amplified fragments were sequenced. Then, the lnpA gene was again amplified with primers having an additional restriction site (underlined; NcoI and XhoI, respectively) (Fwd, AGC ACC CAT GGC CAG CAC CGG CCG CTT CAC GCT GC; Rev, CGC GGC TCG AGG GCT TCA CGC CAT GCG ATG CCG CTG TC), using the genomic DNA as the template, and inserted into the pET28a vector at NcoI/XhoI sites using a DNA ligation kit, Ligation High (Toyobo, Osaka, Japan).
Expression of LNBP.
The constructed plasmid was used to transform E. coli BL21(DE3). The transformants were cultivated at 30°C by shaking them in 1 liter of Luria-Bertani medium (containing 50 μg ml−1 kanamycin) until an optical density at 600 nm of 0.5 was reached. Protein production was then induced by the addition of isopropyl-1-thio-β-d-galactoside to the culture to a final concentration of 0.5 mM, followed by further incubation for 20 h (30°C) with shaking. The cells were harvested by centrifugation at 15,000 × g for 10 min and resuspended in 20 mM MOPS buffer (pH 7.0). The resuspended cells were sonicated (Branson Ultrasonic Corp.), and the cell debris was removed by centrifugation at 17,000 × g for 30 min. The crude enzyme was purified by column chromatography using a column of Ni-nitrilotriacetic acid agarose (QIAGEN, Hilden, Germany) in accordance with the supplier's protocol.
Substrate specificity.
To investigate the substrate specificity, the enzymatic reaction was carried out in a solution containing 0.5 U/ml LNBP, 50 mM donor (Glc1-P or Gal1-P), and 50 mM acceptor (GlcNAc, GalNAc, Glc, or Gal) in 100 mM MOPS buffer (pH 7.0) at 37°C for 2 h. An aliquot of the reaction mixture (1 μl) was then spotted on a thin-layer chromatography plate (5 by 7.5 cm; Silica gel 60F254; Merck, Darmstadt, Germany), and the plate was developed with a solvent system of acetonitrile-water (75:25). Carbohydrates on the plate were visualized by heating them at 120°C after dipping the plate in 5% sulfuric acid in methanol.
RESULTS
Purification of intracellular LNBP of B. bifidum.
We purified LNBP from the cell extract of B. bifidum JCM1254 as shown in Table 1. The purified LNBP appeared as a single band with an apparent molecular mass of 86 kDa on SDS-PAGE and showed a specific activity of 10 U/mg of protein. The protein was considered to take a dimer structure based on its native molecular mass of 140 kDa reported for the partially purified LNBP (13).
TABLE 1.
Purification of LNBP from B. bifidum JCM 1254
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Purification (n-fold) |
|---|---|---|---|---|
| Crude extract | 524 | 37 | 0.07 | 1 |
| 30% saturated (NH4)2SO4 | 181 | 22 | 0.12 | 1.8 |
| Butyl-Toyopearl | 3.4 | 1.9 | 0.56 | 7.8 |
| DEAE-Toyopearl | 0.67 | 1.5 | 2.2 | 30 |
| Mono-Q 1st | 0.23 | 1.0 | 4.6 | 64 |
| Mono-Q 2nd | 0.050 | 0.50 | 10 | 139 |
The purified enzyme phosphorolyzed both LNB and GNB, as previously described with the partially purified LNBP (13). The donor and acceptor specificities in the reverse reaction were also identical to those reported previously (13). The N-terminal and two internal amino acid sequences were determined to be STSGR FTIPS ESNFA EKTAE LARLW GADAV, YGYRL RPEDF VNEGS YNSAW RVPRK, and LGGID FGEPI ADTFP VNEDV TLLRA DGGQV, respectively. On BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) (1), all of the sequences showed high degrees of similarity to a hypothetical protein encoded by the BL1641 gene of Bifidobacterium longum NCC2705 (31) (GenBank accession no. AAN25428.1), the complete genome sequence of which was available, as shown in Fig. 1.
FIG. 1.
Alignment of the amino acid sequences of LNBPs. The origins of the enzymes are indicated in the left column. Conserved amino acid residues are highlighted.
Cloning and expression of LNBP gene of B. longum.
An LNBP gene, lnpA, was successfully amplified from genomic DNA of B. longum JCM1217 as the template with a set of primers designed based on the DNA sequence of B. longum NCC2705 around BL1641. The DNA and amino acid sequences of LNBP from B. longum JCM1217 were deposited in the DNA Databank of Japan (DDBJ) as AB181926. The gene encoded a protein of 752 amino acid residues with a predicted molecular weight of 84,327. The amino acid sequence showed 97% identity to that of the BL1641 gene with the same length and no gaps. On the other hand, the LNBP gene of B. bifidum JCM1254 could not be amplified using the same primer set.
The lnpA gene from B. longum JCM1217 was inserted into an expression vector to form pET28a-lnbp and transformed into E. coli BL21. The transformants harboring the plasmid were cultivated to induce the protein. The recombinant enzyme having an additional His6 sequence at its C terminus was purified. The recombinant LNBP of B. longum showed specific activity (19 U/mg) similar to that of the native LNBP from B. bifidum (10 U/mg). The specificity was tested on thin-layer chromatography, and both the native (B. bifidum) and recombinant (B. longum) LNBPs were shown to use Gal1-P as the donor and GlcNAc/GalNAc as the acceptor with the substrates examined.
Cultivation of B. longum JCM1217 was performed under the same conditions used for B. bifidum, and the strain did not produce LNBP in the glucose medium described in Materials and Methods, suggesting that LNBP is an inducible enzyme for B. longum.
Homology search.
No genes homologous with lnpA were found in other major intestinal bacteria, such as Lactobacillus, Bacteroides, and Escherichia. BLAST searches with the amino acid sequence of LNBP gave hits with only six proteins, including BL1641. One was a hypothetical protein (ZP_00121798.1) in the genomic sequence of another strain of B. longum, DJO10A, with 99% identity. The other three were hypothetical proteins of Clostridium perfringens (33) (locus CPE0573; 47% identity), Propionibacterium acnes (6) (locus PPA0083, 46% identity), Vibrio vulnificus CMCP6 (22) (locus VV21091; 38% identity), and V. vulnificus YJ016 (8) (locus VVA1614; 38% identity). No significant identity to any proteins with known function was found, indicating that LNBP should be classified in a new family.
DISCUSSION
Classification of LNBP.
Enzymes involved in the formation and cleavage of glycosyl linkages are classified into families based on amino acid sequence similarities that are categorized into three classes: glycoside hydrolase (GH) (18), glycosyl transferase (GT) (7), and glycoside lyase (http://afmb.cnrs-mrs.fr/CAZY/). LNBP is one of the carbohydrate-processing phosphorolytic enzymes listed in Table 2 (23). The amino acid sequence of LNBP showed no homology to any other phosphorolytic enzyme. Furthermore, no protein with known function was found to be homologous to LNBP, indicating that it should be classified in a new family.
TABLE 2.
Phosphorolytic enzymes
| EC 2.4.1.x | Name | Mechanism | Product | Family |
|---|---|---|---|---|
| 1 | (Glycogen) phosphorylase | Retention | α-Glc1-P | GT35 |
| 7 | Sucrose phosphorylase | Retention | α-Glc1-P | GH13 |
| 8 | Maltose phosphorylase | Inversion | β-Glc1-P | GH65 |
| 20 | Cellobiose phosphorylase | Inversion | α-Glc1-P | GH94 |
| 30 | 1,3-β-Oligoglucan phosphorylase | Inversion | α-Glc1-P | Not cloned |
| 31 | Laminaribiose phosphorylase | Inversion | α-Glc1-P | Not cloned |
| 49 | Cellodextrin phosphorylase | Inversion | α-Glc1-P | GH94 |
| 64 | Trehalose phosphorylase | Inversion | β-Glc1-P | GH65 |
| 97 | β-1,3-Glucan phosphorylase | Inversion | α-Glc1-P | Not cloned |
| 211 | Lacto-N-biose phosphorylase | Inversion | α-Gal1-P | New family |
| 216 | Trehalose 6-phosphate phosphorylase | Inversion | β-Glc1-P | GH65 |
| 230 | Kojibiose phosphorylase | Inversion | β-Glc1-P | GH65 |
| 231 | Trehalose phosphorylase | Retention | α-Glc1-P | GT4 |
| NDa | Chitobiose phosphorylase | Inversion | α-GlcNAcl-P | GH94 |
ND, not determined.
Phosphorolytic enzymes should initially be categorized into GT families according to the reactions that they catalyze. However, some phosphorolytic enzymes were classified into GH families (GH13 and -65) owing to the presence of homologous hydrolytic enzymes. Furthermore, structural analysis of chitobiose phosphorylase forced recategorization of GT36 into GH94 due to the structural and mechanistic relationships of these phosphorolytic enzymes with glycoside hydrolases of clan GH-L, although none of the members have hydrolytic activity (19). Thus, careful categorization is required to categorize phosphorolytic enzyme families. GT enzymes have been reported to have one of the two clans of fold, GT-A and GT-B (5, 12), whereas GH enzymes take various folds (5). However, prediction of the three-dimensional structure (21) of LNBP did not yield a probable fold, making categorization difficult. At present, we consider that LNBP should be classified in a new family that cannot be categorized rationally as belonging to either GT or GH.
Role of the novel galactose operon involving LNBP.
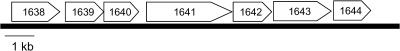
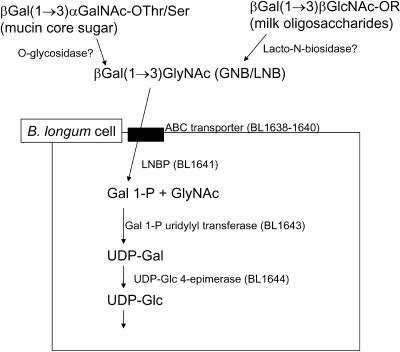
The LNBP gene, BL1641, seems to be located in a putative operon in the B. longum genome, as shown in Fig. 2 (31). In the operon, BL1638-1640 genes are annotated as component proteins of ATP-binding cassette (ABC)-type sugar transporters. BL1642, BL1643, and BL1644 are annotated as mucin desulfatase, galactose-1-phosphate uridylyltransferase (EC 2.7.7.10), and UDP-glucose 4-epimerase (EC 5.1.3.2), respectively. Judging from the members of the operon, LNBP seems to be related to the metabolism of mucin sugars, as shown in Fig. 3, because GNB is the core structure of mucin type sugars (13). In this pathway, Gal1-P formed by the phosphorolysis of LNB/GNB is converted to UDP-glucose to enter other pathways.
FIG. 2.
Putative lacto-N-biose operon found in the genomic sequence of B. longum NCC2705. BL1641 was identified as LNBP in this study. BL1638-1640 genes are annotated as component proteins of the ABC-type sugar transporter. BL1642, BL1643, and BL1644 are annotated as mucin desulfatase, galactose-1-phosphate uridylyltransferase (EC 2.7.7.10), and UDP-glucose 4-epimerase (EC 5.1.3.2), respectively.
FIG. 3.
Proposed scheme of lacto-N-biose metabolism.
Two types of operons of galactose metabolism have been reported (14). The first is the operon with the phosphoenolpyruvate-dependent phosphotransferase system (PTS) transporter. In this pathway, lactose is transported from outside the cell through the membrane by a lactose-specific PTS transporter to form lactose 6-phosphate, followed by hydrolysis with 6-phospho-β-galactosidase to yield glucose and galactose 6-phosphate. The second consists of galactokinase (EC 2.7.1.6), galactose-1-phosphate uridylyltransferase, and UDP-glucose 4-epimerase, accompanied by ABC transporters and α- and/or β-galactosidase. In this pathway, free galactose formed in hydrolysis by galactosidase is phosphorylated into Gal1-P by galactokinase. This is different from the LNB pathway, which does not require galactokinase and starts directly with Gal1-P.
A galactokinase gene is also found in the B. longum genome (BL1210), accompanied by a galactose-1-phosphate uridylyltransferase gene (BL1211). No galactosidase or UDP-glucose-epimerase genes were found near galactokinase. B. longum possesses another UDP-glucose 4-epimerase gene (BL1671) that is not accompanied by enzymes related to galactose metabolism. Based on the above observations, it can be hypothesized that the major galactose operon of B. longum consists of lnpA, expressing an enzyme very specific to LNB and GNB. This seems to constitute a system for intestinal colonization by utilizing mucin-type sugars, as illustrated in Fig. 3.
In addition, this operon may resolve the question of why bifidobacteria are predominant only in breastfed infants (24), as shown in Fig. 3. The major difference between mature bovine milk and human milk is the oligosaccharide content; human milk contains various oligosaccharides with the LNB structure at the nonreducing end (24), whereas mature bovine milk contains no oligosaccharides other than lactose (28). Furthermore, the bovine colostrum contains small amounts of oligosaccharides, but LNB has not been found in their structures (28).
Human milk oligosaccharides have been suggested to be the bifidus factor, but the mechanism and structure necessary for the factor are unknown. In the present study, we showed that LNBP was present in all four strains of bifidobacteria examined for either LNBP activity or the lnbp gene. Taking all the results into account, it is reasonable to hypothesize that the LNB residues in human milk oligosaccharides act as the bifidus factor in breastfed infants. Furthermore, it should be noted that the human colostrum contains more oligosaccharides than the mature milk (24), aiding the rapid colonization of newborn infants.
The results of the present study suggest that LNB is the bifidus factor. Due to the poor availability of human milk oligosaccharides and LNB, it is difficult to examine their function as the bifidus factor. LNBP may be a practical catalyst for producing various derivatives of LNB at reasonable cost. LNB is expected to improve the health of bottle-fed infants.
Acknowledgments
We are grateful to the United Nations University and Kirin Brewery Company for granting a UNU-Kirin Fellowship to J.T. This work was supported in part by a grant from the Bio-oriented Technology Research Advancement Institution.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zheng, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benno, Y., and T. Mitsuoka. 1986. The development of gastrointestinal micro-flora in humans and animals. Bifidobacteria Microflora 5:13-25. [Google Scholar]
- 3.Benno, Y., K. Sawada, and T. Mitsuoka. 1984. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol. Immunol. 28:975-986. [DOI] [PubMed] [Google Scholar]
- 4.Bezkorovainy, A. 1989. Ecology of bifidobacteria, p. 29-72. In A. Bezkorovainy and R. Miller-Catchpole (ed.), Biochemistry and physiology of bifidobacteria. CRC Press, Cleveland, Ohio.
- 5.Bourne, Y., and B. Henrissat. 2001. Glycoside hydrolases and glycosyltransferases: families and functional modules. Curr. Opin. Struct. Biol. 11:593-600. [DOI] [PubMed] [Google Scholar]
- 6.Bruggemann, H., A. Henne, F. Hoster, H. Liesegang, A. Wiezer, A. Strittmatter, S. Hujer, P. Durre, and G. Gottschalk. 2004. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305:671-673. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326:929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow, J. 2002. Probiotics and prebiotics: a brief overview. J. Ren. Nutr. 12:76-86. [DOI] [PubMed] [Google Scholar]
- 10.Cleveland, D. W., S. G. Fischer, M. W. Kirschner, and U. K. Laemmli. 1977. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem. 252:1102-1106. [PubMed] [Google Scholar]
- 11.Collins, M. D., and G. R. Gibson. 1999. Probiotics, prebiotics, and synbiotics: approaches for modulating the microbial ecology of the gut. Am. J. Clin. Nutr. 69:1052S-1057S. [DOI] [PubMed] [Google Scholar]
- 12.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 13.Derensy-Dron, D., F. Krzewinski, C. Brassart, and S. Bouquelet. 1999. β-1,3-Galactosyl-N-acetylhexosamine phosphorylase from Bifidobacterium bifidum DSM 20082: characterization, partial purification and relation to mucin degradation. Biotechnol. Appl. Biochem. 29:3-10. [PubMed] [Google Scholar]
- 14.de Vos, W. M., and E. E. Vaughan. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 15:212-237. [DOI] [PubMed] [Google Scholar]
- 15.Farkas, E., J. Thiem, F. Krzewinski, and S. Bouquelet. 2000. Enzymatic synthesis of Galβ1-3GlcNAc derivatives utilising a phosphorylase from Bifidobacterium bifidum 20082. Synlett 2000:728-730. [Google Scholar]
- 16.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 17.Gyorgy, P., C. S. Rose, and G. F. Springer. 1954. Enzymatic inactivation of bifidus factor and blood group substances. J. Lab. Clin. Med. 43:543-552. [PubMed] [Google Scholar]
- 18.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hidaka, M., Y. Honda, M. Kitaoka, S. Nirasawa, K. Hayashi, T. Wakagi, H. Shoun, and S. Fushinobu. 2004. Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (α/α)6 barrel fold. Structure 12:937-947. [DOI] [PubMed] [Google Scholar]
- 20.Hoskins, L. C., M. Agustines, W. B. Mickee, E. T. Boulding, M. Kriaris, and G. Niedermeyer. 1985. Mucin degradation in human colon ecosystems. J. Clin. Investig. 75:944-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 22.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitaoka, M., and K. Hayashi. 2002. Carbohydrate processing phosphorolytic enzymes. Trends Glycosci. Glycotechnol. 14:35-50. [Google Scholar]
- 24.Kunz, C., S. Rudloff, W. Baier, N. Klein, and S. Strobel. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Nutr. 20:699-722. [DOI] [PubMed] [Google Scholar]
- 25.Larson, G., P. Falk, and L. C. Hoskins. 1988. Degradation of human glycosphingolipids by extracellular glycosidases from mucin-degrading bacteria of the human fecal flora. J. Biol. Chem. 263:10790-10798. [PubMed] [Google Scholar]
- 26.Legendre, N., and P. Matsudaira. 1988. Direct protein microsequencing from immobilon-p transfer. BioTechniques 6:154-159. [PubMed] [Google Scholar]
- 27.Lowry, O. H., and J. A. Lopez. 1946. The determination of inorganic phosphate in the presence of labile phosphate esters. J. Biol. Chem. 162:421-428. [PubMed] [Google Scholar]
- 28.Newburg, D. S., and S. H. Neubauer. 1995. Carbohydrate in milks: analysis, quantities, and significance, p. 273-349. In R. G. Jensen (ed.), Handbook of milk composition. Academic Press, San Diego, Calif.
- 29.Petuely, F. 1957. Bifidusflora bei Fraschenkindern durch bifidogene Substanzen (Bifidusfaktor). Z. Kinderheilkd. 79:174-179. [PubMed] [Google Scholar]
- 30.Rotimi, V. O., and B. I. Duerden. 1981. The development of the bacterial flora in normal neonates. J. Med. Microbiol. 14:51-58. [DOI] [PubMed] [Google Scholar]
- 31.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrezenmeir, J., and M. de Vrese. 2001. Probiotics, prebiotics, and synbiotics: approaching a definition. Am. J. Clin. Nutr. 73:361s-364s. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veerkamp, J. H. 1969. Uptake and metabolism of determinatives of 2-deoxy-2-amino-d-glucose in Bifidobacterium bifidum var. pennsylvanicus. Arch. Biochem. Biophys. 129:248-256. [DOI] [PubMed] [Google Scholar]