Abstract
Background
Homeobox genes of the orthodenticle (otd)/Otx family have conserved roles in the embryogenesis of head and brain. Gene replacement experiments show that the Drosophila otd gene and orthologous mammalian Otx genes are functionally equivalent, in that overexpression of either gene in null mutants of Drosophila or mouse can restore defects in cephalic and brain development. This suggests that otd and Otx genes control a comparable subset of downstream target genes in either organism. Here we use quantitative transcript imaging to analyze this equivalence of otd and Otx gene action at a genomic level.
Results
Oligonucleotide arrays representing 13,400 annotated Drosophila genes were used to study differential gene expression in flies in which either the Drosophila otd gene or the human Otx2 gene was overexpressed. Two hundred and eighty-seven identified transcripts showed highly significant changes in expression levels in response to otd overexpression, and 682 identified transcripts showed highly significant changes in expression levels in response to Otx2 overexpression. Among these, 93 showed differential expression changes following overexpression of either otd or Otx2, and for 90 of these, comparable changes were observed under both experimental conditions. We postulate that these transcripts are common downstream targets of the fly otd gene and the human Otx2 gene in Drosophila.
Conclusion
Our experiments indicate that approximately one third of the otd-regulated transcripts also respond to overexpression of the human Otx2 gene in Drosophila. These common otd/Otx2 downstream genes are likely to represent the molecular basis of the functional equivalence of otd and Otx2 gene action in Drosophila.
Background
Studies on developmental control genes involved in anterior patterning have revealed a set of homologous genes encoding transcription factors that are required for the developmentof the head and brain in diverse animal phyla [1,2,3,4,5]. A striking example for the evolutionary conservation of expression and function of such genes between invertebrates and vertebrates are the homeobox genes of the orthodenticle gene family, which includes the Drosophila orthodenticle (otd) and the murine Otx1 and Otx2 genes [6,7,8,9]. The Drosophila otd gene is expressed in the anterior region of the early embryo in a domain that includes the precursors of the procephalic regions of the head, and it is also expressed in anterior brain regions and in midline CNS structures [6,10,11,12,13,14,15]. Mutational inactivation of otd in Drosophila results in defects in head structures and deletions in anterior parts of the brain as well as in ventral nerve cord defects [6,14,16]. The two otd-related genes in the mouse, Otx1 and Otx2, are also expressed anteriorly in the embryo in nested domains that include the embryonic forebrain and midbrain [17]. Mutational inactivation of these genes results in specific defects in the head and anterior CNS; Otx2 null mice die early in development and fail in specification of the rostral neuroectoderm and proper gastrulation [18,19,20,21]. Otx1 null mice are viable but have spontaneous epileptic seizures and abnormalities affecting the dorsal telencephalic cortex [22].
In addition to the remarkable similarities in expression patterns and mutant phenotypes of the otd/Otx gene family, in vivo gene replacement experiments provide further evidence for conservation of functional properties [3,23,24,25]. In these cross-phylum rescue experiments, human Otx1 or Otx2 genes were overexpressed in Drosophila otd mutants and, conversely, murine Otx1 or Otx2 genes were replaced with the Drosophila otd gene in the mouse. Human Otx1 and Otx2 genes were able to partially rescue the brain and cephalic defects in Drosophila, although Otx2 rescues at a lower frequency than otd, and Otx1 rescues less efficiently still [24,25]. Similarly, the Drosophila otd gene coding sequence introduced into the mice Otx1 locus was able to rescue most of the brain-patterning defects in Otx1 mouse mutants and, when provided with the appropriate Otx2 posttranslational control elements, also in Otx2 mouse mutants [23,26].
Drosophila and vertebrate otd/Otx gene products share structural homology that is confined mainly to the homeodomain. The 60 amino acid residues of the fly otd homeodomain differ from the homeodomains of the human Otx1 and Otx2 protein in only three and two amino acids, respectively. It thus seems likely that most of the conserved functional action of the otd/Otx genes is mediated by the evolutionarily highly conserved homeodomain of the encoded transcription factor [25,27]. Given this highly conserved homeodomain, one might predict that the in vivo functional equivalence of otd/Otx genes demonstrated in the cross-phylum rescue experiments is due to the fact that both otd and Otx genes can control a comparable set of downstream target genes, irrespective of whether the otd/Otx genes are expressed in flies or in mammals [27]. However, currently little is known about the downstream targets of either otd or Otx genes in flies or in mammals, and no information on common targets of otd and Otx genes is available in any species context [27,28].
To address this issue at a genome-wide level we have combined cross-phylum overexpression experiments with expression profiling using oligonucleotide arrays. We sought to identify the common downstream target genes of fly otd and human Otx2 in Drosophila. To this end, we used transgenic flies which carried either the fly otd gene or the human Otx2 gene under the control of a heat-inducible promoter [29,30,31,32,33]. These experiments identified 287 annotated genes that showed highly significant (p ≤ 0.001) changes in expression levels in response to otd overexpression in Drosophila. Among these genes, 93 also showed highly significant differential expression changes in response to Otx2 overexpression. Moreover, the expression levels of 90 of these 93 genes were influenced in the same direction, either upregulated or downregulated, by otd and by Otx2 overexpression. In summary, approximately one third of the candidate otd downstream target genes in Drosophila also respond to overexpression of the human Otx2 gene homolog and nearly all of them display identical patterns of either up- or downregulation under both experimental conditions. From a genome-wide perspective, it is likely that the conserved genetic control of these common otd/Otx2 downstream genes forms the molecular genetic basis for the striking in vivo functional similarity of otd and Otx gene action in Drosophila.
Results
In vivo overexpression and microarray analysis
In this study, transgenic fly strains carrying the otd coding sequence or the human Otx2 coding sequence under the control of the heat-inducible Hsp70 promoter were used [24]. Stage 10-17 embryos were given a 25-minute heat pulse in order to overexpress the otd or Otx2 genes and allowed to recover for 25 minutes (see Materials and methods). Ubiquitous overexpression of otd and Otx2 was verified by whole-mount in situ hybridization with otd- or Otx2-specific antisense RNA probes. These experiments demonstrated that RNA was strongly overexpressed 50 minutes after the onset of heat shock in these strains (data not shown). Wild-type control flies were subjected to the identical heat-shock conditions.
Following ubiquitous overexpression of otd or Otx2, transcript profiles were analyzed using a genome-wide high-density oligonucleotide array and compared to the transcript profiles of heat-shocked wild-type control embryos. The transcripts represented on the oligonucleotide array correspond to probe sets that are complementary to approximately 13,400 annotated Drosophila genes according to Release 1.0 of the Drosophila genome [34]. For each experimental condition, several replicates were carried out (see Materials and methods). The degree of reproducibility within individual replicates is shown in scatter plots for four experimental conditions in Figure 1. A complete description of the microarray content as well as all primary data obtained in each individual microarray experiment are available as Additional data files.
Figure 1.
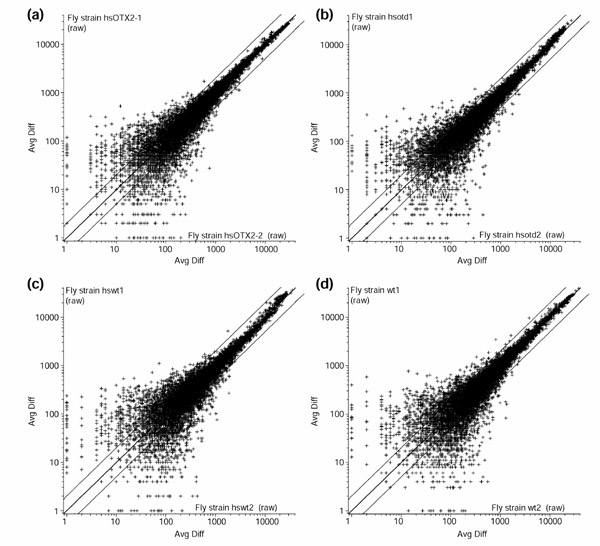
Normalized average difference (Avg Diff) of one pair of replicate arrays for each experimental condition in a log10 scale. (a) heat-shocked Otx2; (b) heat-shocked otd; (c) heat-shocked wild type; (d) wild type. Only probe sets with positive values in both arrays are used. The central line is y = x, and the flanking lines indicate the difference of a factor of two.
Overview of differentially expressed transcripts
An overview of the total number of transcripts that were differentially regulated following otd or Otx2 overexpression is given in Table 1. Two levels of significance for the experimental data are considered in this overview. At a significance level of p ≤ 0.001, a total of 287 genes were found to be differentially regulated following otd overexpression, as compared to heat-shocked wild-type control embryos. This corresponds to 2.1% of the genes represented on the array. At a significance level of p ≤ 0.01, a total of 762 genes were found to be differentially regulated following otd overexpression, as compared to heat-shocked wild-type control embryos. This corresponds to 5.7% of the genes represented on the array. In both cases, approximately a quarter of the differentially regulated transcripts corresponded to known genes, and the rest corresponded to genes that are currently characterized only by sequence information and predicted function (CG-transcripts as described by Celera Genomics [34]).
Table 1.
Numbers of transcripts differentially regulated by overexpression of otd or Otx2
| (a) Differential expression in response to | Total | Named transcripts | CG-transcripts* |
| hsotd | 287 | 63 | 224 |
| hsOtx2 | 682 | 184 | 498 |
| hsotd and hsOtx2 | 93 | 21 | 72 |
| (b) Differential expression in response to | Total | Named transcripts | CG-transcripts* |
| hsotd | 762 | 165 | 597 |
| hsOtx2 | 1395 | 331 | 1064 |
| hsotd and hsOtx2 | 351 | 69 | 282 |
Overview of the numbers of transcripts that were differentially expressed following overexpression of hsp-otd (hsotd) or human hsp-Otx2 (hsOtx2) in Drosophila as a result of heat shock. (a) Number of transcripts that were differentially expressed at a significance level of p ≤ 0.001. (b) Number of transcripts that were differentially expressed at a significance level of p ≤ 0.01. *Genes currently characterized only by sequence information and predicted function (Celera Genomics [34]).
Overexpression of the human Otx2 gene in Drosophila embryos resulted in a larger number of differentially expressed transcripts than did overexpression of the Drosophila otd gene. At a significance level of p ≤ 0.001, a total of 682 genes were found to be differentially expressed following Otx2 overexpression, as compared to heat-shocked wild-type control embryos. This corresponds to 5.1% of the genes represented on the array. At a significance level of p ≤ 0.01, 1,395 genes were found to be differentially expressed following Otx2 overexpression as compared to heat-shocked wild-type control embryos. This corresponds to 10.4% of the genes represented on the array. Again, in both cases, approximately a quarter of the differentially regulated transcripts corresponded to known genes, and the rest were CG-transcripts.
A subset of the transcripts found to be differentially regulated following otd overexpression were also differentially regulated following Otx2 overexpression. Among the transcripts that were differentially expressed at the significance level of p ≤ 0.001, 93 transcripts were found to be differentially regulated following overexpression of either gene. This implies that 32% of the otd-regulated transcripts were also regulated by Otx2. Among the transcripts that were differentially expressed at the significance level of p ≤ 0.01, 351 transcripts were found to be differentially regulated following overexpression of either gene. This implies that 46% of the otd-regulated transcripts were also regulated by Otx2. In the following, only genes that were differentially expressed at the significance level of p ≤ 0.001 are considered further. We propose that these genes are potential direct or indirect downstream targets for the homeodomain transcription factors otd and Otx2.
Functional classification of differentially expressed transcripts
When ubiquitously expressed in the embryo, both otd and Otx2 caused a significant transcriptional response of genes encoding a wide variety of functionally different gene products. A detailed classification of the otd- and Otx2-regulated transcripts into different functional classes was carried out according to Gene Ontology (GO) and is presented in Table 2. (In the GO classification scheme, a given gene can be grouped into more than one functional class [35].) The otd- and Otx2-regulated transcripts fall into 92 GO classes, but only about half of these classes are characterized by more than one regulated transcript.
Table 2.
Classification of transcripts differentially expressed in response to Otx2 and otd overexpression
| Functional class | notd | notd/N (%) | notd/M (%) | nOtx2 | nOtx2/N (%) | nOtx2/M (%) |
| Function unknown (7,108) | 143 | 2.01 | 49.83 | 311 | 4.38 | 45.60 |
| Enzyme (1,872) | 34 | 1.82 | 11.85 | 88 | 4.70 | 12.90 |
| Transcription factor (940) | 23 | 2.45 | 8.01 | 69 | 7.34 | 10.12 |
| Signal transduction (462) | 17 | 3.68 | 5.92 | 24 | 5.19 | 3.52 |
| DNA binding (306) | 14 | 4.58 | 4.88 | 27 | 8.82 | 3.96 |
| Transporter (498) | 12 | 2.41 | 4.18 | 19 | 3.82 | 2.79 |
| Motor (406) | 11 | 2.71 | 3.83 | 22 | 5.42 | 3.23 |
| Protein kinase (365) | 10 | 2.74 | 3.48 | 25 | 6.85 | 3.67 |
| Ligand binding or carrier (581) | 9 | 1.55 | 3.14 | 28 | 4.82 | 4.11 |
| Endopeptidase (413) | 8 | 1.94 | 2.79 | 25 | 6.05 | 3.67 |
| Nucleic acid binding (369) | 8 | 2.17 | 2.79 | 21 | 5.69 | 3.08 |
| Cell adhesion (328) | 8 | 2.44 | 2.79 | 15 | 4.57 | 2.20 |
| Structural protein (335) | 7 | 2.09 | 2.44 | 18 | 5.37 | 2.64 |
| Actin binding (157) | 6 | 3.82 | 2.09 | 10 | 6.37 | 1.47 |
| RNA binding (292) | 4 | 1.37 | 1.39 | 13 | 4.45 | 1.91 |
| Transmembrane receptor (251) | 4 | 1.59 | 1.39 | 9 | 3.59 | 1.32 |
| Chaperone (195) | 3 | 1.54 | 1.05 | 14 | 7.18 | 2.05 |
| Cell cycle regulator (190) | 3 | 1.58 | 1.05 | 12 | 6.32 | 1.76 |
| Ion channel (214) | 3 | 1.40 | 1.05 | 7 | 3.27 | 1.03 |
| Protein phosphatase (91) | 3 | 3.30 | 1.05 | 6 | 6.59 | 0.88 |
| DNA repair protein (65) | 3 | 4.62 | 1.05 | 4 | 6.15 | 0.59 |
| Transcription factor binding (64) | 2 | 3.13 | 0.70 | 11 | 17.19 | 1.61 |
| Cytoskeletal structural protein (121) | 2 | 1.65 | 0.70 | 6 | 4.96 | 0.88 |
| DNA replication factor (42) | 2 | 4.76 | 0.70 | 5 | 11.90 | 0.73 |
| Defense/immunity protein (64) | 2 | 3.13 | 0.70 | 4 | 6.25 | 0.59 |
| G-protein linked receptor (103) | 2 | 1.94 | 0.70 | 3 | 2.91 | 0.44 |
| Receptor (97) | 2 | 2.06 | 0.70 | 2 | 2.06 | 0.29 |
| Cytochrome P450 | 2 | 14.29 | 0.70 | 0 | 0 | 0 |
| Storage protein (25) | 1 | 4.00 | 0.35 | 3 | 12.00 | 0.44 |
| Peptidase (97) | 1 | 1.03 | 0.35 | 3 | 3.09 | 0.44 |
| Lysozyme (8) | 1 | 12.50 | 0.35 | 2 | 25.00 | 0.29 |
| Cyclin-dependent protein kinase (11) | 1 | 9.09 | 0.35 | 2 | 18.18 | 0.29 |
| GABA-B receptor (1) | 1 | 100.00 | 0.35 | 1 | 100.00 | 0.15 |
| Enzyme inhibitor (121) | 1 | 0.83 | 0.35 | 1 | 0.83 | 0.15 |
| Ecdysteroid hormone receptor (2) | 1 | 50.00 | 0.35 | 0 | 0 | 0 |
| 3',5'-cyclic-nucleotide phosphodiesterase (1) | 1 | 100.00 | 0.35 | 0 | 0 | 0 |
| FK506 binding (2) | 1 | 50.00 | 0.35 | 0 | 0 | 0 |
| Peptidylprolyl isomerase (3) | 1 | 33.33 | 0.35 | 0 | 0 | 0 |
| Neurotransmitter transporter (29) | 1 | 3.45 | 0.35 | 0 | 0.00 | 0.00 |
| Steroid hormone receptor (16) | 1 | 6.25 | 0.35 | 0 | 0.00 | 0.00 |
| Acid phosphatase (5) | 1 | 20.00 | 0.35 | 0 | 0.00 | 0.00 |
| Arginine-tRNA ligase (2) | 1 | 50.00 | 0.35 | 0 | 0.00 | 0.00 |
| Carboxypeptidase (1) | 1 | 100.00 | 0.35 | 0 | 0.00 | 0.00 |
| Caspase activator(1) | 1 | 100.00 | 0.35 | 0 | 0.00 | 0.00 |
| Protein tyrosine phosphatase (9) | 0 | 0.00 | 0.00 | 4 | 44.44 | 0.59 |
| Protein serine/threonine kinase (43) | 0 | 0.00 | 0.00 | 4 | 9.30 | 0.59 |
| Chromatin binding (16) | 0 | 0.00 | 0.00 | 4 | 25.00 | 0.59 |
| Ubiquitin conjugating enzyme (12) | 0 | 0.00 | 0.00 | 3 | 25.00 | 0.44 |
| Structural protein of ribosome (136) | 0 | 0.00 | 0.00 | 3 | 2.21 | 0.44 |
| Casein kinase I (6) | 0 | 0.00 | 0.00 | 3 | 50.00 | 0.44 |
| Calcium binding (18) | 0 | 0.00 | 0.00 | 3 | 16.67 | 0.44 |
| Ubiquitin (14) | 0 | 0.00 | 0.00 | 2 | 14.29 | 0.29 |
| Translation factor (70) | 0 | 0.00 | 0.00 | 2 | 2.86 | 0.29 |
| Transcription co-repressor (3) | 0 | 0.00 | 0.00 | 2 | 66.67 | 0.29 |
| GTP binding (14) | 0 | 0.00 | 0.00 | 2 | 14.29 | 0.29 |
| Glutathione transferase (7) | 0 | 0.00 | 0.00 | 2 | 28.57 | 0.29 |
| Furin (2) | 0 | 0.00 | 0.00 | 2 | 100.00 | 0.29 |
| Electron transfer (35) | 0 | 0.00 | 0.00 | 2 | 5.71 | 0.29 |
| Ubiquitinyl hydrolase 1 (2) | 0 | 0.00 | 0.00 | 1 | 50.00 | 0.15 |
| Ubiquitin-specific protease (5) | 0 | 0.00 | 0.00 | 1 | 20.00 | 0.15 |
| Ubiquitin-like conjugating enzyme (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Tubulin-tyrosine ligase (7) | 0 | 0.00 | 0.00 | 1 | 14.29 | 0.15 |
| Transmembrane receptor protein tyrosine phosphatase (4) | 0 | 0.00 | 0.00 | 1 | 25.00 | 0.15 |
| Transmembrane receptor protein tyrosine kinase (7) | 0 | 0.00 | 0.00 | 1 | 14.29 | 0.15 |
| Transcription factor, cytoplasmic sequestering (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Transcription co-activator (2) | 0 | 0.00 | 0.00 | 1 | 50.00 | 0.15 |
| Thioredoxin (4) | 0 | 0.00 | 0.00 | 1 | 25.00 | 0.15 |
| Spermidine synthase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| SNF1A/AMP-activated protein kinase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| SH3/SH2 adaptor protein (2) | 0 | 0.00 | 0.00 | 1 | 50.00 | 0.15 |
| Sarcosine oxidase (2) | 0 | 0.00 | 0.00 | 1 | 50.00 | 0.15 |
| Ribulose-phosphate 3-epimerase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Receptor signaling protein tyrosine phosphatase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Protein tagging (2) | 0 | 0.00 | 0.00 | 1 | 50.00 | 0.15 |
| Prenylated protein tyrosine phosphatase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Phosphoserine phosphatase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Multicatalytic endopeptidase (4) | 0 | 0.00 | 0.00 | 1 | 25.00 | 0.15 |
| mRNA (guanine-N7)-methyltransferase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Mitochondrial processing peptidase(1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| MAP kinase kinase (3) | 0 | 0.00 | 0.00 | 1 | 33.33 | 0.15 |
| Inositol-1,4,5-triphosphate receptor (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Electron transfer flavoprotein (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Effector caspase (3) | 0 | 0.00 | 0.00 | 1 | 33.33 | 0.15 |
| DNA-directed RNA polymerase III (7) | 0 | 0.00 | 0.00 | 1 | 14.29 | 0.15 |
| Cyclin (5) | 0 | 0.00 | 0.00 | 1 | 20.00 | 0.15 |
| CDP-diacylglycerol-serine O-phosphatidyltransferase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Caspase (5) | 0 | 0.00 | 0.00 | 1 | 20.00 | 0.15 |
| cAMP-dependent protein kinase regulator (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| cAMP-dependent protein kinase catalyst (3) | 0 | 0.00 | 0.00 | 1 | 33.33 | 0.15 |
| cAMP-dependent protein kinase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
| Amine oxidase (flavin-containing) (7) | 0 | 0.00 | 0.00 | 1 | 14.29 | 0.15 |
| 3-oxo-5-alpha-steroid 4-dehydrogenase (1) | 0 | 0.00 | 0.00 | 1 | 100.00 | 0.15 |
Genes that were differentially expressed following ubiquitous overexpression of otd or human Otx2, grouped according to Gene Ontology (GO) functional classes. n, Number of transcripts detected that belong to an individual class. N, Number of transcripts represented on the chip for each functional class; the value of N for each functional class is given in parentheses following the class name. n/N × 100, Percentage of transcripts that were differentially regulated for each functional class relative to the total number of transcripts in that class represented on the chip. M, Total number of differentially expressed transcripts (of all classes) following overexpression of otd or human Otx2 (p ≤ 0.001); for otd and Otx2, M is 287 and 682 respectively. n/M × 100, Percentage of transcripts that were differentially regulated in each functional class relative to the the total number of differentially regulated transcripts for otd and Otx2.
In terms of known function, the two classes with the highest absolute and relative numbers of regulated transcripts were 'enzymes' and 'transcription factors'; this was the case for both otd-regulated and Otx2-regulated transcripts. Other functional classes with high numbers of differentially regulated genes were 'signal transduction', 'DNA binding', 'transporter', 'protein kinase', 'motor', 'ligand binding or carrier', and 'endopeptidase'; again this was the case for both otd- and Otx2-regulated transcripts. Indeed, in most cases in which a functional class was characterized by both otd- and Otx2-regulated transcripts, the relative number (n/M; see Table 2) of otd-regulated transcripts was similar to that of Otx2-regulated transcripts. For example, 2.79% of the otd-regulated transcripts versus 2.20% of the Otx2-regulated transcripts were classified under 'cell adhesion', and 3.48% of the otd-regulated transcripts versus 3.67% of the Otx2-regulated transcripts were classified under 'signal transduction'. Approximately half of both the otd-regulated and the Otx2-regulated transcripts belong to the class 'function unknown'.
Quantitative profiling of differentially expressed transcripts
Figure 2 shows the otd-regulated transcripts that correspond to known Drosophila transcripts and presents a quantitative representation of the change in expression levels for these transcripts. For clarity, these transcripts are only grouped into mother classes and not into the detailed GO classes. Most of the 63 known transcripts that were differentially expressed following otd overexpression showed increased expression levels; less than 20% of these transcripts were downregulated. The gene with the highest increase in expression level (78-fold) was otd itself, in accordance with our experimental overexpression protocol. Increases in expression levels above 10-fold were also observed for forkhead domain 96cb (fd96Cb), which encodes a nucleic-acid-binding protein, for patched (ptc), which encodes a protein involved in signal transduction, for picot, which encodes a transporter, and for cortactin and Regulator of cyclin A1 (Rca1), which encode gene products of currently unknown molecular function. Only two transcripts showed increases in the 5-10-fold range, namely sugar transporter1 (sut1) encoding a protein involved in sugar transportation, and scraps (scra) encoding an actin-binding protein. The majority of the upregulated transcripts had increases in the 2-5-fold range. The transcript with the most marked decrease in expression was eyegone (eyg), encoding a transcription factor known to be involved in eye development.
Figure 2.
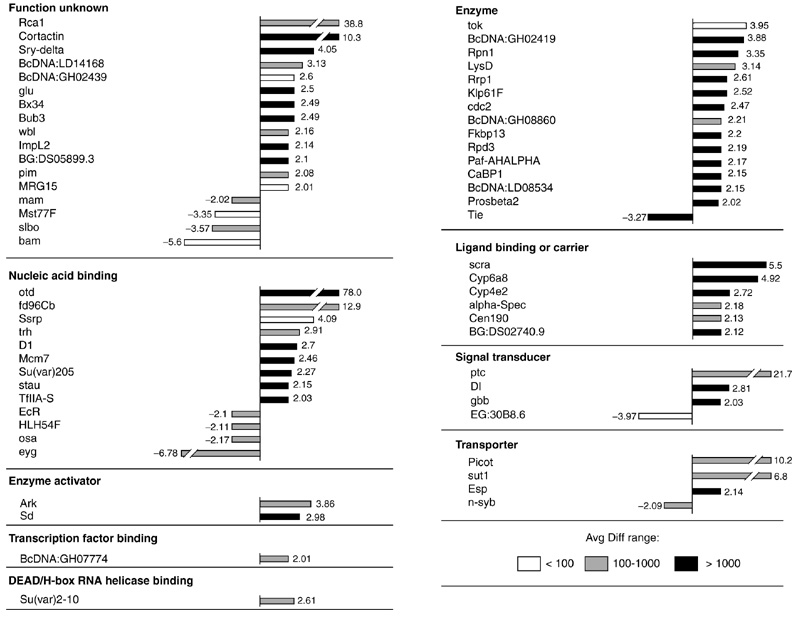
Known transcripts differentially expressed in response to overexpression of otd, grouped according to functional classes. Bars represent the fold change between differentially expressed transcripts in heat-shocked wild-type embryos and heat-shocked otd embryos. Positive values indicate that the relative expression level of a gene is increased (upregulated) following otd overexpression and negative values indicate a decrease (downregulated). Absolute average difference (Avg Diff) values are given for the otd overexpression condition as follows: white bars, Avg Diff < 100; gray bars, Avg Diff from 100-1,000; black bars, Avg Diff > 1,000.
Figure 3 shows the Otx2-regulated transcripts that correspond to known Drosophila genes and presents a quantitative representation of their expression level changes. Again, these transcripts are grouped into mother classes and not into detailed GO classes. As was the case for otd overexpression, most of the known transcripts that were differentially expressed following Otx2 overexpression showed increased expression levels. For example, in the functional class of 'enzyme', 45 out of 49 transcripts were upregulated. In total, less than 13% of the 184 Otx2-regulated known transcripts were downregulated. Increases in expression levels above 10-fold were observed for 23 genes and for 6 of these genes, retained (retn), SMC2, licorne (lic), Rtc1, Hairless (H) and deadhead (dhd), the increases were greater than 50-fold. Twenty-two transcripts showed increases in the 5-10-fold range, and, similarly to the otd overexpression situation, increases of 2-5-fold dominated in most of the functional classes. The transcript with the most marked decrease in expression was once again eyg.
Figure 3.
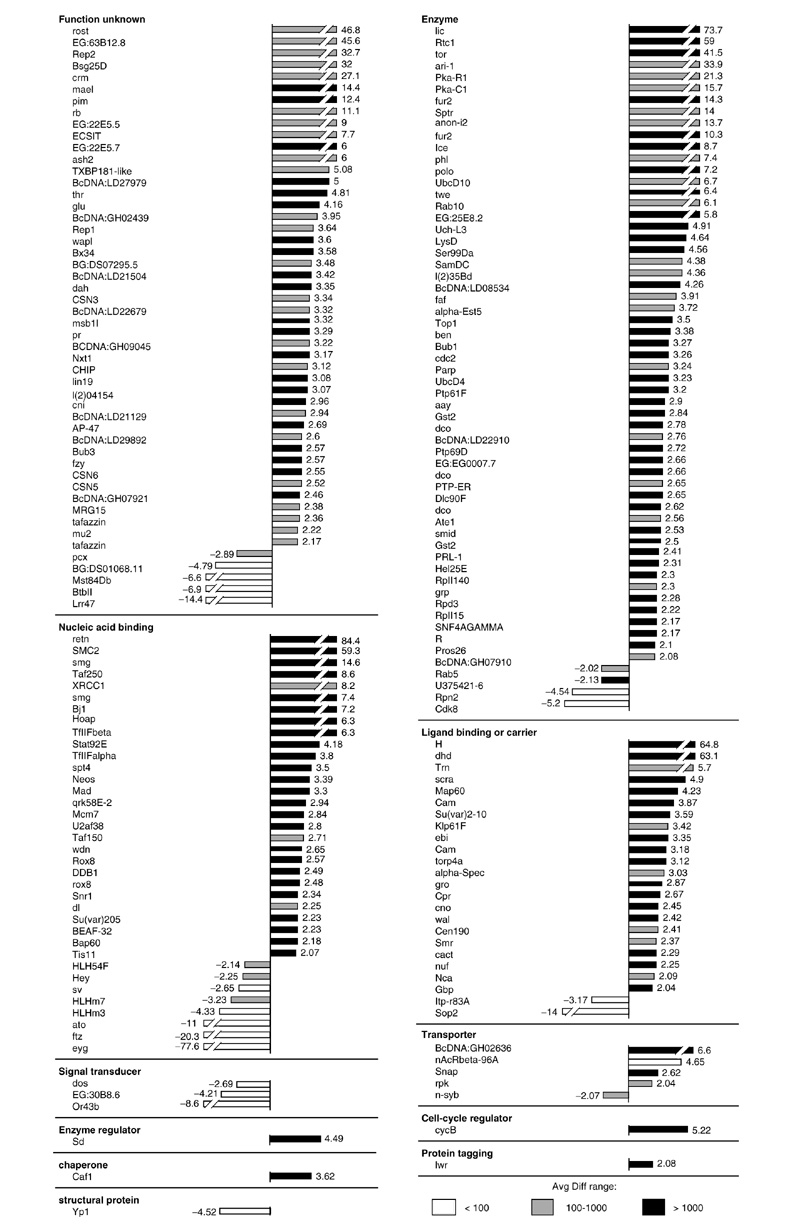
Known transcripts differentially expressed in response to overexpression of Otx2, grouped according to functional classes. Bars represent the fold change between differentially expressed transcripts in heat-shocked wild-type embryos and heat-shocked Otx2 embryos. Positive values indicate that the relative expression level of a gene is increased (upregulated) following Otx2 overexpression and negative values indicate a decrease (downregulated). Avg Diff values are given for the Otx2 overexpression condition as follows: white bars, Avg Diff < 100; gray bars, Avg Diff from 100-1,000; black bars, Avg Diff > 1,000.
Common candidate downstream genes of otd and Otx2
Ninety-three transcripts were differentially expressed in response to both otd overexpression and Otx2 overexpression. This indicates that approximately one third of the otd-regulated genes in Drosophila also respond to overexpression of the human Otx2 gene homolog. Figure 4 shows the expression levels for these transcripts, which are likely to represent the common downstream target genes for otd and Otx2. Twenty-one of these transcripts correspond to known Drosophila genes and 72 correspond to annotated CG-transcripts. The expression levels of all of the known transcripts were influenced in the same manner by overexpression of otd and Otx2, in that a given downstream target gene was either upregulated in both cases or downregulated in both cases. Moreover, for most of these transcripts the absolute expression levels were similar in response to otd and to Otx2. Two marked exceptions were pimple (pim), which was upregulated 12.4-fold following Otx2 overexpression and 2.1-fold following otd overexpression, and eyg, which was downregulated 77.6-fold following Otx2 overexpression (but see PCR data below) and downregulated 6.8-fold following otd overexpression. Similarly, the expression levels of 68 of the CG transcripts were influenced in the same manner by overexpression of otd and Otx2. Only in the three remaining cases were transcripts upregulated by overexpression of one of the otd/Otx transgenes and downregulated by overexpression of the other. Thus, approximately one third of the candidate otd downstream target genes in Drosophila are controlled in a comparable manner by the human Otx2 gene homolog.
Figure 4.
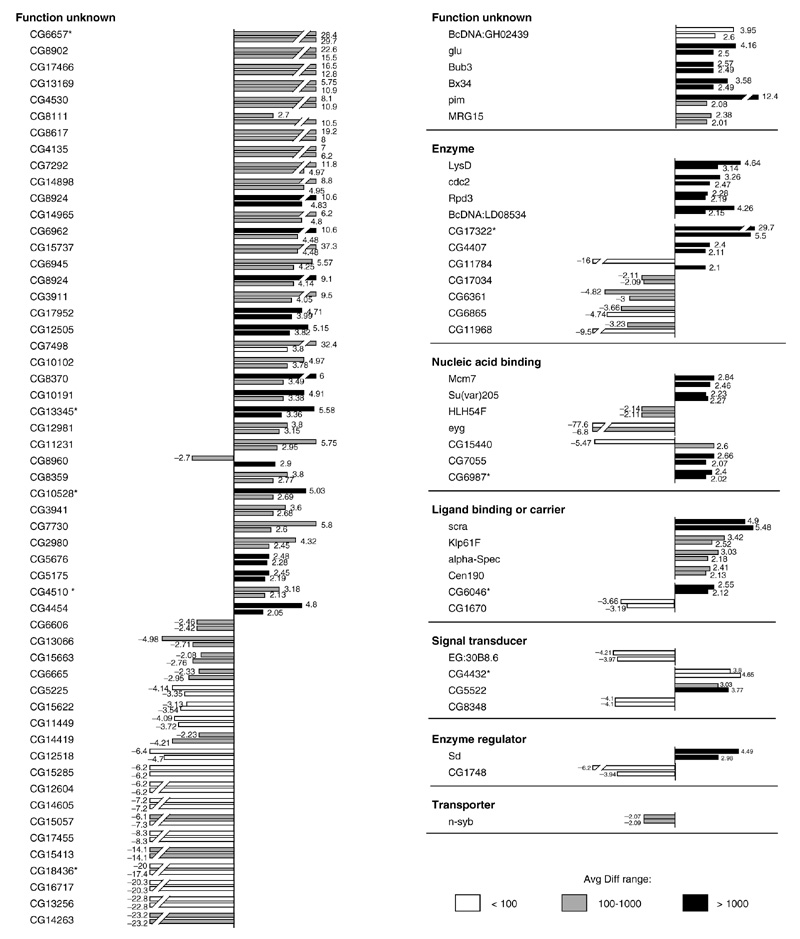
Transcripts differentially expressed in response to overexpression of otd and in response to overexpression of human Otx2, grouped according to functional classes. Bars represent the fold change between differentially expressed transcripts in heat-shocked wild type embryos and heat-shocked otd or heat-shocked Otx2 embryos. The upper bars represent the fold change of differentially expressed transcripts following overexpression of Otx2 and the lower bars represent the fold change of differentially expressed transcripts following overexpression of otd. Positive values indicate that the relative expression level of a gene is increased (upregulated) following otd overexpression and negative values indicate a decrease (downregulated). Avg Diff values are given for the otd overexpression condition as follows: white bars, Avg Diff < 100; gray bars, Avg Diff from 100-1,000; black bars, Avg Diff > 1,000.
There are a number of interesting genes among these common candidate genes. The four known transcripts in class 'ligand binding or carrier', scra, Kinesin-like protein at 61F (Klp61F), alpha-Spectrin (alpha-spec) and Centrosomal protein 190 kD (Cen190), are all involved in actin or microtubule binding or movement [36,37,38,39]. This finding is intriguing as one of the Otx2 downstream genes identified in the mouse is a tropomyosin gene, which also encodes an actin-binding protein [40]. Among the four known transcripts in the class 'nucleic acid binding' are the genes Minichromosome maintenance 7 (Mcm7) and Suppressor of variegation 205 (Su(var)205) [41,42] which encode chromatin-binding proteins, and the genes eyg and HLH54F, which encode transcription factors [43,44]. The four known transcripts in the functional class 'enzymes' are Lysozyme D (LysD), cdc2, Rpd3, and BcDNA:LD08534 [45,46,47,48]. Although the cdc2 gene product is classified as 'enzyme', it also acts at the G2/M transition of the mitotic cell cycle [47]. Moreover, Rpd3 encodes a histone deacetylase which is involved in chromatin structure [46]. In the class 'transporter' the n-synaptobrevin (n-syb) gene, encoding a SNAP receptor, is involved in synaptic-vesicle docking and fusion and is expressed in the embryonic CNS [49]. In the class 'signal transducer', the gene EG:30B8.6 encodes a putative GABA-B receptor [50]. Finally, the gene Segregation distorter (Sd), classified as 'enzyme regulator', encodes a Ran GTPase activator [51]. Among the transcripts of known genes are several genes whose precise functional role is not well defined at the molecular level. These are the Bx34 and MRG15 genes [52,53] which encode components of the nucleus and the gluon, Bub3 and pim genes which are all involved in mitosis. gluon encodes a putative component of the condensin complex, and gluon mutants show peripheral nervous system defects during embryogenesis [54]. The gene product of Bub3 is localized to the kinetochore and may function in the mitotic checkpoint [55]. pim is expressed in the embryonic CNS and encodes a protein implicated in mitotic sister-chromatid separation [56].
Verification of microarray expression data with RT-PCR
To confirm the differences in gene expression levels after heat-shock induced overexpression of otd and human Otx2 as compared to heat-shocked wild-type embryos, quantitative reverse transcription polymerase chain reaction (RT-PCR) was carried out on selected candidate target genes. Changes in expression levels were determined for eight genes that were differentially regulated by otd or human Otx2, namely scra, LysD, glu, Rpd3, pim, n-syb, eyg and otd. The genes wunen (wun) and Scc1, whose expression levels remained unchanged in response to otd or Otx2 overexpression, were used as controls. As indicated in Table 3, these experiments showed that the changes in relative expression level, as measured by RT-PCR, are generally consistent with the data obtained with the oligonucleotide arrays. An exception is the data on the response of the eyg gene to Otx2 overexpression; RT-PCR data indicate a weak downregulation (-1.62) whereas oligonucleotide array data indicate a strong downregulation (-77.6).
Table 3.
Comparison of change folds between oligonucleotide arrays and RT-PCR
| Change fold | |||||||
| Avg Diff | hsotd | hsOtx2 | |||||
| Transcript | hswt | hsotd | hsOtx2 | Array | RT-PCR | Array | RT-PCR |
| scra | 251 | 1375 | 1229 | 5.5 | 1.3 | 4.9 | 1.6 |
| LysD | 525 | 1646 | 2436 | 3.1 | 1.6 | 4.6 | 4.0 |
| glu | 479 | 1196 | 1991 | 2.5 | 1.8 | 4.2 | 10.9 |
| Rpd3 | 1170 | 2562 | 2673 | 2.2 | 2.0 | 2.3 | 2.5 |
| pim | 118 | 246 | 1467 | 2.1 | 1.4 | 12.4 | 8.0 |
| n-syb | 612 | 293 | 296 | -2.1 | -1.5 | -2.1 | -1.5 |
| eyg | 1552 | 229 | 10 | -6.7 | -1.4 | -77.6 | -1.6 |
| wun | 885 | / | 884 | / | / | 1.0 | 1.0 |
| Scc1 | 724 | 723 | / | 1.0 | 1.0 | / | / |
| otd | 84 | 6555 | 108 | 78.0 | 119.4 | 1.3 | 1.5 |
RT-PCR was carried out on cDNA derived from heat-shocked wild type (hswt), heat-shocked otd (hsotd) or heat-shocked Otx2 (hsOtx2) embryos. Change folds determined by RT-PCR are represented as the mean value of eight independent replicates, derived from two different cDNA preparations. wun is used as a control for the comparison of the RT-PCR data between heat-shocked wild type and heat-shocked Otx2. Scc1 is used as a control for the comparison of the RT-PCR data between heat-shocked wild type and heat-shocked otd.
Discussion
Common downstream target genes for otd and Otx
Cross-phylum gene replacement experiments have shown that the fly otd gene and the homologous human Otx genes are functionally equivalent in vivo, in that overexpression of either gene in Drosophila otd null mutants can lead to the restoration of defects in cephalic and brain development [23,24,25,26]. We have used a combination of transgenic overexpression genetics and functional genomics to gain insight into the equivalence of otd and Otx gene expression in Drosophila at a comprehensive, genome-wide level. Using inducible overexpression and quantitative transcript imaging through oligonucleotide arrays representing the total number of 13,400 currently annotated Drosophila genes, we have identified hundreds of candidate downstream genes for both the fly otd gene and the human Otx2 gene. A comparison of these candidate downstream genes reveals that both otd and Otx genes appear to control an overlapping set of genes; we refer to these genes as common downstream genes. The number of identified common downstream genes for otd and Otx2 depends on the statistical level of significance used to determine whether a given gene showed differential expression in response to transgene overexpression. If the analysis is restricted to highly significant (p ≤ 0.001) datasets, we find 93 common downstream genes, equivalent to 32% of the candidate otd downstream genes or approximately 1% of transcripts in the annotated fly genome. If, in contrast, the analysis is based on significant (p ≤ 0.01) datasets, we find 351 common downstream genes, equivalent to 46% of the candidate otd downstream genes or approximately 3% of transcripts in the annotated fly genome. In either case, a substantial, but far from complete, set of the otd regulated genes are common downstream targets of both fly and human transgenes.
It is interesting that, at the genome-wide transcript level, the Otx2 gene does not appear to be able to replace otd action in full; over half of the transcripts that are influenced by otd overexpression are not influenced by Otx2 overexpression. Given the pronounced differences in amino acid sequence between the OTD and OTX2 proteins, this may not be altogether surprising. The OTD and OTX2 proteins consist of 548 and 289 amino acids, respectively. Shared homology between them is restricted to the homeodomain and to a short domain immediately upstream of the homeodomain as well as a tripeptide at the amino terminus [25]. Moreover, as Otx genes cannot completely replace the otd gene in cross-phylum rescue experiments in vivo, a complete correspondence of otd downstream genes and common otd/Otx downstream genes might not be expected [3,24,25]. However, approximately one third of the otd-regulated genes do also respond to Otx2 overexpression. We suggest that these common downstream genes are likely to explain the overlapping roles of the otd/Otx genes in cross-phylum rescue experiments in vivo. These target genes reflect the evolutionarily conserved roles of the members of the otd/Otx gene family in Drosophila. To investigate this further, it will now be important and interesting to carry out similar functional genomic analyses of otd and Otx gene action in a mammalian system such as the mouse [27].
otd overexpression: a genomic perspective on candidate downstream genes
The experiments reported here identify approximately 300 genes that showed highly significant (p ≤ 0.001) changes in expression levels in response to otd overexpression in Drosophila. The genomic perspective of these identified otd downstream target genes reveals several features of otd action at a higher level of insight. First, this finding indicates that the otd gene product, a homeodomain transcription factor, regulates a limited and distinct set of candidate downstream genes. At a significance level of p ≤ 0.001, 287 genes were found to be differentially regulated, corresponding to approximately 2.1% of the transcripts in the annotated fly genome. At a significance level of p ≤ 0.01, 762 genes were found to be differentially regulated, corresponding to approximately 5.7% of the transcripts in the annotated fly genome. This is further evidence for the notion that homeoproteins in Drosophila control only a subset and not the majority of the genes in the genome [30]. Indeed, in similar experiments in which the homeobox gene labial (lab) was overexpressed using the same heat-shock protocol as described here, 6.4% of the genes represented on the array used were shown to be differentially regulated at a significance level of p ≤ 0.01 [30]. (It should however, be noted that the array used in these lab overexpression experiments represents only 10% of the genes in the fly genome.) Thus the relative number of putative otd targets appears to be in the same range as the number of putative lab targets.
Second, these experiments show that the OTD homeodomain transcription factor acts on numerous candidate target genes that also encode transcription factors, consistent with the idea that homeodomain proteins act through a cascade of transcription factors which regulate the expression of their own subset of downstream genes [57]. Currently, we do not know which of the downstream target genes are direct OTD targets and are, thus regulated directly by OTD protein binding to DNA regulatory sequences, and which are indirect targets. At present, little is known about temporal response of putative target genes following pulsed expression of a transcription factor. Some studies have been carried out on the basis of the assumption that direct targets respond immediately whereas indirect targets respond with a delay due to the time required for intermediary gene expression. Nasiadka and Krause used a kinetic approach to identify direct and indirect targets of the ectopically expressed homeodomain transcription factor fushi tarazu (ftz) [58]. Their results show that target genes respond to pulses of ftz expression within two distinct temporal windows. Direct responses (no intermediary gene transcription is required) are 50% complete within about 18 minutes after heat shock. Indirect responses do not reach the same level of response until 26 minutes after heat shock. Assuming that otd expression follows a similar kinetic profile to ftz, it is likely that we have identified primary targets as well as genes whose response was caused by indirect effects requiring intermediate transcription.
Third, these results show that the primary consequence of otd overexpression in Drosophila is the upregulation of its downstream target genes. Indeed more than 80% of the genes that were differentially expressed following otd overexpression showed increased expression levels. This contrasts with the action of the homeotic gene lab; overexpression of lab under comparable conditions resulted in an approximately equal number of upregulated and downregulated target genes [30].
The majority of potential downstream target genes of otd are annotated CG-transcripts and, hence, correspond to predicted genes which have not yet been studied in detail in an in vivo context. This is surprising given the fact that numerous classical genetic screens for genes involved in cephalic and CNS embryogenesis have been carried out [59]. This may indicate that many of the genes involved in those aspects of cephalic and CNS embryogenesis that are under the control of otd in Drosophila have not yet been identified. Alternatively, this finding may reflect specific constraints of the overexpression experiment. For example, the overexpression protocol used makes it difficult to control otd protein concentration and stability. As different levels of a homeoprotein may have different developmental consequences, the relatively high level of OTD protein attained may influence target genes that are not affected by the endogenously attained protein level [60,61]. Moreover, the fact that otd overexpression is not accompanied by simultaneous overexpression of cofactors, which can act together with homeodomain transcription factors to determine their in vivo target specificity, may also lead to nonspecific activation of target genes [62].
Functional genomics of a human transgene overexpressed in Drosophila
In several cases, human transgenes have been overexpressed in Drosophila in order to gain insight into the evolutionary conservation of developmental control gene action [24,25,63,64,65,66]. This has also been the primary goal of the overexpression of human Otx2 in Drosophila reported here. In addition to the identification of common otd/Otx downstream genes, the genomic level of analysis reported here has uncovered remarkable similarities in the activity of the human transgene in the fly as compared to that of its fly homolog. Thus, otd and Otx2 both upregulate most of their target genes upon overexpression. Moreover, the target genes of both transcription factors fall into the same functional categories. For example, the classes 'enzymes' and 'transcription factors' had the highest absolute and relative number of transcripts.
The striking difference in the action of the two transgenes is that overexpression of human Otx2 causes expression changes in many more downstream genes than does overexpression of the fly otd gene. The experiments reported here identify approximately 700 genes that showed highly significant (p ≤ 0.001) changes in expression levels in response to Otx2; this is more than double the number observed in response to otd. It is unlikely that this difference is due to corresponding differences in the expression levels attained for Otx2 versus otd transcripts. Indeed, the transcript abundance of otd was higher than that of Otx2 in these experiments (see Materials and methods). Nevertheless, these data should be interpreted with caution, as several explanations, not mutually exclusive, are possible for the observation that more genes respond to overexpression of Otx2. First, only one single-transgenic strain of otd and only one single-transgenic strain of Otx2 were used. Thus, strain differences or insertion effects might account for the fact that more genes show differential expression following overexpression of Otx2 compared to overexpression of otd. Second, it is conceivable that overexpression of Otx2 affects more downstream genes in Drosophila than otd because the OTX2 transcription factor binds to many more DNA regulatory regions than does OTD. The smaller OTX2 protein might, therefore, have a lower specificity for target gene regulatory regions. Similarly, the OTX2 protein might be more promiscuous than OTD in its interactions with the numerous cofactors that determine target specificity. Third, it has been shown that the DNA-binding specificity of homeoproteins is low in vitro. But given that the homeodomain is conserved and Otx2 rescues the otd phenotype, this suggests that they should recruit a similar subset of cofactors and regulate a common subset of downstream genes, at least in those tissues where otd is endogenously expressed. Furthermore, the Otx2 product, which is not a fly protein, could influence the expression of a small number of transcription factors that are not affected by OTD and which then regulate the expression of their own subset of downstream genes. Whatever the molecular basis for this unexpected difference in the result of Otx2 and otd overexpression may be, its discovery is a further demonstration of the new level of insight that can be attained from a genome-wide functional perspective.
Materials and methods
Embryos
The wild type was Drosophila melanogaster Oregon-R. For overexpression of otd, we used the hsp-otd line 5A generated by Royet and Finkelstein [67]. For overexpression of human Otx2, we used the hsp-Otx2 line generated by Leuzinger et al. [24]. All fly stocks were kept on standard cornmeal/yeast/agar medium at 25°C. Embryos were collected overnight for 12 h on grape juice plates, kept for a further 4 h at 25°C and then subjected to a 37°C heat shock for 25 min, followed by a recovery period of 25 min at 25°C before RNA isolation. Therefore, at the time of RNA isolation these embryos were at embryonic stages 10-17 [29]. Embryos younger than embryonic stage 10 were not used, as heat shock in these earlier stages results in lethality [68]. Embryos used for in situ hybridization studies were collected and heat-shock treated in the same way.
Whole-mount in situ hybridization
For in situ hybridization, digoxigenin-labeled sense and antisense otd/Otx2 RNA probes were generated in vitro, with a DIG labeling kit (Roche Diagnostics) and hybridized to whole-mount embryos following standard procedure [69]. Hybridized transcripts were detected with an alkaline-phosphatase conjugated anti-digoxigenin Fab fragment (RocheDiagnostics) using Nitro blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Sigma) as chromogenic substrates.
High-density oligonucleotide arrays and hybridization
In this study, a custom-designed Drosophila oligonucleotide array (roDROMEGAa, Affymetrix, Santa Clara, CA) was used. It contains 14,090 sequences representing Drosophila-specific transcripts, prokaryotic control sequences and custom-chosen sequences for transgenes such as gal4, gfp, and lacZ. Of the sequences included, 13,998 correspond to Drosophila-specific transcripts annotated by Celera Genome Release 1 [34] and deposited in SWISS-PROT/TrEMBL databases. These 13,998 sequences represent approximately 13,400 genes in the Drosophila genome and therefore some genes are represented by more than one probe set. Each sequence is represented on the array by a set of 14 oligonucleotide probes (25 mers) matching the sequence. To control the specificity of hybridization, the same probes are represented on the array with a single nucleotide mismatch in a central position. As such, each sequence is represented by 14 perfect match and 14 mismatch probes. The average difference (Avg Diff) between the perfect-match hybridization signal and the mismatch signal is proportional to the abundance of a given transcript [32]. RNA was isolated, labeled, and hybridized to the arrays as described [29,30] with minor modifications.
Data analysis
Probe arrays were scanned with a commercial confocal laser scanner (Hewlett-Packard). Pixel intensities were measured, and expression signals were analyzed with commercial software (GENECHIP 3.1, Affymetrix). Data processing was carried out using RACE-A (F. Hoffmann-La Roche), Access 97 and Excel 97 (Microsoft) software. Scatter plots were prepared using GeneSpring™ software version 4.1 (Silicon Genetics, Redwood City, CA). For quantification of relative transcript abundance, Avg Diff value was used [32]. Four replicates were carried out for hsp-otd and hsp-Otx2. Three and five replicates were done for heat-shocked wild type and wild type respectively. All arrays were normalized against the mean of the total sums of Avg Diff values across all 16 arrays. In order to avoid huge fold changes, genes with a normalized Avg Diff below 20 were automatically assigned an Avg Diff of 20 (RACE-A protocol). An unpaired t-test for each individual gene was carried out for the following pairwise comparisons: heat-shocked wild type versus wild type, heat-shocked wild type versus heat-shocked otd, and heat-shocked wild type versus heat-shocked Otx2. For differential transcript imaging, only transcripts that had highly significant or significant changes in Avg Diff (p ≤ 0.001 and p ≤ 0.01, respectively) and whose changes were in the two-fold and above range are presented. Additionally, the higher mean Avg Diff of a pairwise comparison for a given transcript had to be above or equal to 50. To obtain a comprehensive analysis of the number and identity of genes differentially regulated by otd/Otx2, candidates that were already differentially expressed in heat-shocked wild-type embryos compared to non-heat-shocked wild-type controls were excluded from further analysis (data not shown [30]). For a comprehensive list of all genes with their fold changes and significance levels, see Additional data.
RT-PCR
Poly(A)+ RNA (300 ng) was isolated from embryos of wild type, heat-shocked wild type, heat-shocked otd and heat-shocked Otx2 (mRNA isolation kit; Roche Diagnostics) and reverse transcribed with AMV-RT and random hexamers (RT-PCR kit; Roche Diagnostics). PCR was performed with 100 pg template DNA and gene-specific primers (Seq Web, Winsconsin Package Version 10.0, GCG) on a light cycler (LightCycler, Roche Diagnostics). Continuous fluorescence observation of amplifying DNA was possible using SYBR Green I (Roche Diagnostics) After cycling, a melting curve was produced by slow denaturation of the PCR end products, to validate the specificity of amplification. To compare the relative amounts of PCR products we monitored the amplification profile on a graph, displaying the log of the fluorescence against the number of cycles. Relative fold changes for a given gene under both conditions (heat-shocked otd versus heat-shocked wild type or heat-shocked Otx2 versus heat-shocked wild type) were calculated using the fit point method (Light Cycler Manufacturer, Roche).
Quantification of otd and human Otx2 transcripts by RT-PCR
Plasmids containing fly otd or human Otx2 cDNA were linearized with appropriate restriction enzymes and purified. The concentrations of the linearized plasmids were spectrophotometrically quantified using a GeneQuant RNA/DNA calculator (Pharmacia Biotech) and serial dilutions were made. To quantify the concentration of the otd and Otx2 transcripts from heat-shocked otd and heat-shocked Otx2 embryos, a standard curve was established using the serial dilution of the corresponding linearized plasmid on a light cycler (Roche). RT-PCR was carried out when the standard curve was established. Thereafter, the steady-state concentrations of the otd and human Otx2 were calculated in relation to their standard curves, using the second derivative maximum method (Roche). This showed that the concentrations of otd and Otx2 transcripts were 1.5 × 10-6 μg/μl and 3.6 × 10-7 μg/μl, respectively.
Additional data
The following additional data files are available: a list of the genes on the microarray; Primary data (Avg Diff values, both raw and normalized) for each microarray experiment: heat-shocked otd embryos (replicates 1, 2, 3, 4); heat-shocked Otx2 embryos (replicates 1, 2, 3, 4); heat-shocked wild-type embryos (replicates 1, 2, 3); wild-type embryos (replicates 1, 2, 3, 4, 5); normalization factors (as an Excel file) for each replicate; comparisons between pairs of experiments, including the fold change for each gene and the results of a t test: heat-shocked wild-type embryos compared with heat-shocked otd embryos (as an Excel file); heat-shocked wild-type embryos compared with heat-shocked Otx2 embryos (as an Excel file); heat-shocked wild-type embryos compared with wild-type embryos (as an Excel file).
These data have been submitted to the Gene Expression Omnibus at the National Center for Biotechnology Information [70], accession numbers GSM1351-GSM1366 (platform accession GPL70, series accession GSE32).
Supplementary Material
A list of the genes on the microarray
Primary data (Avg Diff values, both raw and normalized) for heat-shocked otd embryos - replicate 1
Primary data (Avg Diff values, both raw and normalized) for heat-shocked otd embryos - replicate 2
Primary data (Avg Diff values, both raw and normalized) for heat-shocked otd embryos - replicate 3
Primary data (Avg Diff values, both raw and normalized) for heat-shocked otd embryos - replicate 4
Primary data (Avg Diff values, both raw and normalized) for heat-shocked Otx2 embryos - replicate 1
Primary data (Avg Diff values, both raw and normalized) for heat-shocked Otx2 embryos - replicate 2
Primary data (Avg Diff values, both raw and normalized) for heat-shocked Otx2 embryos - replicate 3
Primary data (Avg Diff values, both raw and normalized) for heat-shocked Otx2 embryos - replicate 4
Primary data (Avg Diff values, both raw and normalized) for heat-shocked wild-type embryos - replicate 1
Primary data (Avg Diff values, both raw and normalized) for heat-shocked wild-type embryos - replicate 2
Primary data (Avg Diff values, both raw and normalized) for heat-shocked wild-type embryos - replicate 3
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 1
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 2
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 3
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 4
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 5
Normalization factors for each replicate
Heat-shocked wild-type embryos compared with heat-shocked otd embryos
Heat-shocked wild-type embryos compared with heat-shocked Otx2 embryos
Heat-shocked wild-type embryos compared with wild-type embryos
Acknowledgments
Acknowledgements
We thank Jan Mous, Adrian Roth, Michel Tessier, Monika Seiler, and Reto Brem for essential contributions and helpful advice. We are particularly grateful to Clemens Broger and Martin Neeb (F. Hoffman-La Roche) for allowing us to use their RACE-A CHIP analysis software and to Volker Schmid and Natalie Yanze for help with the light cycler. We especially thank Reinhold Koch for contributions to standardizing the description of the statistics. We thank all members of our laboratory, especially Boris Egger, Frank Hirth and Martin Mueller for constant and encouraging discussions, and Lars Kammermeier for help with the photography. This research was supported by grants from the SNSF and by F. Hoffmann-La Roche.
References
- Finkelstein R, Boncinelli E. From fly head to mammalian forebrain: the story of otd and Otx. Trends Genet. 1994;10:310–315. doi: 10.1016/0168-9525(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Thor S. The genetics of brain development: conserved programs in flies and mice. Neuron. 1995;15:975–977. doi: 10.1016/0896-6273(95)90084-5. [DOI] [PubMed] [Google Scholar]
- Sharman AC, Brand M. Evolution and homology of the nervous system: cross-phylum rescues of otd/Otx genes. Trends Genet. 1998;14:211–214. doi: 10.1016/s0168-9525(98)01488-7. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Holland ND. Chordate origins of the vertebrate central nervous system. Curr Opin Neurobiol. 1999;9:596–602. doi: 10.1016/S0959-4388(99)00003-3. [DOI] [PubMed] [Google Scholar]
- Galliot B, Miller D. Origin of anterior patterning. How old is our head? Trends Genet. 2000;16:1–5. doi: 10.1016/s0168-9525(99)01888-0. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Smouse D, Capaci TM, Spradling AC, Perrimon N. The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev. 1990;4:1516–1527. doi: 10.1101/gad.4.9.1516. [DOI] [PubMed] [Google Scholar]
- Simeone A, Gulisano M, Acampora D, Stornaiuolo A, Rambaldi M, Boncinelli E. Two vertebrate homeobox genes related to the Drosophila empty spiracles gene are expressed in the embryonic cerebral cortex. EMBO J. 1992;11:2541–2550. doi: 10.1002/j.1460-2075.1992.tb05319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D'Apice MR, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acampora D, Simeone A. The TINS Lecture. Understanding the roles of Otx1 and Otx2 in the control of brain morphogenesis. Trends Neurosci. 1999;22:116–122. doi: 10.1016/s0166-2236(98)01387-3. [DOI] [PubMed] [Google Scholar]
- Gao Q, Wang Y, Finkelstein R. Orthodenticle regulation during embryonic head development in Drosophila. Mech Dev. 1996;56:3–15. doi: 10.1016/0925-4773(96)00504-7. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Jurgens G. Mediation of Drosophila head development by gap-like segmentation genes. Nature. 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Cadigan KM, Gehring WJ. Three maternal coordinate systems cooperate in the patterning of the Drosophila head. Development. 1994;120:3155–3171. doi: 10.1242/dev.120.11.3155. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott U, Gonzalez-Gaitan M, Jackle H, Technau GM. Number, identity, and sequence of the Drosophila head segments as revealed by neural elements and their deletion patterns in mutants. Proc Natl Acad Sci USA. 1994;91:8363–8367. doi: 10.1073/pnas.91.18.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E, Perrimon N, Finkelstein R. orthodenticle activity is required for the development of medial structures in the larval and adult epidermis of Drosophila. Development. 1992;115:801–811. doi: 10.1242/dev.115.3.801. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Perrimon N. The orthodenticle gene is regulated by bicoid and torso and specifies Drosophila head development. Nature. 1990;346:485–488. doi: 10.1038/346485a0. [DOI] [PubMed] [Google Scholar]
- Klambt C, Jacobs JR, Goodman CS. The midline of the Drosophila central nervous system: a model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nested expression domains of four homeobox genes in developing rostral brain. Nature. 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- Suda Y, Matsuo I, Kuratani S, Aizawa S. Otx1 function overlaps with Otx2 in development of mouse forebrain and midbrain. Genes Cells. 1996;1:1031–1044. doi: 10.1046/j.1365-2443.1996.900288.x. [DOI] [PubMed] [Google Scholar]
- Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122:243–252. doi: 10.1242/dev.122.1.243. [DOI] [PubMed] [Google Scholar]
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Forebrain and midbrain regions are deleted in Otx2-/- mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- Acampora D, Mazan S, Avantaggiato V, Barone P, Tuorto F, Lallemand Y, Brulet P, Simeone A. Epilepsy and brain abnormalities in mice lacking the Otx1 gene. Nat Genet. 1996;14:218–222. doi: 10.1038/ng1096-218. [DOI] [PubMed] [Google Scholar]
- Acampora D, Avantaggiato V, Tuorto F, Barone P, Reichert H, Finkelstein R, Simeone A. Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development. 1998;125:1691–1702. doi: 10.1242/dev.125.9.1691. [DOI] [PubMed] [Google Scholar]
- Leuzinger S, Hirth F, Gerlich D, Acampora D, Simeone A, Gehring WJ, Finkelstein R, Furukubo-Tokunaga K, Reichert H. Equivalence of the fly orthodenticle gene and the human OTX genes in embryonic brain development of Drosophila. Development. 1998;125:1703–1710. doi: 10.1242/dev.125.9.1703. [DOI] [PubMed] [Google Scholar]
- Nagao T, Leuzinger S, Acampora D, Simeone A, Finkelstein R, Reichert H, Furukubo-Tokunaga K. Developmental rescue of Drosophila cephalic defects by the human Otx genes. Proc Natl Acad Sci USA. 1998;95:3737–3742. doi: 10.1073/pnas.95.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyl PP, Signore M, Acampora D, Martinez-Barbera JP, Ilengo C, Annino A, Corte G, Simeone A. Forebrain and midbrain development requires epiblast-restricted Otx2 translational control mediated by its 3' UTR. Development. 2001;128:2989–3000. doi: 10.1242/dev.128.15.2989. [DOI] [PubMed] [Google Scholar]
- Reichert H, Simeone A. Developmental genetic evidence for a monophyletic origin of the bilaterian brain. Philos Trans R Soc Lond B Biol Sci. 2001;356:1533–1544. doi: 10.1098/rstb.2001.0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncinelli E, Morgan R. Downstream of Otx2, or how to get a head. Trends Genet. 2001;17:633–636. doi: 10.1016/s0168-9525(01)02418-0. [DOI] [PubMed] [Google Scholar]
- Leemans R, Egger B, Loop T, Kammermeier L, He H, Hartmann B, Certa U, Hirth F, Reichert H. Quantitative transcript imaging in normal and heat-shocked Drosophila embryos by using high-density oligonucleotide arrays. Proc Natl Acad Sci USA. 2000;97:12138–12143. doi: 10.1073/pnas.210066997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans R, Loop T, Egger B, He H, Kammermeier L, Hartmann B, Certa U, Reichert H, Hirth F. Identification of candidate downstream genes for the homeodomain transcription factor Labial in Drosophila through oligonucleotide-array transcript imaging. Genome Biol. 2001;2:research0015.1–0015.9. doi: 10.1186/gb-2001-2-5-research0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann WEA, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa V, Yamamoto RR, Henderson DS, Glover DM. Mutation of a Drosophila gamma tubulin ring complex subunit encoded by discs degenerate-4 differentially disrupts centrosomal protein localization. Genes Dev. 2000;14:3126–3139. doi: 10.1101/gad.182800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Coyne RS, Dubreuil RR, Goldstein LS, Branton D. Cell shape and interaction defects in alpha-spectrin mutants of Drosophila melanogaster. J Cell Biol. 1993;123:1797–1809. doi: 10.1083/jcb.123.6.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakin L, Reversade B, Virlon B, Rusniok C, Glaser P, Elalouf JM, Brulet P. Gene expression profiles in normal and Otx2-/- early gastrulating mouse embryos. Proc Natl Acad Sci USA. 2000;97:14388–14393. doi: 10.1073/pnas.011513398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto BT. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Feger G. Identification and complete cDNA sequence of the missing Drosophila MCMs: DmMCM3, DmMCM6 and DmMCM7. Gene. 1999;227:149–155. doi: 10.1016/s0378-1119(98)00596-4. [DOI] [PubMed] [Google Scholar]
- Treisman JE. A conserved blueprint for the eye? BioEssays. 1999;21:843–850. doi: 10.1002/(SICI)1521-1878(199910)21:10<843::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regel R, Matioli SR, Terra WR. Molecular adaptation of Drosophila melanogaster lysozymes to a digestive function. Insect Biochem Mol Biol. 1998;28:309–319. doi: 10.1016/s0965-1748(97)00108-2. [DOI] [PubMed] [Google Scholar]
- Wallrath LL. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- Su TT, O'Farrell PH. Size control: cell proliferation does not equal growth. Curr Biol. 1998;8:R687–R689. doi: 10.1016/s0960-9822(98)70436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FlyBase Consortium The FlyBase database of the Drosophila Genome Projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoe M, Schwarz TL, Umbach JA, Gundersen CB, Kidokoro Y. Absence of junctional glutamate receptor clusters in Drosophila mutants lacking spontaneous transmitter release. Science. 2001;293:514–517. doi: 10.1126/science.1061270. [DOI] [PubMed] [Google Scholar]
- Benos PV, Gatt MK, Murphy L, Harris D, Barrell B, Ferraz C, Vidal S, Brun C, Demaille J, Cadieu E, et al. From first base: the sequence of the tip of the X chromosome of Drosophila melanogaster, a comparison of two sequencing strategies. Genome Res. 2001;11:710–730. doi: 10.1101/gr.173801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF. Unmasking a cheating gene. Science. 1999;283:1651–1652. doi: 10.1126/science.283.5408.1651. [DOI] [PubMed] [Google Scholar]
- Zimowska G, Aris JP, Paddy MR. A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci. 1997;110:927–944. doi: 10.1242/jcs.110.8.927. [DOI] [PubMed] [Google Scholar]
- Bertram MJ, Pereira-Smith OM. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266:111–121. doi: 10.1016/s0378-1119(01)00372-9. [DOI] [PubMed] [Google Scholar]
- Prokopenko SN, He Y, Lu Y, Bellen HJ. Mutations affecting the development of the peripheral nervous system in Drosophila : a molecular screen for novel proteins. Genetics. 2000;156:1691–1715. doi: 10.1093/genetics/156.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie KW, Hari KL, Maggert KA, Karpen GH. Centromere proteins and chromosome inheritance: a complex affair. Curr Opin Genet Dev. 1999;9:206–217. doi: 10.1016/S0959-437X(99)80031-8. [DOI] [PubMed] [Google Scholar]
- Philip AV. Mitotic sister-chromatid separation: what Drosophila mutants can tell us. Trends Cell Biol. 1998;8:150. [PubMed] [Google Scholar]
- Kablar B, Vignali R, Menotti L, Pannese M, Andreazzoli M, Polo C, Giribaldi MG, Boncinelli E, Barsacchi G. Xotx genes in the developing brain of Xenopus laevis. Mech Dev. 1996;55:145–158. doi: 10.1016/0925-4773(96)00497-2. [DOI] [PubMed] [Google Scholar]
- Nasiadka A, Krause HM. Kinetic analysis of segmentation gene interactions in Drosophila embryos. Development. 1999;126:1515–1526. doi: 10.1242/dev.126.7.1515. [DOI] [PubMed] [Google Scholar]
- Bate M, Martinez-Arias A. The Development of Drosophila Melanogaster Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1993.
- Hoppler S, Bienz M. Specification of a single cell type by a Drosophila homeotic gene. Cell. 1994;76:689–702. doi: 10.1016/0092-8674(94)90508-8. [DOI] [PubMed] [Google Scholar]
- Cribbs DL, Benassayag C, Randazzo FM, Kaufman TC. Levels of homeotic protein function can determine developmental identity: evidence from low-level expression of the Drosophila homeotic gene proboscipedia under Hsp70 control. EMBO J. 1995;14:767–778. doi: 10.1002/j.1460-2075.1995.tb07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Hartmann B, Hirth F, Walldorf U, Reichert H. Expression, regulation and function of the homeobox gene empty spiracles in brain and ventral nerve cord development of Drosophila. Mech Dev. 2000;90:143–153. doi: 10.1016/s0925-4773(99)00237-3. [DOI] [PubMed] [Google Scholar]
- Yang X, Li DM, Chen W, Xu T. Human homologue of Drosophila lats, LATS1, negatively regulate growth by inducing G(2)/M arrest or apoptosis. Oncogene. 2001;20:6516–6523. doi: 10.1038/sj.onc.1204817. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Goldstein LS. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron. 2001;32:389–401. doi: 10.1016/s0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- Royet J, Finkelstein R. Pattern formation in Drosophila head development: the role of the orthodenticle homeobox gene. Development. 1995;121:3561–3572. doi: 10.1242/dev.121.11.3561. [DOI] [PubMed] [Google Scholar]
- Walter MF, Petersen NS, Biessmann H. Heat shock causes the collapse of the intermediate filament cytoskeleton in Drosophila embryos. Dev Genet. 1990;11:270–279. doi: 10.1002/dvg.1020110405. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- NCBI Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A list of the genes on the microarray
Primary data (Avg Diff values, both raw and normalized) for heat-shocked otd embryos - replicate 1
Primary data (Avg Diff values, both raw and normalized) for heat-shocked otd embryos - replicate 2
Primary data (Avg Diff values, both raw and normalized) for heat-shocked otd embryos - replicate 3
Primary data (Avg Diff values, both raw and normalized) for heat-shocked otd embryos - replicate 4
Primary data (Avg Diff values, both raw and normalized) for heat-shocked Otx2 embryos - replicate 1
Primary data (Avg Diff values, both raw and normalized) for heat-shocked Otx2 embryos - replicate 2
Primary data (Avg Diff values, both raw and normalized) for heat-shocked Otx2 embryos - replicate 3
Primary data (Avg Diff values, both raw and normalized) for heat-shocked Otx2 embryos - replicate 4
Primary data (Avg Diff values, both raw and normalized) for heat-shocked wild-type embryos - replicate 1
Primary data (Avg Diff values, both raw and normalized) for heat-shocked wild-type embryos - replicate 2
Primary data (Avg Diff values, both raw and normalized) for heat-shocked wild-type embryos - replicate 3
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 1
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 2
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 3
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 4
Primary data (Avg Diff values, both raw and normalized) for wild-type embryos - replicate 5
Normalization factors for each replicate
Heat-shocked wild-type embryos compared with heat-shocked otd embryos
Heat-shocked wild-type embryos compared with heat-shocked Otx2 embryos
Heat-shocked wild-type embryos compared with wild-type embryos


