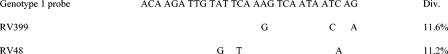Abstract
Several reverse transcription-PCR (RT-PCR) methods have been reported for the detection of rabies and rabies-related viruses. These methods invariably involve multiple transfers of nucleic acids between different tubes, with the risk of contamination leading to the production of false-positive results. Here we describe a single, closed-tube, nonnested RT-PCR with TaqMan technology that distinguishes between classical rabies virus (genotype 1) and European bat lyssaviruses 1 and 2 (genotypes 5 and 6) in real time. The TaqMan assay is rapid, sensitive, and specific and allows for the genotyping of unknown isolates concomitant with the RT-PCR. The assay can be applied quantitatively and the use of an internal control enables the quality of the isolated template to be assessed. Despite sequence heterogeneity in the N gene between the different genotypes, a universal forward and reverse primer set has been designed, allowing for the simplification of previously described assays. We propose that within a geographically constrained area, this assay will be a useful tool for the detection and differentiation of members of the Lyssavirus genus.
Infection with rabies virus requires an accurate and rapid diagnosis, which involves virus detection and genotyping. For suspected human cases, rapid antemortem diagnosis ensures appropriate patient management. Postmortem diagnosis enables the rapid administration of postexposure treatment to all human contacts (7). Furthermore, genetic typing of the isolate increases our epidemiological understanding of the virus (11). The genetic typing of animal isolates (including isolates from bats) allows geographical distributions to be assessed and appropriate control measures to be instigated (9).
Rabies virus and the rabies-related viruses are in the order Mononegavirales, in the Rhabdoviridae family and the Lyssavirus genus. This group of viruses are characterized as enveloped, negative-strand RNA viruses with unsegmented genomes. The genus has been separated into seven genotypes based on the nucleotide sequences and deduced amino acid sequence similarities of the nucleoproteins (N) (4, 5), and these genotypes have subsequently been confirmed by recent studies of the lyssavirus glycoprotein (2) and phosphoprotein (21). The genotypes include classical rabies virus (genotype 1), Lagos bat virus (genotype 2), Mokola virus (genotype 3), Duvenhage virus (genotype 4), European bat lyssavirus (EBLV) types 1 and 2 (genotypes 5 and 6, respectively), and Australian bat lyssavirus (ABLV; genotype 7). In 2003, a new lyssavirus genotype was proposed for a virus isolated from a lesser mouse-eared bat in Kyrghyzstan (1). With the exception of genotype 2, all established lyssavirus genotypes have been associated with clinically indistinguishable fatal diseases in humans. Genotype 1 is found worldwide, with the exception of a few island nations such as Great Britain, Ireland, New Zealand, and Hawaii, the continents of Australia and Antarctica, and an increasing number of Western European countries (2, 31). Genotypes 2 to 4 are widely distributed throughout Africa (18), whereas genotypes 5 and 6 are distributed throughout Europe (22). The United Kingdom is considered “rabies-free,” with only occasional cases of human rabies arising from contact with rabid animals while abroad (15, 24). However, in 1996 a positive case of EBLV type 2 was found in a Daubenton's bat (Myotis daubentonii) in Sussex (30). In September 2002, an EBLV type 2 isolate was detected in a Daubenton's bat in Lancashire (17), and in November of the same year a bat handler in Angus, Scotland, died of EBLV type 2-associated rabies (9). These reports suggest that EBLV-2 may be epizootic in bat populations throughout the United Kingdom. A reverse transcription-PCR (RT-PCR) assay for the detection of rabies and rabies-related viruses developed in our laboratory (12) is a heminested RT-PCR. This technique utilizes a separate RT step, primary amplification, and heminested amplification. The method involves two transfers of material, with an inherent risk of cross-contamination between samples. The amplicon produced is relatively large (586 bp), and although providing a template for sequencing and subsequent phylogenetic analysis, is too large to enable quantitative data to be acquired and renders the assay relatively insensitive compared to other assays, e.g., an ABLV assay (23). The transfer of this assay to a Rapid Cycler (Idaho Technologies, Salt Lake City, Utah) with a nested format decreases the turnaround time of the test but involves a separate RT step, primary amplification, and secondary amplification (13). For an unknown sample, at least seven primers are required if the heminested RT-PCR is applied to amplify the template, and although it is possible to amplify representatives of all seven Lyssavirus genotypes, it is not possible to definitively genotype the virus without further sequencing. The introduction of TaqMan technology (3, 20) further complicated the assay. Again, a separate RT step was required with the JW12 primer followed by amplification with the BB6 primer and three different JW6 primers to yield an ∼500-bp amplicon. Detection of the amplicon with fluorogenic probes required three genotype 1 probes in addition to two probes specific for genotypes 5 and 6. Due to a lack of available dyes at the time, all of the probes in the assay were labeled with the fluorophores FAM and TAMRA, and hence genotypic discrimination was only possible by preparing a panel of reactions, with each containing genotype-specific probes. Due to the size of the amplicon, it would be expected that the assay would be nonquantitative and potentially less sensitive than the heminested assay (25). In addition, since an AB LS50B instrument (Applied Biosystems, Foster City, Calif.) was used to monitor the increase in fluorescence (3), the assay was not done in “real time” and therefore was more time-consuming than, e.g., assays on the LightCycler (Roche Diagnostics Ltd., Lewes, United Kingdom).
The test described in this study addresses all of the disadvantages of the previous assays in that it involves only a single tube for all reactions, reducing the possibility of cross-contamination; the amplicon is small (∼100 bp), allowing for increased sensitivity and relative quantitation; the TaqMan probes are labeled with four different fluorophores, allowing the detection of either genotype 1, 5, or 6 in a single reaction in addition to an internal control transcript (β-actin mRNA), ensuring that the sample was processed correctly; the assay is run in a 96-well standard format for ease of handling and scaling up; there are only two RT-PCR primers (JW12 and N165-146) and only three different TaqMan probes (LysGT1, LysGT5, and LysGT6) involved, reducing the complexity of the assay; the assay is rapid and sensitive, has a dynamic range of 11 log dilutions (for genotype 1), and runs in “real time.”
MATERIALS AND METHODS
RNA samples.
Total RNAs were extracted from field and experimental samples from the Rabies Research and Diagnostic Group archive under category III containment conditions in a dedicated ACDP3/SAPO IV high-security unit by staff who were vaccinated against rabies. Quality standards to ISO9001 were applied. The extractions were performed by using TRIzol (Invitrogen, Paisley, United Kingdom) according to the manufacturer's instructions. In addition, a set of nine samples, previously tested in a ring trial by several regional veterinary laboratories in Germany, was provided by the Institute for Epidemiology, WHO Collaborating Centre for Rabies Surveillance and Research, Friedrich-Loeffler Institute, Federal Research Centre for Animal Health, Wüsterhausen, Germany, and consisted of a brain homogenate of a rabies virus-positive dog diluted in a rabies virus-negative fox brain homogenate. The negative controls in this sample set were uninfected fox brain homogenates. Diagnostic field samples from various animals that were submitted to the Central Veterinary Research Laboratories in Khartoum for investigations of rabies infections (16) were also tested with the real-time TaqMan assay.
Design of TaqMan primer and probe sets.
Primer sequences and locations are indicated in Table 1. The classical rabies virus TaqMan probe, LysGT1, was designed by using 557 partial N gene sequences (402 bases) held in the Veterinary Laboratories Agency-Weybridge database. For EBLV-1 and -2 (lyssavirus genotypes 5 and 6), the probes LysGT5 and LysGT6 were designed starting 2 to 3 nucleotides downstream of the JW12 reverse transcription-PCR primer in the N gene, using sequences derived from 35 genotype 5 and 9 genotype 6 isolates. TaqMan probes were conjugated with the fluorophore and quencher molecules FAM/TAMRA (LysGT1; Sigma-Genosys Ltd., Haverhill, United Kingdom), HEX/Blackhole Quencher 1 (LysGT5; Sigma-Genosys Ltd.), and Cy5/Blackhole Quencher 2 (LysGT6; Proligo, Paris, France). Consensus sequences for the first 402 bases of the N gene sequence were generated for lyssavirus genotypes 1, 5, and 6 by using MegAlign (DNAStar, Madison, Wis.). The consensus sequences were then compared by using the same software to identify conserved regions among the three genotypes in order to design the universal reverse primer N165-146.
TABLE 1.
Oligonucleotide sequences of TaqMan probes and RT-PCR primers
| Name | Role | Length (nt) | Tm (°C) | Sequencea | Positionb |
|---|---|---|---|---|---|
| JW12 | RT-PCR primer | 19 | 49 | ATGTAACACCYCTACAATG | 55-73 |
| N165-146 | PCR primer | 20 | 53 | GCAGGGTAYTTRTACTCATA | 165-146 |
| LysGT1 | Probe | 29 | 62 | ACAAGATTGTATTCAAAGTCAATAATCAG | 81-109 |
| LysGT5 | Probe | 26 | 64 | AACARGGTTGTTTTYAAGGTCCATAA | 80-105 |
| LysGT6 | Probe | 29 | 55 | ACARAATTGTCTTCAARGTCCATAATCAG | 81-109 |
| β act intronic | PCR primer | 22 | 63 | CGATGAAGATCAAG/ATCATTGC | 1051-1072 |
| β act reverse | RT-PCR primer | 17 | 62 | AAGCATTTGCGGTGGAC | 1204-1188 |
| β-Actin | Probe | 32 | 64 | TCCACCTTCCAGCAGATGTGGATCAGCAAG | 1128-1157 |
The β-actin primers and probes were designed by using a consensus sequence derived from β-actin mRNAs corresponding to exons 4 and 5 from the sheep, bat, cow, guinea pig, horse, human, mouse, and rat genes. The forward primer was designed to span the exon boundary between exons 4 and 5, with 8 of 22 bases lying in exon 5. The 5′ terminus of the probe was labeled with ROX and quenched at the 3′ terminus with Deep Dark Quencher 2 (Eurogentec, Liege, Belgium).
TaqMan RT-PCR.
TaqMan RT-PCRs were performed in 50-μl reaction volumes comprised of 20.75 μl of nuclease-free water, 5 μl of 10× PCR buffer (Promega, Madison, Wis.), 12 μl of 25 mM MgCl2, 1 μl of deoxynucleoside triphosphates (a 10 mM concentration of each), 1 μl each of the PCR primers (a 20 μM concentration of each), 1 μl each of the TaqMan probes (a 5 μM concentration of each), 1 μl of Triton X-100 (10% [vol/vol]), 0.25 μl of RNasin (20 to 40 U/μl) (Promega), 0.5 μl of Moloney murine leukemia virus reverse transcriptase (200 U/μl) (Promega), and 0.5 μl of Taq polymerase (5 U/μl) (Promega). One microliter of total RNA was added to this mixture at a concentration of 1 μg/μl. The reactions were carried out in Thermo-Fast 96-well PCR plates or Thermo-tube strips with Ultra Clear caps (ABgene, Epsom, United Kingdom) in an MX3000P multiplex quantitative PCR system (Stratagene, La Jolla, Calif.). The RNAs were reverse transcribed and amplified according to the following heating and cooling program: 1 cycle of 42°C for 30 min and 94°C for 2 min followed by 45 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 20 s. Positive control assays were run using RNAs extracted from mouse neuroblastoma cell cultures infected with lyssavirus strains CVS (challenge virus standard; genotype 1), RV1423 (genotype 5), and RV628 (genotype 6).
For each RT-PCR, a critical threshold cycle number (CT) was determined corresponding to the PCR cycle number at which the fluorescence of the reaction exceeded a value determined to be statistically higher than the background by the software associated with the MX3000P system (Stratagene).
Conventional RT-PCR.
For comparative purposes, separate RT reactions and heminested PCRs (12) were performed with 10-fold serial dilutions of candidate virus strain RNAs corresponding to lyssavirus genotypes 1, 5, and 6. The dilution series were made in nuclease-free water. In brief, 2 μg of total RNA was heat denatured at 100°C for 5 min prior to being reverse transcribed by Moloney murine leukemia virus reverse transcriptase and the primer JW12 at 42°C for 60 min. The cDNA was diluted 1:10 in nuclease-free water, and 5 μl of the mixture was used in a first-round PCR of 46 cycles. One microliter of the first-round PCR product was then subjected to the second-round heminested PCR for 36 cycles. The products of both the first and second rounds of PCR were analyzed in 1.8% agarose gels stained with ethidium bromide.
RESULTS
Comparative sensitivity of TaqMan assay.
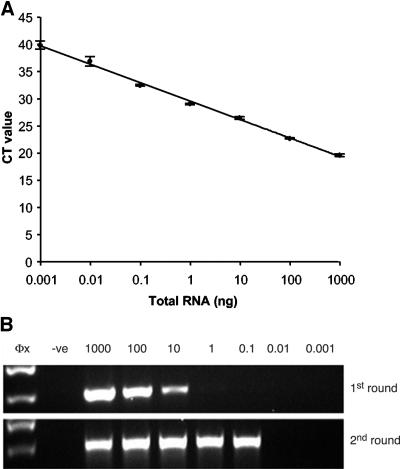
For determination of the relative sensitivity of the single-tube TaqMan RT-PCR assay compared to that previously described (12), a 10-fold serial dilution of total RNA extracted from mouse neuroblastoma cells infected with rabies virus CVS (genotype 1), RV1423 (genotype 5), or RV628 (genotype 6) was prepared in nuclease-free water and tested in both assays. With the heminested PCR assay (12), a product was visible for the first-round PCR for CVS and RV1423 when 1 μg to 10 ng of total RNA was added to the reverse transcription reaction, whereas for RV628, the limit of detection was 100 ng of total RNA. Following the second round of PCR, the limit of detection for CVS was 100 pg of RNA, whereas for RV1423 and RV628, a strong band on the agarose gel was detected if 1 μg to 1 ng of template was added to the reverse transcription reaction. With the single-tube TaqMan RT-PCR assay, all genotypes were detected for the smallest amount of RNA added to the reverse transcription reaction (1 pg), but the linearity of the reaction for genotypes 5 and 6 was only maintained between 10 pg and 1 μg. The efficiencies of the reactions were 92.1%, 98.8%, and 99% for the genotype 1, 5, and 6 TaqMan assays, respectively. A standard curve was generated for the single-tube RT-PCR for genotype 1 and is shown in Fig. 1 (top panel), and the corresponding agarose gel for the same template in the heminested PCR assay (12) is shown in Fig. 1 (bottom panel). The N gene amplicon corresponding to CVS, generated by using the previously described JW12 and JW6 (DPL) primers (12), was cloned into the pGEM-T Easy plasmid vector (Promega) in order to provide a standard template for quantitative purposes. The limit of detection for the PCR component of the assay for genotype 1 with the cloned CVS N gene sequence as the template was determined to be one molecule, as deduced from the amount of plasmid added to the PCR, the molecular weight of the plasmid, and Avogadro's number (data not shown).
FIG. 1.
Comparison of single-tube TaqMan RT-PCR assay for rabies virus (genotype 1) with the heminested PCR method (12), using the same diluted RNA standards. (Top) Inverse linear relationship between RNA concentration and CT value for the single-tube TaqMan RT-PCR assay. The error bars show the standard deviations of the means of three replicates (r2 = 0.994). (Bottom) Agarose gel (1.8%) analysis of diluted total RNA, expressed in nanograms, following first- and second-round heminested PCR. The negative control (−ve) was uninfected mouse brain total RNA.
Detection and genotyping of archival samples.
A total of 62 samples (40 designated genotype 1, 15 designated genotype 5, 6 designated genotype 6, and 1 designated genotype 7) were assayed with the single-tube TaqMan RT-PCR assay, and the results obtained were compared with the genotypes derived by conventional sequencing methods (12, 17). All RNA templates were reverse transcribed and amplified with the primers JW12 and N165-146 to produce an amplicon of the expected size. The CT values of the TaqMan assay for each isolate, corresponding to all of the probes in the reaction, are indicated in Table 2. When no CT is shown for a particular isolate and probe combination, it indicates that for that genotype-specific probe, the fluorescence readings failed to increase during the reaction. In all cases, when the fluorescence readings increased, only one probe hybridized to the template, indicating the genotype and showing that the virus isolate was not mixed. The CT values were found to be quite variable due to the amount of template being added to the TaqMan assay being standardized by reference to the total RNA concentration rather than the viral load. With the exception of RV48, all templates were readily genotyped (genotypes 1, 5, and 6). The RV48 amplicon only weakly hybridized with the genotype 1 probe.
TABLE 2.
List of archival samples tested with the single-tube TaqMan RT-PCR assay
| Isolate | Country of origin | Host species | Genotype |
CT valuea
|
% Divergenceb
|
||||
|---|---|---|---|---|---|---|---|---|---|
| FAM | HEX | Cy5 | GT1 | GT5 | GT6 | ||||
| 8 | Finland | Human | 6 | 15.86 | 11.1 | 26.9 | 0 | ||
| 9 | Germany | Bat | 5 | 15.73 | 19.8 | 0 | 17.6 | ||
| 11 | Germany | Bat | 5 | 13.94 | 19.8 | 0 | 17.6 | ||
| 19 | Denmark | Bat | 5 | 16.46 | 19.8 | 0 | 17.6 | ||
| 29 | The Netherlands | Bat | 6 | 12.12 | 15.3 | 26.9 | 0 | ||
| 31 | The Netherlands | Bat | 5 | 14.12 | 19.8 | 0 | 17.6 | ||
| 32 | The Netherlands | Bat | 5 | 13.62 | 19.8 | 0 | 17.6 | ||
| 33 | The Netherlands | Bat | 5 | 15.67 | 19.8 | 0 | 17.6 | ||
| 37 | The Netherlands | Bat | 5 | 12.95 | 19.8 | 0 | 17.6 | ||
| 38 | The Netherlands | Bat | 5 | 16.57 | 19.8 | 0 | 17.6 | ||
| 44 | USA | Bat | 1 | 11.72 | 3.6 | 18.9 | 12.0 | ||
| 47 | USA | Bat | 1 | 16.19 | 7.4 | 24.8 | 16.6 | ||
| 48 | USA | Bat | 1 | 44.67 | 11.2 | 24.8 | 21.3 | ||
| 49 | USA | Bat | 1 | 10.95 | 7.2 | 18.9 | 21.3 | ||
| 56 | USA | Fox | 1 | 7.39 | 7.3 | 25.7 | 12.0 | ||
| 57 | USA | Skunk | 1 | 7.91 | 3.6 | 31.5 | 16.6 | ||
| 58 | USA | Skunk | 1 | 12.27 | 7.2 | 37.8 | 12.2 | ||
| 66 | Poland | Bat | 5 | 15.08 | 19.8 | 0 | 17.6 | ||
| 68 | Peru | Dog | 1 | 10.12 | 3.6 | 24.8 | 16.6 | ||
| 73 | Belize | Dog | 1 | 8.95 | 3.6 | 24.8 | 12.0 | ||
| 100 | Morocco | Unknown | 1 | 20.27 | 3.6 | 31.5 | 16.6 | ||
| 114 | Chile | Bat | 1 | 15.36 | 7.4 | 18.9 | 12.0 | ||
| 144 | Germany | Bat | 5 | 29.13 | 19.8 | 0 | 17.6 | ||
| 193 | Pakistan | Canine | 1 | 9.25 | 3.6 | 31.5 | 16.6 | ||
| 202 | Turkey | Canine | 1 | 9.72 | 0 | 24.8 | 12.0 | ||
| 227 | Nigeria | Canine | 1 | 8.2 | 3.6 | 24.8 | 12.0 | ||
| 234 | Russia | Canine | 1 | 13.55 | 0 | 24.8 | 12.0 | ||
| 253 | Russia | Wolf | 1 | 12.31 | 3.5 | 25.7 | 7.8 | ||
| 264 | Ukraine | Bat | 5 | 24.78 | 15.3 | 26.9 | 0 | ||
| 266 | France | Bat | 5 | 8.07 | 24.9 | 0 | 17.6 | ||
| 303 | Russia | Raccoon dog | 1 | 32.49 | 0 | 24.8 | 12.0 | ||
| 305 | Georgia | Dog | 1 | 10.27 | 0 | 24.8 | 12.0 | ||
| 307 | Georgia | Cattle | 1 | 8.15 | 0 | 24.8 | 12.0 | ||
| 313 | Germany | Fox | 1 | 9.08 | 0 | 24.8 | 12.0 | ||
| 318 | Germany | Fox | 1 | 25.89 | 3.5 | 19.4 | 7.8 | ||
| 338 | China | Cattle | 1 | 9.88 | 3.6 | 24.8 | 12.0 | ||
| 341 | China | Dog | 1 | 11.09 | 0 | 24.8 | 12.0 | ||
| 342 | China | Cattle | 1 | 11.27 | 3.6 | 24.8 | 12.0 | ||
| 345 | Germany | Bat | 5 | 23.85 | 19.8 | 0 | 17.6 | ||
| 349 | Germany | Bat | 5 | 24.77 | 19.8 | 0 | 17.6 | ||
| 350 | Germany | Bat | 5 | 24.06 | 19.8 | 0 | 17.6 | ||
| 399 | Botswana | Jackal | 1 | 10.46 | 11.6 | 18.9 | 21.6 | ||
| 437 | Estonia | Raccoon dog | 1 | 12.58 | 0 | 24.8 | 12.0 | ||
| 447 | Botswana | Dog | 1 | 9.8 | 3.6 | 31.5 | 16.6 | ||
| 483 | Botswana | Genet | 1 | 13.55 | 11.6 | 18.9 | 21.6 | ||
| 484 | Botswana | Cattle | 1 | 12.62 | 11.6 | 18.9 | 21.6 | ||
| 498 | S. Africa | Cat | 1 | 8.25 | 3.6 | 24.8 | 16.6 | ||
| 519 | Unknown | Cattle | 1 | 9.29 | 0 | 24.8 | 12.0 | ||
| 594 | Switzerland | Bat | 6 | 16.22 | 15.3 | 26.9 | 0 | ||
| 621 | Switzerland | Bat | 6 | 10.78 | 19.8 | 20.3 | 0 | ||
| 628 | England | Bat | 6 | 12.75 | 15.3 | 26.9 | 0 | ||
| 634 | Australia | Bat | 7 | 24.9 | 31.5 | 33.4 | |||
| 758 | Tanzania | Canine | 1 | 10.06 | 0 | 24.8 | 12 | ||
| 759 | Tanzania | Canine | 1 | 29.35 | 0 | 24.8 | 12 | ||
| 889 | Czech Republic | Fox | 1 | 13.95 | 3.7 | 19.9 | 12.5 | ||
| 960 | Estonia | Raccoon dog | 1 | 13.7 | NA | NA | NA | ||
| 1124 | Turkey | Fox | 1 | 16.93 | NA | NA | NA | ||
| 1125 | Turkey | Fox | 1 | 19.08 | NA | NA | NA | ||
| 1176 | Yugoslavia | Fox | 1 | 13.92 | NA | NA | NA | ||
| 1177 | Yugoslavia | Fox | 1 | 17.65 | 3.6 | 31.5 | 16.6 | ||
| 1312 | Poland | Badger | 1 | 13.79 | 0 | 24.8 | 12.0 | ||
| 1333 | Scotland | Human | 6 | 13.05 | 19.8 | 26.9 | 3.8 | ||
For clarity, when no CT value was recorded, this is indicated by the absence of a numerical value for that particular probe.
The divergence was calculated by using the Clustal V component of the MegAlign multiple alignment package (DNASTAR Inc.). NA, sequence data are not available for the probe binding region.
Cross-hybridization was evident at a PCR annealing temperature of 50°C between the genotype 1 probe and the genotype 6 amplicon derived from RV628 (data not shown). The other genotype 6 templates were not assayed at this annealing temperature. At a temperature of 55°C, cross-hybridization was not evident, and this annealing temperature was adopted for all PCRs performed in this study. The RNA from isolate RV634, an Australian bat lyssavirus isolate (genotype 7), was analyzed. An amplicon of the correct size was produced following the single tube TaqMan RT-PCR assay (as determined by agarose gel electrophoresis), but none of the probes was hydrolyzed during the reaction. The isolates tested and the divergence of each template in the region of the three different probes are indicated in Table 2.
Detection and genotyping of unknown samples.
A set of nine coded samples (brain homogenate suspensions), which were previously tested in a ring trial in Germany and were kindly provided by Thomas Müller (Institute for Epidemiology, WHO Collaborating Centre for Rabies Surveillance and Research), were assayed blind for the presence of virus. Following testing, the samples were revealed to be seven mixtures of positive brain material from a single infected dog, diluted in a twofold dilution series in uninfected fox brain material, and two samples of uninfected fox brain homogenate (Table 3). The diagnostic tests employed included the fluorescent antibody test (FAT), the rabies tissue culture inoculation test (RTCIT), and the mouse inoculation test (MIT), all performed as previously described (6, 19, 28), the original first-round RT-PCR as previously described (12), and the new TaqMan RT-PCR methodology (Table 3). A control TaqMan RT-PCR assay for β-actin was included in a duplicate set of samples in addition to the lyssavirus-specific RT-PCR. Seven of the nine samples were shown to be positive for lyssavirus by the PCR-based assays and the MIT. The FAT detected all but the lowest dilution of rabies-infected material, and the RTCIT failed to detect two of the positive samples. The β-actin-positive control RT-PCR was positive for all samples analyzed, did not cause an apparent overall change in the CT values recorded for the rabies virus PCRs, and indicated that there was less total RNA in the 1:8 dilution than in the others (GT1 CT = 18.01; β-actin CT = 17.11). With the single-tube TaqMan RT-PCR method, only the genotype 1 probe hybridized with the amplicons produced, indicating that only classical rabies virus RNA was present in the sample.
TABLE 3.
Comparison of diagnostic methods for rabies used on ring trial material
| Method | Result (+, −, or CT value) at indicated dilution
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1:2 | 1:4 | 1:8 | 1:16 | 1:32 | 1:64 | 1:128 | Negative control 1 | Negative control 2 | |
| FAT | + | + | + | + | + | + | − | − | − |
| RTCIT | + | + | − | + | + | + | − | − | − |
| MIT | + | + | + | + | + | + | + | − | − |
| hnRT-PCR | Weakly + | + | + | + | + | + | + | − | − |
| Q-RT-PCR | |||||||||
| LysGT1 RT-PCR | 16.55 | 16.08 | 18.01 | 12.35 | 13.22 | 11.78 | 15.33 | 0 | 0 |
| LysGT5 RT-PCR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| LysGT6 RT-PCR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| β-Actin RT-PCR | 14.17 | 13.86 | 17.11 | 12.16 | 14.48 | 13.07 | 14.44 | 13.15 | 14.51 |
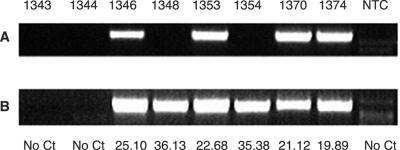
Brain samples from various animals in Sudan suspected of being infected with rabies virus were tested with the single-tube TaqMan RT-PCR assay, and the results were compared with those obtained with the heminested PCR-based assay (12). The CT values generated correlated with the results from the heminested PCR assay, with higher CT values corresponding to those samples with apparently less amplifiable material, as determined by a comparison of the products generated in the heminested PCR assay analyzed by agarose gel electrophoresis with the CT values (Fig. 2). No band or CT value was recorded for sample RV1343 or RV1344 with either the heminested or the real-time RT-PCR assay. All of the viruses identified were genotyped as classical rabies virus, and the host species did not appear to affect the TaqMan assay.
FIG. 2.
Comparison of the conventional heminested RT-PCR (12) and the single-tube RT-PCR methodology (following a 30-min reverse transcription step and 45 rounds of PCR) using 2 μg of total RNA isolated directly from Sudanese brain samples from animals suspected to be infected with rabies. (A) Reverse transcription and first-round PCR with JW12 and JW6 (dpl/e/m). (B) Second-round PCR of products shown in panel A with JW12 and JW10 (dle2/me1/p). The corresponding CT values are shown below the gel. RV1343, goat sample; RV1344, cat sample; RV1346, dog sample; RV1348, goat sample; RV1353, donkey sample; RV1354, donkey sample; RV1370, cow sample; RV1374, cow sample; NTC, no template control.
Effect of genetic divergence at the probe site.
Diversity within the probe site for isolates RV48, RV399, RV483, and RV484 was compared with respect to hybridization with the genotype 1 probe, as these isolates apparently had the most sequence divergence in this area but, with the exception of RV48, still hybridized selectively and efficiently with the probe. Within the binding region of the probe, there were three mismatches in all cases. For RV399, RV483, and RV484, the mismatches were at positions 17, 26, and 28 of the probe sequence, whereas for RV48, they were at positions 11, 14, and 27. A sequence alignment of RV399, RV48, and the genotype 1 probe is shown in Fig. 3.
FIG. 3.
Comparison of the genotype 1 probe sequence with those of RV399, which hybridized with the probe, and RV48, which failed to hybridize or hybridized poorly. Only nucleotide substitutions are shown for the two isolates. The percent divergence was calculated by using the Clustal V component of the MegAlign multiple alignment package (DNASTAR Inc.).
DISCUSSION
A single-tube RT-PCR method has been developed which addresses problems associated with the original heminested PCR assay (12) being relatively insensitive and requiring multiple transfers of material with the concomitant opportunity for cross-contamination. The incorporation of TaqMan technology into a real-time assay not only enables the lyssavirus template to be detected but also allows it to be genotyped. The use of only two universal amplimers (JW12 and N165-146) plus a single probe for each genotype considerably simplifies the TaqMan method, which was developed for use on an Applied Biosystems LS50B instrument (3), although it is acknowledged that the assay is only applicable to three different genotypes of the lyssavirus group. A β-actin TaqMan RT-PCR assay allows the state of the starting material to be ascertained, which may be an important issue for the analysis of small quantities of material transported or stored under less than ideal circumstances (29). This control was not used for normalization purposes in the present study, and it is unlikely that it would be used as such due to the tendency of the primers to amplify genomic DNA in the absence of an upstream RNase-free DNase treatment of the template. However, with the correct enzymatic treatment, this assay could be applied quantitatively in order to estimate the viral load (14, 27).
The single-tube TaqMan RT-PCR assay performed well compared with the gel-based assay with respect to its sensitivity. A direct comparison of the heminested assay and the TaqMan assay revealed that a positive CT value was still obtained when only 1 pg of total RNA was added to the assay, whereas 100 times more RNA was required to generate a visible band on an ethidium bromide-stained agarose gel. In addition, with the cloned classical rabies virus N gene sequence, only one molecule was required in order to produce a positive result in the single-tube TaqMan assay, which is the theoretical minimum amount of template required to produce a positive CT value. With the exception of RV48 (which only poorly hybridized with the LysGT1 probe), all archival samples corresponding to lyssavirus genotypes 1, 5, and 6 were readily detected and correctly genotyped. Positional/combinatorial effects of particular nucleotide substitutions in RV48 appeared to preclude the effective genotyping of this isolate, despite the amplimers binding and a PCR product being generated, as determined by agarose gel electrophoresis. This situation has been suggested previously (14) as a reason for the failure of single nucleotide polymorphisms to generate fluorescent signals in a TaqMan assay when point mutations at the center of the probe have prevented hybridization, despite PCR proceeding as expected. The ABLV isolate RV634 was not detected with the TaqMan assay, despite the fact that an amplicon was produced, indicating that the PCR primers annealed to this template. Since this assay was developed to test viruses likely to be submitted to European laboratories, an inability to detect ABLV was not deemed to be problematic, and as such, this assay will allow the rapid genotyping of suspected rabies samples in the absence of nucleotide sequencing.
Compared to conventional virological methods to detect the viruses employed in the blind ring trial (Table 3), the TaqMan assay performed equivalently or, in the case of the RTCIT (28), was superior, and as such, could work well as an adjunct to the Office International des Epizooties-prescribed assays. The CT values corresponding to the viruses in these samples did not correlate with the apparent dilution of the positive brain material, and we suggest that the reason for this was that the samples contained very high viral loads. With respect to the detection of viruses in samples from Sudan, the host species from which the brain samples were taken did not apparently affect the detection or genotyping of the infecting virus. In addition, the CT values obtained reflected the levels of template amplified in the conventional gel-based assay (12).
The single-tube TaqMan assay is considerably faster to perform than the heminested RT-PCR assay, with the process taking 2 h 11 min, including a 30-min reverse transcription step. This can be reduced to a 2-min reverse transcription step for experimental samples with known levels of template RNA (data not shown), reducing the overall time to perform the assay to 1 h 43 min. Since the results are obtained in “real time,” it is possible to inform competent authorities as the reaction proceeds, more rapidly enabling appropriate decisions to be made with regard to patient management or disease control. The single-tube TaqMan assay has been introduced as a routine test in our laboratory and has been successfully used to detect and characterize viruses from two postmortem field samples from Daubenton's bats found in the United Kingdom. Subsequent sequencing confirmed the presumptive genotyping (genotype 6) conferred by the assay (8, 10).
Acknowledgments
We acknowledge Thomas Müller (Institute for Epidemiology, WHO Collaborating Centre for Rabies Surveillance and Research, Friedrich-Loeffler Institute, Federal Research Centre for Animal Health, Wüsterhausen, Germany) for providing the diagnostic rabies virus samples used in the ring trial assessment and Yahia Beik from the Central Veterinary Research Laboratory, Virology Department, Khartoum, Sudan, for supplying field samples. In addition, we thank all of our collaborators who have, over the years, supplied virus isolates to the Weybridge rabies virus collection.
This work was funded by the Department for Environment, Food and Rural Affairs (Defra), United Kingdom (grant SV3023).
REFERENCES
- 1.Arai, Y. T., I. V. Kuzmin, Y. Kameoka, and A. D. Botvinkin. 2003. New lyssavirus genotype from the lesser mouse-eared bat (Myotis blythi), Kyrghyzstan. Emerg. Infect. Dis. 9:333-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badrane, H., C. Bahloul, P. Perrin, and N. Tordo. 2001. Evidence of two lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J. Virol. 75:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, E. M., J. P. Lowings, J. Smith, P. R. Heaton, and L. M. McElhinney. 2002. A rapid RT-PCR method to differentiate six established genotypes of rabies and rabies-related viruses using TaqMan technology. J. Virol. Methods 105:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourhy, H., B. Kissi, M. Lafon, D. Sacramento, and N. Tordo. 1992. Antigenic and molecular characterization of bat rabies in Europe. J. Clin. Microbiol. 30:2419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourhy, H., B. Kissi, and N. Tordo. 1993. Molecular diversity of the Lyssavirus genus. Virology 194:70-81. [DOI] [PubMed] [Google Scholar]
- 6.Dean, D. J., M. K. Abelseth, and P. Atanasiu. 1996. The fluorescence antibody test, p. 88-93. In F. X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies, 4th ed. World Health Organization, Geneva, Switzerland.
- 7.Department of Health. 2000. Policy and guidance article. Memorandum on rabies: prevention and control, p. 19. Department of Health, London, United Kingdom.
- 8.Fooks, A. R., L. M. McElhinney, D. A. Marston, D. Selden, T. A. Joliffe, P. R. Wakeley, N. Johnson, and S. M. Brookes. 2004. Identification of a European bat lyssavirus type 2 in a Daubenton's bat found in Staines, Surrey, UK. Vet. Rec. 155:434-435. [PubMed] [Google Scholar]
- 9.Fooks, A. R., L. M. McElhinney, D. J. Pounder, C. J. Finnegan, K. Mansfield, N. Johnson, S. M. Brookes, G. Parsons, K. White, P. G. McIntyre, and D. Nathwani. 2003. Case report: isolation of a European bat lyssavirus type 2a from a fatal human case of rabies encephalitis. J. Med. Virol. 71:281-289. [DOI] [PubMed] [Google Scholar]
- 10.Fooks, A. R., D. Selden, S. M. Brookes, N. Johnson, D. A. Marston, T. A. Joliffe, P. R. Wakeley, and L. M. McElhinney. 2004. Identification of a European bat lyssavirus type 2 in a Daubenton's bat found in Lancashire. Vet. Rec. 155:606-607. [PubMed] [Google Scholar]
- 11.Gould, A. R., A. D. Hyatt, R. Lunt, J. A. Kattenbelt, S. Hengstberger, and S. Blacksell. 1998. Characterisation of a novel lyssavirus isolated from Pteropid bats in Australia. Virus Res. 54:165-187. [DOI] [PubMed] [Google Scholar]
- 12.Heaton, P. R., P. Johnstone, L. M. McElhinney, R. Cowley, E. O'Sullivan, and J. E. Whitby. 1997. Heminested PCR assay for the detection of six genotypes of rabies and rabies-related viruses. J. Clin. Microbiol. 35:2762-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heaton, P. R., L. M. McElhinney, and J. P. Lowings. 1999. Detection and identification of rabies and rabies-related viruses using rapid-cycle PCR. J. Virol. Methods 81:63-69. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, G. J., J. S. Smith, C. A. Hanlon, and C. E. Rupprecht. 2004. Evaluation of a TaqMan PCR assay to detect rabies virus RNA: influence of sequence variation and application to quantification of viral loads. J. Clin. Microbiol. 42:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, N., D. W. Lipscomb, R. Stott, G. Gopal Rao, K. Mansfield, J. Smith, L. M. McElhinney, and A. R. Fooks. 2002. Investigation of a human case of rabies in the United Kingdom. J. Clin. Virol. 25:351-356. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, N., L. M. McElhinney, Y. H. Ali, I. K. Saeed, and A. R. Fooks. 2004. Molecular epidemiology of canid rabies in Sudan: evidence for a common origin of rabies with Ethiopia. Virus Res. 104:201-205. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, N., D. Selden, G. Parsons, D. Healy, S. M. Brookes, L. M. McElhinney, A. M. Hutson, and A. R. Fooks. 2003. Isolation of a European bat lyssavirus type 2 from a Daubenton's bat in the United Kingdom. Vet. Rec. 152:383-387. [DOI] [PubMed] [Google Scholar]
- 18.King, A., and J. Crick. 1988. Rabies-related viruses, p. 177-199. In J. B. Campbell and K. M. Charlton (ed.), Rabies. Kluwer, Boston, Mass.
- 19.Koprowski, H. 1996. The mouse inoculation test, p. 80-87. In F. X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies, 4th ed. World Health Organization, Geneva, Switzerland.
- 20.Livak, K. J., S. J. A. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridisation. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 21.Nadin-Davis, S. A., M. Abdel-Malik, J. Armstrong, and A. I. Wandeler. 2002. Lyssavirus P gene characterisation provides insight into the phylogeny of the genus and identifies structural similarities and diversity within the encoded phosphoprotein. Virology 298:286-305. [DOI] [PubMed] [Google Scholar]
- 22.Schneider, L. G., and J. H. Cox. 1994. Bat lyssaviruses in Europe, p. 207-218. In C. E. Rupprecht, B. Dietzschold, and H. Koprowski (ed.), Lyssaviruses. Springer-Verlag, Berlin, Germany.
- 23.Smith, I. L., J. A. Northill, B. J. Harrower, and G. A. Smith. 2002. Detection of Australian bat lyssavirus using a fluorogenic probe. J. Clin. Virol. 25:285-291. [DOI] [PubMed] [Google Scholar]
- 24.Smith, J., L. McElhinney, G. Parsons, N. Brink, T. Doherty, D. Agranoff, M. E. Miranda, and A. R. Fooks. 2003. Case report: rapid ante-mortem diagnosis of a human case of rabies imported into the UK from the Philippines. J. Med. Virol. 69:150-155. [DOI] [PubMed] [Google Scholar]
- 25.Smith, S., L. Vigilant, and P. A. Morin. 2002. The effect of sequence length and oligonucleotide mismatches on 5′ exonuclease assay efficiency. Nucleic Acids Res. 30:e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tordo, N., O. Poch, A. Ermine, G. Keith, and F. Rougeon. 1988. Completion of the rabies virus genome sequence determination: highly conserved domains among the L (polymerase) proteins of unsegmented negative-strand RNA viruses. Virology 165:565-576. [DOI] [PubMed] [Google Scholar]
- 27.Vandesomple, J., A. De Paepe, and F. Speleman. 2002. Elimination of primer dimer artefacts and genomic co-amplification using a two-step SYBR green I real-time RT-PCR. Anal. Biochem. 303:85-98. [DOI] [PubMed] [Google Scholar]
- 28.Webster, W. A., and G. A. Casey. 1996. Virus isolation in neuroblastoma cell culture, p. 96-104. In F. X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies, 4th ed. World Health Organization, Geneva, Switzerland.
- 29.Whitby, J. E., P. Johnstone, and C. Sillero-Zubiri. 1997. Rabies virus in the decomposed brain of an Ethiopian wolf detected by nested reverse transcription-polymerase chain reaction. J. Wildl. Dis. 33:912-915. [DOI] [PubMed] [Google Scholar]
- 30.Whitby, J. E., P. R. Heaton, E. M. Black, M. Wooldridge, L. M. McElhinney, and P. Johnstone. 2000. First isolation of a rabies-related virus from a Daubenton's bat in the United Kingdom. Vet. Rec. 147:385-388. [DOI] [PubMed] [Google Scholar]
- 31.World Health Organization. 2002. World survey for rabies no. 35 for the year 1999. [Online.] http://www.who.int/emc-documents/rabies/whocdscreph200210.html.