Abstract
A cluster of five genes, proposed to be involved in the formation of extracellular polysaccharide (EPS) precursors via the Leloir pathway, have been identified in the acidophilic autotroph Acidithiobacillus ferrooxidans. The order of the genes is luxA-galE-galK-pgm-galM, encoding a LuxA-like protein, UDP-glucose 4-epimerase, galactokinase, phosphoglucomutase, and galactose mutarotase, respectively. The gal cluster forms a single transcriptional unit and is therefore an operon. Two other putative genes of the Leloir pathway, galU, potentially encoding UDP-glucose pyrophosphorylase, and a gene designated galT-like, which may encode a galactose-1-phosphate uridylyltransferase-like activity, were found unlinked in the genome. Using semiquantitative reverse transcription-PCR, the genes of the gal operon were shown to be expressed more during growth in iron medium than in growth in sulfur medium. The functions of galE, pgm, galU, and the galT-like gene were validated by complementation of Escherichia coli mutants and by in vitro enzyme assays. The data suggest that A. ferrooxidans is capable of synthesizing the EPS precursors UDP-glucose and UDP-galactose. In addition, genes rfbA, -B, -C, and -D were identified in the genome of A. ferrooxidans, suggesting that it can also synthesize the EPS precursor dTDP-rhamnose. Since EPSs constitute the major bulk of biofilms, this study may provide an initial model for the metabolic pathways involved in biofilm formation in A. ferrooxidans and aid in understanding the role of biofilms in mineral leaching and the formation of acid mine drainage.
Acidithiobacillus ferrooxidans is an acidophilic, chemolithotrophic, mesophilic, γ-proteobacterium that thrives at pH 2 and functions as part of a consortium of microorganisms for the industrial recovery of metals such as copper and gold (13, 25). In the environment, the microorganism is found in mine drainage, coal wastes, and other acidic sites, especially where pyrite (FeS2) is available as an energy source. A. ferrooxidans can obtain its energy and electron requirements from the oxidation of various forms of reduced sulfur and ferrous iron. It can also fix nitrogen and carbon dioxide.
The attachment and adherence of A. ferrooxidans to mineral surfaces and the subsequent formation of biofilms are prerequisites to mineral dissolution, both in industrial operations and in natural environments (27). Biofilm formation is accompanied by the production of extracellular polysaccharides (EPSs) (28). Whereas the role of biofilm formation by A. ferrooxidans in metal solubilization has been actively studied (8, 28), little is known regarding the underlying genetics, biochemistry, and regulation of EPS formation by this microorganism.
Many organisms use UDP-glucose, UDP-galactose, and dTDP-rhamnose as precursors or building blocks of EPS biosynthesis (33). The galactosides UDP-glucose and UDP-galactose are synthesized from glucose-1-phosphate by two enzymes of the Leloir pathway, GalU (glucose 1-phosphate-pyrophosphorylase) and GalE (UDP-glucose 4-epimerase) (1, 2, 4, 5, 9, 20, 32). Glucose-1-phosphate is also converted to dTDP-rhamnose by the enzymes RfbaA, -B, -C, and -D (14, 17). In addition, glucose-1-phosphate serves as a hub for channeling sugars to the formation of glycogen and connects these pathways with glycolysis, gluconeogenesis, and (ultimately) CO2 fixation.
Since nothing was known about the early steps in EPS biosynthesis in A. ferrooxidans and how these might be connected to general sugar management, we undertook bioinformatic and functional analyses of potential genes involved in both the formation of glucose-1-phosphate and its metabolic connections to the biosynthesis of the EPS precursors UDP-glucose, UDP-galactose, and dTDP-rhamnose.
This study addresses the genetic and biochemical underpinnings of the formation of EPS precursors in A. ferrooxidans. Since EPS typically constitutes the major mass of biofilms, it is hoped that this study will lay the foundation for understanding the formation of biofilms by this organism in both natural habitats and industrial metal recovery operations.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
A. ferrooxidans ATCC 23270 was grown at 30°C, pH 3.5, in minimal-salt medium 9K (iron free) supplemented with elemental sulfur (11, 29). Escherichia coli strains and plasmids used are listed in Table 1. They were grown in Luria broth (LB) or plated on LB solidified with 12 g of agar per liter. Selection for E. coli cells, transformed with the appropriate plasmids, was performed by using LB plates with of 100 μg of ampicillin per ml. MacConkey-Gal plates, containing MacConkey agar base (Difco Laboratories) and 5 g/liter of galactose, and MM-gal, a minimal medium (19) supplemented with 5 g/liter galactose, were used to determine the ability of galactose utilization by these strains. Red colonies in MacConkey-Gal plates and grown in MM-gal were scored as galactose positive (22).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant genotype or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| A. ferrooxidans | ATCC 23270 (type strain) | American Type Culture Collection |
| Escherichia coli | ||
| DH5α | F− Δ(lacZYA-argF) endA1 hsdR17 (rK mK+) supE44 thi-1 recA1 gyrA96 relA1 φ80lacZ ΔM15 | |
| S491 | F−galT22 his lac proA tonA uraP | National Institute of Genetics; SHIGEN |
| HB101 | F−λ−ara-14 galK2 hsdS20 (r-B,m-B) lacY1 leu mtl-1 proA2 recA13 rpsL20 supE44 thi xyl-5 | 1 |
| CSH41 | F+ Δ(lac-pro) galE thi | National Institute of Genetics; SHIGEN |
| JW1091 | λ−lacZ125 relA1 spoT1 trp-49 zbf-507::Tn10 pgm/zbf-507::Tn10 | National Institute of Genetics; SHIGEN |
| FF4001 | MC4100 galU95 | 21 |
| Plasmids | ||
| PKK223-3 | Ampr | Pharmacia Biotech |
| pUC18 | Ampr | Gibco BRL |
| pgalT-1 | Vector pUC18 with sequence of A. ferrooxidans that expresses GalT activity | This study |
| pgalU-1 | Vector pKK 233-3 with galU of A. ferrooxidans | This study |
| ppgm-1 | Vector pKK 233-3 with pgm of A. ferrooxidans | This study |
| pgalE | Vector pKK 233-3 with galE of A. ferrooxidans | This study |
Ampr, ampicillin resistant..
DNA isolation and sequencing, plasmid constructions, and other manipulations.
Chromosomal DNA was isolated from A. ferrooxidans ATCC 23270 as described previously (34). Other DNA manipulations, plasmid preparations, electroporations, and DNA sequencing were performed according to standard procedures as described by Sambrook et al. (26). The expression vector pKK233-3 was digested with EcoRI and subsequently repaired with the Klenow fragment of DNA polymerase I to generate blunt ends, which were dephosphorylated with calf intestinal alkaline phosphatase (New England Bio Labs) prior to being cloned. PCR fragments that correspond to galE, galU, pgm, and luxA genes were amplified with DNA primers whose sequences appear in Table 2. Each PCR fragment and the pKK233-3 vector were ligated using T4 DNA ligase. The constructed plasmids were named pgalE, pgalU, ppgm, and pgalT (Table 1). Restriction enzyme digestion EcoRI (Invitrogen), Klenow (large) fragment of DNA polymerase I (Invitrogen), T4 DNA ligase (Promega), and calf intestinal alkaline phosphatase were used under conditions recommended by the suppliers.
TABLE 2.
Primers used for PCR experiments and gene cloning and RT-PCR experiments
| Gene | Positiona | Primer used for: |
|---|---|---|
| PCR | ||
| luxA | A | 5′ ATGCGGCATTTATTTTGGAGG 3′ |
| B | 5′ CGGTGACCAGCACCTGC 3′ | |
| galE | A | 5′ ATGCAGGTGTTGGTCACCGGCG 3′ |
| B | 5′ CAAAGTTCATAAGCCATTCTGCCACG 3′ | |
| pgm | A | 5′ ATGGCGGTATTGCAGATTG 3′ |
| B | 5′ TCAGGTGATGACAGTGGGG 3′ | |
| galU | A | 5′ ATGGCTGAAGTGCGCAAGGC 3′ |
| B | 5′ ACCCACCACCGGACGGGC 3′ | |
| RT-PCR | ||
| rbn | 1 | 5′ ATGTCCCGAAAGCGATGGT 3′ |
| luxA | 2 | 5′ TAAATGCCGCATCCACCCCG 3′ |
| 3 | 5′ ATGCGGCATTTATTTTGGAGG 3′ | |
| galE | 4 | 5′ CGGTGACCAGCACCTGC 3′ |
| 5 | 5′ ATGCAGGTGTTGGTCACCGGCG 3′ | |
| galK | 6 | 5′ TCACAAGGCAAAGCCCTGGGTG 3′ |
| 7 | 5′ ATGAACTTTGCTCATCATATTCCTGC 3′ | |
| pgm | 8 | 5′ TGCAATACCGCCATGGCA 3′ |
| 9 | 5′ ATGGCGGTATTGCAGATTG 3′ | |
| galM | 10 | 5′ GCCAATGGTATCGAGCAGT 3′ |
| 11 | 5′ GACAACGGCATGGGCTAC 3′ | |
| mgt | 12 | 5′ CACTTCCGTGGGCTC 3′ |
Position relative to genes shown in Fig. 1A and B.
Isolation of a galT-like complementing gene.
DNA of A. ferrooxidans was partially digested with Sau3A (Gibco BRL) according to the manufacturer's directions, yielding fragments with an average size of approximately 1 to 3 Kb. These fragments were cloned into pUC18, and the resulting library was used to transform E. coli strain S491 (Table 1). After overnight incubation at 37°C on MacConkey-Gal plates, red colonies were picked and grown in MM-gal with 100 μg of ampicillin/ml (21). A colony, galT-1, was selected and DNA was prepared for DNA sequencing.
Isolation of RNA and RT-PCR.
Total RNA was isolated from cells of A. ferrooxidans grown in 9K medium supplemented with S or FeSO4 to mid-log phase by the method of Hagen and Young (12) as modified by Guacucano et al. (10). Reverse transcription-PCR (RT-PCR), with various control reactions, was carried out as previously described (10). The sequences of the RT and PCR primers used are provided in Table 2.
Preparation of cell extracts.
Fifty milliliters of a fresh LB culture of E. coli grown for 12 h (late log phase) were harvested by centrifugation (6,000 rpm for 20 min; 4°C), and the cell pellet was suspended and washed twice with 0.05 M potassium phosphate, pH 7. Cells were suspended in 20 ml of 0.05 M potassium phosphate, pH 7, and disrupted by sonication (sonic power, 375 W; output control, 10) at 4°C for a total of 6 min (12 30-s sonication pulses and 30 s of rest in an ice bath with a Bronson 450 sonifier) (7). Cell debris was removed by centrifugation (6,000 rpm for 20 min; 4°C). The protein concentration in the supernatant was determined using the Bio-Rad protein assay based on the method of Bradford (3)
Enzyme assays.
All enzyme assays were performed at 30°C in a total volume of 1 ml with freshly prepared cell extracts. The formation of NADPH (α-phosphoglucomutase activity and galactose 1-phosphato-uridiltransferase activity) or NADH (UDP-galactose 4-epimerase activity and UDP-glucose pyrophosphorylase activity), products of the coupled reaction, were determined by measuring the increase in absorbance at 340 nm at different times (7). The blank consisted of the reaction mixture without cell extract.
α-Phosphoglucomutase (EC 2.7.5.1) activity was measured in a reaction mixture consisting of 179 mM glycylglycine (pH 7.4), 0.67 mM β-NADP, 0.02 mM glucose 1,6-diphosphate, 30 mM MgCl2, 43 mM l-cysteine, 1 U glucose 6-phosphate dehydrogenase, and 300 μl of cell extract. The reaction was initiated by the addition of 5.0 mM α-glucose 1-phosphate (7).
UDP-galactose 4-epimerase (EC 5.1.3.2) activity was measured in a reaction mixture consisting of 400 mM glycylglycine-NaOH buffer (pH 8.5), 5 mM MgCl2, 0.5 mM NAD, 0.015 U UDP-glucose dehydrogenase, and 300 μl of cell extract. The reaction was started by the addition of 0.2 mM UDP-galactose (7).
UDP-glucose pyrophosphorylase (EC 2.7.7.9) activity was measured in a reaction mixture consisting of 50 mM Tris-HCl buffer (pH 7.5), 8 mM MgCl2, 1.58 mg cysteine hydrochloride (pH 7.5), 0.5 mM NAD, 1.25 mM UTP, 0.015 U UDP-glucose dehydrogenase, and 300 μl of cell extract. The reaction was initiated by the addition of 1 mM α-glucose 1-phosphate (7).
Galactose 1-phosphate-uridiltransferase (EC 2.7.7.12) activity was measured in a reaction mixture consisting of 100 mM TEA buffer (pH 7.8) containing 10 mM MgCl2, 1 mM β-NADP, 0.25 mM glucose 1,6-diphosphate, 5 U of glucose 6-phosphate dehydrogenase, 3 of phosphoglucomutase, and 300 μl of cell extract. The reaction was started by the addition of 1 mM UDP-glucose and 1 mM galactose 1-phosphate (15).
Bioinformatic analysis.
Known metabolic pathways involved in galactose catabolism were obtained from BIOCYC (www.biocyc.org), KEGG (www.genome.ad.jp/kegg/), and ERGO (http://ergo.integratedgenomics.com/ERGO/). Amino acid sequences derived from genes identified as being involved in galactose metabolism were used as query sequences to search the partial genome sequence of A. ferrooxidans ATCC 23270 in the TIGR (www.tigr.org/) and ERGO databases using TBlastN and BlastP, respectively. When a prospective candidate gene was identified in TIGR or ERGO, its predicted amino acid sequence was then used to formulate a BlastP (www.ncbi.nlm.nih.gov) search of the nonredundant database at NCBI. Only bidirectional best hits were accepted as evidence for putative orthologs. Candidate genes and their translated proteins were further characterized employing the following bioinformatic tools: Block Maker (http://blocks.fhcrc.org/blocks/make_blocks.html), Pfam, Prosite, and domain predictions (http://motif.genome.jp/).
Nucleotide sequence accession numbers.
The following nucleotide sequences reported in this paper have been assigned GenBank accession numbers: AY751082 (galT-like) and AY789510 to AY789512 for galE, galU, and pgm, respectively.
RESULTS
Identification and characterization of candidate genes involved in the formation of EPS precursors. The following candidate genes and their predicted protein products, potentially involved in the biosynthesis of glucose-1-phosphate and its conversion to the EPS precursors (UDP-glucose, UDP-galactose, and dTDP-rhamnose), were identified in the genome sequence of the type strain of A. ferrooxidans ATCC 23270 by bioinformatic analysis: galM (galactose mutarotase; EC 5.1.3.3), galK (galactokinase; EC 2.7.1.6), galU (glucose 1-phosphate-pyrophosphorylase; EC 2.7.7.9), two copies of galE (UDP-glucose 4-epimerase; EC 5.1.3.2), pgm (phosphoglucomutase; EC 5.4.2.2), rfbA (dTDP glucose pyrophosphorylase; EC 2.7.7.24), rfbB (dTDP glucose-4,6-dehydratase; EC 4.2.1.4.6), rfbC (dTDP 4-hydrorhamnose-3,5-epimerase; EC 5.1.3.13), and rfbD (dTDP hydrorhamnose reductase; EC 1.1.1.133) (Table 3). The similarity of these candidate genes with their database matches, the predicted presence of diagnostic functional motifs and domains, and their suggested cluster of orthologous genes (COG) designations are listed in Table 3. No candidate gene with statistically significant similarity to galT was detected in the genome of A. ferrooxidans by bioinformatic analysis. However, a potential gene encoding a GalT-complementing activity (galactose 1-phosphate-uridyltransferase; EC 2.7.7.12) was experimentally isolated (see below).
TABLE 3.
List of candidate genes proposed to be involved in the formation of EPS precursors in A. ferrooxidans and their proposed activities
| Genea | Proposed enzyme activityb | Best BlastP hitc | % Simd | Scoree | E valuef | Motif(s)g |
|---|---|---|---|---|---|---|
| galE | UDP-glucose epimerase (EC 5.1.3.2) | Mycobacterium avium | 43 | 93 | 7e−18 | pfam 1370; COG 1087 |
| galK | Galactokinase (EC 2.7.1.6) | Streptomyces coelicor | 48 | 106 | 5e−22 | PD 339735 |
| pgm | Phosphoglucomutase (EC 5.4.2.2) | Synechococcus elongatus | 77 | 711 | 0.0 | pfam 2878; COG 0033 |
| galM | Aldose-1 epimerase (EC 5.1.3.3) | Sinorhizobium meliloti | 52 | 33.9 | 1.1 | |
| galT-like | Possible galactose-1-phosphate urydylyltransferase (EC 2.7.7.10) | Kinecococcus radiotolerans | 42 | 32 | 4.3 | COG 1085 |
| galU | UDP-glucose pyrophosphorylase (EC 2.7.7.9) | Burkholderia fungorum | 73 | 371 | 2e−73 | pfam 0483; COG 0451; PD 01252 |
| luxA-like | Possible coenzyme F420-dependent reductase | Bradyrhizobium japonicum | 44 | 157 | 3e−37 | pfam 0296; COG 2141 |
| rfbA | d-TDP-glucose pyrophosphorylase (EC 2.7.7.24) | Azotobacter vinelandii | 80 | 406 | 1e−112 | pfam 0483; COG 1209 |
| rfbB | d-TDP-glucose4,6dehydratase (EC 4.2.1.46) | Pseudomonas putida | 72 | 443 | 1e−123 | pfam 1370; COG 1088 |
| rfbC | d-TDP-4-dehydrorhamnose 3,5-epimerase (EC 5.1.3.13) | Pseudomona stutzeri | 79 | 245 | 3e−64 | pfam 0908; COG 1898 |
| rfbD | d-TDP-4-dehydrorhamnose reductase (EC 1.1.1.133) | Synechococcus elongatus | 63 | 261 | 1e−68 | pfam 4321; COG 1091 |
Proposed gene name.
Proposed enzyme activity.
Organism with the best BlastP hit to the candidate gene.
Percentage of similarity (% Sim) of candidate gene to that found in the organism listed in row (c).
Score of BlastP match.
E value of BlastP match.
Motif and domains identified in the candidate proteins. Pfam, protein families; PD, Prodom (protein domains).
Organization and expression of candidate genes potentially involved in the metabolism of glucose-1-phosphate.
Candidate genes lux A-like, galE, galK, pgm, and galM proposed to be involved in glucose-1-phosphate metabolism were found to be organized in a gene cluster (Fig. 1A). The cluster was flanked by the potential genes rbn and mgt, potentially encoding an oligoribonuclease and magnesium transporting ATPase, respectively. A possible pseudo-tRNA gene was detected between galM and mgt. RT-PCR experiments showed that luxA-like, galE, galK, pgm, and galM are cotranscribed, demonstrating that they form an operon, whereas the flanking rbn and mgt genes did not constitute part of this operon (Fig. 1B).
FIG. 1.
Organization and cotranscription of the gal operon of A. ferrooxidans. (A) The gal operon consists of luxA-like, galE, galK, pgm, and galM flanked by the genes rbn and mgt. A potential pseudo-tRNA is located between galM and mgt. The direction of transcription is indicated by the block arrows. Locations of PCR primers are shown below the operon. (B) Determination of cotranscription of the genes comprising the gal operon by RT-PCR. Gel electrophoresis of DNA fragments amplified by RT-PCR using purified RNA as a substrate (a) or PCR using genomic DNA as a substrate (b). Arrows indicate the predicted sizes of the amplified DNA fragments in base pairs. (C) A comparison of the organization of the gal operon of A. ferrooxidans with the gal operons of E. coli (18), Lactobacillus casei, and Klebsiella pneumoniae. The organization of these operons was derived from the Integrated Genomics web site (www.integratedgenomics.com).
A comparison of the organization of the A. ferrooxidans gal operon with other known or putative gal operons indicates a considerable degree of conservation of genes, although not necessarily preservation of gene order (Fig. 1C). This evidence supports the proposed function of the A. ferrooxidans gal operon and permits a focus to be placed on identifying genetic regulatory features in future studies.
Expression of the gal operon during growth in sulfur and iron.
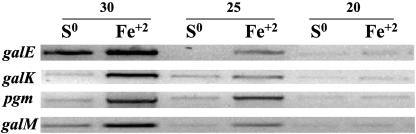
Several genes have been demonstrated to be differentially expressed in A. ferrooxidans grown in either iron or sulfur (16). With the goal of determining whether the oxidizable energy source also influenced the expression of genes potentially involved in galactose metabolism, a comparison was made by semiquantitative PCR of the expression of genes in the gal operon when A. ferrooxidans was grown in the presence of iron or sulfur. As shown in Fig. 2, four of the five genes of the gal operon (galE, galK, pgm, and galM) showed higher levels of transcription when cells were grown in iron rather than in sulfur-containing medium. The fifth gene of the gal operon, the luxA-like gene, was not tested.
FIG. 2.
Expression of several genes of the Gal cluster of A. ferrooxidans grown in the presence of sulfur (S0) or iron (Fe2+) as determined by semiquantitative PCR using 20, 25, or 30 cycles, respectively. PCR primers used are shown in Table 2.
Analysis of the lux A-like gene.
The predicted protein potentially derived from lux A-like exhibits similarity with the LuxA family of bacterial luciferase-like monooxygenases and contains the pfam00296 domain characteristic of this family (6, 31). It also contains COG2141, characteristic of the coenzyme F420-dependent N5,N10-methylene tetrahydromethanopterin reductase and related flavin-dependent oxidoreductases. For example, the domain is found in glucose-6-phosphate dehydrogenase from Rhizobium sp. (30). The A. ferrooxidans LuxA-like protein exhibited weak similarity (47%) with this protein (BlastP score = 79; E value = 1e−13), which initiates the pentose phosphate pathway (23, 24). Therefore, a possible function of the LuxA-like protein could be to connect the Leloir pathway with the pentose phosphate pathway. Supporting this conjecture is the presence of the putative pgm gene in the same operon as luxA-like, encoding a PGM protein involved in the synthesis of glucose-6-phosphate. The latter is a major point of entry of carbon into the pentose phosphate pathway.
An alternative hypothesis is that lux A-like encodes the missing GalT function typically found in the gal operon but not detected in the A. ferrooxidans gal operon. However, in an in vitro enzyme assay, LuxA-like did not exhibit galactose 1-phosphate-uridyltransferase activity, diminishing the likelihood of this conjecture.
Identification of a galT-like gene.
Since no candidate galT gene was detected in the genome of A. ferrooxidans by bioinformatic analysis, it was decided to explore the possibility of the existence of GalT activity by direct experimental techniques. DNA of A. ferrooxidans was partially digested with Sau3A (1- to 3-Kb fragments) and was inserted into pUC18. The resulting recombinant library was used to transform a strain of E. coli (S491) lacking galT. A plasmid that was able to complement the missing GalT function was isolated and designated GalT-1. The insert in GalT-1 was sequenced, and the resulting sequence was used to identify a corresponding open reading frame in the partial genome sequence of A. ferrooxidans (TIGR) using BlastN. This open reading frame was designated galT-like.
Further analysis of the partial genome sequence revealed that galT-like was embedded in a gene cluster that potentially encodes other genes that may be involved in galactose metabolism, although their bioinformatic identification by BlastP was below a statistically significant threshold and could only be detected by Psi-BLAST (analysis not shown). The predicted GalT-like product was 120 amino acids long, which is shorter than most GalT proteins (340 amino acids). It also lacked the pfam01230 domain that includes the characteristic histidine triad of the GalT family (18).
Complementation of E. coli mutants with genes from A. ferrooxidans.
E. coli strains CSH41(lacking galE), JW1091(lacking pgm), FF4001(lacking galU), and S491(lacking galT) were independently transformed with pKK233-3 containing cloned candidate genes from A. ferrooxidans potentially encoding GalE, PGM, GalU, and GalT, respectively, and giving rise to the recombinant plasmids pgalE-1, ppgm-1, pgalU-1, and pgalT-1, respectively. In the first three cases, the cloned genes were prepared by PCR of A. ferrooxidans genomic DNA using primers designed from the predicted start sites to the predicted stop sites as identified by bioinformatic analysis of the genome sequence. Expression of these cloned inserts was presumed to occur from the vector promoter. Each of these recombinant plasmids was capable of complementing the respective E. coli mutant, as shown by comparing growth with that of wild-type E. coli DH5α (Fig. 3).
FIG. 3.
Growth at 37°C in minimal medium supplemented with 0.5% galactose of E. coli mutants with or without vectors containing A. ferrooxidans genes. □, wild-type E. coli DH5α; ▪, mutant E. coli strain CSH41 (lacking galE), JW1091 (lacking pgm), FF4001 (lacking galU), orS491(galT); ♦, CSH41 (lacking galE) with pgalE-1; X, JW1091 (lacking pgm) with ppgm-1; ▴, FF4001 (lacking galU) with pgalU-1; and ○, S491 (lacking galT) with pgalT-1.
In contrast, pgalT-1 was derived by complementation of E. coli S491 (lacking galT) using a randomly prepared library of A. ferrooxidans genomic DNA in pUC18. It was capable of complementing E. coli S491 (lacking galT) (Fig. 3), but growth of the recombinant strain was slower than that of E. coli DH5α. It is possible that the proposed galT was transcribed from an A. ferrooxidans promoter, resulting in lower levels of transcription or, alternatively, that the A. ferrooxidans GalT was less efficient than E. coli GalT for reasons of translation inefficiency, instability, or reduced enzyme activity.
Assays of enzyme activities from E. coli harboring cloned A. ferrooxidans genes.
E. coli strains CSH41(lacking galE), JW1091(lacking pgm), FF4001(galU), and S491(lacking galT) harboring the respective complementing plasmids pgalE-1, ppgm-1, pgalU-1, and pgalT-1 were grown overnight, and the enzymatic activities of GalE, PGM, GalU and GalT were analyzed as described in Materials and Methods (Table 4). These activities were compared to those derived from the appropriate E. coli mutant strain and from wild-type strain DH5α (Table 4). E. coli mutant strains harboring the proposed galE, pgm, and galU of A. ferrooxidans exhibited more activity for each of the respective enzymes than E. coli DH5α, possibly because of the presence of more enzyme mass due to expression of a multicopy plasmid. E. coli S491 harboring the potential galT-like gene of A. ferrooxidans also exhibited slightly more activity for GalT than did E. coli DH5α. Enzyme activities are measured in late-log cells where the growth of E. coli S491 containing galT-like was similar to that of E. coli DH5α (Fig. 3), which suggests that the initial reduction in growth rate observed for E. coli S491 containing galT-like could reflect the less-efficient induction of the A. ferrooxidans galT-like gene in a heterologous host than that of the native gene and not to some intrinsic property of the GalT-like enzyme.
TABLE 4.
Enzyme activity measurements
| Strain (relevant mutation) | Enzyme activitya
|
||
|---|---|---|---|
| Negative controlb | Positive controlc | + Complementing plasmidd | |
| CSH41 (galE) | 0 | 68 ± 5 | 109 ± 2 (pgalE-1) |
| JW1091 (pgm) | 27 ± 1 | 60 ± 12 | 121 ± 7 (ppgm-1) |
| FF4001 (galU) | 0.5 ± 0.1 | 19 ± 6 | 45 ± 7 (pgalU-1) |
| S491 (galT) | 5 ± 1.0 | 10 ± 2 | 18 ± 1 (pgalT-1) |
Enzyme activity expressed as the number of nanomoles per minute per milligram of total cell protein. Each value is the average of three measurements (± standard deviation). All strains were grown in 50 ml of LB medium at 37°C for 12 h with 0.5% galactose at 30°C.
Enzyme activities in mutant strains transformed with the vector pKK233-3 but lacking the A. ferrooxidans insert.
Enzyme activities of E. coli DH5α (wild type for each of the respective enzyme activities).
Enzyme activities of mutant strains complemented with the plasmid indicated in brackets carrying the respective A. ferrooxidans insert.
DISCUSSION
Inspection of the partial genome sequence of A. ferrooxidans provided by the Institute for Genome Research and by Integrated Genomics reveals the presence of a number of potential genes that exhibit significant similarity to genes known to be involved in the metabolism of glucose-1-phosphate and its conversion to the EPS precursors UDP-glucose, UDP-galactose, and dTDP-rhamnose via the Leloir pathway (Table 3). Figure 4 shows a working model for how these genes and their products might be connected in the formation of EPS precursors in A. ferrooxidans.
FIG. 4.
Schematic representation of proposed pathways (solid arrows) and potential genes involved in the biosynthesis of glucose-1-phosphate (circled) and its role in the production of the precursors of EPS formation in A. ferrooxidans. Dotted arrows indicate suggested connections between glucose-1-phosphate and glycogen anabolism and catabolism, pentose phosphate pathway, glycolysis, gluconeogenesis, and the formation of EPS via glycosyltransferases (GTs).
The genes that have been identified include galE, galK, pgm, and galM, organized in a gene cluster that is cotranscribed (Fig. 1A and B) and thus constituting an operon (gal operon) in A. ferrooxidans. This gal operon exhibits conservation of gene organization with other well-described gal operons (Fig. 1C). Two of the genes of the gal operon, galE and pgm, were shown to complement E. coli mutants lacking these functions (Fig. 3) and also to encode GalE and Pgm activities, respectively, as determined by enzyme assays (Table 4). Another gene that has been identified in this study that probably plays a role in glucose-1-phosphate metabolism but does not form part of the A. ferrooxidans gal operon is a potential galU. The putative GalU protein has significant sequence similarity to GalU proteins in other organisms and exhibits GalU enzymatic activity in vitro (Table 4). It also complements an E. coli mutant lacking galU (Fig. 3).
Semiquantitative PCR experiments demonstrate that the genes of the A. ferrooxidans gal operon are expressed more when the microorganism is grown in the presence of Fe2+ as an energy source than with S0 (Fig. 2). It has been established that the energy substrate used by A. ferrooxidans influences the quantity and chemical composition of the EPS formed (8, 26). It was proposed that variations in EPS formation demonstrate the need for different mechanisms of adhesion of the microorganism to diverse substrates encountered in the environment. We speculate that the differential expression of genes of the gal operon reported here reflect this need and suggest how it might be accomplished.
A galT gene could not be detected initially in the genome sequence of A. ferrooxidans by bioinformatic analysis. However, an A. ferrooxidans sequence (galT-like) complementing growth of E. coli S491 (Fig. 3) and providing GalT enzymatic activity (Table 4) was experimentally identified from a shotgun plasmid library of A. ferrooxidans DNA cloned into E. coli S491. The sequence of galT-like revealed very weak similarity with known galT genes (Table 3), although it was considerably shorter than typical galT and did not exhibit the GalT prosite motif.
Putative genes rfbA, -B, -C, and -D potentially encoding enzymes that convert glucose-1-phosphate to dTDP-rhamnose were identified by bioinformatic analysis in the genome of A. ferrooxidans and exhibit significant sequence similarity to enzymes in other organisms (Table 3). However, their function awaits experimental validation.
More troublesome is the identification of the function of the luxA-like gene that forms part of the A. ferrooxidans gal operon. It exhibits weak similarity to an F420-dependent glucose-6-phosphate dehydrogenase that catalyzes the conversion of glucose-6-phosphate to 6-phosphogluconolactone. pgm also forms part of the gal operon; its role is the conversion of glucose-1-phosphate to glucose-6-phosphate, which could then be converted to 6-phosphogluconolactone by the hypothetical product of the luxA-like gene and subsequently enter the pentose phosphate pathway. Thus, a speculative function for the LuxA-like product could be to connect glucose-1-phosphate metabolism with the pentose phosphate pathway (Fig. 4). If LuxA-like exhibited reverse activity, it could catalyze the conversion of 6-phosphogluconolactone to glucose-6-phosphate, and then it could help channel products of CO2 fixation towards the formation of EPS precursors.
Taking these results as a whole, including sequence similarities, genetic organization, and (in several instances) experimental validation, it appears that A. ferrooxidans has the genetic potential to encode the EPS precursors UDP-glucose, UDP-galactose, and dTDP-rhamnose using glucose-1-phosphate as a central starting point (Fig. 4). Glucose-1-phosphate serves as a hub of carbohydrate metabolism, channeling sugars to the formation of extracellular polysaccharides (EPS) and glycogen and connecting these pathways with glycolysis, gluconeogenesis, perhaps the pentose phosphate pathway, and ultimately CO2 fixation (Fig. 4). Future work will be directed towards an examination of the mechanisms involved in the regulation of the genes involved in glucose-1-phosphate formation in A. ferrooxidans and the pathways involved in the formation of EPS from the EPS precursors suggested in this study. It is anticipated that this information will provide a working model for understanding the formation of biofilms by A. ferrooxidans and help in understanding the role of this microorganism in mineral leaching and the formation of acid mine drainage.
Acknowledgments
The work was supported by Fondecyt grant no. 1010623. M.B. was supported by a scholarship from Deutscher Akademischer Austauschdienst (DAAD). Sequencing of A. ferrooxidans at TIGR was supported by the U.S. Department of Energy (DOE).
We thank the National Institute of Genetics (SHIGEN) for providing E. coli strains S491, CSH41, and JW1091 and Katja Bettenbrock and Ernesto Garcia for E. coli strain FF4001. We thank the Institute of Genome Research (TIGR) and Integrated Genomics, Inc. (IG), for the use of their partial sequences of the A. ferrooxidans genome.
REFERENCES
- 1.Bettenbrock, K., and C. Alpert. 1998. The gal genes for the Leloir pathway of Lactobacillus casei 64H. Appl. Environ. Microbiol. 64:2013-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boels, I., A. Ramos, M. Kleerebezem, and W. De Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Broadbent, J. R., D. McMahon, D. L. Welker, C. J. Ober, and S. Moineau. 2003. Biochemistry, genetics, and applications of exopolysaccharide production in Streptococcus thermophilus: a review. J. Dairy Sci. 86:407-423. [DOI] [PubMed] [Google Scholar]
- 5.Chatterje, S., Y. N. Zhou, S. Roy, and S. Adhya. 1997. Interaction of Gal repressor with inducer and operator: induction of Gal transcription from repressor-bound DNA. Proc. Natl. Acad. Sci. USA 94:2957-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czyz, A., B. Wróbel, and G. Wegrzyn. 2000. Vibrio harveyi bioluminescence plays a role in stimulation of DNA repair. Microbiology 146:283-288. [DOI] [PubMed] [Google Scholar]
- 7.Degeest, B., and L. Vuyst. 2000. Correlation of activities of the enzymes α-phosphoglucomutase, UDP-galactose 4-epimerase, and UDP-glucose pyrophosphorylase with exopolysaccharide biosynthesis by Streptococcus thermophilus LY03. Appl. Environ. Microbiol. 66:3519-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gehrke, T., J. Telegdi, D. Thierry, and W. Sand. 1998. Importance of extracellular polymeric substances from Thiobacillus ferrooxidans for bioleaching. Appl. Environ. Microbiol. 64:2743-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrich, J. A., and W. McClure. 1992. Regulation of open complex formation at the Escherichia coli galactose operon promoters. J. Mol. Biol. 224:15-29. [DOI] [PubMed] [Google Scholar]
- 10.Guacucano, M., G. Levican, D. S. Holmes, and E. Jedlicki. 2000. An RT-PCR artifact in the characterization of bacterial operons. Electron. J. Biotechnol. 3:213-216. [Google Scholar]
- 11.Guiliani, N., A. Bengrine, F. Borne, M. Chippaux, and V. Bonnefoy. 1997. Alanyl-tRNA synthetase gene of the extreme acidophilic chemolithoautotrophic Thiobacillus ferrooxidans is highly homologous to alaS genes from all living kingdoms but cannot be transcribed from its promoter in Escherichia coli. Microbiology 143:2179-2187. [DOI] [PubMed] [Google Scholar]
- 12.Hagen, F., and E. Young. 1978. Effect of RNase III on efficiency of translation of bacteriophage T7 lysozyme mRNA. J. Virol. 29:793-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes, D. S. 1999. Biotechnology: miners in miniature. Chem. Ind. 1:20-24. [Google Scholar]
- 14.Laws, A., Y. Gu, and V. Marshall. 2001. Biosynthesis, characterization, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol. Adv. 18:597-625. [DOI] [PubMed] [Google Scholar]
- 15.Levander, F., M. Svensson, and P. Rådström. 2002. Enhanced exopolysaccharide production by metabolic engineering of Streptococcus thermophilus. Appl. Environ. Microbiol. 68:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levican, G., P. Bruscella, M. Guacunano, C. Inostroza, V. Bonnefoy, D. S. Holmes, and E. Jedlicki. 2002. Characterization of the petI and res operons of Acidithiobacillus ferrooxidans. J. Bacteriol. 184:1498-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Looijesteijn, P., I. Boels, M. Kleerebezem, and M. Hugenholtz. 1999. Regulation of exopolysaccharide production by Lactococcus lactis subs. cremoris by the sugar source. Appl. Environ. Microbiol. 65:5003-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchler-Bauer, A., J. B. Anderson, C. De Weese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, and C. J. Lanczycki. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1978. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 20.Moller, T., T. Franch, C. Udesen, K. Gerdes, and P. Valentin-Hansen. 2002. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 16:1696-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mollerach, M., R. Lopez, and E. Garcia. 1998. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J. Exp. Med. 188:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng, H-L., T-F. Fu, S.-F. Liu, and H.-Y. Chang. 1992. Cloning and expression of the Klebsiella pneumoniae galactose operon. J. Biochem. 112:604-608. [DOI] [PubMed] [Google Scholar]
- 23.Purwantini, E., and L. Daniels. 1998. Molecular analysis of the gene encoding F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J. Bacteriol. 180:2212-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purwantini, E., T. Gillis, and L. Daniels. 1997. Presence of F420-dependent glucose-6-phosphate dehydrogenase in Mycobacterium and Nocardia species, but absence from Streptomyces and Corynebacterium species and methanogenic Archaea. FEMS Microbiol. Lett. 146:129-134. [DOI] [PubMed] [Google Scholar]
- 25.Rawlings, D., and T. Kusano. 1994. Molecular genetics of Thiobacillus ferrooxidans. Microbiol. Rev. 58:39-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Sand, W., T. Gehrke, P.-G. Jozsa, and A. Schippers. 1999. Direct versus indirect bioleaching, p. 27-47. In R. Amils and A. Ballester (ed.), Biohydrometallurgy and the environment toward the mining of the 21st century. Elsevier, Madrid, Spain.
- 28.Sand, W., and T. Gehrke. 1999. Analysis and function of the EPS from the strong acidophile Thiobacillus ferrooxidans. pg. 127-141. In J. Wingender, T. R. Neu and H.-C. Fleming (ed.), Microbial extracellular polymeric substances: characterization, structure and function. Springer, Berlin, Germany.
- 29.Schrader, J., and D. Holmes. 1988. Phenotypic switching of Thiobacillus ferrooxidans. J. Bacteriol. 170:3915-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Streit, W. R., R. A. Schmitz, X. Perret, C. Staehelin, W. J. Deakin, C. Raasch, H. Liesegang, and W. J. Broughton. 2004. An evolutionary hot spot: the pNGR234b replicon of Rhizobium sp. strain NGR234. J. Bacteriol. 186:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szpilewska, H., A. Czyz, and G. Wegrzyn. 2003. Experimental evidence for the physiological role of bacterial luciferase in the protection of cells against oxidative stress. Curr. Microbiol. 47:379-382. [DOI] [PubMed] [Google Scholar]
- 32.Weickert, M., and S. Adhya. 1993. The galactose regulon of Escherichia coli. Mol. Microbiol. 10:245-251. [DOI] [PubMed] [Google Scholar]
- 33.Whitfield, C., and A. Paiment. 2003. Biosynthesis and assembly of group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr. Res. 338:2491-2502. [DOI] [PubMed] [Google Scholar]
- 34.Yates, J. R., and D. S. Holmes. 1986. Two families of repeated DNA sequences in Thiobacillus ferrooxidans. J. Bacteriol. 169:1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]