Abstract
The ergot alkaloids are a family of indole-derived mycotoxins with a variety of significant biological activities. Aspergillus fumigatus, a common airborne fungus and opportunistic human pathogen, and several fungi in the relatively distant taxon Clavicipitaceae (clavicipitaceous fungi) produce different sets of ergot alkaloids. The ergot alkaloids of these divergent fungi share a four-member ergoline ring but differ in the number, type, and position of the side chains. Several genes required for ergot alkaloid production are known in the clavicipitaceous fungi, and these genes are clustered in the genome of the ergot fungus Claviceps purpurea. We investigated whether the ergot alkaloids of A. fumigatus have a common biosynthetic and genetic origin with those of the clavicipitaceous fungi. A homolog of dmaW, the gene controlling the determinant step in the ergot alkaloid pathway of clavicipitaceous fungi, was identified in the A. fumigatus genome. Knockout of dmaW eliminated all known ergot alkaloids from A. fumigatus, and complementation of the mutation restored ergot alkaloid production. Clustered with dmaW in the A. fumigatus genome are sequences corresponding to five genes previously proposed to encode steps in the ergot alkaloid pathway of C. purpurea, as well as additional sequences whose deduced protein products are consistent with their involvement in the ergot alkaloid pathway. The corresponding genes have similarities in their nucleotide sequences, but the orientations and positions within the cluster of several of these genes differ. The data indicate that the ergot alkaloid biosynthetic capabilities in A. fumigatus and the clavicipitaceous fungi had a common origin.
The ergot alkaloids are a complex family of indole-derived mycotoxins with varied and significant biological activities (6, 7, 14, 25). Fungi derived from two relatively divergent ascomycete orders (Hypocreales and Eurotiales) produce ergot alkaloids. Several fungi in the family Clavicipitaceae (order Hypocreales), including the ergot fungi in the genus Claviceps and certain grass endophytes in the genera Epichloë (including their imperfect relatives in the genus Neotyphodium) and Balansia, produce ergot alkaloids in association with their grass hosts (3, 6, 7, 14, 25). The rather distantly related imperfect fungus Aspergillus fumigatus, a common saprophyte and opportunistic human pathogen with close relatives in the order Eurotiales, produces ergot alkaloids in broth culture (4, 23) and in association with its airborne conidia (13).
Ergot alkaloid-producing members of the Clavicipitaceae (clavicipitaceous fungi) produce a wide variety of clavine and lysergyl-derived ergot alkaloids but commonly accumulate ergopeptines (nonribosomally synthesized peptides containing lysergic acid and three amino acids) or simple amides of lysergic acid, such as ergine and ergonovine (6, 7, 8, 14). A. fumigatus produces several clavine ergot alkaloids, including festuclavine and fumigaclavines A, B, and C (4, 13, 23). Most of the clavine and lysergyl-derived ergot alkaloids contain the same four-member ergoline ring system, but they differ in the number, type, and position of the side chains (6, 7, 8, 14).
Festuclavine, one of the ergot alkaloids produced by A. fumigatus, was first described from Claviceps purpurea (6, 7) and also has been detected in Neotyphodium spp. (15, 19). However, none of the fumigaclavines produced by A. fumigatus has been detected in a member of the Clavicipitaceae. Likewise, the end products of the Claviceps spp. and Neotyphodium spp. pathways (ergopeptines and lysergic acid amides) have not been found in A. fumigatus. This distribution of alkaloids is consistent with the hypothesis that early steps of the ergot alkaloid pathways are shared by these diverse fungal species but the later portions of the pathways are different in A. fumigatus and the clavicipitaceous fungi.
The ergot alkaloid pathway is relatively long and may have alternate branches or spurs in some ergot alkaloid producers (7, 8, 14, 15). Functions of three genes involved in the ergot alkaloid pathway have been demonstrated by gene knockout. The dmaW gene encoding dimethylallyltryptophan synthase (DMAT synthase) was knocked out in Neotyphodium sp. strain Lp1, an endophyte of perennial ryegrass (29). Loss of all ergot alkaloids in the knockout mutant indicated that dmaW controls the determinant step in the ergot alkaloid pathway. A gene encoding lysergyl peptide synthetase 1 (LPS1), one of two components of the lysergyl peptide synthetase complex responsible for assembly of ergopeptines from lysergic acid and three amino acids (20, 28), was also knocked out in Neotyphodium sp. strain Lp1 (16). LPS1-deficient knockout mutants failed to produce ergopeptines and simple amides of lysergic acid but still produced clavine ergot alkaloids (15, 16). A gene encoding lysergyl peptide synthetase 2 (LPS2), the lysergic acid-activating component of the lysergyl peptide synthetase complex, has been cloned and knocked out in C. purpurea, resulting in the loss of ergopeptines from the knockout mutant (5).
Genes encoding DMAT synthase, LPS1, and LPS2 are clustered within ∼32 kb of each other in the genome of C. purpurea (5, 25). Moreover, at least six additional genes that encode activities likely to be required for ergot alkaloid production are interspersed in this cluster with the ergot alkaloid biosynthesis genes (5, 25, 26). Analysis of mRNAs from five of the clustered genes indicate they are expressed in cultures grown under conditions conducive to ergot alkaloid production but are not expressed under repressive conditions (5). Such clustering and common regulation of secondary metabolite biosynthetic pathway genes are common in fungi (9, 27).
Our objective in this study was to determine whether the ergot alkaloids of A. fumigatus had a common biosynthetic origin with those of the clavicipitaceous fungi by identification and functional analysis of the dmaW gene controlling the determinant step in the pathway and by comparison of sequences flanking dmaW in A. fumigatus and C. purpurea.
MATERIALS AND METHODS
Fungi and culture conditions.
A. fumigatus isolate WVU1943 (=FGSC A1141) from a parakeet lung is described in the accompanying paper (13) and was manipulated in a class II biosafety cabinet. Isolates were maintained on potato dextrose agar (PDA) (20 g/liter dehydrated mashed potatoes [Idaho Spuds; Pillsbury, Minneapolis, MN], 20 g/liter glucose, 15 g/liter agar [Bacto agar; Difco, Detroit, MI]) at room temperature or at 37°C. Cultures for analysis of ergot alkaloids were grown on PDA at 37°C. For preparation of DNA or protoplasts, the fungus was cultured overnight in 15 ml of potato dextrose broth (Difco) in a deep petri dish (depth, 20 mm; diameter, 100 mm; Fisher Scientific, Pittsburgh, PA) on an orbital shaker at 37°C and 150 rpm. Cultures were inoculated by lightly dusting the medium with conidia that had adhered to a petri dish lid formerly covering a mature culture. In mock inoculations, an average of 1.2 × 107 conidia per petri dish was deposited by this inoculation method.
Analysis of the A. fumigatus genome for ergot alkaloid biosynthesis genes.
A. fumigatus preliminary sequence data were obtained from The Institute for Genomic Research website (http://www.tigr.org). All A. fumigatus sequences described in this paper are located on assembly 92 (chromosome 2, formerly assembly 72) between nucleotides 2894958 and 2926335. The presence of sequences in the A. fumigatus genome that are similar to known or putative ergot alkaloid biosynthesis genes was assessed by application of the tblastn algorithm (1) at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov) with the deduced protein sequences of DMAT synthase of Claviceps fusiformis (24) and LPS1 of Neotyphodium lolii (16) or C. purpurea (26) as query sequences. The nature of genes linked to the ergot alkaloid biosynthesis genes or candidate genes was investigated by retrieving contiguous 2-kb segments of DNA from the A. fumigatus genome and querying the nonredundant protein database of the National Center for Biotechnology Information with these sequences by application of the blastx search algorithm (1).
Knockout and complementation of dmaW in A. fumigatus.
Fungal DNA for amplification by PCR was isolated by using the Gene Clean Spin protocol (Bio 101, Vista, CA). An internal fragment of A. fumigatus dmaW (encoding amino acids 136 to 352 of the predicted 497-amino-acid product) was amplified by PCR with primers dmaW forward (5′-TTGATCTGGAGTGGTTCCGC-3′) and dmaW reverse (5′-CGTTCATGCCGAAGGTTGTG-3′). A 50-μl PCR mixture contained 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% (vol/vol) Triton X-100, 1.5 mM MgCl2, 200 μM of each deoxyribonucleotide triphosphate, 1 μM of each primer, and 2.5 U of Taq DNA polymerase (Promega, Madison, WI), which was added once the thermocycler reached 95°C in the initial denaturing period. The reaction began with an initial denaturation step consisting of 2.5 min at 95°C, which was followed by 35 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. The 651-bp dmaW PCR product was ligated into pCR2.1 (Invitrogen, Carlsbad, CA) based on T/A overhangs and was transformed into chemically competent Escherichia coli cells. The resulting 4.5-kb gene knockout construct, pDMAT1, was linearized at a unique PpuMI site in the dmaW coding sequences prior to transformation of A. fumigatus.
Protoplasts of A. fumigatus were prepared by the method of Murray et al. (11), except that 5 mg/ml of Driselase (InterSpex, Foster City, CA), 1.3 mg/ml Novozyme 234 (InterSpex), and 25 μg/ml chitinase (Sigma, St. Louis, MO) were used as lysing enzymes. Cotransformation of the protoplasts with PpuMI-linearized pDMAT1 and the hygromycin resistance-conferring plasmid pMOcosX (12), which had been linearized at a unique NotI site, was performed as previously described (16), but with 2 μg of each DNA in 10 μl of water. Considering the relative differences in the sizes of the cotransformed molecules, the equal masses introduced corresponded to an approximately twofold molar excess of the gene knockout construct (pDMAT1, 4.5 kb) relative to the selectable marker (pMOcosX, 8.8 kb). The transformation mixture was divided into six aliquots and plated on regeneration medium containing 300 μg/ml hygromycin (Calbiochem, La Jolla, CA) as previously described (16). Transformation plates were incubated at 37°C for 2 days, and hygromycin-resistant colonies were transferred onto PDA plates containing 400 μg/ml hygromycin.
Transformants were screened for homologous recombination of pDMAT1 with the native dmaW gene by PCR (using the conditions described above, except that the annealing temperature was 57°C) with primer UF (5′-TGTAAAACGACGGCCAGTGAAT-3′), which annealed to vector sequences near the universal primer annealing site of pCR2.1, and primer 3S (5′-AAGTAAGTCCCGAGCTGTTCAT-3′), which annealed to dmaW sequences near the 3′ end of the gene and flanking the intended site of integration (Fig. 1A). Transformants yielding a fragment of the expected size for a gene knockout at dmaW (1.2 kb) were purified to nuclear homogeneity by culturing them from individual, germinated conidia. Single-spore colonies were again screened by PCR (using the conditions described above but with an annealing temperature of 60°C) with primers UR (5′-AGCTATGACCATGATTACGCCA-3′), which annealed to vector sequences near the reverse primer annealing site of pCR2.1, and primer 5S (5′-TAGTGAGATACACATTCGAGCC-3′), which annealed to dmaW sequences near the 5′ end of the gene and flanking the intended site of integration (Fig. 1A). In this way, both the 5′ and 3′ borders of the integration site were analyzed by PCR. Candidate gene knockout strains also were screened with primers 5S and 3S under the PCR conditions described above with an annealing temperature of 57°C. Finally, transformants producing fragments of the expected sizes in the PCR analyses were analyzed further by Southern hybridization as described below.
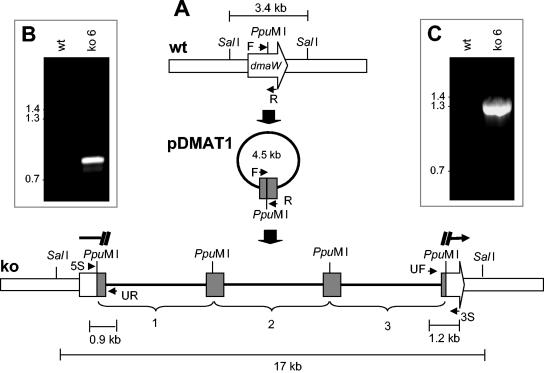
FIG. 1.
(A) Diagrammatic representation of homologous recombination at dmaW. In the wild-type (wt) diagram, F and R are annealing sites for primers dmaW forward and dmaW reverse, respectively, which amplify an internal portion of dmaW. The gene knockout construct pDMAT1, containing the internal fragment of dmaW, was linearized at a unique PpuMI site internal to dmaW prior to transformation. The diagram labeled ko represents the dmaW locus with three copies of pDMAT1 integrated in tandem, disrupting dmaW. UF, annealing site for a primer derived from universal primer sequences in the vector; 3S, annealing site for a primer flanking the 3′ border of the site of integration; UR, annealing site for a primer derived from reverse primer sequences in the vector; 5S, annealing site for a primer flanking the 5′ border of the site of integration. (B) PCR from primers UR and 5S amplified the expected 852-bp band from a strain carrying the disrupted dmaW allele but not from the wild-type isolate. (C) PCR from primers UF and 3S amplified the 1.2-kb product only from the strain with a disrupted dmaW gene. Although additional amplicons can be predicted from duplicate annealing sites for primers UF and UR, which result from the multiple integrations of pDMAT1, these theoretical amplicons are much too large to be amplified with the PCR conditions and reagents used in this study. The relative mobilities of relevant fragments (in kilobases) of BstEII-digested bacteriophage lambda are indicated on the left in panels B and C.
Fungal DNA for Southern blot hybridization was digested with SalI, size fractionated by electrophoresis through 0.8% agarose, and transferred to a Biodyne A nylon membrane (Pall-Gelman Sciences, Ann Arbor, MI) by capillary action with 20× SSC (1× SSC is 150 mM NaCl plus 15 mM trisodium citrate) as the transfer medium. A dmaW probe was labeled with digoxigenin (DIG)-dUTP by PCR amplifying wild-type genomic DNA with primers dmaW forward and dmaW reverse (see above). The PCR conditions were the same as those described above except that a 1× DIG DNA labeling mixture (Roche Applied Science, Indianapolis, IN) was used instead of the standard deoxynucleoside triphosphates. Hybridization was performed at 65°C in 5× SSC, 0.1% (wt/vol) N-lauroylsarcosine, 0.2% (wt/vol) sodium dodecyl sulfate, 1% (wt/vol) blocking reagent (Roche), 10 mM maleic acid, and 15 mM NaCl (pH 7.5). The final washes were with 0.5× SSC-0.1% sodium dodecyl sulfate at 65°C. Bound probe was detected with anti-DIG antibody, followed by chemiluminescence performed by the standard method (DIG Manual; Roche Applied Science, Indianapolis, IN).
An ergot alkaloid-deficient dmaW knockout strain was complemented with a full-length dmaW-containing fragment produced by PCR with primers 5′-GAGAGTGGTGTTGAGAGCTGCC-3′ and 5′-CCTTCAGGCTTCATGAACGGAC-3′, which primed amplification of a 3.4-kb fragment containing the entire dmaW coding sequence along with 784 bp of the 5′ flanking sequence and 1,123 bp of the 3′ flanking sequence. The PCR conditions were the same as those described above except that the annealing temperature was 57°C and the extension time at 72°C was 3 min 20 s. The PCR product was cotransformed into a dmaW knockout strain along with phleomycin resistance-conferring plasmid pBC-phleo (22) (Fungal Genetics Stock Center, University of Missouri-Kansas City, Kansas City, MO). Selection was on complete regeneration medium (16) containing phleomycin at a concentration of 100 μg/ml.
Analysis of ergot alkaloids.
Ergot alkaloids from conidia produced on PDA were extracted with 80% methanol and analyzed by high-performance liquid chromatography with fluorescence detection as described in the accompanying paper (13).
RESULTS
Identification and analysis of a dmaW homolog in A. fumigatus.
A search of the A. fumigatus genome database revealed an excellent match for C. fusiformis dmaW, which encodes DMAT synthase, the enzyme that catalyzes the first step in the ergot alkaloid pathway in clavicipitaceous fungi. The deduced A. fumigatus DMAT synthase was 50% identical to the DMAT synthase from C. fusiformis (24), 53% identical to the DMAT synthase from C. purpurea (26), and 59% identical to the DMAT synthase from Neotyphodium sp. strain Lp1 (29). The A. fumigatus dmaW sequence contained a single intron at a position identical to the position of the single introns in the dmaW genes isolated from the Claviceps and Neotyphodium spp. (data not shown).
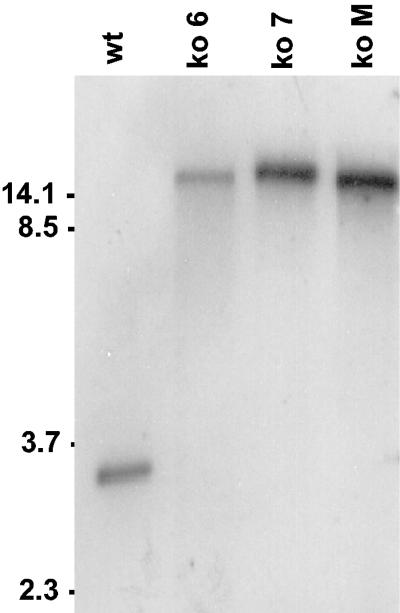
A gene knockout construct, designed to integrate at dmaW by a single crossover (Fig. 1), was targeted to the dmaW locus in 3 of 25 hygromycin-resistant transformants analyzed (Fig. 1 and 2). PCR analysis of the 5′ and 3′ junctions of the native dmaW gene and the integrated gene knockout construct yielded fragments of the expected length for homologous recombination of such a construct at dmaW (Fig. 1B and C). PCR amplification of the gene knockout strains with primers 5S and 3S, spanning the site of recombination, failed to yield the 1.2-kb product amplified from the wild-type isolate (data not shown). Southern hybridization of DNA from the putative knockout strains digested with SalI, which had sites flanking the targeted integration site and not within the knockout construct, indicated that three or more copies of the knockout construct integrated in tandem at dmaW in these transformants (Fig. 2).
FIG. 2.
Southern blot of SalI-digested DNA from nontransformed A. fumigatus (wt) and three dmaW knockout mutants probed with a dmaW fragment derived from primers dmaW forward and dmaW reverse (see Fig. 1A). The relative intensities of hybridization reflect minor differences in loading of the samples. The relative mobilities of relevant fragments (in kilobases) of BstEII-digested bacteriophage lambda are indicated on the left.
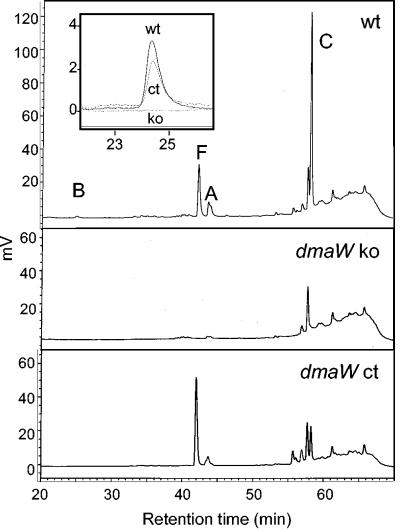
The dmaW knockout strains did not produce any of the known ergot alkaloids (Fig. 3). Additional peaks, including uncharacterized molecules eluting at 55.5 min and 56 min (Fig. 3) and a minor peak eluting at 32.8 min (evident only when the traces were viewed at much higher sensitivity), also were eliminated by knockout of dmaW. The peak eluting at 32.8 min is proposed to contain one or more of the isomers of chanoclavine based on a measured molecular mass of 256 and coelution with a chanoclavine standard. The molecules that eluted later were not characterized further.
FIG. 3.
High-performance liquid chromatography traces of nontransformed A. fumigatus (wt), a dmaW knockout strain (dmaW ko6), and strain dmaW ko6 complemented by ectopic integration of a DNA fragment containing the gene (dmaW ct). The insert shows overlaid traces for nontransformed A. fumigatus (solid line; wt), strain dmaW ko6 (dotted line; ko), and the complemented strain (dashed line; ct) at a higher sensitivity to demonstrate the presence or absence of fumigaclavine B. B, fumigaclavine B; F, festuclavine; A, fumigaclavine A; C, fumigaclavine C.
Transformation of strain dmaW ko6 with a DNA fragment containing the entire dmaW coding sequences plus 784 bp of 5′ flanking sequences and 1,123 bp of 3′ flanking sequences restored the ability to produce ergot alkaloids (Fig. 3). The complementing fragment was determined to have integrated ectopically, because the gene knockout construct was retained at the native dmaW locus (data not shown). All known A. fumigatus ergot alkaloids were produced in the complemented strains (Fig. 3).
Genes clustered with dmaW in A. fumigatus.
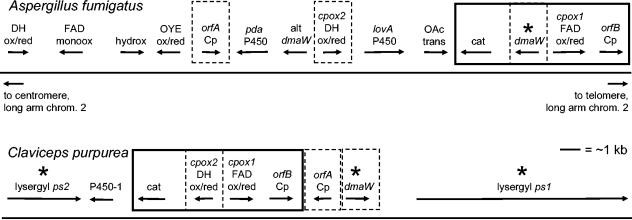
Database searches with five C. purpurea gene sequences, which in addition to dmaW have been proposed to be involved in ergot alkaloid biosynthesis (but which have not yet been functionally analyzed), identified potential homologs clustered with each other and with the dmaW homolog of A. fumigatus (Fig. 4). These genes included two putative oxidoreductase-encoding genes identified as cpox1 and cpox2, as well as a catalase-encoding gene and two unidentified open reading frames (labeled orfA Cp and orfB Cp), whose sequences and close linkage with dmaW are conserved in C. purpurea (25, 26) and C. fusiformis (10). Despite the similarity in the composition of the clusters of genes associated with dmaW in A. fumigatus and C. purpurea, the physical arrangement in terms of the orientation and order of genes within the cluster was different in the two fungi (Fig. 4).
FIG. 4.
Map of the genes clustered with dmaW of A. fumigatus compared to the cluster surrounding dmaW of C. purpurea (redrawn from data in reference 25). The arrows above the chromosomes indicate the positions of genes and the orientations of transcription. Abbreviations: Cp, C. purpurea; ox/red, oxidoreductase; cat, catalase; OAc trans, O-acetyltransferase; P450, cytochrome P450 monooxygenase; lovA P450, molecule similar to cytochrome P450 involved in lovastatin biosynthesis; pda P450, molecule similar to cytochrome P450 involved in pisatin demethylase activity; DH, short-chain alcohol dehydrogenase; alt, alternate; OYE, old yellow enzyme; hydrox, hydroxylase; monoox, monooxygenase; ps, peptide synthetase; FAD, flavin adenine dinucleotide. Genes that are found in the same relative position and orientation in the two clusters are enclosed in a solid boxes. Genes previously proposed to be involved in ergot alkaloid production that are present in both clusters but differ in their relative positions and orientations are enclosed in dashed boxes. Genes for which a function has been demonstrated by gene knockout (5, 16, 29) are indicated with asterisks. The scale applies to both clusters. The A. fumigatus sequences illustrated span from nucleotide 2894958 (left end of diagram) to nucleotide 2926335 (right end of diagram) of assembly 92 (chromosome 2; formerly assembly 72) of the A. fumigatus genome. The identifiers for the A. fumigatus sequences (from left to right) are m19874, m19875, m19876, m19877, m19878, m19880, m19881, m19882, m19883, m19884, m19885, m19886, m19888, and m19891. Data are available from The Institute for Genomic Research (www.tigr.org). The GenBank accession numbers for the C. purpurea sequences are as follows: lysergyl ps2, AJ438610; cytochrome P450-1, no accession number available (described by Correia et al. [5]); catalase, AJ703808; cpox2 (short-chain alcohol dehydrogenase oxidoreductase), CAB39316; cpox1 (flavin adenine dinucleotide oxidoreductase), AJ703809; orfB, AY836772; orfA, AY836771; dmaW, CAC37397; and lysergyl ps1, CAB39315.
Analysis of additional sequences clustered near dmaW of A. fumigatus revealed several other genes that may encode enzymes in the ergot alkaloid pathway (Fig. 4). These sequences include sequences similar to the sequences encoding cytochrome P450 monooxygenases, a flavin adenine dinucleotide-dependent monooxygenase, and short-chain alcohol dehydrogenases. Additional sequences whose deduced functions are consistent with roles in formation of the side chains that are different in the three fumigaclavines also were identified. These sequences include sequences encoding several monooxygenases or hydroxylases and an O-acetyltransferase and a divergent dmaW-like (prenyl transferase) gene. The predicted product of the divergent dmaW-like gene exhibited 25% identity with the deduced product of A. fumigatus dmaW required for ergot alkaloid biosynthesis and 26%, 25%, and 27% identity with the deduced products of the dmaW genes of C. purpurea, C. fusiformis, and Neotyphodium sp. strain Lp1, respectively.
Distal to orfB Cp in A. fumigatus is a metalloproteinase-encoding gene and a retrotransposon-rich region, after which the assembly ends (within 36 kb). Proximal to the A. fumigatus cluster are several genes encoding unknown hypothetical proteins, a few additional genes encoding deduced products also potentially involved in alkaloid biosynthesis, and interspersed genes for which no role in ergot alkaloid biosynthesis has been deduced.
DISCUSSION
Our results demonstrate that the genome of A. fumigatus contains a homolog of dmaW that is required for ergot alkaloid biosynthesis. The loss of all known ergot alkaloids upon inactivation of dmaW in A. fumigatus indicates that this gene controls the determinant step in the ergot alkaloid pathway of A. fumigatus, as it does in the clavicipitaceous fungus Neotyphodium sp. strain Lp1 (29). Moreover, in the A. fumigatus genome, dmaW is clustered with genes that are similar to and extend the ergot alkaloid gene cluster observed in the ergot fungus C. purpurea. Collectively, the data indicate a common biosynthetic and genetic origin for the ergot alkaloids of A. fumigatus and of the ergot alkaloid-producing members of the Clavicipitaceae.
Each of the three gene knockout strains investigated contained three or more copies of the knockout construct integrated at dmaW. The multiple integrations may have resulted from either or both of two modifications of the more typical gene knockout strategy that we used in this experiment. First, the knockout construct was introduced by cotransformation at two times the molar concentration of the selectable marker construct. Second, the knockout was created by integrating an internal fragment of the gene via a single crossover, a strategy that has resulted in multiple integrations in previous studies with other fungi (2, 17, 18, 21).
This approach was chosen for two reasons. The knockout construct can be easily prepared in a single cloning step, which should facilitate similar analyses of many of the genes in the cluster. Also, since we intended to complement the mutation in a subsequent step, any phenotype associated with ectopic integration of the cotransformed selectable marker construct would have been revealed at that stage. The observations that three independent transformants containing the integration at dmaW lacked ergot alkaloids and that complementation restored ergot alkaloid production indicate that the strategy provides a reasonable means for rapid analysis of gene function.
A. fumigatus produces at least four ergot alkaloids, all belonging to the clavine class of ergot alkaloids (4, 13, 23). Analysis of the peaks eliminated in the gene knockout compared to the wild-type strains in the present study indicated that conidia of A. fumigatus also contain small quantities of chanoclavine. This result is not surprising since chanoclavine is an early pathway intermediate in the biosynthesis of all known ergot alkaloids and it accumulates to various degrees in different ergot alkaloid producers (7, 8, 14, 15, 25).
Among the ergot alkaloids produced by A. fumigatus, only festuclavine and chanoclavine have been detected in any of the clavicipitaceous ergot alkaloid producers that have been studied (6, 7, 15, 19). Conversely, the ergot alkaloid-producing members of the Clavicipitaceae produce numerous ergot alkaloids that are not found in A. fumigatus. These include the ergopeptines (tripeptide derivatives of lysergic acid) and simple amides of lysergic acid (such as ergonovine and ergine). The production of both the ergopeptine and simple amide classes of ergot alkaloids is dependent upon the activities or products of lysergyl peptide synthetases (5, 15, 16). Based on the ergot alkaloid profiles and sequences clustered with dmaW in A. fumigatus and C. purpurea, we hypothesize that the early stages of the pathway are shared by A. fumigatus and C. purpurea but that later steps in the pathways differ in the two fungi.
The ergot alkaloid pathways of A. fumigatus and C. purpurea probably share several steps in addition to the determinant step catalyzed by DMAT synthase. The cluster of genes immediately around dmaW in A. fumigatus contains five hypothetical genes that are highly similar to genes previously hypothesized to be involved in the early stages of the ergot alkaloid pathway of C. purpurea (5, 25, 26). These genes include the following: a sequence labeled orfB, which encodes a product similar to a methyl transferase, an activity required for the second step in the pathway; two genes (cpox1 and cpox2) encoding oxidoreductase that may catalyze early steps in the pathway (7, 14, 25); a sequence labeled orfA, for which a biochemical function has not been proposed; and a catalase-encoding gene. A function for the catalase in ergot alkaloid biosynthesis is not obvious. One possibility is that the catalase acts peroxidatively to oxidize the primary alcohol of chanoclavine to the corresponding aldehyde, which is a step required prior to cyclization of the fourth ring of the basic four-member ergoline ring system (7, 8, 14). Interestingly, the orientation and position of three members of this putative core set of genes differ in the gene clusters of the two fungi, suggesting that numerous recombination events occurred after divergence of the lineages of these two fungi. Alternatively, if the cluster had been acquired by horizontal gene transfer, then the dissimilarities indicate that any such transfer was ancient and not recent.
We propose that after a shared early stage, the ergot alkaloid pathways of A. fumigatus and the clavicipitaceous fungi diverge and yield different end products. Cultures of A. fumigatus do not contain detectable levels of the later products of the pathway, such as lysergic acid or lysergyl derivatives such as ergopeptines (13). Consistent with this hypothesis, searches with fragments of genes encoding LPS1 (required for the assembly of the ergopeptine class of ergot alkaloids) from N. lolii (16) or C. purpurea (26) and LPS2 (required for activating lysergic acid for peptide formation) from C. purpurea (5) produced only low-level matches to miscellaneous peptide synthetase genes in the A. fumigatus genome. The degree of identity observed was typical of the degrees of identity among peptide synthetases catalyzing the assembly of unrelated nonribosomally synthesized peptides in fungi (27), and none of the low-level matches was located near the dmaW gene.
Conversely, the A. fumigatus cluster contained genes not found in the C. purpurea cluster. Some of these hypothetical genes encode functions that we hypothesize to be involved in the later steps of the A. fumigatus pathway leading from festuclavine to the fumigaclavines, which are not produced by C. purpurea. For example, the product of the gene in the A. fumigatus cluster predicted to encode an O-acetyltransferase may function in acetylating fumigaclavine B to fumigaclavine A. The unusual, alternate dmaW gene (labeled alt dmaW in Fig. 4) may control the prenylation of fumigaclavine A to fumigaclavine C. The lack of ergot alkaloids in the mutant containing a knockout at the typical dmaW gene demonstrates that the highly divergent dmaW-like prenyl transferase gene found in the cluster is not functionally redundant with dmaW and thus may catalyze another prenylation step.
The ergot alkaloid gene cluster of C. purpurea and the cluster of genes containing dmaW in A. fumigatus appear to have a common origin. The cluster of genes in A. fumigatus lacks the large peptide synthetase genes found in the C. purpurea cluster, revealing several additional genes potentially involved in ergot alkaloid production. Systematic gene knockouts and their characterization should allow definition of the early (and probably shared) steps of the ergot alkaloid pathway, several of which have been difficult to characterize biochemically in C. purpurea (7, 8, 14). Ergot alkaloid-deficient mutants, such as the dmaW knockout, may be useful for assessing the contribution of ergot alkaloids to virulence to animals or potential contributions of the alkaloids to the success of A. fumigatus.
Acknowledgments
This work was supported by USDA-NRI grant 2001-35319-10930 and Hatch funds. Sequencing of the A. fumigatus genome was funded by National Institute of Allergy and Infectious Disease grant U01 AI 48830 to David Denning and William Nierman, the Wellcome Trust, and Fondo de Investigaciones Sanitarias.
We thank Caroline Machado, Christopher Schardl, and Mark Farman (University of Kentucky) for access to data and helpful suggestions and Brian Tapper (AgResearch Limited, New Zealand) and Miroslav Flieger (Czech Academy of Sciences) for the chanoclavine standard. Preliminary sequence data for the A. fumigatus genome were obtained from The Institute for Genomic Research (http://www.tigr.org). The assistance of Bill Nierman and Natalie Federova with questions concerning the data is greatly appreciated.
Footnotes
This paper is published with the approval of the Director of the West Virginia Agricultural and Forestry Experiment Station as Scientific Article no. 2896.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apel, P. A., D. G. Panaccione, F. R. Holden, and J. D. Walton. 1993. Cloning and targeted gene disruption of XYL1, a β1,4-xylanase gene from the maize pathogen Cochliobolus carbonum. Mol. Plant-Microbe Interact. 6:467-473. [DOI] [PubMed] [Google Scholar]
- 3.Clay, K., and C. Schardl. 2002. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 160:S199-S127. [DOI] [PubMed] [Google Scholar]
- 4.Cole, R. J., J. W. Kirksey, J. W. Dorner, D. M. Wilson, J. C. Johnson, Jr., A. N. Johnson, D. M. Bedell, J. P. Springer, K. K. Chexal, J. C. Clardy, and R. H. Cox. 1977. Mycotoxins produced by Aspergillus fumigatus species isolated from molded silage. J. Agric. Food Chem. 25:826-830. [DOI] [PubMed] [Google Scholar]
- 5.Correia, T., N. Grammel, I. Ortel, U. Keller, and P. Tudzynski. 2003. Molecular cloning and analysis of the ergopeptine assembly system in the ergot fungus Claviceps purpurea. Chem. Biol. 10:1281-1292. [DOI] [PubMed] [Google Scholar]
- 6.Flieger, M., M. Wurst, and R. Shelby. 1997. Ergot alkaloids—sources, structures, and analytical methods. Folia Microbiol. 42:3-30. [DOI] [PubMed] [Google Scholar]
- 7.Floss, H. G. 1976. Biosynthesis of ergot alkaloids and related compounds. Tetrahedron 32:873-912. [Google Scholar]
- 8.Gröger, D., and H. G. Floss. 1998. Biochemistry of ergot alkaloids—achievements and challenges. Alkaloids 50:171-218. [Google Scholar]
- 9.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 10.Machado, C. 2004. Studies of ergot alkaloid biosynthesis genes in clavicipitaceous fungi. Ph.D. thesis. University of Kentucky, Lexington.
- 11.Murray, F. R., G. C. M. Latch, and D. B. Scott. 1992. Surrogate transformation of perennial ryegrass, Lolium perenne, using genetically modified Acremonium endophyte. Mol. Gen. Genet. 233:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Orbach, M. J. 1994. A cosmid with a HyR marker for fungal library construction and screening. Gene 150:159-162. [DOI] [PubMed] [Google Scholar]
- 13.Panaccione, D. G., and C. M. Coyle. 2005. Abundant respirable ergot alkaloids from the common airborne fungus Aspergillus fumigatus. Appl. Environ. Microbiol. 71:3106-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panaccione, D. G., and C. L. Schardl. 2003. Molecular genetics of ergot alkaloid biosynthesis, p. 399-424. In J. F. White, Jr., C. W. Bacon, N. L. Hywel-Jones, and J. W. Spatafora (ed.), The clavicipitalean fungi: evolutionary biology, chemistry, biocontrol, and cultural impacts. Marcel-Dekker, New York, N.Y.
- 15.Panaccione, D. G., B. A. Tapper, G. A. Lane, E. Davies, and K. Fraser. 2003. Biochemical outcome of blocking the ergot alkaloid pathway of a grass endophyte. J. Agric. Food Chem. 51:6429-6437. [DOI] [PubMed] [Google Scholar]
- 16.Panaccione, D. G., R. D. Johnson, J. Wang, C. A. Young, P. Damrongkool, B. Scott, and C. L. Schardl. 2001. Elimination of ergovaline from a grass-Neotyphodium endophyte symbiosis by genetic modification of the endophyte. Proc. Natl. Acad. Sci. USA 98:12820-12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panaccione, D. G., J. S. Scott-Craig, J.-A. Pocard, and J. D. Walton. 1992. A cyclic peptide synthetase gene required for pathogenicity of the fungus Cochliobolus carbonum on maize. Proc. Natl. Acad. Sci. USA 89:6590-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panaccione, D. G., M. McKiernan, and R. M. Hanau. 1988. Colletotrichum graminicola transformed with homologous and heterologous benomyl resistance genes retains expected pathogenicity to corn. Mol. Plant-Microbe Interact. 1:113-120. [Google Scholar]
- 19.Porter, J. K., C. W. Bacon, J. D. Robbins, and D. Betowski. 1981. Ergot alkaloid identification in Clavicipitaceae systemic fungi of pasture grasses. J. Agric. Food Chem. 29:653-657. [Google Scholar]
- 20.Riederer, B., M. Han, and U. Keller. 1996. d-Lysergyl peptide synthetase from the ergot fungus Claviceps purpurea. J. Biol. Chem. 271:27524-27530. [DOI] [PubMed] [Google Scholar]
- 21.Scott-Craig, J. S., D. G. Panaccione, F. Cervone, and J. D. Walton. 1991. Endopolygalacturonase is not required for pathogenicity of Cochliobolus carbonum on maize. Plant Cell 2:1191-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silar, P. 1995. Two new easy to use vectors for transformation. Fungal Genet. Newsl. 42:73. [Google Scholar]
- 23.Spilsbury, J. F., and S. Wilkinson. 1961. The isolation of festuclavine and two new clavine alkaloids from Aspergillus fumigatus Fres. J. Chem. Soc. 5:2085-2091. [Google Scholar]
- 24.Tsai, H.-F., H. Wang, J. C. Gebler, C. D. Poulter, and C. L. Schardl. 1995. The Claviceps purpurea gene encoding dimethylallyltryptophan synthase, the committed step for ergot alkaloid biosynthesis. Biochem. Biophys. Res. Commun. 216:119-125. [DOI] [PubMed] [Google Scholar]
- 25.Tudzynski, P., T. Correia, and U. Keller. 2001. Biotechnology and genetics of ergot alkaloids. Appl. Microbiol. Biotechnol. 57:593-605. [DOI] [PubMed] [Google Scholar]
- 26.Tudzynski, P., K. Holter, T. Correia, C. Arntz, N. Grammel, and U. Keller. 1999. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol. Gen. Genet. 261:133-141. [DOI] [PubMed] [Google Scholar]
- 27.Walton, J. D., D. G. Panaccione, and H. Hallen. 2004. Peptide synthesis without ribosomes, p. 127-162. In J. Tkacz and L. Lange (ed.), Advances in fungal biotechnology for industry, agriculture, and medicine. Kluwer Academic, New York, N.Y.
- 28.Walzel, B., B. Riederer, and U. Keller. 1997. Mechanism of alkaloid cyclopeptide synthesis in the ergot fungus Claviceps purpurea. Chem. Biol. 4:223-230. [DOI] [PubMed] [Google Scholar]
- 29.Wang, J., C. Machado, D. G. Panaccione, H.-F. Tsai, and C. L. Schardl. 2004. The determinant step in ergot alkaloid biosynthesis by an endophyte of perennial ryegrass. Fungal Genet. Biol. 41:189-198. [DOI] [PubMed] [Google Scholar]