Abstract
Determination of the hepatitis C virus (HCV) genotype has become accepted as the standard procedure in laboratory practice. Genotype assignment helps in disease prognosis and assists in establishing the appropriate duration of treatment. More than 10 types and 70 subtypes of HCV have been described. In Russia the most common subtypes are 1a, 1b, 2a, and 3a, and the types 4 and 5 are relatively rare. The “gold standard” for testing is gene sequencing. However, a variety of other assays had been developed to provide more rapid and cheaper forms of testing. The aim of this study was to determine the HCV genotype by minisequencing followed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. Fragments of 5′ untranslated region of the HCV genome were amplified. Three oligonucleotide primers were designed to detect two sets of genotype-specific single nucleotide polymorphisms. The primer extension reaction was performed using modified thermostable DNA polymerase and in the presence of dideoxynucleosides. The molecular weights of the reaction products were analyzed with MALDI-TOF mass spectrometer. The HCV genotype was determined by registering the particles of the expected molecular weights. The method was used to genotype HCV from HCV-positive blood sera or plasma. The 1a, 1b, 2a, 3a, and 4 genotype HCVs were determined in the samples examined. The data were confirmed by direct sequencing. Thus, we propose a new accurate and efficient method for HCV genotyping based on minisequencing followed by mass spectrometry.
Laboratory diagnosis and monitoring of viral hepatitis are an important research area in the infectious pathology of the liver. Traditionally, diagnosis and monitoring represent a complicated problem, which is far from being solved. An important task in the clinical laboratory diagnosis of hepatitis C virus (HCV) is the detection of the serological and/or genetic markers of the virus: namely, antibodies to HCV and genomic RNA of HCV.
HCV has extremely variable genomic RNA. Several distinct types differ by as much as 33% over the entire genome (14). At present, according to the classification of Simmonds (18), 11 types and 70 subtypes of the virus were discriminated (18). Discovery of all of the HCV genotypes is of interest for epidemiological studies, while in clinical practice it is sufficient to distinguish the major genotypes of HCV.
Recently, geographical distributions of the various genotypes have been determined. Thus, in Japan, Taiwan, and partially in China, genotypes 1b, 2a, and 2b are predominantly registered. Type 1b was even termed Japanese. Rare types of viruses, i.e., types 4, 5, and 6, are found considerably more often in African countries than in European countries and America. The dominating genotype in the United States is type 1a, also known as American. In European countries, the dominating HCV genotype also appears to be 1a, with the share of genotype 1b increasing appreciably (22). In the Russian Federation, the most common genotypes of the virus are 1b, 1a, 2a, and 3a, with their ratio constantly shifting (10, 12).
Besides the particular geographical distributions, HCV genotypes also vary in the disease outcome and the response to therapy. Many studies have indicated an association between HCV type and both the responsiveness to interferon treatment and the degree of clinical progression of the HCV infection (6, 8, 9).
Gene sequencing is the reference method for identifying different HCV genotypes. Usually, the analysis of sequences of HCV NS5, core, E1, and 5′ untranslated region (5′ UTR) is used for HCV genotyping (1, 2, 3, 16). However, direct sequencing is expensive, time-consuming, and does not identify mixed infections when two different HCV genotypes are present. Several other methods for typing HCV, such as restriction fragment length polymorphism analysis (5), type-specific amplification (10, 17), reverse hybridization with type-specific probes (21), and cleavage fragment length polymorphism (Third Wave Technologies, Inc., Madison, Wisconsin) analysis (13) have been developed. Two of them were realized as commercial kits, the InnoLiPA HCV kit (Innogenetics, Belgium) and the Invader HCVG ASRs kit (Third Wave Technologies, Inc., Madison, Wisconsin).
As clinically, in almost all cases, it is enough to distinguish the HCV genomes on the level of types (and not subtypes), using a single amplification for diagnostic detection of HCV RNA and for genotype determination would be the most efficient way of clinical HCV genotyping. The high degree of conservation in the 5′ UTR has made it the target of choice for reverse transcription-PCR-based detection assays. Several methods exploit differences in the 5′ UTR sequences for genotyping: for example, direct sequencing (7), restriction fragment length polymorphism (5), line probe assay (21), and single-strand conformation polymorphism (11) analyses.
The two existing commercial kits for HCV genotyping, the InnoLiPA HCV kit (Innogenetics, Belgium) and the TRUGENE HCV 5′NC genotyping kit (Visible Genetics, Toronto, Canada) are based on the analysis of the 5′ UTR. Though any genotyping methods based on the 5′ UTR in some cases are not reliable at the subtype level, these methods produce accurate results at the genotype level. This inherent disadvantage of the 5′ UTR for HCV genotyping is not crucial for clinical purposes. Clinically, it is not necessary to know precisely the HCV subtype and in almost all cases it is enough to determine genotype at the type level only.
Most of the existing methods for HCV genotyping are based on the detection of the genotype-specific polymorphisms in the RNA fragments. Currently, scientific laboratories spend considerable effort in the development of new optimal approaches for determining single nucleotide polymorphisms (SNPs). One of the newly emerged technologies is mass spectrometry of the minisequencing reaction products using the matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) (20). The method is based on the selective enzymatic extension of the oligonucleotide primers and, in terms of the accuracy of SNP identification, is comparable to the direct determination of the nucleotide sequence of the loci of interest. At the same time, the MALDI-TOF technique is considerably cheaper in application than the direct sequencing procedure.
In the present study, we have implemented the detection of the genotype-specific point mutations in the 5′ UTR of HCV using minisequencing and determination of the molecular weights of the reaction products on the Reflex-IV (Bruker Daltonics, Germany) MALDI-TOF MS. For genotyping of HCV RNA, this approach is used for the first time.
MATERIALS AND METHODS
Clinical samples.
The materials for the study were HCV-positive samples of blood serum or plasma, taken from the laboratory bank (10). Prior to being used, all the specimens were kept at a temperature of −110°C.
Isolation of HCV RNA and RT-PCR.
HCV RNA was isolated from serum or plasma samples by using a QIAamp viral RNA kit (QIAGEN, Netherlands) according to the manufacturer's instruction. The 163-bp fragments of the 5′-UTR HCV were amplified using a PolyHepC kit (Lytech Ltd, Russia) according to the manufacturer's instruction. The amplification products were analyzed by means of electrophoresis in a 2% agarose gel.
Dephosphorylation of amplification products.
Dephosphorylation of the 5′-end phosphate groups of deoxynucleoside triphosphate in the postamplification reaction mixture was carried out during incubation with 0.5 U of shrimp alkaline phosphatase (Fermentas, Lithuania) for 20 min at 37°C, followed by inactivation of the enzyme by heating for 10 min at 85°C.
Minisequencing reaction.
To identify the type-specific nucleotide contexts in the 5′-UTR of the HCV genome, we used three oligonucleotide primers: HC1, 5′ GGGCGTGCCCCCGC 3′, HC2, 5′ GAGTACACCGGAATCGC 3′, and HC21, 5′ TGAGTACACCGGAATTGC 3′.
The thermocyclic minisequencing reaction was carried out in the reaction mixture of 66 mM Tris-HCl, pH 9.0; 16.6 mM (NH4)2SO4; 2.5 mM MgCl2; 0.2 mM dATP; 0.2 mM dTTP; 0.2 mM dCTP; 0.2 mM ddGTP; 20 pmol each primer; and 2 units TermiPol DNA polymerase (Solis Biodyne, Estonia), using HCV RNA amplified fragments as the matrix. Thus, we used amplicons for analysis because it is necessary to produce PCR and minisequencing reaction in different places in the laboratory (rooms or PCR-boxes) to avoid amplicon contamination. The minisequencing products were obtained according to the following procedure: 94°C for 20 s, 58°C for 20 s, and 72°C for 15 s, in 70 cycles.
Purification of the minisequencing products.
The minisequencing products were purified by means of the SpectroCLEAN kit (Sequenom) according to the manufacturer's instructions. The kit's intrinsic sorbent in the amount of 8 μg was diluted in 15 μl of superpure water (Merck, Germany) after which the suspension obtained was placed in a test tube containing the minisequencing reaction products. The test tube's contents were then thoroughly mixed and incubated at room temperature for 15 min. Centrifuging for 5 min at 1,000 rpm precipitated the sorbent. The supernatant was used for the mass spectral analysis.
Registration of the HCV genotyping results.
The sample's aliquot (0.2 to 1 μl) at the concentration of 10 to 30 pmol/μl of oligonucleotides, obtained after the purification procedure, was applied to the matrix dried on the target AnchorChip (400 μm, Bruker Daltonic, Germany), prepared from the saturated solution of 3-hydroxypicolinic acid (Fluka, Germany) in 50% acetonitrile (Merck, Germany), adding 10 g/liter dibasic ammonium citrate (Fluka, Germany) and air dried. All the solvents used including water (Merck, Germany) were of analytical grade only or, at least, specific for mass spectrometry.
The mass spectra were obtained on the Reflex IV (Bruker Daltonics, Germany) MALDI-TOF MS in the linear mode using a nitrogen laser with a wavelength of 337 nm and pulse frequency of 9 Hz in the mode of positive ions. The analyzer's delay time was 200 ns. The voltage at the amplifier's electrode was 20.0 kV, at the accumulating electrode 17.1 kV, and at the focusing lens 9.4 kV. The mass spectrometer's parameters were optimized for the range of the m/z values from 1,000 to 10,000, using peptide mass spectra for the calibration. The mass spectra of the oligonucleotides obtained were additionally calibrated using internal standards. Each mass spectrum was obtained at 30 laser pulses at constant laser power and constant threshold value in order to enhance the resolution. From the mass spectra of the products of the reaction, we inferred the particular genotypes in the initial HCV samples (Table 1).
TABLE 1.
Expected masses of the minisequencing reaction products depending on the HCV genotypea
| HCV genotype | Product mass in extension of primer HC1b (Da) | Product mass in extension of primer HC2c (Da) | Product mass in extension of primer HC21d (Da) |
|---|---|---|---|
| 1a | 5,198 (+dA+dA+ddG) | *5,205 | 6,456 (+dC+dA+ddG) |
| 1b | 4,571 (+ddG) | *5,205 | 6,456 (+dC+dA+ddG) |
| 2a | 5,198 (+dA+dA+ddG) | *5,205 | 6,142 (+dC+ddG) |
| 3a | 4,571 (+ddG) | 5,838 (+dT+ddG) | *5,524 |
| 4 | 5,198 (+dA+dA+ddG) | 5,823 (+dC+ddG) | *5,524 |
| 5 | 4,571 (+ddG) | *5,205 | 6,142 (+dC+ddG) |
*, no enzymatic extension with this probe. The terminating synthesis of nucleotides (ddG) is displayed in bold type.
HC1 is 4,242 Da.
HC2 is 5,205 Da.
HC21 is 5,524 Da.
DNA sequence analysis.
The amplification products (163 bp) were purified on the Wizard PCR Preps DNA Purification System (Promega) according to the instruction manual. The nucleotide sequence of the 5′ UTR of HCV was determined by the modified Sanger method using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit and the ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Hitachi, Japan) according to the manufacturer's instruction.
Nucleotide sequence accession numbers.
The following sequences from GenBank were included in the analysis: genotype 1a, hepatitis C virus, complete genome, NC_004102; genotype 1b, hepatitis C virus strain HCV-N, complete genome, AF139594; genotype 2a, hepatitis C virus clone pJ6CF, complete genome, AF177036; genotype 3a, hepatitis C virus complete genome sequence, D17763; type 4, hepatitis C virus strain GE 139 5′ noncoding region type 4, AF021899; and type 5, hepatitis C virus type 5 gene, 5′ untranslated region, L29612.
RESULTS
The alignment of the nucleotide sequences of the amplified fragment of the 5′ UTR of HCV is presented in Fig. 1. Using the alignment, we chose the two SNP sites that allow differentiation of the HCV genotypes most commonly found in the territory of the Russian Federation. The choice of the two particular sites was determined by the degree of conservation of the 5′ UTR of HCV in the scope of the alignment. Although it is not possible to differentiate all of the known HCV genotypes using just these two SNP sites, analysis of the two SNP sites is of substantial clinical importance.
FIG. 1.
Alignment of the nucleotide sequences of the analyzed 5′ UTR fragments of HCV RNA belonging to various genotypes. The sites of annealing of the nucleotide primers participating in the minisequencing reaction are given in italics and underlined. The points of nucleotide polymorphisms are in bold capital letters.
Using the alignment, we selected three oligonucleotide primers (Fig. 1), extending which at the 3′ end allows determination of the genotype-specific nucleotide contexts of the HCV RNA. The primers for genotyping were selected to ensure that the primer extension reaction products produced in the analysis of RNA of various genotypes of HCV were unambiguously differentiated. It was necessary to select two primers in the same place (primers HC2 and HC21). One of them was designed to anneal with the nucleotide sequences of 1a, 1b, 2a, and 5 HCV genotypes and the other with the 3a and 4 genotypes. The expected masses of the reaction products of the primer extension for each genotype studied are shown in Table 1.
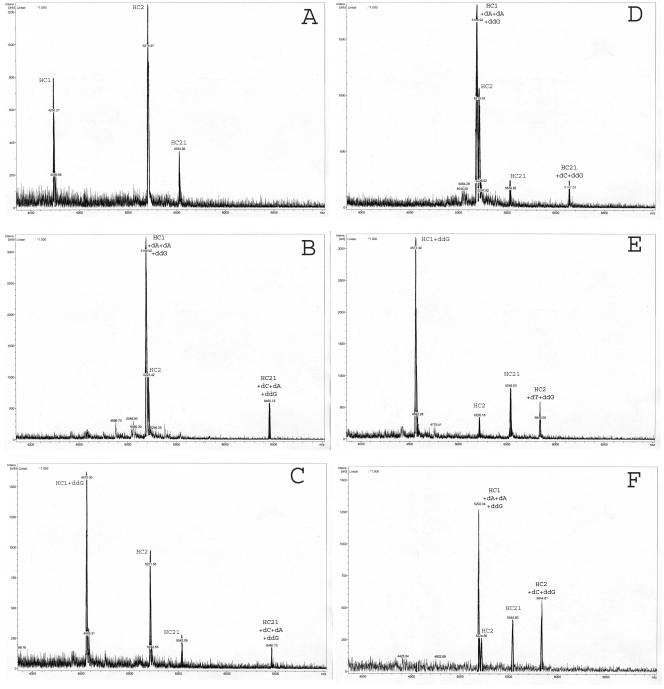
The conditions of the multiprimer minisequencing reaction were optimized for the simultaneous extension of the three oligonucleotide primers at the two sites of nucleotide polymorphism. This makes possible identification of both mono- and mixed infections (when more than one genotype of the virus is present in the clinical specimen). Examples of the mass spectra of the reaction products for each particular genotype are shown in Fig. 2.
FIG. 2.
Mass spectra of the minisequencing reaction products of the analyzed HCV genotypes. A. Nonextended oligonucleotide primers. B. The products of the primer extension in analysis of HCV genotype 1a. C. The products of the primer extension in analysis of HCV genotype 1b. D. The products of the primer extension in analysis of HCV genotype 2a. E. The products of the primer extension in analysis of HCV genotype 3a. F. The products of the primer extension in analysis of HCV genotype 4.
The proposed method of HCV RNA typing was validated on the RNA-positive samples from the laboratory bank of blood sera or plasma. The specimens from the bank were previously tested by allele-specific amplification for determination of the viral genotype. Of the 69 samples studied, viral genotype 1a was revealed in 4.5% of cases, 1b in 48%, 2a in 4.5%, 3a in 29%, and 4 in 1.5%. Mixed infection was registered in eight (12%) of the samples (Table 2). Of these, a combination of 3a plus 4 was observed in three samples, 3a plus 1b in two samples, 1a plus 1b in two samples, and 1a plus 3a in one sample. In two samples (3%) a discrepancy between the HCV genotype determination by the two methods was registered. Thus, comparative analysis of the minisequencing and of allele-specific amplification shows a high percentage of concordance, 97%, including the determination of the mixed infections. In other words, the standard sequencing of the amplified HCV fragments confirms the correctness of the genotype determination using the minisequencing technology paired with MALDI-TOF MS on the Reflex-IV.
TABLE 2.
HCV typing results for 69 HCV-positive sera obtained by the minisequencing reaction followed by mass spectrometry, allele-specific amplification, and direct sequencinga
| HCV genotype | No. of isolates with HCV genotype according to:
|
||
|---|---|---|---|
| Minisequencing reaction followed by mass spectrometry | Allele-specific amplification | Direct sequencing | |
| 1a | 3 | 4 | 3 |
| 1b | 34 | 33 | 34 |
| 2a | 3 | 3 | 3 |
| 3a | 20 | 21 | 20 |
| 4 | 1 | 0 | 1 |
| Mixed | 8 | 8 | * |
Chi-square for the correlations between allele-specific amplification and direct sequencing/minisequencing for the 1b and 3a genotypes was 27 (P < 0.001), corresponding to 97% concordance. Minisequencing and direct sequencing show 100% concordance by genotype. *, the correctness of the genotype determination has not been confirmed by sequencing in cases of mixed infection.
DISCUSSION
The highly conserved 5′ UTR of HCV can be routinely used to identify HCV RNA in sera and to determine HCV genotypes. Some commercial kits are based on the analysis of the sequence variability of the 5′ UTR of HCV for genotyping (InnoLiPA and TRUGENE). The current limitations of using the 5′ UTR for HCV genotyping are related to the low discriminating power of the 5′ UTR sequence for determination of the particular subtype (4, 14, 19). The high level of conservation found in this region does not allow accurate distinction of the HCV genotypes at the subtype level (as in the some cases with 2a and 2c or 1a and 1b).
However, these inherent disadvantages of using the 5′ UTR for HCV genotyping are not crucial for actual clinical purposes. As practice shows, any HCV typing system based on the 5′ UTR sequences (including our method) is reliable at the genotype level (e.g., type 1, 2, 3, or 4). That is quite enough for clinical studies. At the same time, it should be mentioned that HCV typing systems based on NS5′ or core sequence regions are characterized by sensitivities slightly less than that of the 5′ UTR-based methods.
In our study we propose the classical three-step minisequencing reaction for the identification of the genotype-specific nucleotide polymorphism in the 5′ UTR of HCV. The “know-how” for the approach was using the MALDI-TOF MS for analysis of the minisequensing reaction products. The selectivity of the proposed technology is based on the selective enzymatic extension of the oligonucleotide primers for 1, 2, or 3 nucleotide links and also depends on the nucleotide sequence at the point of polymorphism. The resolution of the Reflex-IV MALDI-TOF MS allows accurate discrimination between the oligonucleotides 10 to 30 bases in size, which is entirely satisfactory for the task.
MS setups are widely used in organic and analytical chemistry to detect and to identify substances of low molecular weights. In molecular biology modifications of the TOF MS (electrospray, MALDI) are used to also detect high-molecular-weight material such as proteins, nucleic acids, and oligosaccharides. The first application of MS in molecular biology was checking the sequences of newly synthesized oligonucleotides. MS analysis is considerably less time- and cost-consuming than the widespread sequencing procedures. The most important advantage of MS is the direct analysis of molecules, with no labels to be introduced and no separation steps followed by the image processing.
The technological approach outlined in the present article (minisequencing reaction followed by MS analysis) for identification of the SNPs in the 5′ UTR of HCV allows unambiguous determination of the viral genotypes most commonly found in the territory of the Russian Federation. The high accuracy of the proposed technology (as verified by direct sequencing) also allows discrimination of the minor nucleotide changes distinguish genotypes from each other (for example, A or G at position −99). The technology presented here is characterized by higher precision than the reverse hybridization with type-specific probes used in the InnoLiPA HCV kit. At the same time the method is less time- and cost-consuming than the sequencing procedures realized in the TRUGENE HCV 5′NC genotyping kit. Of course, using the technology of minisequencing based on the MALDI-TOF MS analysis for routine genotyping requires specific training and experience of working with MS. Similar training and experience are also required, however, for work with any device for direct sequencing (ABI Prism Genetic Analyzer or any other).
In conclusion, considering the clinical importance of genotyping in the treatment and management of hepatitis C, the method proposed herein for HCV RNA typing can be efficiently used for the discrimination of the major HCV genotypes most commonly found in the territory of the Russian Federation, namely, genotypes 1a, 1b, 2a, and 3a.
Acknowledgments
We gratefully acknowledge I. Y. Torshin's assistance with the writing of the manuscript.
REFERENCES
- 1.Bukh, J., R. H. Purcell, and, R. H. Miller. 1992. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc. Natl. Acad. Sci. USA 89:4942-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukh, J., R. H. Purcell, and, R. H. Miller. 1994. Sequence analysis of the core gene of the 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, S. W., F. McOmish, E. C. Holmes, B. Dow, J. F. Peuhterer, E. Follett, P. L. Yap, and, P. Simmonds. 1992. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J. Gen. Virol. 73:1131-1141. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z., and, K., E. Weck. 2002. Hepatitis C Virus Genotyping: Interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J. Clin. Microbiol. 40:3127-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson, F., P., J. C. Simmonds, L. M. Ferguson, B. C. Jarvis, E. A. Dow, C. R. Follett, T. Seed, C. Krusius, G. A. Lin, et al. 1995. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J. Gen. Virol. 76:1197-1204. [DOI] [PubMed] [Google Scholar]
- 6.Davis, G. L., and J. Y. Lau. 1997. Factors predictive of a beneficial response to therapy of hepatitis C. J. Hepatol. 26:122S-127S. [DOI] [PubMed] [Google Scholar]
- 7.Gelmer, J. J., P. N. Rys, J. N. Thorvilson, and D. H. Persing. 1999. Determination of hepatitis C virus genotype by direct sequences analysis of products generated with the Amplicor HCV test. J. Clin. Microbiol. 37:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi, J., Y., E. Kishihara, Y. Yoshimura, K. Tani, H. Yamaji, H. Ikematsu, and S. Kashiwagi. 1995. Relationship of genotype to level of Hepatitis C viraemia determined by competitive polymerase chain reaction. J. Infect. 30:235-239. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi, J., M. Ohmiya, Y. Kishihara, Y. Tani, N. Kinukawa, H. Kematsu, and, S. Kashiwagi. 1994. A statistical analysis of predictive factors of response to human lymphoblastoid interferon in patients with chronic hepatitis C. Am. J. Gastroenterol. 89:2151-2156. [PubMed] [Google Scholar]
- 10.Ilina, E. N., E. K. Artemov, V. M. Govorun, L. M. Ivanova, I. and O. Ivanikov. 2002. Genotyping of hepatitis C virus RNA by allele-specific amplification. Kremlin Med. Clin. Rev. 1:38-41. (In Russian.) [Google Scholar]
- 11.Lareu, R. R., R. N. Swanson, and S. A. Fox. 1997. Rapid and sensitive genotyping of hepatitis C virus by single-strand conformation polymorphism. J. Virol. Methods 64:11-18. [DOI] [PubMed] [Google Scholar]
- 12.L'vov, D. K., E. I. Samokhvalov, S. Mishiro, et al. 1997. Regularities in the spread of hepatitis C virus and its genotypes in Russian and countries within the former USSR. Vopr. Virusol. 42:157-161. (In Russian.) [PubMed] [Google Scholar]
- 13.Marshall, D. J., L. M. Heisler, V. Lyamichev, C. Murvine, D. M. Olive, and G. D. Ehrich. 1997. Determination of hepatitis C virus genotypes in the United States by cleavage fragment length polymorphism analysis. J. Clin. Microbiol. 35:3156-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolte, F., S., A., M. Green, K., R. Fiebelkorn, A. M. Caliendo, C. Sturchio, A. Grunwald, and M. Healy. 2003. Clinical evaluation of two methods for genotyping hepatitis C virus based on analysis of the 5′ noncoding region. J. Clin. Microbiol. 41:1558-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto, H., M. Kojima, S. Okada, K. Yamamoto, H. Iizuka, T. Tanaka, S. Fukuda, F. Tsuda, and S. Mishiro. 1992. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology 188:331-341. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto, H., M. Kojima, M. Sakamoto, H. Iizuka, S. Hadiwandowo, S. Suwignyo, Y. Miyakawa, and M. Mayumi. 1994. The entire nucleotide sequence and classification of a hepatitis C virus isolate of a novel genotype from an Indonesian patient with chronic liver disease. J. Gen. Virol. 75:629-635. [DOI] [PubMed] [Google Scholar]
- 17.Okamoto, H., Y. Sugiyama, S. Okada, K. Kurai, Y. Akahane, Y. Sugai, T. Tanaka, K. Sato, F. Tsuda, Y. Miyakawa, et al. 1992. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J. Gen. Virol. 73:673-679. [DOI] [PubMed] [Google Scholar]
- 18.Simmonds, P., A. Alberti, H. J. Alter, F. Bonino, D. W. Bradley, C. Brechot, J. T. Brouwer, S. W. Chan, K. Chayama, D. S. Chen, et al. 1994. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology 19:1321-1324. [PubMed] [Google Scholar]
- 19.Smith, D. B., J. Mellor, L. M. Jarvis, F. Davidson, J. Kolberg, M. Urdea, P. L. Yap, P. Simmonds, and the International HCV Collaborative Study Group. 1995. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J. Gen. Virol. 76:1749-1761. [DOI] [PubMed] [Google Scholar]
- 20.Storm, N., B. Darnhofer-Demar, D. van den Boom, and C. P. Rodi. 2002. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol. Biol. 212:241-262. [DOI] [PubMed] [Google Scholar]
- 21.Stuyver, L., R. Rossau, A. Wyseur, M. Duhamel, B. Vanderborght, H. Heuverswyn, and, G. Maertens. 1993. Typing of hepatitis C virus isolates and characterization of new subtype using a line probe assay. J. Gen. Virol. 74:1093-1102. [DOI] [PubMed] [Google Scholar]
- 22.Zein, N. N. 2000. Clinical significance of hepatitis C genotypes. Clin. Microbiol. Rev. 13:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]