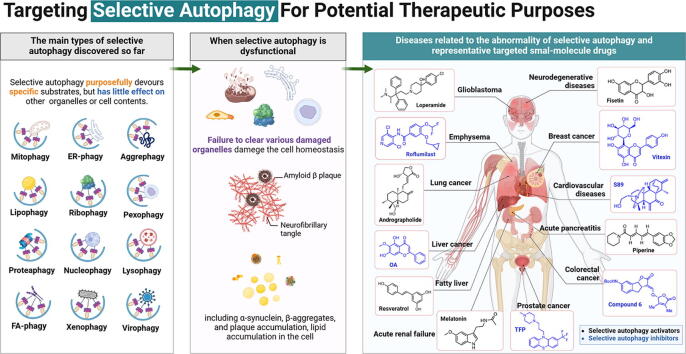
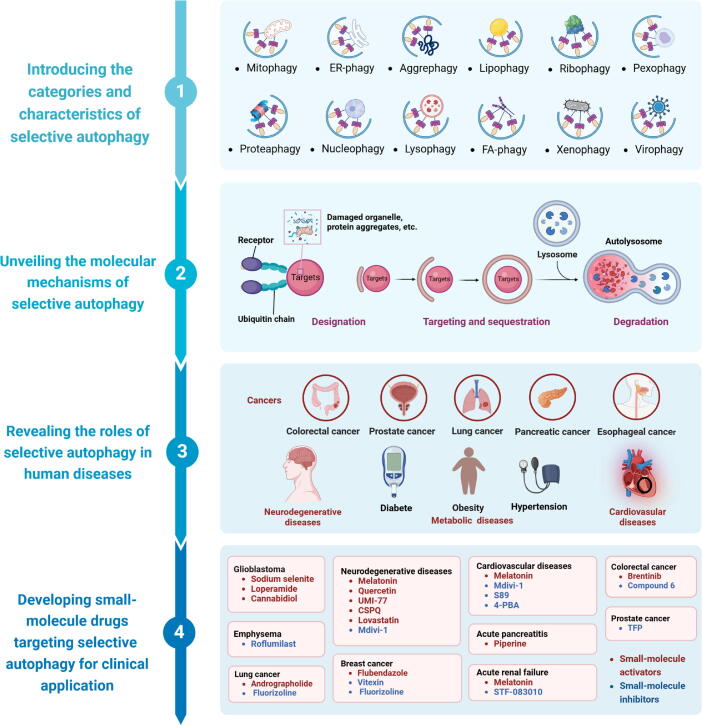
Graphical abstract
Keywords: Autophagy, Selective autophagy, Molecular mechanism, Human disease, Small-molecule drug, Clinically relevant implementation
Highlights
-
•
Selective autophagy is characterized by its ability to specifically degrade a certain substrate, rather than causing widespread cell autophagy.
-
•
The mechanisms underlying selective autophagy can be broadly dispersed in three steps: designation, targeting and sequestration, and degradation.
-
•
Numerous human diseases and their progression are closely linked to aberrant selective autophagy.
-
•
Deciphering the molecular mechanisms underlying selective autophagy provides a theoretical framework for treating relevant clinical disorders.
-
•
Regulation of selective autophagy by discovering and developing small-molecule agents has great clinical application prospects for the treatment of related diseases.
Abstract
Background
Autophagy is an evolutionarily conserved turnover process for intracellular substances in eukaryotes, relying on lysosomal (in animals) or vacuolar (in yeast and plants) mechanisms. In the past two decades, emerging evidence suggests that, under specific conditions, autophagy can target particular macromolecules or organelles for degradation, a process termed selective autophagy. Recently, accumulating studies have demonstrated that the abnormality of selective autophagy is closely associated with the occurrence and progression of many human diseases, including neurodegenerative diseases, cancers, metabolic diseases, and cardiovascular diseases.
Aim of Review
This review aims at systematically and comprehensively introducing selective autophagy and its role in various diseases, while unravelling the molecular mechanisms of selective autophagy. By providing a theoretical basis for the development of related small-molecule drugs as well as treating related human diseases, this review seeks to contribute to the understanding of selective autophagy and its therapeutic potential.
Key Scientific Concepts of Review
In this review, we systematically introduce and dissect the major categories of selective autophagy that have been discovered. We also focus on recent advances in understanding the molecular mechanisms underlying both classical and non-classical selective autophagy. Moreover, the current situation of small-molecule drugs targeting different types of selective autophagy is further summarized, providing valuable insights into the discovery of more candidate small-molecule drugs targeting selective autophagy in the future. On the other hand, we also reveal clinically relevant implementations that are potentially related to selective autophagy, such as predictive approaches and treatments tailored to individual patients.
Introduction
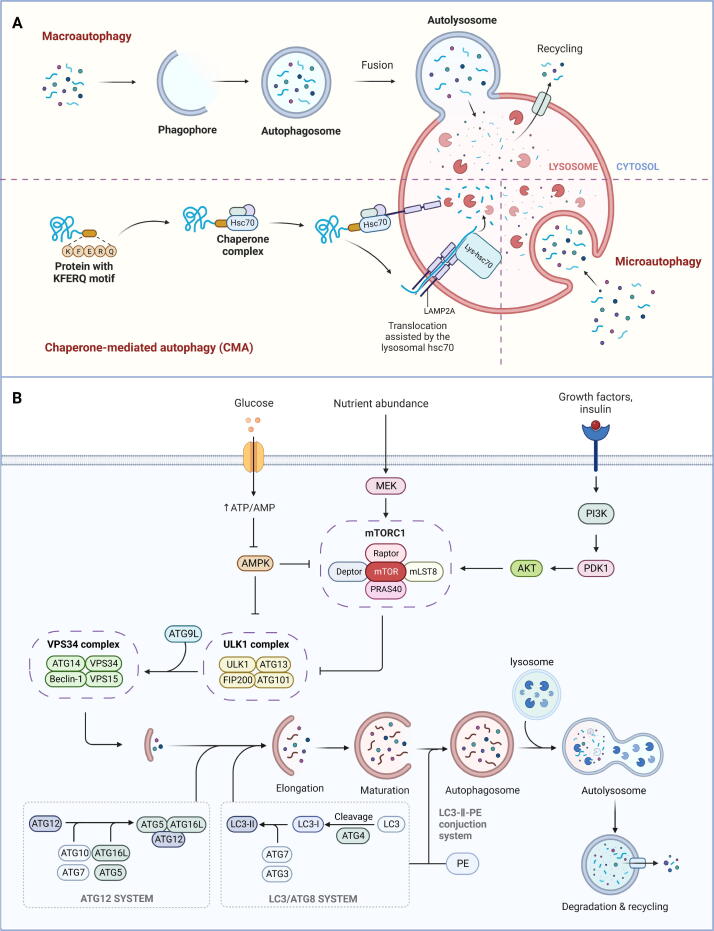
Autophagy stands as a subcellular degradation pathway that is pivotal for upholding the homeostasis of eukaryotic cells and overall cell health. It was first discovered by Ashford and Porten in human liver cells in 1962 [1]. In mammalian cells, autophagy is usually categorized into three distinct types based on the pathways through which substrate (also called cargo) enters lysosomes: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (Fig. 1A) [2], [3]. Eukaryotic cells exhibit two principal pathways for protein degradation: the ubiquitin–proteasome system (UPS) and the autophagy-lysosome pathway (ALP) [4]. Compared to the UPS, which is prevalent in eukaryotes and archaea, autophagy exhibits stronger degradation ability. Its substrates include not only biological macromolecules such as soluble proteins but also larger cellular structures such as damaged organelles [5]. The process of autophagy generally conforms to the “cargo-ligand-receptor” model, which comprises four main processes: induction of autophagy, formation of autophagosome, fusion between autophagosome and lysosome, and subsequent content degradation [6]. Autophagy is known to be a highly evolutionarily conserved process, and its occurrence and development are governed by a cascade of autophagy-related genes (ATGs). Currently, more than 30 autophagy-specific genes [7] and more than 50 related genes have been identified. The proteins encoded by these ATGs significantly contribute to various phases of autophagy initiation and progression (Fig. 1B). Moreover, numerous studies have substantiated the connection between autophagy dysregulation and the pathogenesis of various human diseases, highlighting it as a pivotal research focus in recent years [8].
Fig. 1.
Overview of autophagy in mammalian cells. (A) The classification of autophagy based on the pathways substrate (also called cargo) entering lysosomes. (B) The main molecular mechanisms of autophagy discovered so far.
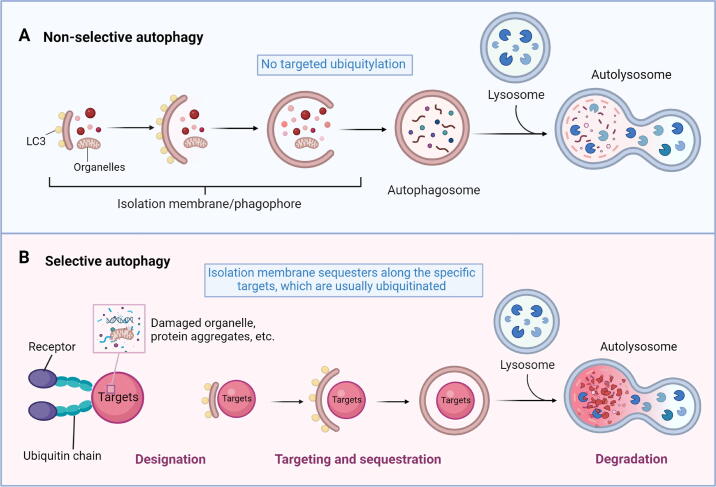
It has long been assumed that autophagy is a non-specific process. However, as research has progressed, it has become evident that autophagy can specifically degrade some biological macromolecules or organelles under certain conditions, a phenomenon referred to as selective autophagy [9]. Unlike non-selective autophagy (Fig. 2A), selective autophagy is characterized by its ability to specifically degrade a certain substrate rather than inducing widespread cell autophagy (Fig. 2B). Nowadays, extensive research is being conducted on various forms of selective autophagy, including mitophagy [10], endoplasmic reticulum (ER)-phagy/reticulophagy [11], aggrephagy [12], lipophagy [13], pexophagy [14], ribophagy [15], and others. In addition to the organelles inside the cell, invasive substances from outside of the cell, such as viruses and bacteria, can also be degraded by selective autophagy [16]. However, while the molecular mechanism of generalized autophagy is relatively well studied, the detailed mechanism of selective autophagy remains unclear and requires further investigation. At present, the existing data accumulated by related studies prove that selective autophagy exerts a substantial physiological impact across various facets of organisms. And its aberrant function is directly linked to the onset and advancement of numerous severe human diseases, encompassing neurodegenerative diseases, cancers, metabolic diseases, and so on [17], [18]. Therefore, elucidating the molecular mechanism of selective autophagy is poised to provide a novel theoretical foundation for addressing related diseases therapeutically. The development of new small-molecule drugs targeting selective autophagy is currently a research hotspot, as these drugs are potentially more specific to their targets and have fewer side effects. Owing to its high specificity, drug development aimed at selective autophagy pathways holds significant promise for therapeutic interventions, making it a field of high clinical relevance.
Fig. 2.
Comparative model of non-selective autophagy and selective autophagy. (A) Non-selective autophagy allows cells to survive through nutrient starvation until the next nutrient source is available. Once cells sense lack of nutrient, an isolation membrane is mostly formed at ER-mitochondria contact sites, LC3-II labelled membranes elongate as they engulf materials and eventually closes to form autophagosomes. Autophagosomes then fuse with lysosomes to degrade their contents. (B) In selective autophagy, many cargoes are ubiquitinated and specifically recognized by receptors, which does not happen in non-selective autophagy.
Here, we summarize the presently discovered categories of selective autophagy and delve into the underlying mechanisms governing this process, providing a comprehensive understanding of selective autophagy. Additionally, we elucidate the diseases associated with different types of selective autophagy, categorizing them into several classifications, such as neurodegenerative diseases, cancers, cardiovascular diseases, and metabolic diseases. Moreover, existing small-molecule drugs for regulating selective autophagy are also described to give inspiration to related drug development. This article aspires to contribute to the identification and development of novel drugs targeting selective autophagy, presenting novel perspectives for the treatment of associated diseases.
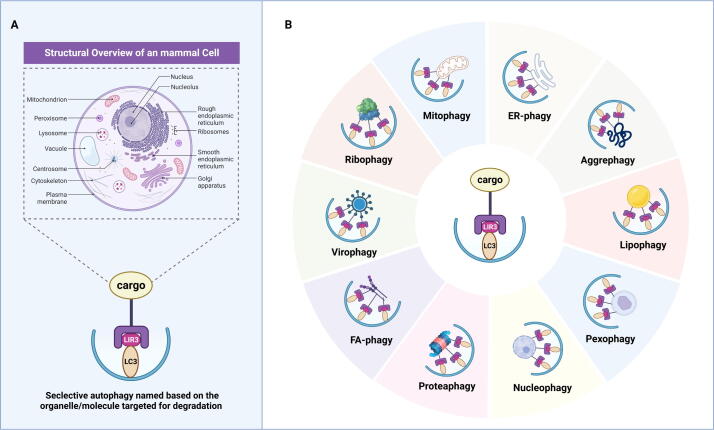
The categories and characteristics of selective autophagy
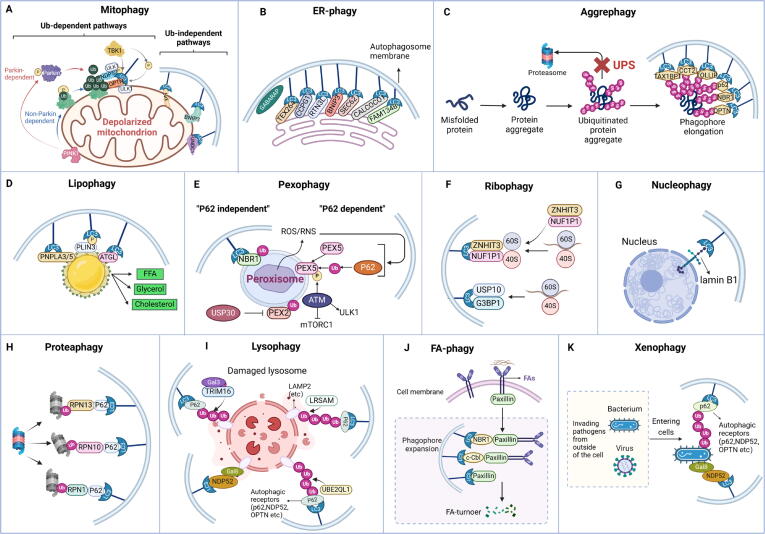
In contrast to non-selective autophagy, which is characterized by “eating oneself,” selective autophagy is a deliberate “phagocytosis” process that selectively targets and engulfs different substrates, including mitochondria, peroxisomes, ribosomes, ERs, lysosomes, nuclei, proteasomes, lipid droplets (LDs), and so forth (Fig. 3A). In this section, we provide a summary of the types of selective autophagy that are relatively well studied at present, outlining their distinct mechanistic features (Fig. 3B).
Fig. 3.
The classifications of selective autophagy. (A) The regular cargoes of selective autophagy inside mammal cells. (B) The representative types of selective autophagy based on its targeting cargoes.
Mitophagy
Mitochondria, as the main source of ATP, hold critical significance in numerous intracellular processes and play a pivotal role in instigating programmed cell death. However, damaged mitochondria will significantly destroy the metabolic homeostasis of cells, leading to the excessive generation of reactive oxygen species (ROS) and cellular demise. Hence, quantitative and quality control to maintain mitochondrial homeostasis is a prerequisite for the therapeutic intervention of diverse diseases. Mitophagy, a subtype of selective autophagy, is essential for preserving the functional integrity of mitochondria and cellular homeostasis by specifically eliminating dysfunctional mitochondria from the cytoplasm.
At present, the mechanisms of mitophagy are categorized into two groups: the ubiquitin (Ub)-dependent pathway and the Ub-independent pathway. And the most extensively studied mitophagy regulation pathway is the PTEN-induced putative kinase 1 (PINK1)/Parkin pathway in the Ub-dependent pathway. The initiation of PINK1-Parkin pathway-mediated mitophagy is usually associated with changes in mitochondrial membrane potential (MMP). In mammalian cells, PINK1-Parkin pathway-mediated mitophagy can be triggered by inhibitors of the respiratory chain, protein toxicity, and mitochondrial ROS [19]. PINK1 denotes a mitochondrial serine/threonine kinase encoded by nuclear DNA and transported to the mitochondria for function [20]. Under physiological circumstances, PINK1 is synthesized within the cytoplasm and transported to the inner mitochondrial membrane (IMM). The changes of MMP prevent PINK1 from entering the IMM, leading to its accumulation on the outer mitochondrial membrane (OMM) and subsequent recruitment of the E3 ubiquitin ligase Parkin to the mitochondrial surface [21]. Subsequently, the E3 ubiquitin ligase activity of Parkin was triggered by PINK1 by a series of modifications, including the phosphorylation of Parkin and ubiquitin [22]. Activated Parkin can cause polyubiquitination of various mitochondrial outer membrane proteins and thus be recognized by a variety of autophagy adaptor proteins, including sequestosome 1 (SQSTM1/p62), optineurin (OPTN), nuclear dot protein 52 (NDP52), and neighbor of BRCA1 gene 1 (NBR1) [23], all of which contain the LC3-interacting region (LIR). By engaging in interactions between the LIR motifs and microtubule-associated protein 1 light chain 3 (LC3), the autophagy vesicles wrap the damaged mitochondria to form mitochondrial autophagosomes, which subsequently undergo fusion with lysosomes to degrade the damaged mitochondria [23]. In addition, autophagy receptors, such as OPTN and NDP52, can be directly recruited by PINK1 into mitochondria via ubiquitin phosphorylation, thus promoting the occurrence of mitophagy [24]. In the non-ubiquitination pathway, the OMM proteins such as BCL2-interacting protein 3 like (BNIP3L)/Nip3-like protein X (NIX), BCL2-interacting protein 3 (BNIP3), and FUN14 domain-containing protein 1 (FUNDC1) can directly bind to LC3, thereby promoting mitochondria to be wrapped by the autophagosomal membrane (Fig. 4A). Recently, the membrane scaffold protein prohibitin 1 (PHB1) and prohibitin 2 (PHB2) have also been recognized as novel mitophagy receptors, suggesting the existence of additional potential mitophagy receptors [25], [26].
Fig. 4.
Autophagy induction mechanisms of specific selective autophagy types. (A) The mechanisms of mitophagy are divided into two categories: the Ub-dependent pathway and the Ub-independent pathway. And the most extensively studied mitophagy regulation pathway is the PINK1/Parkin pathway in the Ub-dependent pathway. (B) The autophagic receptors of ER-phagy found in mammals up to now are SQSTM1/p62, BNIP3, FAM134B, TEX264, RTN3L, SEC62, CCPG1, CALCOCO, and so on. (C) When neurodegenerative diseases occur, the UPS pathway is disrupted, resulting in the inability to degrade misfolded proteins (such as Aβ and Tau in AD), which leads to irreversible damage. Therefore, the promotion of aggrephagy holds promise as a novel therapeutic approach for this kind of disease. (D) Lipophagy, which decomposes triglycerides into FFAs, is an important metabolic pathway for the body to reduce lipid toxicity. (E) Some proteins on the peroxisome membrane are ubiquitinated. Then ubiquitinated membrane proteins are combined with LC3 mediated by NBR1, p62, or other proteins to form autophagosomes, which are further fused with lysosomes and degraded. (F) NUFIP1 acts as a major receptor of ribophagy by specific binding to LC3 with the assistance of ZNHIT3. USP10 and G3BP1 were found to be mammalian homologs of ribophagy receptors in yeast, indicating their potential role in mammalian ribophagy. (G) Nucleophagy is programmed to selectively remove nuclear components through the process of autophagy. Lamin B1 and chromatin can be degraded by autophagy mechanisms under aging exposure. (H) The inactivated proteasome subunit is labeled by ubiquitin and then recognized by autophagic receptors. The proteasome subunits, RPN1, RPN10, and RPN13, have been identified as important ubiquitin receptors in mammals. (I) Lysophagy is essential for the quality control of lysosomes. Lysophagy factors, such as UBE2QL1, LRSAM1, and TRIM16, are demanded to ubiquitinate lysosomal membrane proteins. The ubiquitinated proteins then recruit autophagy receptors, leading to the induction of lysophagy. (J) FAs serve as selective targets for autophagy, with the autophagy receptors NBR1, c-Cbl, and SQSTM1/p62 identified as crucial mediators in this process by facilitating the targeting of FAs to autophagosomes. (K) Xenophagy can remove harmful invading pathogens from outside of the cell, such as bacteria and viruses, to maintain cell homeostasis.
As the most widely studied type of selective autophagy, abnormal or damaged mitophagy has been implicated in numerous human diseases, a more comprehensive exploration of the role of mitophagy in diseases will be provided in a subsequent section.
ER-phagy/Reticulophagy
ER assumes a pivotal role in numerous biological processes within the body, such as calcium storage and lipid biosynthesis, as well as the maturation and transport of both secreted and membrane proteins. However, disruption of the environmental equilibrium within the ER and impairment of its functionality lead to the accumulation of numerous unfolded or misfolded proteins within its lumen. This accumulation ultimately disrupts ER homeostasis, initiates ER stress, along with the unfolded protein response (UPR), and may even trigger a comprehensive remodeling of the ER. ER-phagy, also known as reticulophagy, is a lysosomal-mediated process that specifically removes damaged ER. Its primary function is to degrade redundant ER membranes and insoluble or toxic protein aggregates, thus controlling ER volume and maintaining cell homeostasis. Additionally, ER-phagy has a hidden function in expanding ER capacity by releasing the vesicle membrane that has been engulfed. This expansion of ER capacity is beneficial to the aggregation of newly synthesized ER enzymes and the reduction of the buildup of unfolded proteins, thereby inhibiting the aggregation of misfolded proteins. Essentially, ER-phagy can modulate the steady-state capacity of the ER, making it an indispensable process for preserving cellular homeostasis.
It is noteworthy that the targeted autophagic degradation of the ER occurs due to the specific recognition by certain autophagy-related proteins, such as ATG8 in yeast, LC3 in mammals, or other effector factors [27]. These proteins selectively identify ER-phagy receptors, triggering the initiation of autophagosome formation. Therefore, understanding alterations in ER-phagy receptors is imperative for comprehending the mechanisms underlying ER-phagy-related diseases. Among the recognized mammalian ER-phagy receptors are SQSTM1/p62 [28], BNIP3 [29], family with sequence similarity 134 member B (FAM134B, also known as RETREG1) [30], testis expressed 264 (TEX264) [31], reticulon-3L (RTN3L) [32], SEC62 Homolog, preprotein translocation factor (SEC62) [33], cell cycle progression protein 1 (CCPG1) [34], calcium binding and coiled-coil domain protein 1 (CALCOCO1) [35], and so on (Fig. 4B). Originally recognized as a tumor suppressor gene named JK1, FAM134B underwent its initial characterization. However, a recent study revealed its role as a protein interacting with ATG8/LC3 and shown that the deletion of FAM134B led to observable ER swelling in neuronal cells [30]. Likewise, numerous studies have demonstrated the pivotal role played by different specific ER-phagy receptor proteins in preserving ER homeostasis [32], [35], [36], [37]. These investigations have elucidated that different ER-phagy receptors are responsible for distinct stress conditions, and the expression levels of diverse ER-phagy receptors in different tissues also exhibit significant variation [38]. Consequently, the different physiological functions and regulatory mechanisms of these receptors remain an area requiring in-depth investigation.
On balance, ER-phagy is currently a focal point of research, yet numerous pressing issues persist. Urgent challenges include deciphering the precise molecular mechanism governing ER-phagy, comprehending its pathophysiological functions, exploring specific regulatory methods (e.g., post-translational modifications), identifying novel ER-phagy receptors, and more. Despite notable progress in understanding the molecular mechanisms of ER-phagy, fundamental questions surrounding how various physiological and pathological signals influence its specific mechanisms, the exact process of ER fragmentation, and the subsequent sequestration into autophagosomes and lysosomes remain pivotal problems that are unanswered. Uncovering the physiological and pathological processes related to ER-phagy-induced diseases and gene mutations is essential, offering potential avenues for disease treatment. As scientific exploration deepens, unraveling further aspects of ER-phagy will furnish novel insights and strategies for preventing and treating associated diseases.
Aggrephagy
Numerous diseases arise due to genetic mutations or the buildup of abnormal proteins. The targeted degradation of these pathogenic proteins is crucial for maintaining cellular homeostasis and can significantly impact disease development or treatment. In this context, autophagy, as an important mechanism for cytoplasmic cleaning, has attracted widespread attention from scientists. Of particular interest is aggrephagy, an important branch of cellular autophagy, which has shown unique potential for regulating protein aggregates and removing misfolded proteins. As one of the important types of selective autophagy in cells, it is primarily responsible for degrading protein aggregates and assumes a decisive function in the quality surveillance system for abnormal proteins. Therefore, regulating the degradation of some aggregated or misfolded proteins through aggrephagy holds promise for potential applications in the treatment of related diseases.
Mechanistically, the ubiquitination of the aggregates is essential for facilitating their degradation. Additionally, SQSTM1/p62, NBR1, toll interacting protein (TOLLIP), Tax1 binding protein 1 (TAX1BP1), and OPTN are shown to be cargo receptors specific to aggrephagy, acting as bridging connectors between polyubiquitinated substrates and LC3 on autophagosomes (Fig. 4C) [39]. The ubiquitin-binding domain (UBD) and LIR motif are essential for the function of aggrephagy receptors. In both SQSTM1/p62 and NBR1, the UBD situated in the C-terminal region distinctly recognizes Lys63-linked polyubiquitin substrates, resulting in the formation of complexes [40]. Simultaneously, the LIR motif within SQSTM1/p62 and NBR1 facilitates the transportation of complexes, resulting from the union of SQSTM1/p62 or NBR1 with polyubiquitinated aggregates, to autophagosomes. TOLLIP interacts with and colocalizes alongside ubiquitin and LC3, signifying TOLLIP's role as a receptor for Ub-ATG8 [41]. Elevated TOLLIP expression facilitates the effective degradation of polyglutamine (polyQ) proteins associated with HD. Conversely, the absence of TOLLIP induces cytotoxic effects in response to the overexpression of polyQ proteins [42]. Similarly, the deficiency of TAX1BP1 leads to the accumulation of protein aggregates within the brain, which leads to the deterioration of neurodegenerative diseases such as Huntington's disease (HD) [43]. Moreover, recent studies have identified a novel aggregate receptor, chaperonin containing TCP1 subunit 2 (CCT2), which operates independently of ubiquitin-binding receptors and CMA. Unlike SQSTM1/p62, NBR1, and TAX1BP1, which promote the elimination of soluble protein aggregates, CCT2 assists in the autophagic degradation of solid protein aggregates characterized by low motility. CCT2 operates selectively, specifically promoting the degradation of solid aggregates [44].
Lipophagy
LDs are large accumulations of neutral lipids found in adipocytes, consisting mainly of a monolayer structure of triglycerides and sterol esters. LDs are cellular organelles responsible for storing intracellular neutral lipids, with numerous studies linking them to obesity and various diseases [45]. Beyond adipocytes, LDs have been identified in diverse cell types, including hepatocytes, smooth muscle cells, and glial cells. Since autophagy was first reported as a process involving LD decomposition in 2009, lipophagy has garnered significant attention as a novel process of lipid metabolism based on LD decomposition [46]. Lipophagy represents a distinct form of autophagy capable of selectively identifying LDs and integrating them into autophagosomes efficiently. After fusion with lysosome, the lysosomal enzymes are used to decompose triacylglycerol (TAG) to produce free fatty acids (FFAs), providing a substrate for fatty acid oxidation (FAO), which is an important metabolic pathway for the body to reduce lipid toxicity (Fig. 4D) [47].
Adipocyte triglyceride lipase (ATGL) has been shown to initiate the hydrolysis of TAG to release free fatty acids, indicating that ATGL may play an important role in regulating lipophagy [48]. Mechanistically, ATGL promotes the activity of Sirtuin 1 (SIRT1), which is a prerequisite for ATGL-induced initiation of lipophagy to control hepatic LD catabolism [49]. Therefore, ATGL is identified as a key player in the selective autophagic degradation of lipophagy. In a mouse model subjected to a high-fat diet (HFD), another member of the same lipase family, patatin-like phospholipase domain-containing protein 8 (PNPLA8), was proven to engage with LC3, instigating lipophagy and thus ameliorating the symptoms of NAFLD [50]. Furthermore, PNPLA3 and PNPLA5 are essential components for the induction of lipophagy in starved human hepatocytes [51], [52]. Perilipin (PLIN) proteins are the main cytosolic lipid droplet-related proteins in many diseases, and they are considered to participate in the establishment and stability of LDs. Demonstrably, the mammalian target of rapamycin complex 1 (mTORC1) orchestrates the regulation of hepatic lipophagy by modulating PLIN3 phosphorylation, indicating a pivotal role for PLIN3 as a significant lipophagy receptor [53]. Studies have demonstrated that the lipid transfer protein oxysterol-binding protein-related protein 8 (ORP8), which is located on LDs and acts as the receptor for autophagic turnover of LDs, promotes the envelopment of LDs by autophagosomal membranes [54]. Notably, this function of ORP8 is detached from its lipid transport activity. Instead, it is accomplished through its direct interaction with LC3 anchored in the phagophore [55]. Mucolipin TRP cation channel 1 (MCOLN1), identified as a lysosomal Ca2+ channel that controls lysosome-plasma membrane fusion, has recently been demonstrated to participate in the extracellular efflux of FFAs produced from lipophagy via lysosomal exocytosis [13]. In conclusion, the investigation into lipid autophagy receptors, particularly the elucidation of mechanisms involving lipophagy, has significantly advanced our understanding of cellular lipid metabolism and its implications for health and disease.
Pexophagy
Peroxisomes are organelles with a single-membrane structure found in eukaryotic cells. Named for their abundance of catalase and various peroxidases, peroxisomes play a crucial role in fatty acid oxidation, phospholipid synthesis, and oxidative stress [56]. Peroxisome autophagy, also called pexophagy, is the primary mechanism for peroxisome degradation [57]. When various factors trigger pexophagy, specific proteins on the peroxisome membrane undergo ubiquitination. Subsequently, ubiquitinated membrane proteins bind to LC3 mediated by NBR1, SQSTM1/p62, or other proteins to form autophagosomes, which are further fused with lysosomes and degraded (Fig. 4E) [58].
There are numerous proteins present on the peroxisome membrane, collectively referred to as peroxisomal membrane proteins (PMPs). Among these, the most thoroughly studied PMP related to pexophagy is peroxisomal biogenesis factor 5 (PEX5) [59]. During the occurrence of oxidative stress, PEX5 locates an important protein kinase, ataxia telangiectasia-mutated kinase (ATM), onto the peroxidase body membrane, thus promoting the initiation of pexophagy [60]. Secondly, the peroxisomal biogenesis factor 2 (PEX2), serving as peroxisomal E3 ubiquitin ligase, undergoes ubiquitination to designate peroxisomes for pexophagy [61]. Specifically, this leads to widespread ubiquitination of peroxisomes and their subsequent degradation through an autophagic process mediated by NBR1, indicating that pexophagy occurrence can be regulated by controlling the ubiquitination process [62]. The deubiquitinating enzyme ubiquitin-specific protease 30 (USP30) can inhibit pexophagy by antagonizing PEX2, thus maintaining the balance of the number of peroxisomes in cells [63]. In addition to oxidative stress and ubiquitination, amino acid starvation emerges as another prevalent factor inducing pexophagy. It was revealed that pexophagy was promoted under amino acid-deficient condition in human cervical cancer HeLa cells [14]. Importantly, mitochondria and peroxisomes are closely related metabolic organelles in origin and function. Studies have proved that BNIP3L/NIX can regulate not only mitophagy but also pexophagy, revealing the dual functions of BNIP3L/NIX and highlighting the interrelationship between different selective autophagy pathways [64]. In summary, pexophagy, as the main degradation mode of peroxisomes, together with the biosynthesis process of peroxisomes, maintains the dynamic balance of peroxisome number in cells. Disturbance of this equilibrium can give rise to peroxisomal dysfunction and contribute to the onset of various diseases.
Ribophagy
Ribosomes are intracellular ribonucleoprotein particles primarily composed of ribosomal RNA (rRNA) and proteins and mainly function as molecular machines for intracellular protein synthesis. Through electron microscopy, ribosomes have been observed inside autophagosomes [15]. Previously, it was widely believed that ribosomes enclosed within autophagosomes underwent non-selective bulk degradation. Nevertheless, recent emerging findings show that there is a link between ribosomes and selective autophagy. Ribophagy represents a distinctive form of autophagy dedicated to the selective degradation of ribosomes, which was first discovered in 2008 [65]. In mammals, the inactivation of mTORC1 is engaged in the selective degradation of ribosomes [66]. Ribophagy relies on the ability of nuclear FMRP interacting protein 1 (NUFIP1) to interact with LC3B and enhance cell viability, indicating that NUFIP1 acts as a ribosome receptor for starvation-induced ribophagy [66]. And the binding partner of NUFIP1, zinc finger HIT domain-containing protein 3 (ZNHIT3), which transports ribosomes to autolysosomes by directly associating with LC3B, undergoes relocation from the nucleus to autophagosomes, lysosomes, and ribosomes following mTORC1 inhibition. Specifically, inhibiting mTORC1 causes changes in ribosomal stability and promotes the binding of ribosomes to NUFIP1-ZNHIT3 (Fig. 4F). It was reported that Ubp3, a ubiquitin protease, and Bre5, a cofactor, participated in the ribophagy of yeast, mainly targeting the 60S subunit and not affecting the 40S subunit. Recently, ubiquitin-specific protease 10 (USP10) and GTPase activating protein (SH3 domain) binding protein 1 (G3BP1) were found to be mammalian homologues of Ubp3 and Bre5, respectively [67]. In addition, due to the close structural correlation between ribosomes, ER, and mitochondria, when ER-phagy and mitophagy occur, ribosomes can be degraded through the bypass autophagy pathway [68].
Ribophagy holds great promise as a therapeutic target for various diseases, indicating drugs targeting ribophagy may become a therapeutic strategy for various diseases in the future [69]. However, there is a limited understanding of ribosome turnover and its implications for cellular homeostasis, development, and the pathogenesis of human diseases [70]. The signaling pathways and regulatory pathways of ribophagy are still unclear, and the existence of other receptors mediating the occurrence of ribophagy needs further clarification [71].
Nucleophagy
Nucleophagy, a specific category of autophagy directing the cell nucleus toward autophagic degradation, has not only been demonstrated as a model system for investigating selective macroautophagy but has also been associated with various disease conditions [72]. Despite the nucleus serving as the command center of the cell by protecting our genetic information and regulating gene expression, the mechanisms and implications of nuclear autophagy remain poorly understood [73].
During the initiation stage of nuclear autophagy, the nuclear lamina protein lamin B1 directly interacts with LC3 found within the nucleus, facilitating autophagy membrane transport and substrate delivery, and binds to the lamin-associated domains on chromatin [74]. The degradation of lamin B1 is achieved through its transport from nuclear to cytoplasmic and then to the lysosome, where it is degraded (Fig. 4G). Interestingly, detailed investigations into the initiation mechanisms of nucleophagy revealed a potential association between the regulation of nucleophagy and lipid metabolism. Studies have found that lamin gene expression abnormalities or mutations can cause a variety of physiological and pathological processes, such as autosomal dominant familial partial lipodystrophy type 2 (FPLD2), fat storage disorders, and other diseases [75]. Furthermore, investigations have revealed that the inner nuclear membrane (INM) exhibits distinct lipid composition and lipid metabolism functions, further supporting this conclusion [76].
Nuclear abnormalities are common in progeria syndromes, carcinogenic damage, and degenerative diseases. Preventing premature aging and maintaining cell homeostasis depend significantly on the selective autophagy of organelles. We aim to achieve significant advancements in understanding the mechanism of nucleophagy and its physiological and pathological significance in the future, which will help to clarify the pathogenesis of many human diseases and foster the formulation of efficacious treatment strategies involving nucleophagy.
Proteaphagy
In UPS, multiple ubiquitin molecules form covalent connections with the target protein, marking it for degradation by 26S proteasomes [77]. Notably, the proteasomes themselves are also subject to degradation. The lysosomal degradation of proteasomes was first discovered in 1995 when researchers observed that rats administered leupeptin, an inhibitor of lysosomal proteases, showed an accumulation of proteasomes in their lysosomes [78]. In 2015, it was demonstrated for the first time in Arabidopsis that proteasomes can be degraded through the ATG8-mediated autophagy pathway. In this process, the inactivated proteasome subunit is labeled by ubiquitin and then recognized by the ubiquitin receptor ribophorin 10 (RPN10), which further mediates the degradation of the inactivated proteasome by binding to ATG8, thus maintaining the stability of the intracellular proteasome library. This process was later named proteaphagy [79]. However, no interaction between RPN10 and LC3/ATG8 was found in human and yeast cells, suggesting that there may be other molecules mediated proteaphagy in other organisms. In HeLa cells, the ubiquitinated proteasome was degraded by the SQSTM1/p62-mediated autophagy pathway [80]. Moreover, crucial ubiquitin receptors in mammals have been identified as the proteasome subunits ribophorin 1 (RPN1), RPN10, and ribophorin 13 (RPN13) (Fig. 4H) [81]. Under conditions of amino acid starvation, these three subunits become poly-ubiquitinated, making them easier to recognize by SQSTM1/p62. By interacting with LC3, SQSTM1/p62 facilitates the transport of inactive 26S proteasomes to the growing phagophore, eventually leading to their turnover through autophagy [82].
Lysophagy
Interestingly, in addition to the proteasome itself, which can be degraded by selective autophagy, the lysosome, a key organelle in autophagy, can also be targeted for degradation by autophagy. Lysosomes are organelles that decompose biological macromolecules, including proteins, nucleic acids, and polysaccharides, and contain many hydrolytic enzymes. When lysosomes are damaged or unstable, a large amount of hydrolase will be released into the cytosol, posing a threat to cell health [83]. Therefore, the removal of damaged lysosomes is also extremely important for the maintenance of cellular homeostasis. Compromised lysosomes can be engulfed by autophagosomes in a phenomenon recognized as lysophagy. Under circumstances where lysosomal membranes are impaired, or even in regular conditions, lysophagy-related factors encompassing ubiquitin conjugating enzyme E2Q family-like 1 (UBE2QL1), leucine-rich repeat and sterile alpha motif-containing protein 1 (LRSAM1), and tripartite motif containing 16 (TRIM16) are demanded for the ubiquitination of lysosomal membrane proteins [84], [85]. Ubiquitinated proteins subsequently attract autophagy receptors, leading to the induction of lysophagy (Fig. 4I) [86]. Moreover, Galectin-3, which is typically distributed within the cytoplasm and nucleus, can be mobilized to compromised lysosomes. The TRIM16-galectin-3 complex serves as a foundation for the assembly of autophagic initiation proteins, consequently prompting the formation of phagophores [87]. Conversely, galectin-8 establishes a direct interaction with the autophagic receptor NDP52, irrespective of ubiquitin, facilitating the recruitment of LC3-positive phagophores for the mediation of lysophagy [88].
FA-phagy
Focal adhesions (FAs) are structures located beneath the cell membrane, composed of integrins situated on the cellular membrane and actin in the cell [89]. They assume a significant function in fostering adhesion between cells as well as establishing a connection between cells and the extracellular matrix. FAs can be decomposed and reassembled in the process of cell movement and morphological changes, thus regulating cell adhesion and movement. Significantly, autophagy has been demonstrated to degrade FAs in diverse cell types, which is called FA-phagy. The FA protein paxillin engages with processed LC3 via a preserved LIR motif in the N-terminal end of paxillin, culminating in FA-phagy and subsequent disassembly of FAs, which can reduce the migration and invasion of tumor cells [90]. Furthermore, the depletion of essential autophagy genes such as ATG5 or ATG7 has been proven to increase both the number and size of FAs, providing further support for the concept that autophagy negatively regulates FAs [90]. Multiple studies have demonstrated that FAs serve as selective targets for autophagy, with the autophagy cargo receptors NBR1, casitas B-lineage lymphoma (c-Cbl), and SQSTM1/p62 identified as crucial mediators in this process by facilitating the targeting of FAs to autophagosomes (Fig. 4J) [91], [92]. The exploration of FA-phagy as a significant mechanism for FAs degradation has enriched our understanding of cellular dynamics and regulatory pathways governing cell adhesion and movement. The intricate interplay between autophagy and FAs unveils a previously unrecognized facet of cellular homeostasis, shedding light on the adaptability and plasticity of cell-matrix interactions.
Xenophagy and virophagy
Selective autophagy can not only target various intracellular organelles as degradation goods but also identify harmful invading pathogens, such as bacteria and viruses, from outside of the cell to maintain cellular homeostasis. Xenophagy is an important autophagy mechanism used by the host to clear intracellular pathogens and assumes a pivotal function in resisting external pathogen infection. Similar to various forms of selective autophagy, xenophagy brings the cargo specifically to the autophagosomal membrane by means of autophagy receptors (SQSTM1/p62, NDP52, OPTN, and NBR1) (Fig. 4K) [93]. But the current research progress is limited, and it is not yet clear how xenophagy is initiated. Recent studies have highlighted the essential role of the V-ATPase-ATG16L1 axis in initiating xenophagy [94]. Studies have found that bacteria can evade heterologous autophagy by inhibiting the initial signaling pathway of autophagosomes, pretending to be cellular components to avoid autophagy recognition, blocking the formation of autophagosomes, inhibiting the fusion of autophagosomes and lysosomes, etc., so as to achieve the purpose of intracellular survival [95]. Therefore, further study of the specific mechanism of xenophagy can offer the possibility of regulating intracellular xenophagy utilizing small-molecule drugs and provide new approaches for treating associated diseases. The xenophagic disposal of viruses, known as virophagy, is a subtype of xenophagy identified in various viral infections, such as human immunodeficiency virus-1 (HIV-1) [96] and the coronavirus disease 2019 (COVID-19) [97]. At the cellular level, virophagy can selectively target viruses to degrade them, which is beneficial to cell survival. However, some invading viruses have evolved strategies to escape, manipulate, and even inhibit the autophagy mechanism, which is beneficial to virus replication [98].
Selective autophagy is a process characterized by distinct recognition mechanisms for different degradation targets compared with general autophagy, while sharing key protein molecules in subsequent autophagic processes. For example, the three processes, ubiquitination labeling of target goods in the early stage, fusion of autophagosomes with lysosomes, and final degradation in lysosomes, have the same demand for some receptor proteins [27]. In recent years, new types of selective autophagy, such as ferritinophagy [99], have been discovered, leading to the ongoing development of a comprehensive selective autophagy system. From mitophagy to xenophagy, the diverse categories of selective autophagy collectively contribute to maintaining cellular homeostasis by selectively eliminating specific cellular components. Deciphering the intricate roles of selective autophagy pathways in cellular quality control and stress responses necessitates a comprehensive understanding of the underlying molecular mechanisms.
Molecular mechanisms of selective autophagy
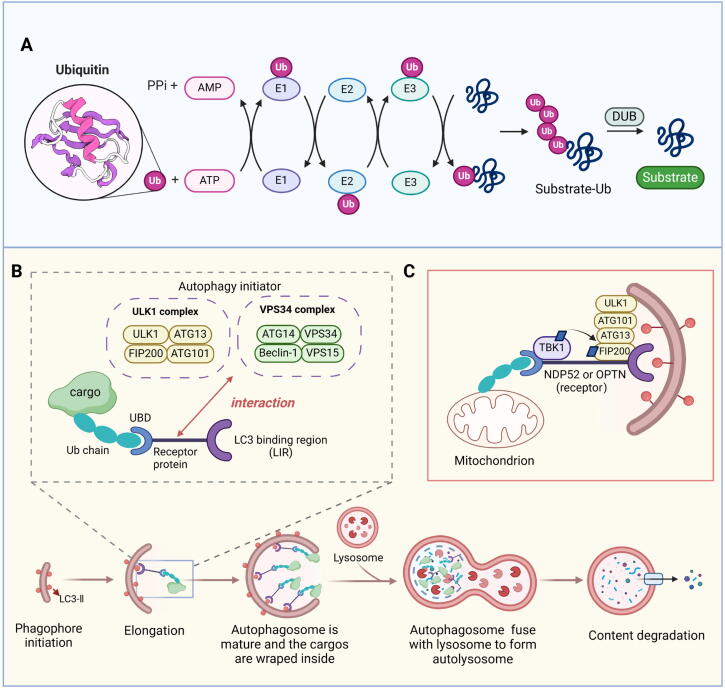
The ongoing investigation into the molecular mechanisms of selective autophagy remains a focal point of research, and this process requires the synergistic effect of receptors, chaperones, and autophagy proteins [100]. Current research suggests that the mechanisms underlying selective autophagy can be broadly dispersed into three specific processes: designation, targeting and sequestration, and degradation.
Designation
The determination of selective autophagy cargoes primarily relies on targeted ubiquitination. Ub, a small protein composed of 76 amino acids, is ubiquitously present in all tissues of eukaryotic organisms [101]. It functions as a modulator by covalently attaching to cellular proteins, facilitated by an enzymatic cascade involving three classes of enzymes referred to as E1 (activation), E2 (conjugation), and E3 (ligation) (Fig. 5A) [102]. The diversity of ubiquitin linkage patterns and proteins that interact with ubiquitin allows Ub to carry out various functions. Numerous studies have demonstrated that the targets of selective autophagy are usually ubiquitinated prior to their degradation [39]. With the identification of autophagy receptors, a clearer and more detailed selective autophagy mechanism is being unveiled.
Fig. 5.
An overview of the main molecular mechanisms in selective autophagy. (A) Ubiquitination plays an important role in the identification of selective autophagy cargo. (B) Selective autophagy induction via the recruitment of the autophagy initiation complex. (C) A typical example of parsimonious model of selective autophagy (mitophagy), involving Ub, autophagy receptor and autophagy initiation complex.
There have been many studies on how Ub is affixed to its substrates, including conjugation of a single ubiquitin monomer (monoubiquitination) or sequential conjugation of several ubiquitin moieties (polyubiquitination) of varying length [103]. The presence of a distinct ubiquitin-linkage type specific to autophagy has been a topic of prolonged debate [104]. Some studies suggested that maybe the monoubiquitination of proteins or organelles is sufficient for their specific encapsulation into autophagosomes, indicating that the linkage types may not significantly affect selectivity [105]. Importantly, aside from Ub systems, two ubiquitin-like systems (UBLs) are required for autophagy: the ATG5-ATG12-ATG16L system and the LC3 system. The two UBL conjugation systems are highly conserved and function during autophagosome formation during both selective and non-selective autophagy. Once a target protein is modified by ubiquitination, the monomer or polymer of ubiquitin linked to the target protein can be recognized and bound by various UBDs, protein domains capable of regulating the process of ubiquitination modification. The specific binding between UBD and Ub determines the specificity of ubiquitination substrate function [106]. Furthermore, current research has demonstrated that fluidity is the key determinant of selective autophagy. Recognition in selective autophagy necessitates that the condensates exhibit semi-liquid characteristics, and solid-like aggregates may not qualify as optimal autophagy cargoes [107].
Collectively, ubiquitin serves as a signal for cargo targeting and determines the formation position of autophagosome. In principle, the degradation agent inducing the proximity between ubiquitin ligase and its substrate can initiate the selective autophagy process and the degradation of substrate [108]. However, more efforts are needed to identify ligases suitable for this purpose. Understanding the relationship between the diversity of Ub chain and autophagy will empower researchers to identify ligases suitable for targeting, thus paving the way for developing selective degradation agents.
Targeting and sequestration
Recently, considerable progress has been made in the recognition of ubiquitin-dependent selective autophagy receptors [109], including SQSTM1/p62, NBR1, OPTN, and NDP52. These receptors possess the ability to simultaneously bind substrate and ubiquitin, initiating the pathway leading to autophagy and recruiting the autophagosomal membrane. They selectively interact with cargoes and guide them to the elongated autophagosomal membrane [39]. In mammals, autophagy receptors containing the LIR can usually bind to LC3 on the isolation membrane, and most autophagy receptors contain both UBDs and LIR [110]. In this way, ubiquitin substrates binding to autophagy receptors are labeled for selective autophagic degradation. Notably, these autophagy receptors participate in the removal of diverse substrates, including protein aggregates, organelles, and pathogens [111].
SQSTM1/p62 was the first selective autophagy receptor discovered. With its ubiquitin-binding motif, SQSTM1/p62 collects ubiquitinated protein aggregates or other cellular components into autophagosomes through specific binding with LC3, bringing about their degradation [112]. SQSTM1/p62 is renowned for its function of scavenging protein aggregates through aggrephagy. However, recent evidence has demonstrated its involvement as a receptor in other selective autophagy types as well, such as mitophagy [113] and lipophagy [112]. Following the identification of SQSTM1/p62 as an autophagy receptor, the evolutionarily related NBR1 emerged as the second mammalian autophagy receptor [114]. The list was subsequently expanded to include NDP52 [115], OPTN [116], TAX1BP1 [117], etc. These autophagy receptors play a crucial role in the selective autophagy of diverse intracellular components [111]. Intriguingly, different kinds of selective autophagy have distinctive representative receptors involved (Table 1) [118]. The receptor-mediated mechanism stands out as one of the earliest-established explanations for the selectivity observed in autophagy.
Table 1.
Receptors involved in mammalian selective autophagy.
| Pathway | Substrate | Mammalian autophagy receptors | Related diseases | Refs. |
|---|---|---|---|---|
| Ub-dependent mitophagy |
Mitochondria | SQSTM1/p62, OPTN, NBR1, NDP52, TAX1BP1 | Neurodegenerative diseases (particularly PD), amyotrophic lateral sclerosis, cancer, metabolic diseases, heart defects | [24] |
| Ub-independent mitophagy |
Mitochondria | NIX, BNIP3, FUNDC1, FKBP8, BCL2L13 | Neurodegenerative diseases, cancer, heart defects | [260] |
| ER-phagy | Endoplasmic reticulum |
FAM134B, SEC62, RTN3, BNIP3, TEX264, RTN3L, SQSTM1/p62, CCPG1 | Neurodegenerative diseases, cancer, renal diseases | [223] |
| Pexophagy |
Peroxisome | NBR1, SQSTM1/p62 | Peroxisomal disorders (for example, Zellweger syndrome) | [14] |
| Aggrephagy | Protein aggregates | SQSTM1/p62, NBR1, OPTN, TAX1BP1, TOLLIP, CCT2 |
Neurodegenerative diseases (such as HD) | [44] |
| Lipophagy |
Lipid droplets (LDs) |
SQSTM1/p62, ATGL, AIP4, ORP8, PLIN2/3, PNPL3/5/7/8, |
Liver diseases, obesity, cancer, atherosclerosis | [49] |
| Ribophagy | Ribosomes | NUFIP1 | May exacerbate disease- related protein dyshomeostasis | [66] |
| FA-phagy | Focal adhesions (FAs) | SQSTM1/p62, NBR1, c- Cbl, |
Cancer, vascular diseases (including intracranial aneurysms) |
[261] |
Another mechanism that contributes to the selectivity of cargo is the recruitment of specific autophagy initiation complexes, including the ATG1/UNC-52-like kinase 1 (ULK1) complex and the vesicular protein sorting 34 (VPS34) complex, which participate in the initiation of autophagy (Fig. 5B). In the absence of growth factors, acetylation of ULK1 in the FIP200-ATG13-ULK1 complex activates its kinase activity and promotes autophagy. On the contrary, in conditions of nutrient deficiency, reduced acetylation of components of the VPS34 complex, such as VPS34 and Beclin 1, due to acetyltransferase inactivation, leads to increased activity of the VPS34 complex and the initiation of autophagy [119]. The ULK1 complex can be assembled in ER tubulovesicular regions marked by ATG9 vesicle to initiate autophagy, emphasizing the potential for recruitment of autophagy initiation complexes to promote autophagy [120]. In addition, the ULK1 complex is attracted by damaged mitochondria via the receptor proteins OPTN and NDP52, contributing to a concise model of selective autophagy. This underscores the significance of coordinating ULK1 complex localization by autophagy receptors and TBK1 as pivotal factors driving the formation of targeted autophagosomes (Fig. 5C) [121].
Degradation
Similar to conventional autophagy, the degradation of selective autophagy occurs following the fusion of autophagosome and lysosome. Autophagosomes undergo fusion with lysosomes when they are fully mature, resulting in the formation of autolysosomes. Lysosome-related proteins involved in the maturation stage of autolysosomes include lysosomal associated membrane protein 1 (LAMP1), lysosomal associated membrane protein 2 (LAMP2), UV radiation resistance associated gene (UVRAG), and so on [122]. Finally, the membrane of autolysosome ruptures, and its contents are degraded by lysosomal hydrolase. Of note, amino acids and some proteins generated during the degradation process serve as a source of nutrition, energy, or are recycled to support cellular functions [123].
Collectively, the whole mechanism underlying selective autophagy is highly specific. Especially, the labeling of substrates and the recognition of autophagy receptors must be extremely selective to avoid the elimination of other normal cell components. With the in-depth study of selective autophagy, it is anticipated that additional receptors will be found, and their functions in specific cell conditions will be thoroughly analyzed. These advancements will hopefully provide us with new therapeutic targets and strategies to treat diseases caused by intracellular waste accumulation.
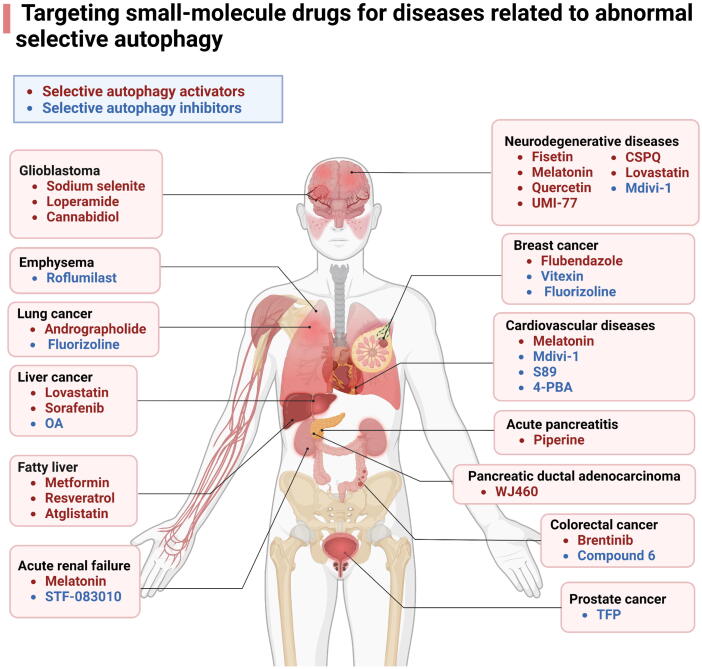
The roles of selective autophagy in human diseases
With the advancement of life sciences, selective autophagy, recognized as a pivotal mechanism in cellular self-regulation, has garnered substantial interest. Here, we provide a detailed discussion on the function of selective autophagy in various diseases and highlight its potential as a target for treatment (Table 2, Table 3). By intervening in the selective autophagy pathway, the regulation of key processes encompassing intracellular metabolism, immune response, and cell death may provide new ideas for disease treatment.
Table 2.
Selective autophagy and associated proteins involved in neurodegenerative diseases.
| Neurodegenerative diseases | Forms of selective autophagy involved | Key protein /pathway | Molecular mechanism | Refs. |
|---|---|---|---|---|
| AD | Aggrephagy | Tau, Aβ | The misfolded protein (Aβ and Tau) aggregates can be reduced by aggrephagy, which relieves the symptoms of AD | [125] |
| Mitophagy | PINK1 | Impaired PINK1 is observed in neuronal cells in AD patients | [129] | |
| Lysophagy | TRIM16 | TRIM16-mediated lysophagy suppresses high-glucose-accumulated neuronal Aβ | [85] | |
| PD | Mitophagy | PINK1, Parkin | Inhibiting mitophagy causes the deterioration of PD | [131] |
| ER-phagy | α-synuclein | Increased α-synuclein, can trigger ER stress, leading to ER-phagy in nerve cells and deteriorating PD |
[134] | |
| Aggrephagy | α-synuclein | α-synuclein accumulation can be reduced by aggrephagy | [139] | |
| HD | Aggrephagy | mHTT | mHTT clearance can be facilitated by aggrephagy receptors | [142] |
| Mitophagy | DRP1, SIRT3 |
mHTT interacts with Drp1 , leading to mitochondrial Abnormality; SIRT3 plays a neuroprotective role |
[143], [144] | |
| ALS | Mitophagy | OPTN, TBK1, SQSTM1 | Dysfunction of ALS-related proteins lead to abnormal quality control of mitochondria, accelerating neuronal death |
[147] |
| Aggrephagy | SOD1 |
SQSTM1/p62 interacted with ALS mutants of SOD1 to promote its degradation | [150] |
Table 3.
Cancers, metabolic diseases and cardiovascular diseases that are linked with selective autophagy.
| Diseases | Forms of selective autophagy | Key protein /pathway | References |
|---|---|---|---|
| Breast cancer | Mitophagy, ER-phagy | DRP1, CALCOO1 | [151] |
| CRC | Mitophagy, ER-phagy | Akt/mTOR pathway, TEX264, FAM134B, CALCOO1 | [152], [157] |
| Cervical cancer | Mitophagy, ER-phagy | Parkin, SEC62 | [36], [153]. |
| Ovarian cancer | Mitophagy | CRL4 | [155] |
| AML | Mitophagy | OPTN | [156]. |
| Prostate cancer | ER-phagy | SEC62 | [262] |
| NSCLC | ER-phagy | SEC62 | [36]. |
| ESCC | ER-phagy | FAM134B | [158] |
| HCC | ER-phagy | FAM134B | [159] |
| Pancreatic cancer | ER-phagy | FAM134B | [160] |
| Gastric cancer | Xenophagy | LC3 | [177] |
| Metabolic syndrome | Lipophagy | FGF21 | [184] |
| Obesity cardiomyopathy | Mitophagy, Lipophagy | DRP1, Parkin, FUNDC1 | [181], [188], [263] |
| Diabetes | ER-phagy, Lipophagy | PTP1B | [182] |
| NAFLD | Lipophagy | FGF21 | [184] |
Neurodegenerative diseases
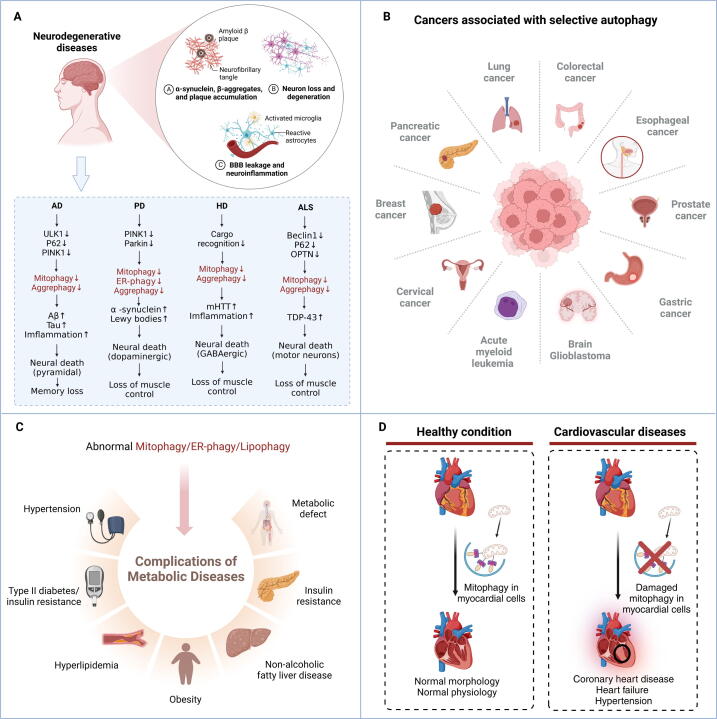
Neurodegenerative diseases, characterized by the degeneration of a large number of specific neurons, are a class of progressive, disabling, and even fatal complex diseases. An evident feature of neurodegenerative diseases is the abnormal accumulation of proteins, exerting toxic effects on neurons and ultimately leading to neuronal death and corresponding clinical symptoms [124]. There is a significant association between autophagy and neurodegenerative diseases. The modulation of selective autophagy presents promising research and application prospects for intervening in neurodegenerative diseases. In this section, we will delve into the involvement of selective autophagy in several major neurodegenerative diseases, including Alzheimer's disease (AD), Parkinson's disease (PD), HD, and Amyotrophic lateral sclerosis (ALS) (Fig. 6A).
Fig. 6.
The diseases associated with abnormal selective autophagy. (A) Abnormal selective autophagy in neurodegenerative diseases. (B) An overview of cancers that are linked with abnormal selective autophagy. (C) Metabolic diseases that are connected with abnormal selective autophagy. (D) The impact of damaged mitophagy on cardiovascular diseases.
AD is a degenerative disorder affecting the central nervous system, predominantly manifesting in the elderly and those approaching old age. Pathologically, it is identified by the presence of misfolded protein aggregates within the brain, such as amyloid β (Aβ) and hyperphosphorylated Tau (p-Tau) protein deposition, which causes progressive dementia [125]. The pathological proteins associated with AD, namely Aβ and p-Tau, can become aggregate. Under normal physiological conditions, the misfolded proteins containing the KFERQ motif are recognized by HSC70 and subsequently recruited directly into lysosomes via LAMP2a for degradation through CMA. The ubiquitinated misfolded proteins undergo degradation within the proteasome in UPS. Nevertheless, in AD, the conventional proteolytic pathways are compromised due to their vulnerability to AD aggregates, resulting in obstruction. Aggrephagy emerges as the only viable pathway to eliminate misfolded protein aggregates [126]. In aggrephagy, ubiquitinated aggregates are identified by the aggrephagy receptors SQSTM1/p62, OPTN, and NBR1, leading to their recruitment into LC3-containing autophagosomes. Subsequently, these autophagosomes fuse with lysosomes, forming autolysosomes where the AD aggregates undergo degradation [127]. Therefore, eliminating misfolded protein aggregates by modulating aggrephagy with small-molecule drugs represents an effective approach for preventing and treating AD.
The accumulation of damaged mitochondria in the brain is a hallmark of neurodegenerative diseases, with AD being one of the most prominent examples. Impairment of mitophagy leads to notable disruptions in mitochondrial transport and dynamics within neurons, exacerbating the pathological changes observed in AD [128]. Therefore, enhancing mitophagy can inhibit the aggregation of Aβ and microtubule-associated proteins and reverse the cognitive defects in AD model. Some regulatory factors of mitophagy pathway, such as PINK1, ULK1, MCL-1, phosphatidylinositol-binding clathrin assembly protein (PICALM), presenilin 1 (PS1), and Bcl2-associated athanogene 3 (BAG-3), are found to be lowly expressed or impaired in AD patients [129]. Restoration of these genes and/or drugs targeting mitophagy has shown promise in inhibiting disease progression in preclinical AD models. Given the ongoing challenges in developing anti-AD drugs, approaches targeting broader aspects of AD pathology, such as defective mitophagy, may hold therapeutic potential. Recent research has demonstrated that high-glucose-accumulated neuronal Aβ was suppressed by TRIM16-mediated lysophagy, implicating that other types of selective autophagy may also fulfill a crucial function in AD [85]. Therefore, further exploration of the relationship between selective autophagy and AD is warranted.
PD is a progressive neurodegenerative disorder with pathological features involving the depletion of dopaminergic neurons in the substantia nigra pars compacta (SNc). It was found that mitophagy in mouse neurons only occurred in non-dopaminergic neurons (tyrosine hydroxylase (TH) negative), suggesting that dopaminergic neurons in SNc may lead to the deterioration of PD by inhibiting mitophagy [130]. Mutations in PINK1 and Parkin genes can induce PD by causing impairments in the specific elimination of damaged mitochondria, leading to their accumulation [131]. Current therapeutic strategies primarily focus on blocking mitochondrial USP30, which antagonizes Parkin by removing ubiquitin from the mitochondrial surface. This intervention enhances mitochondrial ubiquitination and facilitates the recruitment of cargo receptors, ultimately reinstating the degradation of impaired mitochondria [132].
Increasing research has linked abnormal ER-phagy to a variety of neurodegenerative diseases, including PD [133]. Both increased α-synuclein, an abnormally folded protein, and decreased dopaminergic neurons in PD can trigger ER stress [134]. ER stress can normally facilitate the correct folding of unfolded and misfolded proteins through the UPR. However, excessive ER stress can lead to ER damage, causing autophagy and apoptosis in nerve cells [135]. Autophagy, including selective autophagy, has been demonstrated to be involved in the pathology of PD in cellular and animal models [136]. The abnormal folding of α-synuclein, which is unable to be degraded by the proteasome, accumulates in the ER, triggering ER stress and autophagy to remove the injured ER [137]. Notably, inhibition of ER stress has been shown to play a neuroprotective role in PD [138]. In summary, in the development of neurodegenerative diseases, ER-phagy may act as a neuroprotective mechanism to remove the damaged ER to maintain the homeostasis of nerve cells and prevent the occurrence of lesions. Similarly, PD is characterized by the accumulation of α-synuclein in cells, indicating that eliminating these abnormally aggregated proteins by regulating aggrephagy could also be a reliable and potential treatment for neurodegenerative diseases [139].
As a rare autosomal dominant genetic disease, HD is also known as progressive chorea. Patients generally develop symptoms in middle age, mainly manifested as involuntary movement, cognitive impairment, intellectual retardation, and emotional disorders [140]. The pathogenesis is related to mutations in the Huntington (HTT) gene that encodes HTT protein. Compared to normal HTT, mutant HTT (mHTT) proteins are more likely to form aggregates and introduce other proteins into the cell, thereby affecting the normal function of the cell [141]. Following the initial identification of SQSTM1/p62 as a selective autophagy receptor capable of aiding in the removal of mHTT aggregates [142], numerous receptors for aggrephagy have been identified, with OPTN, TPLLIP, and TAX1BP1 documented as mediators of mHTT aggregate degradation [41], [43], making the regulation of aggrephagy a potential therapeutic direction. Of note, through its interaction with mitochondrial dynamin-related protein 1 (Drp1), mHTT increases its activity, which causes aberrant distribution and excessive mitochondrial fragmentation [143]. Mitochondrial Sirtuin 3 (SIRT3) has been found to play a neuroprotective role in HD, suggesting a correlation between mitochondrial abnormalities and HD [144]. Recent investigations have indicated that the onset of HD is triggered by the inability to effectively clear dead or dying mitochondria [145]. These examples illustrate the association between impaired mitophagy and the development of HD.
ALS is a devastating neuromuscular disorder marked by the progressive degeneration of motor neurons in the spinal cord and brain. The degeneration of these motor neurons results in neuromuscular denervation, sporadic skeletal muscle wasting, and, ultimately, paralysis and fatality [146]. Existing treatment strategies for ALS are relatively limited in effectiveness, and there is currently no cure for this deadly disease. Several genes associated with ALS have been found to be involved in autophagy, including OPTN, TBK1, and SQSTM1/p62. They are related to autophagy to different extent, particularly the clearance of damaged mitochondria and protein aggregates. For example, dysfunction of ALS-related proteins OPTN and TBK1 will lead to abnormal quality control of mitochondria, and the damaged mitochondria cannot be removed in time, which will accelerate neuronal death [147]. Current research suggests that autophagy induction can serve as a treatment strategy for most neurodegenerative diseases. However, extensive autophagy is harmful for maintaining intracellular homeostasis. Therefore, more precise targeted selective autophagy, such as mitophagy, has the potential to become an effective and less toxic treatment approach.
Studies have shown that the dysfunction of autophagy receptors in aggrephagy may contribute to the pathogenesis of ALS [148]. The mutations in the gene encoding Cu/Zn superoxide dismutase 1 (SOD1) are responsible for the neuropathological manifestations observed in certain cases of familial ALS. There are several studies that indicate the interaction of SQSTM1/p62 with ALS mutants of SOD1, with the ubiquitin-association domain of SQSTM1/p62 being essential for this interaction [149]. Accordingly, manipulating the associated autophagy pathway has been contemplated as a therapeutic strategy for addressing this ailment. In the treatment of ALS, these cargoes, including protein aggregates, are targeted toward undergoing selective autophagy to degradation [148]. More recently, tripartite motif containing 44 (TRIM44) has been identified as a link between the UPS and SQSTM1/p62-dependent aggrephagy, facilitating the removal of misfolded proteins. Also, investigating the interplay between these two degradation pathways might uncover novel mechanisms for addressing diseases associated with aggrephagy, including neurodegenerative conditions and cancers [150].
Cancers
Cancer stands as a leading global cause of mortality, and accumulating evidence establishes a significant association between the development of numerous cancers and selective autophagy (Fig. 6B). Similar to non-selective autophagy, mitophagy typically assumes a dual role, acting as a double-edged sword in various cancers. In most cancer cells, the activation of mitophagy can effectively inhibit the proliferation of cancer cells and thus block the occurrence and progression of malignant tumors. For instance, flubendazole can induce Drp1-mediated mitophagy in breast cancer cells [151]. Methanol extracted from the immature fruit of Poncirus trifoliata can promote apoptosis of colorectal cancer (CRC) cells by inducing mitophagy [152]. Moreover, research has found that inhibiting histone deacetylase (HDAC) can activate mitophagy by mediating the acetylation of Parkin, thereby inhibiting the proliferation of cervical cancer cells [153]. In addition, drug resistance has always been one of the challenges in clinical cancer therapy. Fortunately, mitophagy can inhibit the metabolic adaptation in cancer cells, providing new strategies to combat the drug resistance of cancer [154]. Moreover, targeting Cullin RING E3 ubiquitin ligase 4 (CRL4) inhibits the growth of chemotherapy-resistant ovarian cancer (OC) through inducing mitophagy, demonstrating activation of mitophagy is a promising therapeutic approach to overcome OC chemotherapy resistance [155]. However, the induction of mitophagy may also promote cancer progression in some other cancer cells. For instance, it has been found that inhibiting the mitophagy receptor OPTN may be an effective treatment for malignant tumors, such as acute myeloid leukemia (AML) [156]. Currently, there is a dearth of comprehensive and rigorous investigations into the dual nature of mitophagy in cancer, and resolving this issue remains a focus for future research.
In the progressive stage of malignant tumors, autophagy can help cancer cells fight against nutritional deficiency and hypoxia, thus promoting tumor metastasis [18]. For example, the expression of SEC62 is upregulated in prostate cancer, non-small cell lung cancer (NSCLC), cervical cancer, and other cancers, and the ER-phagy mediated by SEC62 makes tumor cells show stronger tolerance, drug resistance, and migration ability, indicating that SEC62 can promote the formation of malignant tumors [36]. However, it has been observed that ER-phagy can also induce cancer cell death. Mutated FAM134B acts as a “double-edged sword”, exerting a tumor suppressor function in CRC [157]. Moreover, the mutation or overexpression of FAM134B is also related to the occurrence of other tumors, encompassing esophageal squamous cell carcinoma (ESCC) [158], hepatocellular carcinoma (HCC) [159], and pancreatic cancer [160]. Similar to FAM134B, TEX264 was found as a marker protein in CRC cells [161], while the CALCOCO1 expression is probably upregulated in CRC and breast cancer [162]. Moreover, the knockdown of endoplasmic reticulum-Golgi intermediate-compartment 3 (ERGIC3) suppresses lung cancer via ER-phagy [163]. In conclusion, ER-phagy exhibits an extremely intricate dual role in the context of cancer therapy, and modulating its activity, either inhibiting or activating it, holds promise as a novel approach for the treatment of malignant tumors.
Research has unveiled that pexophagy can be induced by hypoxia, while hypoxia is pervasive in malignant tumors [164]. Hence, we posit that pexophagy is intricately linked to the onset and progression of cancer. It was found that the upregulation of PEX2 expression was observed in liver cancer tissues, and conversely, silencing PEX2 expression markedly suppressed the proliferation of liver cancer cells. As expected, knocking down PEX10 or PEX12 also got the same result [165]. Furthermore, inhibition of ATM expression also suppresses the proliferation, migration, and invasion of CRC cells [166]. The altered expression of these pexophagy-related molecules in tumors probably means that tumor occurrence and growth are significantly influenced by pexophagy.
While nucleophagy has not received as much attention as general autophagy, abnormalities in various nuclear components have been demonstrated to be closely linked to cancer [167]. The interaction between LC3 and lamin B1 in nucleophagy leads to the downregulation of lamin B1 during carcinogenic damage. Disrupting LC3-lamin B1 interaction or autophagy can prevent the loss of lamin B1 and alleviate oncogene-induced aging within human primary cells. Thus, selective autophagy of the nucleus serves as a protective mechanism, safeguarding cells from the initiation of tumorigenesis [168]. Genetic changes, such as chromatin deletions and trans-localizations, have the potential to induce uncontrolled cell proliferation and evade the immune system, transforming healthy cells into malignant ones [169]. Furthermore, alterations in the composition of the nuclear envelope are crucial for cancer, since many components of the nuclear envelope play crucial roles in cellular functions that impact carcinogenesis and tumor growth [170]. Therefore, it is imperative to explore the impact of degrading nuclear components through nucleophagy on the initiation and advancement of cancer. Recently, it has been found that in cancer-associated fibroblasts, ribophagy mediated by NUFIP1 promotes the growth of pancreatic cancer cells by secreting nucleosides, which brings new inspiration for the treatment of pancreatic cancer [171].
For a long time, it has been known that bacterial effectors interact with and activate host oncoproteins, leading to cell cycle disruption and, ultimately, carcinogenesis [172]. Accordingly, xenophagy can operate as a shield, impeding tumor growth by eliminating bacterial infection [173]. One significant factor that increases the risk of gastric cancer is the infection of Helicobacter pylori. It may promote the occurrence of gastric cancer through a variety of mechanisms, including chronic inflammation, production of carcinogens, influence of host cell signaling pathway, and even interference with the immune system [174]. Research has shown that xenophagy within cells is notably inhibited by the highly pathogenic Helicobacter pylori strain GC026, suggesting that damaged xenophagy may contribute to the carcinogenesis of gastric cancer [175]. Except for Helicobacter pylori, another common bacterium that accumulates in malignant tumor lesions is Salmonella [176]. As Salmonella accumulates in malignant lesions, cancer cells induce a heightened level of xenophagy via LC3 processing to eliminate the bacteria [177]. In a nutshell, targeting xenophagy is an anti-cancer strategy with great potential.
Notably, FAs are significantly associated with the invasion and metastasis of tumor cells. To illustrate, a reduction in FAs typically correlates with heightened cellular migration and the progression of cancer metastasis. As the central protein of FA, focal adhesion kinase (FAK) promotes the turnover and cell migration of FA, and FAK inhibitors were found to have anti-breast cancer effects due to their stability to FAs [178]. Moreover, since FAs are crucial for maintaining the integrity of blood vessels, their deficiency can result in bleeding, a commonly observed phenomenon in various vascular disorders, including intracranial aneurysms [179]. Additionally, inhibiting FA-phagy, thereby stabilizing FAs, has shown promising effects in mitigating breast cancer metastasis [92]. These findings indicate that targeting FA-phagy is a prospective cancer therapeutic strategy.
In general, the prospect of targeting selective autophagy for cancer therapy is promising, offering a new avenue for exploring treatment approaches that are both more effective and tailored to individual needs. Ongoing exploration in this field is expected to drive advancements in cancer treatment, ultimately leading to improved therapeutic outcomes for patients.
Metabolic diseases
Metabolic diseases such as diabetes and obesity are often accompanied by intracellular energy imbalances, and selective autophagy helps maintain cellular health by removing excessive or damaged organelles [180] (Fig. 6C). This process has been fully embodied in mitophagy, where selective autophagy helps maintain the energy production efficiency of cells by removing damaged mitochondria [19]. Studies have shown that Drp1 is essential for mediated mitophagy in high-obesity cardiomyopathy, making it a promising target to relieve obesity cardiomyopathy [181].
The protein-tyrosine phosphatase 1B (PTP1B) is recognized to be located in the ER and participates in the negative regulation of islet signal transduction. It has been proven that ER stress upregulates PTP1B and impairs the glucose uptake function of cells [182]. Nonetheless, the intervention with ER-phagy can effectively remove the damaged ER and aggregated protein, reduce the occurrence of endoplasmic reticulum stress, and thus maintain the glucose uptake function of cells. Accordingly, regulating ER-phagy may become a new method to treat insulin resistance-related diseases, such as metabolic syndrome and type 2 diabetes.
When lipophagy is impaired, excessive storage of LDs has the potential to lead to various diseases, such as non-alcoholic fatty liver disease (NAFLD), obesity, and other metabolic diseases. Dysregulation of lipophagy appears to be a contributing factor in conditions such as fatty liver diseases (FLDs), presenting a significant risk factor for HCC development. Dysfunctional lipophagy has also been linked to the progression of disease in individuals with NAFLD [183]. In addition, nonalcoholic steatohepatitis (NASH), an advanced and more severe stage of NAFLD that poses a growing threat to global human health, is also closely related to the accumulation of LDs in hepatocytes [184]. The buildup of LDs in hepatocytes is a consequence of disrupted lipid metabolism, closely linked to a metabolic syndrome characterized by obesity, insulin resistance, dyslipidemia, and hypertension [185]. It has been found that targeting lipophagy can effectively prevent abnormal lipid metabolism. Accordingly, lipophagy modulators, such as fibroblast growth factor 21 (FGF21) [186], are regarded as potential targets for future rational therapies aimed at addressing NASH through the manipulation of lipophagy. Even though inducing lipophagy to improve these liver diseases is an appealing hypothesis, it necessitates further investigation because potential therapeutic advantages may be attributed to the modulation of additional pathways, as none of these strategies exclusively focus on lipophagy.
Cardiovascular diseases
Major cardiovascular diseases, including coronary heart disease, heart failure, and hypertension, pose significant global public health challenges. And their in-depth molecular mechanism research and the development of innovative treatment strategies are driving cutting-edge science in the cardiovascular field [112]. It is worth noting that these diseases can often be linked to abnormal selective autophagy, especially mitophagy (Fig. 6D).
Recent investigations have revealed a robust association between mitophagy and the occurrence of cardiovascular diseases [187]. The absence of FUNDC1, a receptor protein that mediates mitophagy, exacerbates myocardial remodeling, decreased myocardial function, mitochondrial abnormalities, and cell death caused by HFD. At the same time, the levels of IP3R3 increase and intracellular calcium significantly overload in myocardial cells, indicating that FUNDC1 and its interacting proteins can serve as prevention and treatment targets for obesity cardiomyopathy, which provides important scientific ideas for the development of related drugs for obesity cardiomyopathy [188]. The latest research reveals that nitric oxide (NO) promotes mitophagy mediated by MCM8 and E3 ubiquitin ligase TRIM21, thus maintaining normal coronary artery function and cardiovascular homeostasis [189]. This study not only reveals a novel mitophagy mechanism that does not depend on PINK1/Parkin and Drp1 but also provides important targets for the treatment of vasculitis diseases. In summary, in-depth research on the relationship between selective autophagy and cardiovascular diseases provides important scientific ideas for drug development related to cardiovascular diseases and is anticipated to pave the way for their clinical treatment and prognosis.
Other human diseases
Nephrin, a transmembrane protein located on the glomerular hiatal membrane, undergoes processing and modification in the ER before translocating to the cell membrane. It is essential for preserving glomerular selective permeability and normal function [190]. Mutations in nephrin can result in the occurrence of proteinuria or congenital nephrotic syndrome [190]. Missense mutants of nephrin in humans give rise to impaired glycosylation and enhanced binding of the mutants to ER chaperones and calnexin [191]. Furthermore, nephrin mutants accumulate in the ER, which activates the activating transcription factor-6 (ATF6) signaling pathway of the UPR and enhances the expression of ER chaperone [191]. Simultaneously, nephrin mutants enhance the ubiquitination of cells, thereby initiating ER-phagy and reducing the number of mutants entering the plasma membrane [192]. In renal diseases such as proteinuria or congenital nephrotic syndrome, enhancing ER-phagy may effectively remove misfolded and aggregated proteins and damaged ER in pathological cells, offering a potential treatment strategy for these diseases. Consequently, ER-phagy may emerge as a novel therapeutic approach for renal diseases. Additionally, FAM134B mutations can cause hereditary sensory and autonomic neuropathy type II (HSANII), an autosomal recessive genetic disorder characterized by impaired pain perception and ulceration of hands and feet [193]. It is implied that nerve cells are likely to be extremely sensitive to ER-phagy, and this disease progression may be effectively halted by targeted regulation of ER-phagy levels [194].
Peroxisome biogenesis disorder (PBD) is the most typical disease linked to inordinate pexophagy. It was previously believed that it is the mutation of the PEX gene involved in peroxisome biogenesis that makes the process of peroxisome biogenesis unable to proceed normally, leading to the deletion of mature peroxisomes and finally resulting in PBD. However, this view has changed recently because it has been discovered that the most common genes mutated in PBD are three genes that make up the AAA ATPase complex (ATPases associated with diverse cell activities), namely PEX1, PEX6, and PEX26, accounting for 48.5 %, 13.1 %, and 3.4 %, respectively. The AAA ATPase complex is responsible for detaching ubiquitinated PEX5 from the membrane of peroxisome, thus preventing the autophagy process of peroxisome, which is then preserved. However, the mutant complex loses this ability, and the peroxisome will continue the autophagy process, leading to a reduction in the quantity of peroxisomes [195]. Therefore, it is now believed that at least 65 % of PBD is related to excessive pexophagy [196]. This discovery provides a new therapeutic direction for PBD, namely, inhibiting pexophagy by targeting drugs to increase the number of peroxisomes. Correspondingly, the number of peroxisomes in fibroblasts of patients with PEX1 deletion (PEX1 null) or mutation (PEX1-G843D) increased significantly after being stimulated by three different autophagy inhibitors (LY294002, bafilomycin A1, or chloroquine). The number of peroxisomes in PEX1-G843D cells even reached the level observed in wild-type cells [195]. Consequently, targeted inhibition of pexophagy is expected to emerge as a new therapeutic target for PBD.
Overall, approaches targeting selective autophagy for therapeutic interventions hold promise for alleviating these disorders. Currently, there is a thorough effort in the in-depth investigation of small-molecule drugs targeting selective autophagy for underlying therapeutic applications. Despite persisting unresolved challenges in the selective autophagy domain, the targeting of this process through small-molecule drugs has demonstrated considerable promise as a viable treatment modality for diseases.
Small-molecule drugs targeting selective autophagy
Up until now, numerous compounds with therapeutic promise have been documented in scientific literature and patents as potential regulators of the autophagy process. Nonetheless, a significant proportion of autophagy modulators are derived from natural products or compounds originally intended for the targeting of diverse proteins or pathways. And the precise mechanisms by which these compounds interact with autophagic targets to induce or inhibit autophagy remain incompletely elucidated. As autophagy is a complex process, and given the involvement of multiple pathways in signal transduction, it is noteworthy that autophagy modulators lacking specificity may exert effects on multiple targets. Pharmacologically targeting non-selective autophagy can yield both beneficial and detrimental therapeutic effects, as it serves as a modulator across various cellular events. Targeting selective autophagy, on the other hand, might minimize adverse drug reactions. Despite considerable advancements in exploring selective autophagy, the pace of developing small-molecule modulators tailored to various types of selective autophagy is still slow, with the current species of such modulators being extremely rare. Due to the absence of specific and powerful autophagy modulators, our comprehension of the correlation between autophagy and diseases remains constrained. Therefore, it is attractive and imperative to find new selective modulators that target autophagy-related proteins. In addition, it is expected that selective autophagy modulators will be applied to clinical treatment, aiming for maximum clinical benefit with minimal adverse effects in the future. Consequently, the quest for new selective autophagy modulators has emerged as a focal point of research. Here, we present a comprehensive summary of the recent advancements in selective autophagy modulators, which are divided into activators and inhibitors (Fig. 7). Also, the challenges of targeting selective autophagy with small-molecule drugs are discussed in this part.
Fig. 7.
Small-molecule drugs targeting selective autophagy for treating related diseases.
Small-molecule activators of selective autophagy
With the increase of age, the autophagy function of cells steadily weakens, contributing to a shortened lifespan and establishing a detrimental cycle. Hence, augmenting autophagy levels has emerged as a pivotal area of investigation. Autophagy presents a crucial role in aging-related diseases, such as neurodegenerative diseases, cardiovascular diseases, immune diseases, inflammatory diseases, and even cancers [197]. Various mouse models have underscored the intimate association between disease occurrence and the downregulation or defects in autophagy. Conversely, drugs that promote autophagy can effectively improve or delay diseases in animal models, especially neurodegenerative diseases accompanied by the misfolding of proteins and abnormal protein accumulation [197]. At present, numerous investigations have explored small-molecule activators targeting macroautophagy, but there are still few studies on selective autophagy activators with high specificity, such as the development of specific protein autophagy activators, ER-phagy activators, mitophagy activators, and so on. For example, rapamycin, recognized as the most widely employed autophagy activator, exerts its effects by inhibiting the mTOR pathway, consequently inducing extensive non-selective autophagy. It has also been proven that rapamycin can prolong the life span of female Drosophila and mice by promoting autophagy and improve related aging problems at the same time [198]. Investigations into non-selective autophagy activators suggest that promoting selective autophagy holds promise for treating related diseases. In this part, the small-molecule activators of selective autophagy found in current research were clearly introduced and summarized (Table 4).
Table 4.
Small-molecule activators of selective autophagy.
| Names | Chemical structure | Selective autophagy types of action | Targets | Indications | Ref. |
|---|---|---|---|---|---|
| Fisetin | 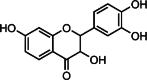 |
Mitophagy | PINK1 | Neuroinflammation, cognitive impairment | [199], [264] |
| Melatonin | 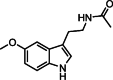 |
Mitophagy | Mcoln1, OPA1, Sirt3 |
AD, ischemia–reperfusion injury, atherosclerosis, sepsis-induced acute kidney injury | [200], [201], [203] |
| Quercetin | 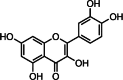 |
Mitophagy | NLRP3 | Inflammation, neuronal damage |
[204] |
| UMI-77 |  |
Mitophagy | MCL-1, |
AD | [205] |
| Lovastatin |  |
Mitophagy | SHP2, GOLM1 |
PD, HCC | [209] |
| Andrographolide |  |
Mitophagy | Parkin | PD | [210] |
| Flubendazole |  |
Mitophagy | EVA1A | Breast cancer | [151] |
| WJ460 |  |
Mitophagy | Myoferlin | PDAC | [212] |
| Sorafenib |  |
ER-phagy | FAM134B | HCC | [213] |
| Flufenamic acid |  |
Lipophagy | AKR1C3 | HCC | [214] |
| Brentinib |  |
ER-phagy | FAM134B, ORP8/USP5 | CRC | [215]. |
| Sodium selenite | 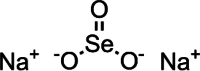 |
Mitophagy | Damaged mitochondrial | Glioblastoma | [216] |
| Loperamide | 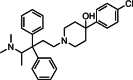 |
ER-phagy | ATF4 | Glioblastoma | [217] |
| Cannabidiol | 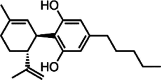 |
Mitophagy | TRPV4 | Glioblastoma | [218] |
| Metformin | 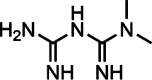 |
Lipophagy | SIRT1 | Hepatosteatosis, NAFLD | [219] |
| Resveratrol | 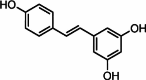 |
Lipophagy | SIRT1 | NAFLD | [221] |
| Atglistatin | 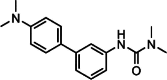 |
Lipophagy | ATGL | Chronic inflammation and metabolic abnormalities induced by HFD | [222] |
| Piperine | 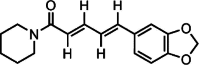 |
ER-phagy | FAM134B, CCPG1 | Acute pancreatitis | [223] |
Some natural or synthetic compounds have demonstrated the capacity to regulate mitophagy. For instance, fisetin, the active component of the traditional Chinese medicine Cotinus coggygria, blocked NLRP3 inflammasome activation via promoting mitophagy through the PINK1-Parkin pathway, which can effectively ameliorate cognitive impairment [199]. Melatonin has been proven to produce a variety of beneficial effects based on promoting mitophagy, including the amelioration of cognitive deficiencies in AD [200], the reduction of cardiac ischemia–reperfusion injury [201], and the prevention of atherosclerosis development [202]. More recently, melatonin has emerged as a potent mitigator of sepsis-induced acute kidney injury by augmenting mitophagy via SIRT3-mediated mitochondrial transcription factor A (TFAM) deacetylation, revealing a new mechanism of melatonin as a mitophagy activator that was previously overlooked [203]. Additionally, quercetin, a natural flavonoid, has been shown to prevent neuronal damage by promoting mitophagy and suppressing the activation of NLRP3 inflammatory corpuscles mediated by mitochondrial ROS in microglia, offering a potential innovative therapeutic approach for diseases related to neuroinflammation [204]. For small-molecule synthetic compounds, targeting the MCL-1 protein to stimulate mitophagy is a promising method to treat AD. In a comprehensive screening encompassing over 2,000 FDA-approved drugs or candidate drugs, researchers identified a small-molecule compound named UMI-77 capable of safely and effectively inducing mitophagy. Importantly, UMI-77 has been proven to be an effective selective mitophagy activator, which can significantly reduce the level of inflammatory factors in mouse brain and clear the Aβ deposition, reversing the pathological features of AD [205]. Generally, research concerning modulators of mitophagy is relatively extensive among various forms of selective autophagy. Numerous activators have been reported in previous studies, such as pifithrin-α, resveratrol, rotenone, ST-539, etc. [19], [206], [207]. However, there remains a paucity of studies on specific modulators of other types of selective autophagy, necessitating further efforts and development. Besides AD, another neurodegenerative disease garnering significant attention is PD. And the misfolding and aggregation of α-synuclein represent crucial pathogenic mechanisms in PD. A research team found that the Cu2-xSe antioxidative nanoparticles facilitate the selective degradation of α-synuclein aggregates in neurons, leading to improved motor and memory abilities in mice with PD [208]. This finding reveals the significance and innovativeness of regulating the selective autophagy of α-synuclein in neurons using nanoparticles for PD therapy, paving the way for related drug discovery. Additionally, triggering Src homology phosphotyrosyl phosphatase 2 (SHP2)-mediated mitophagy by repurposing lovastatin can reduce the damage to dopaminergic neurons and improve MPTP-induced Parkinson-like symptoms in mice, providing a novel therapeutic approach for delaying the progress of PD [209]. Recent research has demonstrated that andrographolide reduces the activation of NLRP3 inflammatory corpuscles in microglia and saves the loss of dopaminergic neurons in PD. Specifically, this natural product may promote mitophagy through Parkin-dependent pathway [210]. In summary, these examples underscore the promise of small-molecule selective autophagy activators in the treatment of neurodegenerative diseases.
In cancer, tumor metabolism profoundly influences the proliferation, growth, migration, invasion, and other key biological processes of tumor cells. In recent years, cholesterol metabolism has emerged as a focal point in tumor metabolism research. Cholesterol can inhibit autophagic degradation of receptor tyrosine kinases (RTKs) by suppressing selective autophagy, which is mediated by Golgi membrane protein 1 (GOLM1) through the mTOR pathway. Therefore, the use of lovastatin can indirectly promote autophagy, enhancing the efficacy of tyrosine kinase inhibitors (TKIs) in HCC cells and impeding tumor growth [211]. Flubendazole, a widely-used class of broad-spectrum anthelmintic drugs, has been shown to promote mitochondrial dysfunction and initiate Drp1-mediated mitophagy by targeting EVA1A in breast cancer cells. Flubendazole-induced excessive mitophagy leads to mitochondrial damage and cellular dysfunction, thereby inhibiting the proliferation and migration of breast cancer cells [151]. Furthermore, a recently identified compound, WJ460, which triggered mitophagy through targeting the membrane-anchored protein myoferlin, was demonstrated to promote mitophagy and thus initiate ferroptosis in pancreatic ductal adenocarcinoma (PDAC) cells [212]. Sorafenib (a ferroptosis inducer) has been proven to be effective in activating ER-phagy mediated by receptor protein FAM134B, which significantly enhances the sensitivity of HCC cells to ferroptosis without affecting macroautophagy [213]. Recent discoveries have revealed that AKR1C3 inhibits lipophagy to facilitate the formation of lipid droplets. Therefore, the AKR1C3 inhibitor flufenamic acid can effectively promote lipophagy in HCC xenograft tumors, thus reducing the accumulation of LDs and sensitizing drug-resistant HCC cells to sorafenib [214]. In addition, an ALK inhibitor called brentinib has been identified to trigger FAM134B-mediated ER-phagy and induce ER stress by enhancing the interaction between ORP8 and ubiquitin-specific protease 5 (USP5), therefore exerting an inhibition effect on CRC cells [215]. This finding shed light on the prospect of discovering small-molecule compounds targeting ER-phagy for cancer treatment. Glioblastoma, the most common primary malignant brain tumor, is associated with low survival rates and restricted therapeutic modalities. Sodium selenite has garnered considerable research attention due to its ability to induce lethal mitophagy in human glioma cells, which brings about irreversible cell death in gliomas [216]. Exposure of glioblastoma cells to the anti-diarrhea drug loperamide induced ER stress, increased the expression of ATF4, and induced FAM134B and TEX264 mediated ER-phagy, ultimately culminating in autophagic cell death [217]. Recent studies have also proved that cannabidiol can inhibit human gliomas by activating TRPV4 to induce fatal mitophagy [218]. These discoveries offer a novel approach for glioma treatment. Besides, it is worthwhile to investigate whether the autophagy induced by these compounds is selective. Further research and clinical trials will help reveal the specific mechanism and effect of selective autophagy activators in cancer treatment. If these activators can be successfully used in cancer treatment, it will definitely provide patients with a brand-new therapeutic choice. However, additional research is imperative to assess their safety and efficacy, ensuring their feasibility and optimal efficacy in clinical applications.
Obesity and liver diseases are intimately linked to disturbances in lipid metabolism, highlighting the compromised functionality of lipophagy. Metformin mitigates hepatic lipid accumulation by reinstating SIRT1-mediated autophagy initiation, particularly lipophagy, through a pathway independent of AMP-activated protein kinase [219]. Resveratrol, a type of traditional Chinese herbal extract, has shown promise in numerous clinical trials for reducing steatosis primarily by enhancing autophagy, particularly lipophagy [220]. More recently, another study illustrated for the first time that the combination of metformin and resveratrol provides robust protective effects against hepatosteatosis via activating autophagy through the cAMP/AMPK/SIRT1 signaling pathway, thereby identifying a potential treatment direction for NAFLD [221]. The pharmacological inhibitor of ATGL, Atglistatin, in combination with CARD9 KO, was discovered to restore the imbalanced lipolysis and enhance the impaired lipophagy, providing protection against chronic inflammation and metabolic abnormalities induced by HFD [222].
In fact, in many other kinds of diseases, promoting selective autophagy also plays an important part. In terms of inflammatory diseases, piperine demonstrates efficacy in reducing ER stress by enhancing the ER-phagy dependent on FAM134B and CCPG1, thereby mitigating pancreatic injury and holding promise for acute pancreatitis treatment [223]. It has been reported that resveratrol can induce xenophagy, thereby enhancing the clearance of invasive bacteria in intestinal epithelial cells and macrophages and offering a potential treatment strategy for life-threatening infections by enhancing intracellular bacterial clearance [224]. Furthermore, as widely known, the COVID-19 pandemic, resulting from the infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019, has significantly affected global public health and economic activities. Notably, through inhibiting the SUMO modification of angiotensin converting enzyme 2 (ACE2), the selective autophagy mediated by cargo receptor TOLLIP can be enhanced to protect the body, thus reducing the infection of SARS-CoV-2 [225].
Collectively, selective autophagy activators have surfaced as a promising approach for relieving various diseases. However, these drugs designed to target selective autophagy often encounter challenges such as inadequate targeting and safety concerns, which limit their clinical efficacy. Unlike subcellular organs such as nucleus and mitochondria, ER is a transit point for intracellular substances. While the ER targeting system can significantly improve the accumulation of drugs in ER in a short time, the retention time remains insufficient, and a large part of drugs will still be excreted to the extracellular space. At present, there remains a dearth of research into extending the duration of drug accumulation within the ER following targeting. Therefore, it is necessary to integrate pharmaceutical methods such as targeting nanoparticles and monoclonal antibodies to improve the targeting of small-molecule drugs and increase their aggregation in target organelles. For instance, long-chain lipophilic molecules modified by naphthalene sulfonamide groups possess the ability to self-assemble into spherical nanoparticles capable of encapsulating the Bcl-2 inhibitor obatoclax. The drug delivery system inhibits Bcl-2 by targeting ER and promotes ER-phagy and mitochondrial damage, leading to apoptosis of human cervical cancer HeLa cells [226]. On the other hand, the development of selective autophagy inhibitors is equally important and urgent, as upregulated selective autophagy has also been linked to numerous pathophysiological conditions. Interestingly, enhancing autophagy strategy proves to be more effective in promoting ER-targeted anti-tumor and anti-metastatic therapies compared to inhibiting autophagy strategy, which may account for the phenomenon that the research and development efforts focused on small-molecule drugs targeting selective autophagy activation rather than inhibition [227].
Small-molecule inhibitors of selective autophagy
The downregulation of selective autophagy has the potential to modulate autophagic flux, leading to reduced clearance of toxic aggregates and damaged organelles, as well as improved cellular function and homeostasis. Furthermore, selective autophagy inhibition may represent a valuable therapeutic approach to target diseases associated with excessive autophagic flux, such as neurodegenerative disorders and cancer. Consequently, the development of selective autophagy inhibitors represents an innovative research frontier with profound implications for therapeutic intervention. While the number of selective autophagy modulators exhibiting high specificity remains limited, investigations into specific types of selective autophagy inhibitors are underway with vigor (Table 5). Prominent examples include the research on inhibitors targeting mitophagy, ER-phagy, lipophagy, etc.
Table 5.
Small-molecule inhibitors of selective autophagy.
| Names | Chemical structure | Selective autophagy types of action | Targets | Indications | Ref. |
|---|---|---|---|---|---|
| Mdivi-1 | 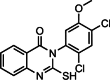 |
Mitophagy | Drp1 | Cardiovascular diseases, neurodegenerative diseases such as AD and PD. | [230], [265] |
| S89 |  |
Mitophagy | MFN1 | Cardiovascular diseases | [266] |
| Oroxylin A |  |
Mitophagy | PINK9 | HCC | [231] |
| Fluorizoline |  |
Mitophagy | PHB1, PHB2 | Breast cancer, lung cancer | [232] |
| Roflumilast |  |
Mitophagy | Drp1, PINK1 | Emphysema | [234] |
| Compound 6 | 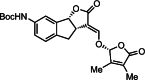 |
Mitophagy | Parkin | Colorectal cancer | [233] |
| Vitexin |  |
ER-phagy | FAM134B | Breast cancer | [235] |
| Trifluoperazine | 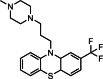 |
ER-phagy | SEC62 | Prostate cancer | [236] |
| STF-083010 | 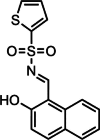 |
ER-phagy | IRE1 | Acute renal failure | [237] |
| 4-PBA |  |
ER-phagy | HDAC | Myocardial ischemia–reperfusion injury, ophthalmic diseases such as glaucoma |
[238] |
It is well known that mitochondrial division, facilitated by the interplay of the division proteins fission 1 (Fis1) and Drp1, is a prerequisite for mitophagy. Accordingly, mitochondrial division inhibitor 1 (Mdivi-1), an allosteric regulator of Drp1, stands out as a typical inhibitor of selective autophagy targeting mitochondria and hardly interferes with other types of selective autophagy [228]. Besides, the protective effect of Mdivi-1 on the heart has been confirmed in a wide range of cardiovascular diseases [229], and its therapeutic potential for neurodegenerative diseases has also been verified in several models of AD and PD [230]. In addition, oroxylin A, a novel CDK9 inhibitor from Scutellaria baicalensis, exhibits the capacity to inhibit PINK1-Parkin pathway-mediated mitophagy, thereby overcoming drug resistance in HCC and demonstrating potent therapeutic promise for this condition [231]. Mitophagy destruction emerges as a promising strategy for HCC treatment, and oroxylin A stands out as a potential candidate for developing novel mitophagy inhibitors [231]. A prohibitin-binding compound, fluorizoline, was shown to inhibit mitophagy induced by carbonyl cyanide m-chlorophenyl hydrazone (CCCP) and a combination of 1 μM oligomycin and 1 μM antimycin A through directly targeting PHB1 and PHB2 in both HeLa cells and A549 cells [232]. This discovery positions fluorizoline as a promising and innovative modulator of mitophagy, with potential applications as an anti-cancer agent. Compound 6, a newly discovered analogue of strigolactones, demonstrates potent and selective cytotoxicity against CRC cells while sparing normal cells. And compound 6 plays an anti-cancer role as an effective mitophagy inhibitor, selectively enhancing autophagy flux and impeding autophagy-lysosome fusion in HCT116 cells [233]. Additionally, roflumilast, a phosphodiesterase 4 inhibitor, has been identified as capable of suppressing cigarette smoke extract-induced mitophagy by inhibiting the expression of phosphorylated Drp1 and PINK1, thus alleviating smoking-associated lung diseases like emphysema [234].
An effective strategy to target ER-phagy involves focusing on autophagy receptors unique to ER, such as FAM134B. Vitexin has been identified as a pharmacological agent that can destroy the FAM134B-BiP complex, inhibit ER-phagy, and effectively suppress the progress of breast cancer in vivo [235]. Moreover, in tumor cells that overexpress SEC62, trifluoperazine treatment can downregulate the ER membrane protein SEC62 and prevent the occurrence of ER-phagy, thus inhibiting the potential for prostate cancer growth and metastasis [236]. In addition, STF-083010 is a specific inhibitor of inositol-requiring enzyme-1 (IRE1), which inhibits apoptosis induced by ER stress and probably induces ER-phagy, thus alleviating acute renal failure in rats [237]. Recent research indicates that 4-phenylbutyrate (4-PBA), a drug that has been extensively utilized in clinics, can inhibit ER-phagy and improve myocardial ischemia–reperfusion injury [238]. Moreover, evidence supports the efficacy of 4-PBA in the treatment of glaucoma and various other ophthalmic conditions [220]. Ginkgolide injections have been widely used in clinical practice, with Ginkgolide B and bilobalide accounting for 34 % and 48 %, respectively. Studies have demonstrated their capacity to effectively inhibit ER stress and selective autophagy, thus conferring protective effects against cerebral ischemia–reperfusion injury [217].
Nevertheless, there remains a scarcity of selective and highly specific inhibitors targeting pexophagy, aggrephagy, ribophagy, and other forms of selective autophagy, reflecting the nascent nature of this field and the ongoing research. Targeting these types of selective autophagy activators has research prospects and potential for treating diseases. And further endeavors are required to develop selective autophagy-targeted small molecular drugs to provide reference and improve the efficacy of treatments for various diseases.
Chloroquine (CQ) and hydroxychloroquine (HCQ) stand out as the sole compounds targeting autophagy that have entered clinical studies [239]. The mechanism of action is to cause lysosome deacidification, thereby hindering the fusion of the autophagosome with the lysosome and impeding the cargo degradation process. More than 50 anti-tumor clinical trials of CQ or HCQ have been initiated to date [240], [241]. Nevertheless, the issue of specificity of CQ inhibitors remains a challenge. At higher concentrations, they have been observed to induce DNA damage and impede angiogenesis. Even when an anti-tumor effect is evident, it may not solely result from autophagy inhibition. Furthermore, while CQ primarily targets the lysosome, its specificity is constrained as it encompasses many other functions beyond its implication in autophagy. In contrast, a novel inhibitor of lysosomal autophagy, lys05, which is a water-soluble analog of HCQ, shows better specificity, efficiently accumulates in lysosomes in tumor cells, and reduces lysosomal acidity, leading to sustained inhibition of autophagy and tumor growth [242]. The progression of clinical trials involving drugs targeting autophagy or selective autophagy holds the potential to expand treatment options for patients and deepen our understanding of the pathophysiological mechanisms underlying various diseases.
On balance, manipulating the selective autophagy pathway through small-molecule drugs has surfaced as a promising therapeutic approach for targeting various diseases, ranging from cancer to neurodegenerative disorders. Recent progress has marked significant strides in the development of selective autophagy modulators, yet substantial efforts are requisite to translate these advancements into clinical efficacy. The key challenge is to identify selective autophagy targets and develop potent and specific small-molecule drugs that can interfere with their activity. In this regard, high-throughput screening approaches, coupled with structural and chemical biology techniques, have provided powerful tools for the design, discovery, and optimization of selective autophagy modulators. Nevertheless, significant knowledge gaps persist regarding the interaction between the selective autophagy machinery and key substrates, particularly protein aggregates, leading to challenges in comprehending how substrates evade autophagic degradation. Addressing these gaps could facilitate the rational development of drugs aimed at addressing deficiencies in cargo removal. Presently, pioneering methodologies that strive to artificially guide protein aggregates towards autophagic degradation, such as AUTAC, ATTEC, and AUTOTAC, are undergoing investigation [243]. And some therapeutic intervention may be of great significance before massive aggregations of pathogenic proteins have occurred. In summary, the discovery of novel targets for selective autophagy and the development of highly specific and potent small-molecule modulators present novel avenues for disease treatment, opening opportunities for innovative drug development. However, although autophagy is intricately linked to a variety of human diseases, the widespread non-selective autophagy that is usually induced can also cause many off-target effects, posing significant challenges for the clinical application of related drugs [244]. Therefore, there is a pressing requirement for the development of more precise, specific, and safe selective autophagy drugs, which can reduce side effects and increase the possibility of small-molecule targeted drugs being put into clinical application.
Conclusions and perspectives
Conceptual innovation by targeting selective autophagy
Selective autophagy acts as an exceptionally specific quality control system that surveils cellular components and earmarks redundant or damaged elements for autophagic degradation. Over the last decade, human genetic research has increasingly shown that selective autophagy is related to diseases, especially neurodegenerative diseases, cancers, inflammatory diseases, and autoimmune diseases, suggesting the potential of targeting selective autophagy in the treatment of diseases. However, it is difficult to dissect how selective autophagy occurs in vivo, because most organelles are relatively difficult to be observed and operated experimentally in mammals. Surprisingly, zebrafish (Danio rerio) have gained a lot of popularity in recent years as a model for studying autophagic processes in vivo due to their accessibility to light, capacity to be genetically altered, and potential of translation. So far, zebrafish have already been used to investigate many types of selective autophagy, including mitophagy, xenophagy, lipophagy, and aggrephagy [245]. Probably the most comprehensively investigated instance of selective autophagy is the PINK1-Parkin pathway in mitophagy, which has been described in detail in our previous article [19], [21], [246]. Nonetheless, further research into other forms of selective autophagy is imperative to gain a more profound comprehension of the mechanisms behind selective autophagy.
Clinically relevant implementation benefiting healthcare
Exploring the mechanism behind selective autophagy in diseases not only enhances our understanding of cellular processes but also holds promise for clinical applications. Firstly, the identification of disease-specific, selective autophagy-related biomarkers represents a crucial step in disease prediction and prognosis. These biomarkers, which can be obtained from blood, urine, tissue samples, etc., may serve as indicators of disease progression and treatment response. For instance, recent studies have demonstrated the potential of selective autophagy-related biomarkers in predicting the progression of various diseases [247]. Secondly, molecular imaging techniques such as magnetic resonance imaging (MRI) and positron emission tomography (PET) scanning can be utilized to evaluate the extent of disease involvement and monitor treatment response in patients. By employing these techniques, clinicians can better assess the efficacy of selective autophagy-based therapies and make informed adjustments regarding treatment strategies [248].
Moreover, personalized medicine strategies should be implemented to optimize therapeutic outcomes. Understanding the correlation between genotype and phenotype is essential for tailoring treatment to individual patients. By considering the genetic background and clinical manifestations of patients, personalized treatment regimens can be developed to target selective autophagy pathways effectively [249]. Second, drug screening and targeted therapy approaches should be refined to accommodate patient-specific variations. This involves the development of screening methods for different patient subgroups and the selection of the most appropriate treatment regimen based on individual patient characteristics [250]. Additionally, combination therapy strategies represent a promising approach to enhance treatment efficacy while minimizing side effects. By combining small-molecule drugs targeting selective autophagy with other therapeutic modalities such as chemotherapy, radiotherapy, and immunotherapy, synergistic effects can be achieved, leading to improved clinical outcomes [251]. These integrated approaches hold significant potential for the development of more effective and personalized treatment strategies for a wide range of diseases in clinical practice.
Regarding clinical drug development, many drugs have been found to be related to autophagy but often lack specificity for autophagy. Some drugs may cause excessive autophagy, thus causing various off-target effects while producing therapeutic effects. And some drugs even have both induced and inhibitory effects on autophagy. These defects bring great challenges to the clinical application of targeted autophagy drugs. One typical example is crizotinib, which induces autophagy through inhibition of the STAT3 pathway in multiple lung cancer cell lines, but at the same time, it can induce pulmonary toxicity by blocking autophagy flux in alveolar epithelial cells [252], [253]. Consequently, it is of great significance to develop selective autophagy drugs with higher targeting properties that have broad clinical application prospects.
From underlying mechanisms to potential therapies: The roadmap
All in all, this review firstly introduces the categories and characteristics of selective autophagy and also comprehensively dissects the underlying molecular mechanisms of each type of selective autophagy. Next, following the roles and significance selective autophagy plays in various human diseases, we discuss possible clinical therapeutic approaches represented by targeted small-molecule drug development, completing the extension from potential mechanisms to potential therapies (Fig. 8).
Fig. 8.
From underlying mechanisms to potential therapies: The roadmap.
Despite the great promise of clinical application for targeting selective autophagy, potential therapies targeting selective autophagy, represented by the development of small-molecule drugs, are still faced with some challenges. Firstly, the complexity of intracellular autophagy pathway makes the selectivity and specificity of drugs a difficult problem. Fortunately, by modifying specific organelle targeting groups on the surface of drugs or carriers, it has become a basic strategy for the design of selective autophagy-targeting drugs [254]. For instance, it is promising to modify drugs targeting ER-phagy by adding groups with endoplasmic reticulum affinity, such as methyl benzene sulfonyl group, chloropropane group, or dansyl group, to increase the drug selectivity to ER and the residence time in ER [255]. Also, nanoparticles, polymers, liposomes, etc. can also be used as drug delivery systems to improve the stability of drugs and enhance their distribution and release in the body. Secondly, the safety of drugs targeting selective autophagy needs to be further confirmed. For example, CCCP and oligomycin A are two widely used selective inducers of mitophagy, which drive mitophagy by causing mitochondrial damage. However, their severe cytotoxicity limits their clinical application [256]. Pre-clinical and clinical trials and evaluations need to be further strengthened to ensure the safety and efficacy of these drugs.
Apart from small-molecule drugs, research on protein peptides, antibody therapy that directly target selective autophagy is at an early stage and is still evolving. A highly potent GABARAP-selective inhibitory peptide has been identified within the 270/480 kDa ankyrin-G, and an exceptionally powerful pan-Atg8 inhibitory peptide has been observed in the 440 kDa ankyrin-B. These peptides demonstrated remarkable efficacy in blocking autophagy when expressed in cultured cells [257]. Furthermore, the latest research has shown that certain RNA and antibody molecules can also function as tools and drugs to regulate selective autophagy [258]. For example, AUTAC, AUTOTAC, and the recently discovered ATNC are all antibody-based selective degraders for intracellular targets, suggesting that antibody drugs developed based on selective autophagy have the potential to downregulate some biomarkers, thereby inhibiting the occurrence and progression of disease [259].
In a nutshell, selective autophagy stands as a vital mechanism for cellular self-repair and metabolic balance, contributing to the preservation of cell health and vitality. Its pivotal role in the development of various diseases underscores its importance. The integration of Artificial Intelligence (AI) could reveal a potential link between selective autophagy and disease development, opening new possibilities for identifying potential therapeutic targets and biomarkers. The discovery and development of modulators for selective autophagy hold substantial clinical promise, offering a key direction for current drug development. Looking forward, as the basic research of selective autophagy continues to deepen, an increasing number of selective autophagy modulators will be discovered and applied to disease treatment. Additionally, modern techniques, including high-throughput screening and computer-aided drug design, will further expedite the identification and creation of selective autophagy modulators, offering potential therapeutic approaches for a wide range of human diseases.
Compliance with Ethics Requirement
This review does not involve any studies with human or animal subjects.
CRediT authorship contribution statement
Wei Ma: Writing – original draft, Visualization, Validation. Yingying Lu: Writing – original draft, Visualization, Validation. Xin Jin: Writing – original draft. Na Lin: Conceptualization, Writing – review & editing, Supervision. Lan Zhang: Conceptualization, Funding acquisition, Supervision. Yaowen Song: Conceptualization, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 22277102).
Biographies

Wei Ma received her Ph.D. degree in oncology from China Medical University in 2019. Now, she is a lecturer and an attending physician of the First Hospital of China Medical University. Her research interests include breast oncology.

Yingying Lu received her bachelor's degree in Tianjin University of Traditional Chinese Medicine in 2022. Now, she is a postgraduate student at School of Life Science and Engineering in Southwest Jiaotong University. Her research interests include are the development of autophagy targeting drugs and related anti-tumor drugs.

Xin Jin received her Master's degree in Imaging Medicine and Nuclear Medicine from China Medical University in 2013. She is a lecturer and an attending physician of the First Hospital of China Medical University. Her research interests include the application of ultrasound and artificial intelligence in the diagnosis and efficacy evaluation of gynecological tumors.

Na Lin received his Ph.D. degree from China Medical University in 2017. She is an Associate Professor of China Medical University since 2021. Her research interests are in pathogenesis and treatment of hematological malignancies.

Lan Zhang received his Ph.D. degree in Medicinal Chemistry from Shenyang Pharmaceutical University in 2015. Subsequently, he joined the faculty of State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, as a postdoctoral fellow. Currently, he is worked as an associate professor at the School of Life Science and Engineering, Southwest Jiaotong University. His research interests include the design and discovery of targeted small-molecule drugs, drug repurposing studies.

Yaowen Song received her Ph.D. degree from China Medical University in 2023. She is a lecturer and an attending physician of the First Hospital of China Medical University since 2016. Her research interests include oncobiology and oncology therapeutics.
Contributor Information
Na Lin, Email: salinaofcmu@163.com.
Lan Zhang, Email: zhanglanx_9@126.com.
Yaowen Song, Email: ywsong@cmu.edu.cn.
References
- 1.Levine B., Klionsky D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Wang L., Klionsky D.J., Shen H.M. The emerging mechanisms and functions of microautophagy. Nat Rev Mol Cell Biol. 2022 doi: 10.1038/s41580-022-00529-z. [DOI] [PubMed] [Google Scholar]
- 3.Kaushik S., Cuervo A.M. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19(6):365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Le WD. Autophagy and Ubiquitin-Proteasome System. Advances in Experimental Medicine and Biology. 2019;1206527-50. doi: Doi: 10.1007/978-981-15-0602-4_25. [DOI] [PubMed]
- 5.Pohl C., Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366(6467):818–822. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 6.Fleming A., Bourdenx M., Fujimaki M., Karabiyik C., Krause G.J., Lopez A., et al. The different autophagy degradation pathways and neurodegeneration. Neuron. 2022;110(6):935–966. doi: 10.1016/j.neuron.2022.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geng J., Baba M., Nair U., Klionsky D.J. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol. 2008;182(1):129–140. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klionsky D.J., Petroni G., Amaravadi R.K., Baehrecke E.H., Ballabio A., Boya P., et al. Autophagy in major human diseases. Embo j. 2021;40(19):e108863. doi: 10.15252/embj.2021108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reggiori F., Komatsu M., Finley K., Simonsen A. Autophagy: more than a nonselective pathway. Int. J Cell Biol. 2012;2012219625 doi: 10.1155/2012/219625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickles S., Vigié P., Youle R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr Biol. 2018;28(4):R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang J.R., Lingeman E., Luong T., Ahmed S., Muhar M., Nguyen T., et al. A genome-wide ER-phagy screen highlights key roles of mitochondrial metabolism and ER-resident UFMylation. Cell. 2020;180(6) doi: 10.1016/j.cell.2020.02.017. 1160-77.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakashima A., Shima T., Tsuda S., Aoki A., Kawaguchi M., Furuta A., et al. Aggrephagy deficiency in the placenta: a new pathogenesis of preeclampsia. Int J Mol Sci. 2021;22(5) doi: 10.3390/ijms22052432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui W., Sathyanarayan A., Lopresti M., Aghajan M., Chen C., Mashek D.G. Lipophagy-derived fatty acids undergo extracellular efflux via lysosomal exocytosis. Autophagy. 2021;17(3):690–705. doi: 10.1080/15548627.2020.1728097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germain K., Pexophagy K.PK. A Model for Selective Autophagy. Int J Mol Sci. 2020;21(2) doi: 10.3390/ijms21020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An H., Harper J.W. Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol. 2018;20(2):135–143. doi: 10.1038/s41556-017-0007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y.J., Kim J.K., Jung C.H., Kim Y.J., Jung E.J., Lee S.H., et al. Chemical modulation of SQSTM1/p62-mediated xenophagy that targets a broad range of pathogenic bacteria. Autophagy. 2022;18(12):2926–2945. doi: 10.1080/15548627.2022.2054240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W., Ocak U., Gao L., Tu S., Lenahan C.J., Zhang J., et al. Selective autophagy as a therapeutic target for neurological diseases. Cell Mol Life Sci. 2021;78(4):1369–1392. doi: 10.1007/s00018-020-03667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichimiya T., Yamakawa T., Hirano T., Yokoyama Y., Hayashi Y., Hirayama D., et al. Autophagy and Autophagy-Related Diseases: A Review. Int J Mol Sci. 2020;21(23) doi: 10.3390/ijms21238974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y., Li Z., Zhang S., Zhang T., Liu Y., Zhang L. Cellular mitophagy: Mechanism, roles in diseases and small molecule pharmacological regulation. Theranostics. 2023;13(2):736–766. doi: 10.7150/thno.79876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han R., Liu Y., Li S., Li X.J., Yang W. PINK1-PRKN mediated mitophagy: differences between in vitro and in vivo models. Autophagy. 2023;19(5):1396–1405. doi: 10.1080/15548627.2022.2139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eiyama A., Okamoto K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr Opin Cell Biol. 2015;3395–101 doi: 10.1016/j.ceb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 23.Onishi M., Yamano K., Sato M., Matsuda N., Okamoto K. Molecular mechanisms and physiological functions of mitophagy. Embo j. 2021;40(3):e104705. doi: 10.15252/embj.2020104705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter B., Sliter D.A., Herhaus L., Stolz A., Wang C., Beli P., et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A. 2016;113(15):4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S., Ma J., Yin P., Liang F. The landscape of mitophagy in sepsis reveals PHB1 as an NLRP3 inflammasome inhibitor. Front Immunol. 2023;141188482 doi: 10.3389/fimmu.2023.1188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Falco F., Gentile I., Cerino P., Cutarelli A., Catoi C., Roperto S. Prohibitin 2 is Involved in Parkin-Mediated Mitophagy in Urothelial Cells of Cattle Infected with Bovine Papillomavirus. Pathogens. 2020;9(8) doi: 10.3390/pathogens9080621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamark T., Johansen T. Mechanisms of Selective Autophagy. Annu Rev Cell Dev Biol. 2021;37143–69 doi: 10.1146/annurev-cellbio-120219-035530. [DOI] [PubMed] [Google Scholar]
- 28.Kim H.Y., Yoon H.S., Heo A.J., Jung E.J., Ji C.H., Mun S.R., et al. Mitophagy and endoplasmic reticulum-phagy accelerated by a p62 ZZ ligand alleviates paracetamol-induced hepatotoxicity. Br J Pharmacol. 2022 doi: 10.1111/bph.16004. [DOI] [PubMed] [Google Scholar]
- 29.Bozi L.H.M., Takano A.P.C., Campos J.C., Rolim N., Dourado P.M.M., Voltarelli V.A., et al. Endoplasmic reticulum stress impairs cardiomyocyte contractility through JNK-dependent upregulation of BNIP3. Int J Cardiol. 2018;272194–201 doi: 10.1016/j.ijcard.2018.08.070. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X., Wang X., Ding X., Du M., Li B., Weng X., et al. FAM134B oligomerization drives endoplasmic reticulum membrane scission for ER-phagy. EMBO J. 2020;39(5):e102608. doi: 10.15252/embj.2019102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An H., Ordureau A., Paulo J.A., Shoemaker C.J., Denic V., Harper J.W. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol Cell. 2019;74(5):891–908.e10. doi: 10.1016/j.molcel.2019.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H., Voeltz G.K. Reticulon-3 Promotes Endosome Maturation at ER Membrane Contact Sites. Dev Cell. 2021;56(1):52–66.e7. doi: 10.1016/j.devcel.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y., Han Y., Yang S., Cao J., Jiang M., Liang J. Endoplasmic reticulum-resident protein Sec62 drives colorectal cancer metastasis via MAPK/ATF2/UCA1 axis. Cell Prolif. 2022;55(12):e13253. doi: 10.1111/cpr.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith M.D., Harley M.E., Kemp A.J., Wills J., Lee M., Arends M., et al. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell. 2018;44(2) doi: 10.1016/j.devcel.2017.11.024. 217-32.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nthiga T.M., Kumar Shrestha B., Lamark T., Johansen T. The soluble reticulophagy receptor CALCOCO1 is also a Golgiphagy receptor. Autophagy. 2021;17(8):2051–2202. doi: 10.1080/15548627.2021.1940610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fumagalli F., Noack J., Bergmann T.J., Cebollero E., Pisoni G.B., Fasana E., et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016;18(11):1173–1184. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 37.Nthiga T.M., Kumar Shrestha B., Sjøttem E., Bruun J.A., Bowitz Larsen K., Bhujabal Z., et al. CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. EMBO J. 2020;39(15):e103649. doi: 10.15252/embj.2019103649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Q., Xiao Y., Chai P., Zheng P., Teng J., Chen J. ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr Biol. 2019;29(5) doi: 10.1016/j.cub.2019.01.041. 846-55.e6. [DOI] [PubMed] [Google Scholar]
- 39.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20(3):233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi C.S., Kehrl J.H. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3(123) doi: 10.1126/scisignal.2000751. ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu K., Psakhye I., Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158(3):549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 42.Pokatayev V., Yang K., Tu X., Dobbs N., Wu J., Kalb R.G., et al. Homeostatic regulation of STING protein at the resting state by stabilizer TOLLIP. Nat Immunol. 2020;21(2):158–167. doi: 10.1038/s41590-019-0569-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarraf S.A., Shah H.V., Kanfer G., Pickrell A.M., Holtzclaw L.A., Ward M.E., et al. Loss of TAX1BP1-Directed Autophagy Results in Protein Aggregate Accumulation in the Brain. Mol Cell. 2020;80(5) doi: 10.1016/j.molcel.2020.10.041. 779-95.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma X., Lu C., Chen Y., Li S., Ma N., Tao X., et al. CCT2 is an aggrephagy receptor for clearance of solid protein aggregates. Cell. 2022;185(8) doi: 10.1016/j.cell.2022.03.005. [DOI] [PubMed] [Google Scholar]
- 45.Robichaud S., Fairman G., Vijithakumar V., Mak E., Cook D.P., Pelletier A.R., et al. Identification of novel lipid droplet factors that regulate lipophagy and cholesterol efflux in macrophage foam cells. Autophagy. 2021;17(11):3671–3689. doi: 10.1080/15548627.2021.1886839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q., Zhao Y., Guo H., Li Q., Yan C., Li Y., et al. Impaired lipophagy induced-microglial lipid droplets accumulation contributes to the buildup of TREM1 in diabetes-associated cognitive impairment. Autophagy. 2023;19(10):2639–2656. doi: 10.1080/15548627.2023.2213984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zechner R., Madeo F., Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol. 2017;18(11):671–684. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- 49.Sathyanarayan A., Mashek M.T., Mashek D.G. ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Rep. 2017;19(1):1–9. doi: 10.1016/j.celrep.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim K.Y., Jang H.J., Yang Y.R., Park K.I., Seo J., Shin I.W., et al. SREBP-2/PNPLA8 axis improves non-alcoholic fatty liver disease through activation of autophagy. Sci Rep. 2016;635732 doi: 10.1038/srep35732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Negoita F., Blomdahl J., Wasserstrom S., Winberg M.E., Osmark P., Larsson S., et al. PNPLA3 variant M148 causes resistance to starvation-mediated lipid droplet autophagy in human hepatocytes. J Cell Biochem. 2019;120(1):343–356. doi: 10.1002/jcb.27378. [DOI] [PubMed] [Google Scholar]
- 52.Dupont N., Chauhan S., Arko-Mensah J., Castillo E.F., Masedunskas A., Weigert R., et al. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol. 2014;24(6):609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Macia M., Santos-Ledo A., Leslie J., Paish H.L., Collins A.L., Scott R.S., et al. A Mammalian Target of Rapamycin-Perilipin 3 (mTORC1-Plin3) Pathway is essential to Activate Lipophagy and Protects Against Hepatosteatosis. Hepatology. 2021;74(6):3441–3459. doi: 10.1002/hep.32048. [DOI] [PubMed] [Google Scholar]
- 54.Wang Z., Zhang H. Join the club: ORP8 is a lipophagy receptor. Protein Cell. 2023 doi: 10.1093/procel/pwad005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pu M., Zheng W., Zhang H., Wan W., Peng C., Chen X., et al. ORP8 acts as a lipophagy receptor to mediate lipid droplet turnover. Protein Cell. 2022 doi: 10.1093/procel/pwac063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Houten S.M., Wanders R.J.A., Ranea-Robles P. Metabolic interactions between peroxisomes and mitochondria with a special focus on acylcarnitine metabolism. Biochim Biophys Acta. 2020;1866(5) doi: 10.1016/j.bbadis.2020.165720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker C.L., Pomatto L.C.D., Tripathi D.N., Davies K.J.A. Redox Regulation of Homeostasis and Proteostasis in Peroxisomes. Physiol Rev. 2018;98(1):89–115. doi: 10.1152/physrev.00033.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Werner A., Herzog B., Voigt O., Valerius O., Braus G.H., Pöggeler S. NBR1 is involved in selective pexophagy in filamentous ascomycetes and can be functionally replaced by a tagged version of its human homolog. Autophagy. 2019;15(1):78–97. doi: 10.1080/15548627.2018.1507440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demers ND, Riccio V, Jo DS, Bhandari S, Law KB, Liao W, et al. PEX13 prevents pexophagy by regulating ubiquitinated PEX5 and peroxisomal ROS. Autophagy. 20231-22. doi: Doi: 10.1080/15548627.2022.2160566. [DOI] [PMC free article] [PubMed]
- 60.Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T., et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015;17(10):1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sargent G., van Zutphen T., Shatseva T., Zhang L., Di Giovanni V., Bandsma R., et al. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol. 2016;214(6):677–690. doi: 10.1083/jcb.201511034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng P., Wu X., Erramilli S.K., Paulo J.A., Knejski P., Gygi S.P., et al. A peroxisomal ubiquitin ligase complex forms a retrotranslocation channel. Nature. 2022;607(7918):374–380. doi: 10.1038/s41586-022-04903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Riccio V., Demers N., Hua R., Vissa M., Cheng D.T., Strilchuk A.W., et al. Deubiquitinating enzyme USP30 maintains basal peroxisome abundance by regulating pexophagy. J Cell Biol. 2019;218(3):798–807. doi: 10.1083/jcb.201804172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilhelm L.P., Zapata-Muñoz J., Villarejo-Zori B., Pellegrin S., Freire C.M., Toye A.M., et al. BNIP3L/NIX regulates both mitophagy and pexophagy. EMBO J. 2022;41(24):e111115. doi: 10.15252/embj.2022111115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kraft C., Deplazes A., Sohrmann M., Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 66.Wyant G.A., Abu-Remaileh M., Frenkel E.M., Laqtom N.N., Dharamdasani V., Lewis C.A., et al. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science. 2018;360(6390):751–778. doi: 10.1126/science.aar2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yao R.Q., Ren C., Xia Z.F., Yao Y.M. Organelle-specific autophagy in inflammatory diseases: a potential therapeutic target underlying the quality control of multiple organelles. Autophagy. 2021;17(2):385–401. doi: 10.1080/15548627.2020.1725377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beese C.J., Brynjólfsdóttir S.H., Frankel L.B. Selective Autophagy of the Protein Homeostasis Machinery: Ribophagy, Proteaphagy and ER-Phagy. Front Cell. Dev Biol. 2019:7373. doi: 10.3389/fcell.2019.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.An H., Harper J.W. Ribosome Abundance Control Via the Ubiquitin-Proteasome System and Autophagy. J Mol Biol. 2020;432(1):170–184. doi: 10.1016/j.jmb.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li W., He P., Huang Y., Li Y.F., Lu J., Li M., et al. Selective autophagy of intracellular organelles: recent research advances. Theranostics. 2021;11(1):222–256. doi: 10.7150/thno.49860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin M., Klionsky D.J. Finding a ribophagy receptor. Autophagy. 2018;14(9):1479–1480. doi: 10.1080/15548627.2018.1483672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bo Otto F., Thumm M. Nucleophagy-Implications for Microautophagy and Health. Int J Mol Sci. 2020;21(12) doi: 10.3390/ijms21124506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Papandreou M.E., Tavernarakis N. Nucleophagy: from homeostasis to disease. Cell Death Differ. 2019;26(4):630–669. doi: 10.1038/s41418-018-0266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dou Z., Xu C., Donahue G., Shimi T., Pan J.A., Zhu J., et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527(7576):105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan W., Zhang M., Wang C., Li B., Li L., Ye F., et al. Resveratrol attenuates high-fat diet-induced hepatic lipotoxicity by upregulating Bmi-1 expression. J Pharmacol Exp Ther. 2022;381(2):96–105. doi: 10.1124/jpet.121.001018. [DOI] [PubMed] [Google Scholar]
- 76.Lee J.M., Lee Y.K., Mamrosh J.L., Busby S.A., Griffin P.R., Pathak M.C., et al. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011;474(7352):506–510. doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee D., Hong J.H. Physiological overview of the potential link between the UPS and Ca(2+) signaling. Antioxidants (Basel) 2022;11(5) doi: 10.3390/antiox11050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cuervo A.M., Palmer A., Rivett A.J., Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227(3):792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- 79.Marshall R.S., Li F., Gemperline D.C., Book A.J., Vierstra R.D. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in arabidopsis. Mol Cell. 2021;81(9):2053. doi: 10.1016/j.molcel.2021.03.026. [DOI] [PubMed] [Google Scholar]
- 80.Cohen-Kaplan V., Livneh I., Avni N., Fabre B., Ziv T., Kwon Y.T., et al. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc Natl Acad Sci U S A. 2016;113(47):E7490–E7499. doi: 10.1073/pnas.1615455113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hamazaki J., Hirayama S., Murata S. Redundant roles of Rpn10 and Rpn13 in recognition of ubiquitinated proteins and cellular homeostasis. PLoS Genet. 2015;11(7):e1005401. doi: 10.1371/journal.pgen.1005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marshall R.S., Vierstra R.D. Dynamic regulation of the 26S proteasome: from synthesis to degradation. Front Mol Biosci. 2019:640. doi: 10.3389/fmolb.2019.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Settembre C., Fraldi A., Medina D.L., Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14(5):283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kravic B., Behrends C., Meyer H. Regulation of lysosome integrity and lysophagy by the ubiquitin-conjugating enzyme UBE2QL1. Autophagy. 2020;16(1):179–180. doi: 10.1080/15548627.2019.1687217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chae C.W., Yoon J.H., Lim J.R., Park J.Y., Cho J.H., Jung Y.H., et al. TRIM16-mediated lysophagy suppresses high-glucose-accumulated neuronal Aβ. Autophagy. 2023;19(10):2752–2768. doi: 10.1080/15548627.2023.2229659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Papadopoulos C., Kravic B., Meyer H. Repair or Lysophagy: Dealing with Damaged Lysosomes. J Mol Biol. 2020;432(1):231–1229. doi: 10.1016/j.jmb.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 87.Wang K, Fu S, Dong L, Zhang D, Wang M, Wu X, et al. Periplocin suppresses the growth of colorectal cancer cells by triggering LGALS3 (galectin 3)-mediated lysophagy. Autophagy. 20231-19. doi: Doi: 10.1080/15548627.2023.2239042. [DOI] [PMC free article] [PubMed]
- 88.Bell S.L., Lopez K.L., Cox J.S., Patrick K.L., Watson R.O. Galectin-8 senses phagosomal damage and recruits selective autophagy adapter TAX1BP1 to control mycobacterium tuberculosis infection in macrophages. MBio. 2021;12(4):e0187120. doi: 10.1128/mBio.01871-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burridge K. Focal adhesions: a personal perspective on a half century of progress. FEBS J. 2017;284(20):3355–3361. doi: 10.1111/febs.14195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sharifi M.N., Mowers E.E., Drake L.E., Collier C., Chen H., Zamora M., et al. Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of Paxillin with LC3. Cell Rep. 2016;15(8):1660–1672. doi: 10.1016/j.celrep.2016.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kenific C.M., Debnath J. NBR1-dependent selective autophagy is required for efficient cell-matrix adhesion site disassembly. Autophagy. 2016;12(10):1958–11199. doi: 10.1080/15548627.2016.1212789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang C.H., Bijian K., Qiu D., Su J., Saad A., Dahabieh M.S., et al. Endosomal sorting and c-Cbl targeting of paxillin to autophagosomes regulate cell-matrix adhesion turnover in human breast cancer cells. Oncotarget. 2017;8(19):31199–31214. doi: 10.18632/oncotarget.16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gomes L.C., Dikic I. Autophagy in antimicrobial immunity. Mol Cell. 2014;54(2):224–233. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 94.Xu Y., Zhou P., Cheng S., Lu Q., Nowak K., Hopp A.K., et al. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell. 2019;178(3) doi: 10.1016/j.cell.2019.06.007. 552-66.e20. [DOI] [PubMed] [Google Scholar]
- 95.Kwon D.H., Song H.K. A structural view of xenophagy, a battle between host and microbes. Mol Cells. 2018;41(1):27–34. doi: 10.14348/molcells.2018.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brest P., Benzaquen J., Klionsky D.J., Hofman P., Mograbi B. Open questions for harnessing autophagy-modulating drugs in the SARS-CoV-2 war: hope or hype? Autophagy. 2020;16(12):2267–2270. doi: 10.1080/15548627.2020.1779531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mijaljica D., Klionsky D.J. Autophagy/virophagy: a “disposal strategy” to combat COVID-19. Autophagy. 2020;16(12):2271–3222. doi: 10.1080/15548627.2020.1782022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liang S., Wu Y.S., Li D.Y., Tang J.X., Liu H.F. Autophagy in Viral Infection and Pathogenesis. Front Cell. Dev Biol. 2021:9766142. doi: 10.3389/fcell.2021.766142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qin X., Zhang J., Wang B., Xu G., Yang X., Zou Z., et al. Ferritinophagy is involved in the zinc oxide nanoparticles-induced ferroptosis of vascular endothelial cells. Autophagy. 2021;17(12):4266–4285. doi: 10.1080/15548627.2021.1911016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Faruk M.O., Ichimura Y., Komatsu M. Selective autophagy. Cancer Sci. 2021;112(10):3972–13398. doi: 10.1111/cas.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee T.Y., Chen S.A., Hung H.Y., Ou Y.Y. Incorporating distant sequence features and radial basis function networks to identify ubiquitin conjugation sites. PLoS One. 2011;6(3):e17331. doi: 10.1371/journal.pone.0017331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Varshavsky A. The Ubiquitin System, Autophagy, and Regulated Protein Degradation. Annu Rev Biochem. 2017;86123–28 doi: 10.1146/annurev-biochem-061516-044859. [DOI] [PubMed] [Google Scholar]
- 103.Shaid S., Brandts C.H., Serve H., Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20(1):21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kwon Y.T., Ciechanover A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem Sci. 2017;42(11):873–886. doi: 10.1016/j.tibs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 105.Kim P.K., Hailey D.W., Mullen R.T., Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105(52):20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 107.Yamasaki A., Alam J.M., Noshiro D., Hirata E., Fujioka Y., Suzuki K., et al. Liquidity is a critical determinant for selective autophagy of protein condensates. Mol Cell. 2020;77(6) doi: 10.1016/j.molcel.2019.12.026. 1163-75.e9. [DOI] [PubMed] [Google Scholar]
- 108.Yamanaka S., Horiuchi Y., Matsuoka S., Kido K., Nishino K., Maeno M., et al. A proximity biotinylation-based approach to identify protein-E3 ligase interactions induced by PROTACs and molecular glues. Nat Commun. 2022;13(1):183. doi: 10.1038/s41467-021-27818-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Deng Z., Lim J., Wang Q., Purtell K., Wu S., Palomo G.M., et al. ALS-FTLD-linked mutations of SQSTM1/p62 disrupt selective autophagy and NFE2L2/NRF2 anti-oxidative stress pathway. Autophagy. 2020;16(5):917–931. doi: 10.1080/15548627.2019.1644076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rogov V., Dötsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53(2):167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 112.Jeong S.J., Zhang X., Rodriguez-Velez A., Evans T.D., Razani B. p62/SQSTM1 and Selective Autophagy in Cardiometabolic Diseases. Antioxid Redox Signal. 2019;31(6):458–471. doi: 10.1089/ars.2018.7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Geisler S., Holmström K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 114.Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 115.Thurston T.L., Ryzhakov G., Bloor S., von Muhlinen N., Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10(11):1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 116.Wild P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R., et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333(6039):228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Newman A.C., Scholefield C.L., Kemp A.J., Newman M., McIver E.G., Kamal A., et al. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-κB signalling. PLoS One. 2012;7(11):e50672. doi: 10.1371/journal.pone.0050672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sun D., Wu R., Zheng J., Li P., Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28(4):405–415. doi: 10.1038/s41422-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu Y., Wan W. Acetylation in the regulation of autophagy. Autophagy. 2023;19(2):379–387. doi: 10.1080/15548627.2022.2062112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Karanasios E., Walker S.A., Okkenhaug H., Manifava M., Hummel E., Zimmermann H., et al. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016;712420 doi: 10.1038/ncomms12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vargas J.N.S., Wang C., Bunker E., Hao L., Maric D., Schiavo G., et al. Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol Cell. 2019;74(2) doi: 10.1016/j.molcel.2019.02.010. 347-62.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saftig P., Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10(9):623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 123.Yao Z., Delorme-Axford E., Backues S.K., Klionsky D.J. Atg41/Icy2 regulates autophagosome formation. Autophagy. 2015;11(12):2288–2299. doi: 10.1080/15548627.2015.1107692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hou Y., Dan X., Babbar M., Wei Y., Hasselbalch S.G., Croteau D.L., et al. Ageing as a risk factor for neurodegenerative disease. Nat Rev Neurol. 2019;15(10):565–581. doi: 10.1038/s41582-019-0244-7. [DOI] [PubMed] [Google Scholar]
- 125.Malampati S., Song J.X., Chun-Kit Tong B., Nalluri A., Yang C.B., Wang Z., et al. Targeting aggrephagy for the treatment of Alzheimer's disease. Cells. 2020;9(2) doi: 10.3390/cells9020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hyttinen J.M., Amadio M., Viiri J., Pascale A., Salminen A., Kaarniranta K. Clearance of misfolded and aggregated proteins by aggrephagy and implications for aggregation diseases. Ageing Res Rev. 2014;1816–28 doi: 10.1016/j.arr.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 127.Eshraghi M., Ahmadi M., Afshar S., Lorzadeh S., Adlimoghaddam A., Rezvani Jalal N., et al. Enhancing autophagy in Alzheimer's disease through drug repositioning. Pharmacol Ther. 2022;237108171 doi: 10.1016/j.pharmthera.2022.108171. [DOI] [PubMed] [Google Scholar]
- 128.Xie C., Zhuang X.X., Niu Z., Ai R., Lautrup S., Zheng S., et al. Amelioration of Alzheimer's disease pathology by mitophagy inducers identified via machine learning and a cross-species workflow. Nat Biomed Eng. 2022;6(1):76–93. doi: 10.1038/s41551-021-00819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chu C.T. Mechanisms of selective autophagy and mitophagy: implications for neurodegenerative diseases. Neurobiol Dis. 2019;12223–34 doi: 10.1016/j.nbd.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Katayama H., Hama H., Nagasawa K., Kurokawa H., Sugiyama M., Ando R., et al. Visualizing and modulating mitophagy for therapeutic studies of neurodegeneration. Cell. 2020;181(5) doi: 10.1016/j.cell.2020.04.025. 1176-87.e16. [DOI] [PubMed] [Google Scholar]
- 131.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20(9):1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 132.Bingol B., Tea J.S., Phu L., Reichelt M., Bakalarski C.E., Song Q., et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510(7505):370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 133.Matus S., Castillo K., Hetz C. Hormesis: protecting neurons against cellular stress in Parkinson disease. Autophagy. 2012;8(6):997–1001. doi: 10.4161/auto.20748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kelly L.P., Carvey P.M., Keshavarzian A., Shannon K.M., Shaikh M., Bakay R.A., et al. Progression of intestinal permeability changes and alpha-synuclein expression in a mouse model of Parkinson's disease. Mov Disord. 2014;29(8):999–1009. doi: 10.1002/mds.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Esmaeili Y., Yarjanli Z., Pakniya F., Bidram E., Łos M.J., Eshraghi M., et al. Targeting autophagy, oxidative stress, and ER stress for neurodegenerative disease treatment. J Control Release. 2022;345147–75 doi: 10.1016/j.jconrel.2022.03.001. [DOI] [PubMed] [Google Scholar]
- 136.Rakowski M., Porębski S., Grzelak A. Nutraceuticals as Modulators of Autophagy: Relevance in Parkinson's Disease. Int J Mol Sci. 2022;23(7) doi: 10.3390/ijms23073625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ghavami S., Shojaei S., Yeganeh B., Ande S.R., Jangamreddy J.R., Mehrpour M., et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog Neurobiol. 2014;11224–49 doi: 10.1016/j.pneurobio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 138.Zeng X.S., Jia J.J., Kwon Y., Wang S.D., Bai J. The role of thioredoxin-1 in suppression of endoplasmic reticulum stress in Parkinson disease. Free Radic Biol Med. 2014;6710–8 doi: 10.1016/j.freeradbiomed.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 139.Lee J., Sung K.W., Bae E.J., Yoon D., Kim D., Lee J.S., et al. Targeted degradation of ⍺-synuclein aggregates in Parkinson's disease using the AUTOTAC technology. Mol Neurodegener. 2023;18(1):41. doi: 10.1186/s13024-023-00630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim A., Lalonde K., Truesdell A., Gomes Welter P., Brocardo P.S., Rosenstock T.R., et al. New Avenues for the Treatment of Huntington's Disease. Int J Mol Sci. 2021;22(16) doi: 10.3390/ijms22168363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tabrizi S.J., Ghosh R., Leavitt B.R. Huntingtin lowering strategies for disease modification in Huntington's disease. Neuron. 2019;101(5):801–819. doi: 10.1016/j.neuron.2019.01.039. [DOI] [PubMed] [Google Scholar]
- 142.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shirendeb U.P., Calkins M.J., Manczak M., Anekonda V., Dufour B., McBride J.L., et al. Mutant huntingtin's interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington's disease. Hum Mol Genet. 2012;21(2):406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Naia L., Carmo C., Campesan S., Fão L., Cotton V.E., Valero J., et al. Mitochondrial SIRT3 confers neuroprotection in Huntington's disease by regulation of oxidative challenges and mitochondrial dynamics. Free Radic Biol Med. 2021;163163–79 doi: 10.1016/j.freeradbiomed.2020.11.031. [DOI] [PubMed] [Google Scholar]
- 145.Franco-Iborra S., Plaza-Zabala A., Montpeyo M., Sebastian D., Vila M., Martinez-Vicente M. Mutant HTT (huntingtin) impairs mitophagy in a cellular model of Huntington disease. Autophagy. 2021;17(3):672–689. doi: 10.1080/15548627.2020.1728096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hardiman O., Al-Chalabi A., Chio A., Corr E.M., Logroscino G., Robberecht W., et al. Amyotrophic lateral sclerosis. Nat Rev Dis Primers. 2017;317071 doi: 10.1038/nrdp.2017.71. [DOI] [PubMed] [Google Scholar]
- 147.Ma Q, Xin J, Peng Q, Li N, Sun S, Hou H, et al. UBQLN2 and HSP70 participate in Parkin-mediated mitophagy by facilitating outer mitochondrial membrane rupture. EMBO Rep. 2023e55859. doi: Doi: 10.15252/embr.202255859. [DOI] [PMC free article] [PubMed]
- 148.Vicencio E., Beltrán S., Labrador L., Manque P., Nassif M., Woehlbier U. Implications of Selective Autophagy Dysfunction for ALS Pathology. Cells. 2020;9(2) doi: 10.3390/cells9020381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gal J., Ström A.L., Kwinter D.M., Kilty R., Zhang J., Shi P., et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009;111(4):1062–1073. doi: 10.1111/j.1471-4159.2009.06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lyu L., Chen Z., McCarty N. TRIM44 links the UPS to SQSTM1/p62-dependent aggrephagy and removing misfolded proteins. Autophagy. 2022;18(4):783–798. doi: 10.1080/15548627.2021.1956105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhen Y., Yuan Z., Zhang J., Chen Y., Fu Y., Liu Y., et al. Flubendazole induces mitochondrial dysfunction and DRP1-mediated mitophagy by targeting EVA1A in breast cancer. Cell Death Dis. 2022;13(4):375. doi: 10.1038/s41419-022-04823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim S.-Y., Yi H.-K., Yun B.-S., Lee D.-Y., Hwang P.H., Park H.-R., et al. The extract of the immature fruit of Poncirus trifoliata induces apoptosis in colorectal cancer cells via mitochondrial autophagy. Food Sci Human Wellness. 2020;9(3):237–244. doi: 10.1016/j.fshw.2020.05.001. [DOI] [Google Scholar]
- 153.Sun X., Shu Y., Ye G., Wu C., Xu M., Gao R., et al. Histone deacetylase inhibitors inhibit cervical cancer growth through Parkin acetylation-mediated mitophagy. Acta Pharm Sin B. 2022;12(2):838–852. doi: 10.1016/j.apsb.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang M.M., Xu F.J., Su Y., Geng Y., Qian X.T., Xue X.L., et al. A new strategy to fight metallodrug resistance: mitochondria-relevant treatment through mitophagy to inhibit metabolic adaptations of cancer cells. Angew Chem Int Ed Engl. 2022;61(27):e202203843. doi: 10.1002/anie.202203843. [DOI] [PubMed] [Google Scholar]
- 155.Meng Y., Qiu L., Zeng X., Hu X., Zhang Y., Wan X., et al. Targeting CRL4 suppresses chemoresistant ovarian cancer growth by inducing mitophagy. Signal Transduct Target Ther. 2022;7(1):388. doi: 10.1038/s41392-022-01253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Meyer LM, Koschade SE, Vischedyk JB, Thoelken M, Gubas A, Wegner M, et al. Deciphering the mitophagy receptor network identifies a crucial role for OPTN (optineurin) in acute myeloid leukemia. Autophagy. 20231-15. doi: Doi: 10.1080/15548627.2023.2230839. [DOI] [PMC free article] [PubMed]
- 157.Islam F., Gopalan V., Lam A.K. RETREG1 (FAM134B): A new player in human diseases: 15 years after the discovery in cancer. J Cell Physiol. 2018;233(6):4479–4489. doi: 10.1002/jcp.26384. [DOI] [PubMed] [Google Scholar]
- 158.Haque M.H., Gopalan V., Chan K.W., Shiddiky M.J., Smith R.A., Lam A.K. Identification of Novel FAM134B (JK1) Mutations in Oesophageal Squamous Cell Carcinoma. Sci Rep. 2016;629173 doi: 10.1038/srep29173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang Z.Q., Chen J., Huang W.Q., Ning D., Liu Q.M., Wang C., et al. FAM134B induces tumorigenesis and epithelial-to-mesenchymal transition via Akt signaling in hepatocellular carcinoma. Mol Oncol. 2019;13(4):792–810. doi: 10.1002/1878-0261.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Michaud D.S., Ruan M., Koestler D.C., Pei D., Marsit C.J., De Vivo I., et al. Epigenome-Wide Association Study Using Prediagnostic Bloods Identifies New Genomic Regions Associated With Pancreatic Cancer Risk. JNCI Cancer Spectr. 2020;4(5):pkaa041. doi: 10.1093/jncics/pkaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liu X.X., Liu F.J. Novel bioinformatic identification of differentially expressed tissue-specific and cancer-related proteins from the Human Protein Atlas for biomarker discovery. Genet Mol Res. 2015;14(2):4557–4565. doi: 10.4238/2015.May.4.14. [DOI] [PubMed] [Google Scholar]
- 162.Yu G., Xiong D., Liu Z., Li Y., Chen K., Tang H. Long noncoding RNA LINC00052 inhibits colorectal cancer metastasis by sponging microRNA-574-5p to modulate CALCOCO1 expression. J Cell Biochem. 2019;120(10):17258–17272. doi: 10.1002/jcb.28988. [DOI] [PubMed] [Google Scholar]
- 163.Hong S.H., Chang S.H., Cho K.C., Kim S., Park S., Lee A.Y., et al. Endoplasmic reticulum-Golgi intermediate compartment protein 3 knockdown suppresses lung cancer through endoplasmic reticulum stress-induced autophagy. Oncotarget. 2016;7(40):65335–65347. doi: 10.18632/oncotarget.11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Feng X., Zhang H., Meng L., Song H., Zhou Q., Qu C., et al. Hypoxia-induced acetylation of PAK1 enhances autophagy and promotes brain tumorigenesis via phosphorylating ATG5. Autophagy. 2021;17(3):723–742. doi: 10.1080/15548627.2020.1731266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cai M., Sun X., Wang W., Lian Z., Wu P., Han S., et al. Disruption of peroxisome function leads to metabolic stress, mTOR inhibition, and lethality in liver cancer cells. Cancer Lett. 2018;42182–93 doi: 10.1016/j.canlet.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 166.Liu R., Tang J., Ding C., Liang W., Zhang L., Chen T., et al. The depletion of ATM inhibits colon cancer proliferation and migration via B56γ2-mediated Chk1/p53/CD44 cascades. Cancer Lett. 2017;39048–57 doi: 10.1016/j.canlet.2016.12.040. [DOI] [PubMed] [Google Scholar]
- 167.Denais C., Lammerding J. Nuclear mechanics in cancer. Adv Experiment Med Biol. 2014;773435–70 doi: 10.1007/978-1-4899-8032-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhao L., Li W., Luo X., Sheng S. The multifaceted roles of nucleophagy in cancer development and therapy. Cell Biol Int. 2021;45(2):246–257. doi: 10.1002/cbin.11504. [DOI] [PubMed] [Google Scholar]
- 169.Xiong W., Gao Y., Wei W., Zhang J. Extracellular and nuclear PD-L1 in modulating cancer immunotherapy. Trends Cancer. 2021;7(9):837–846. doi: 10.1016/j.trecan.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 170.Wu F., Yang J., Liu J., Wang Y., Mu J., Zeng Q., et al. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal Transduct Target Ther. 2021;6(1):218. doi: 10.1038/s41392-021-00641-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yuan M., Tu B., Li H., Pang H., Zhang N., Fan M., et al. Cancer-associated fibroblasts employ NUFIP1-dependent autophagy to secrete nucleosides and support pancreatic tumor growth. Nat Cancer. 2022;3(8):945–960. doi: 10.1038/s43018-022-00426-6. [DOI] [PubMed] [Google Scholar]
- 172.Ammanathan V., Vats S., Abraham I.M., Manjithaya R. Xenophagy in cancer. Semin Cancer Biol. 2020:66163–66170. doi: 10.1016/j.semcancer.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 173.Sui X., Liang X., Chen L., Guo C., Han W., Pan H., et al. Bacterial xenophagy and its possible role in cancer: A potential antimicrobial strategy for cancer prevention and treatment. Autophagy. 2017;13(2):237–247. doi: 10.1080/15548627.2016.1252890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sharafutdinov I., Tegtmeyer N., Linz B., Rohde M., Vieth M., Tay A.C., et al. A single-nucleotide polymorphism in Helicobacter pylori promotes gastric cancer development. Cell Host Microbe. 2023;31(8) doi: 10.1016/j.chom.2023.06.016. 1345-58.e6. [DOI] [PubMed] [Google Scholar]
- 175.Castaño-Rodríguez N., Kaakoush N.O., Goh K.L., Fock K.M., Mitchell H.M. Autophagy in helicobacter pylori infection and related gastric cancer. Helicobacter. 2015;20(5):353–369. doi: 10.1111/hel.12211. [DOI] [PubMed] [Google Scholar]
- 176.Gurbatri C.R., Arpaia N., Danino T. Engineering bacteria as interactive cancer therapies. Science. 2022;378(6622):858–864. doi: 10.1126/science.add9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wen X., Klionsky D.J. How bacteria can block xenophagy: an insight from Salmonella. Autophagy. 2020;16(2):193–1114. doi: 10.1080/15548627.2019.1666580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tiede S., Meyer-Schaller N., Kalathur R.K.R., Ivanek R., Fagiani E., Schmassmann P., et al. The FAK inhibitor BI 853520 exerts anti-tumor effects in breast cancer. Oncogenesis. 2018;7(9):73. doi: 10.1038/s41389-018-0083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jiang P., Wu J., Chen X., Ning B., Liu Q., Li Z., et al. Quantitative proteomics analysis of differentially expressed proteins in ruptured and unruptured cerebral aneurysms by iTRAQ. J Proteomics. 2018;18245–52 doi: 10.1016/j.jprot.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 180.Alim Al-Bari A., Ito Y., Thomes P.G., Menon M.B., García-Macia M., Fadel R., et al. Emerging mechanistic insights of selective autophagy in hepatic diseases. Front Pharmacol. 2023;141149809 doi: 10.3389/fphar.2023.1149809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tong M., Mukai R., Mareedu S., Zhai P., Oka S.I., Huang C.Y., et al. Distinct roles of DRP1 in conventional and alternative mitophagy in obesity cardiomyopathy. Circ Res. 2023;133(1):6–21. doi: 10.1161/circresaha.123.322512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Panzhinskiy E., Hua Y., Culver B., Ren J., Nair S. Endoplasmic reticulum stress upregulates protein tyrosine phosphatase 1B and impairs glucose uptake in cultured myotubes. Diabetologia. 2013;56(3):598–607. doi: 10.1007/s00125-012-2782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Carotti S., Aquilano K., Zalfa F., Ruggiero S., Valentini F., Zingariello M., et al. Lipophagy impairment is associated with disease progression in NAFLD. Front Physiol. 2020;11850 doi: 10.3389/fphys.2020.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schuster S., Cabrera D., Arrese M., Feldstein A.E. Triggering and resolution of inflammation in NASH. Nat Rev Gastroenterol Hepatol. 2018;15(6):349–364. doi: 10.1038/s41575-018-0009-6. [DOI] [PubMed] [Google Scholar]
- 185.Fotbolcu H., Zorlu E. Nonalcoholic fatty liver disease as a multi-systemic disease. World J Gastroenterol. 2016;22(16):4079–4090. doi: 10.3748/wjg.v22.i16.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Byun S., Seok S., Kim Y.C., Zhang Y., Yau P., Iwamori N., et al. Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase. Nat Commun. 2020;11(1):807. doi: 10.1038/s41467-020-14384-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mao Y., Ren J., Yang L. FUN14 Domain containing 1 (FUNDC1): a promising mitophagy receptor regulating mitochondrial homeostasis in cardiovascular diseases. Front Pharmacol. 2022;13887045 doi: 10.3389/fphar.2022.887045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ren J., Sun M., Zhou H., Ajoolabady A., Zhou Y., Tao J., et al. FUNDC1 interacts with FBXL2 to govern mitochondrial integrity and cardiac function through an IP3R3-dependent manner in obesity. Sci Adv. 2020;6(38) doi: 10.1126/sciadv.abc8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lin M., Xian H., Chen Z., Wang S., Liu M., Liang W., et al. MCM8-mediated mitophagy protects vascular health in response to nitric oxide signaling in a mouse model of Kawasaki disease. Nat Cardiovasc Res. 2023;2(8):778–792. doi: 10.1038/s44161-023-00314-x. [DOI] [PubMed] [Google Scholar]
- 190.Yoshida S., Wei X., Zhang G., O'Connor C.L., Torres M., Zhou Z., et al. Endoplasmic reticulum-associated degradation is required for nephrin maturation and kidney glomerular filtration function. J Clin Invest. 2021;131(7) doi: 10.1172/jci143988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Drozdova T., Papillon J., Cybulsky A.V. Nephrin missense mutations: induction of endoplasmic reticulum stress and cell surface rescue by reduction in chaperone interactions. Physiol Rep. 2013;1(4):e00086. doi: 10.1002/phy2.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nishibori Y., Liu L., Hosoyamada M., Endou H., Kudo A., Takenaka H., et al. Disease-causing missense mutations in NPHS2 gene alter normal nephrin trafficking to the plasma membrane. Kidney Int. 2004;66(5):1755–1765. doi: 10.1111/j.1523-1755.2004.00898.x. [DOI] [PubMed] [Google Scholar]
- 193.Kurth I., Pamminger T., Hennings J.C., Soehendra D., Huebner A.K., Rotthier A., et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet. 2009;41(11):1179–1181. doi: 10.1038/ng.464. [DOI] [PubMed] [Google Scholar]
- 194.Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522(7556):354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 195.Law K.B., Bronte-Tinkew D., Di Pietro E., Snowden A., Jones R.O., Moser A., et al. The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy. 2017;13(5):868–884. doi: 10.1080/15548627.2017.1291470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nazarko T.Y. Pexophagy is responsible for 65% of cases of peroxisome biogenesis disorders. Autophagy. 2017;13(5):991–994. doi: 10.1080/15548627.2017.1291480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wong S.Q., Kumar A.V., Mills J., Lapierre L.R. Autophagy in aging and longevity. Hum Genet. 2020;139(3):277–290. doi: 10.1007/s00439-019-02031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Juricic P., Lu Y.-X., Leech T., Drews L.F., Paulitz J., Lu J., et al. Long-lasting geroprotection from brief rapamycin treatment in early adulthood by persistently increased intestinal autophagy. Nature Aging. 2022;2(9):824–836. doi: 10.1038/s43587-022-00278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ding H., Li Y., Chen S., Wen Y., Zhang S., Luo E., et al. Fisetin ameliorates cognitive impairment by activating mitophagy and suppressing neuroinflammation in rats with sepsis-associated encephalopathy. CNS Neurosci Ther. 2022;28(2):247–258. doi: 10.1111/cns.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen C., Yang C., Wang J., Huang X., Yu H., Li S., et al. Melatonin ameliorates cognitive deficits through improving mitophagy in a mouse model of Alzheimer's disease. J Pineal Res. 2021;71(4):e12774. doi: 10.1111/jpi.12774. [DOI] [PubMed] [Google Scholar]
- 201.Zhang Y., Wang Y., Xu J., Tian F., Hu S., Chen Y., et al. Melatonin attenuates myocardial ischemia-reperfusion injury via improving mitochondrial fusion/mitophagy and activating the AMPK-OPA1 signaling pathways. J Pineal Res. 2019;66(2):e12542. doi: 10.1111/jpi.12542. [DOI] [PubMed] [Google Scholar]
- 202.Ma S., Chen J., Feng J., Zhang R., Fan M., Han D., et al. Melatonin ameliorates the progression of atherosclerosis via mitophagy activation and NLRP3 inflammasome inhibition. Oxid Med Cell Longev. 2018 doi: 10.1155/2018/9286458. 20189286458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tang H., Yang M., Liu Y., Zhu X., Liu S., Liu H., et al. Melatonin alleviates renal injury by activating mitophagy in diabetic nephropathy. Front Endocrinol (Lausanne) 2022:13889729. doi: 10.3389/fendo.2022.889729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Han X., Xu T., Fang Q., Zhang H., Yue L., Hu G., et al. Quercetin hinders microglial activation to alleviate neurotoxicity via the interplay between NLRP3 inflammasome and mitophagy. Redox Biol. 2021:44102010. doi: 10.1016/j.redox.2021.102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cen X., Chen Y., Xu X., Wu R., He F., Zhao Q., et al. Pharmacological targeting of MCL-1 promotes mitophagy and improves disease pathologies in an Alzheimer's disease mouse model. Nat Commun. 2020;11(1):5731. doi: 10.1038/s41467-020-19547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Luo H., Krigman J., Zhang R., Yang M., Sun N. Pharmacological inhibition of USP30 activates tissue-specific mitophagy. Acta Physiol (Oxf) 2021;232(3):e13666. doi: 10.1111/apha.13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Xu J., Sun L., He M., Zhang S., Gao J., Wu C., et al. Resveratrol Protects against Zearalenone-Induced Mitochondrial Defects during Porcine Oocyte Maturation via PINK1/Parkin-Mediated Mitophagy. Toxins (Basel) 2022;14(9) doi: 10.3390/toxins14090641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu H., Zheng Q., Yuan J., Gao Y., Wang T., Zhang H., et al. Modulating SQSTM1/p62-dependent selective autophagy of neurons by activating Nrf2 with multifunctional nanoparticles to eliminate α-synuclein aggregates and boost therapy of Parkinson’s disease. Nano Today. 2023:49101770. [Google Scholar]
- 209.Liu W., Wang M., Shen L., Zhu Y., Ma H., Liu B., et al. SHP2-mediated mitophagy boosted by lovastatin in neuronal cells alleviates parkinsonism in mice. Signal Transduct Target Ther. 2021;6(1):34. doi: 10.1038/s41392-021-00474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ahmed S., Kwatra M., Ranjan Panda S., Murty U.S.N., Naidu V.G.M. Andrographolide suppresses NLRP3 inflammasome activation in microglia through induction of parkin-mediated mitophagy in in-vitro and in-vivo models of Parkinson disease. Brain Behav Immun. 2021:91142–91158. doi: 10.1016/j.bbi.2020.09.017. [DOI] [PubMed] [Google Scholar]
- 211.Shao W.Q., Zhu W.W., Luo M.J., Fan M.H., Li Q., Wang S.H., et al. Cholesterol suppresses GOLM1-dependent selective autophagy of RTKs in hepatocellular carcinoma. Cell Rep. 2022;39(3) doi: 10.1016/j.celrep.2022.110712. [DOI] [PubMed] [Google Scholar]
- 212.Rademaker G., Boumahd Y., Peiffer R., Anania S., Wissocq T., Liégeois M., et al. Myoferlin targeting triggers mitophagy and primes ferroptosis in pancreatic cancer cells. Redox Biol. 2022:53102324. doi: 10.1016/j.redox.2022.102324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Liu Z., Ma C., Wang Q., Yang H., Lu Z., Bi T., et al. Targeting FAM134B-mediated reticulophagy activates sorafenib-induced ferroptosis in hepatocellular carcinoma. Biochem Biophys Res Commun. 2022:589247–589253. doi: 10.1016/j.bbrc.2021.12.019. [DOI] [PubMed] [Google Scholar]
- 214.Wu C., Dai C., Li X., Sun M., Chu H., Xuan Q., et al. AKR1C3-dependent lipid droplet formation confers hepatocellular carcinoma cell adaptability to targeted therapy. Theranostics. 2022;12(18):7681–7698. doi: 10.7150/thno.74974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhang Z., Gao W., Zhou L., Chen Y., Qin S., Zhang L., et al. Repurposing Brigatinib for the Treatment of Colorectal Cancer Based on Inhibition of ER-phagy. Theranostics. 2019;9(17):4878–4892. doi: 10.7150/thno.36254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kim E.H., Sohn S., Kwon H.J., Kim S.U., Kim M.J., Lee S.J., et al. Sodium selenite induces superoxide-mediated mitochondrial damage and subsequent autophagic cell death in malignant glioma cells. Cancer Res. 2007;67(13):6314–6324. doi: 10.1158/0008-5472.Can-06-4217. [DOI] [PubMed] [Google Scholar]
- 217.Zielke S., Kardo S., Zein L., Mari M., Covarrubias-Pinto A., Kinzler M.N., et al. ATF4 links ER stress with reticulophagy in glioblastoma cells. Autophagy. 2021;17(9):2432–2448. doi: 10.1080/15548627.2020.1827780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Huang T., Xu T., Wang Y., Zhou Y., Yu D., Wang Z., et al. Cannabidiol inhibits human glioma by induction of lethal mitophagy through activating TRPV4. Autophagy. 2021;17(11):3592–3606. doi: 10.1080/15548627.2021.1885203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Song Y.M., Lee Y.H., Kim J.W., Ham D.S., Kang E.S., Cha B.S., et al. Metformin alleviates hepatosteatosis by restoring SIRT1-mediated autophagy induction via an AMP-activated protein kinase-independent pathway. Autophagy. 2015;11(1):46–59. doi: 10.4161/15548627.2014.984271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Liu C., Liao J.Z., Li P.Y. Traditional Chinese herbal extracts inducing autophagy as a novel approach in therapy of nonalcoholic fatty liver disease. World J Gastroenterol. 2017;23(11):1964–1973. doi: 10.3748/wjg.v23.i11.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Afshari H., Noori S., Zarghi A. A novel combination of metformin and resveratrol alleviates hepatic steatosis by activating autophagy through the cAMP/AMPK/SIRT1 signaling pathway. Naunyn Schmiedebergs Arch Pharmacol. 2023 doi: 10.1007/s00210-023-02520-7. [DOI] [PubMed] [Google Scholar]
- 222.Getiye Y., Rice T.A., Phillips B.D., Carrillo D.F., He G. Dysregulated lipolysis and lipophagy in lipid droplets of macrophages from high fat diet-fed obese mice. J Cell Mol Med. 2022;26(18):4825–4836. doi: 10.1111/jcmm.17513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Huang W., Zhang J., Jin W., Yang J., Yu G., Shi H., et al. Piperine alleviates acute pancreatitis: A possible role for FAM134B and CCPG1 dependent ER-phagy. Phytomedicine. 2022:105154361. doi: 10.1016/j.phymed.2022.154361. [DOI] [PubMed] [Google Scholar]
- 224.Al Azzaz J., Rieu A., Aires V., Delmas D., Chluba J., Winckler P., et al. Resveratrol-Induced Xenophagy Promotes Intracellular Bacteria Clearance in Intestinal Epithelial Cells and Macrophages. Front Immunol. 2018:93149. doi: 10.3389/fimmu.2018.03149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jin S., He X., Ma L., Zhuang Z., Wang Y., Lin M., et al. Suppression of ACE2 SUMOylation protects against SARS-CoV-2 infection through TOLLIP-mediated selective autophagy. Nat Commun. 2022;13(1):5204. doi: 10.1038/s41467-022-32957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Pandey S., Patil S., Ballav N., Basu S. Spatial targeting of Bcl-2 on endoplasmic reticulum and mitochondria in cancer cells by lipid nanoparticles. J Mater Chem B. 2020;8(19):4259–4266. doi: 10.1039/d0tb00408a. [DOI] [PubMed] [Google Scholar]
- 227.Shen X, Deng Y, Chen L, Liu C, Li L, Huang Y. Modulation of Autophagy Direction to Enhance Antitumor Effect of Endoplasmic-Reticulum-Targeted Therapy: Left or Right? Adv Sci (Weinh). 2023e2301434. doi: Doi: 10.1002/advs.202301434. [DOI] [PMC free article] [PubMed]
- 228.Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14(2):193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Aishwarya R., Alam S., Abdullah C.S., Morshed M., Nitu S.S., Panchatcharam M., et al. Pleiotropic effects of mdivi-1 in altering mitochondrial dynamics, respiration, and autophagy in cardiomyocytes. Redox Biol. 2020:36101660. doi: 10.1016/j.redox.2020.101660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Liu X., Song L., Yu J., Huang F., Li Y., Ma C. Mdivi-1: a promising drug and its underlying mechanisms in the treatment of neurodegenerative diseases. Histol Histopathol. 2022;37(6):505–512. doi: 10.14670/hh-18-443. [DOI] [PubMed] [Google Scholar]
- 231.Yao J., Wang J., Xu Y., Guo Q., Sun Y., Liu J., et al. CDK9 inhibition blocks the initiation of PINK1-PRKN-mediated mitophagy by regulating the SIRT1-FOXO3-BNIP3 axis and enhances the therapeutic effects involving mitochondrial dysfunction in hepatocellular carcinoma. Autophagy. 2022;18(8):1879–1897. doi: 10.1080/15548627.2021.2007027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Núñez-Vázquez S., Saura-Esteller J., Sánchez-Vera I., Guilbaud E., Cosialls A.M., Pons G., et al. The prohibitin-binding compound fluorizoline inhibits mitophagy in cancer cells. Oncogenesis. 2021;10(9):64. doi: 10.1038/s41389-021-00352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yang S.T., Fan J.B., Liu T.T., Ning S., Xu J.H., Zhou Y.J., et al. Development of strigolactones as novel autophagy/mitophagy inhibitors against colorectal cancer cells by blocking the autophagosome-lysosome fusion. J Med Chem. 2022 doi: 10.1021/acs.jmedchem.2c00275. [DOI] [PubMed] [Google Scholar]
- 234.Kyung S.Y., Kim Y.J., Son E.S., Jeong S.H., Park J.W. The phosphodiesterase 4 inhibitor roflumilast protects against cigarette smoke extract-induced mitophagy-dependent cell death in epithelial cells. Tuberc Respir Dis. 2018;81(2):138–147. doi: 10.4046/trd.2017.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chipurupalli S., Ganesan R., Martini G., Mele L., Reggio A., Esposito M., et al. Cancer cells adapt FAM134B/BiP mediated ER-phagy to survive hypoxic stress. Cell Death Dis. 2022;13(4):357. doi: 10.1038/s41419-022-04813-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zimmermann J.S.M., Linxweiler J., Radosa J.C., Linxweiler M., Zimmermann R. The endoplasmic reticulum membrane protein Sec62 as potential therapeutic target in SEC62 overexpressing tumors. Front Physiol. 2022:131014271. doi: 10.3389/fphys.2022.1014271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Liu L., Xu L., Zhang S., Wang D., Dong G., Chen H., et al. STF-083010, an inhibitor of XBP1 splicing, attenuates acute renal failure in rats by suppressing endoplasmic reticulum stress-induced apoptosis and inflammation. Exp Anim. 2018;67(3):373–382. doi: 10.1538/expanim.17-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Jian L., Lu Y., Lu S., Lu C. Chemical chaperone 4-phenylbutyric acid reduces cardiac ischemia/reperfusion injury by alleviating endoplasmic reticulum stress and oxidative stress. Med Sci Monit. 2016:225218–225227. doi: 10.12659/msm.898623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Levy J.M.M., Towers C.G., Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17(9):528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rangwala R., Chang Y.C., Hu J., Algazy K.M., Evans T.L., Fecher L.A., et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Barnard R.A., Wittenburg L.A., Amaravadi R.K., Gustafson D.L., Thorburn A., Thamm D.H. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy. 2014;10(8):1415–1425. doi: 10.4161/auto.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mei L., Chen X., Wei F., Huang X., Liu L., Yao J., et al. Tethering ATG16L1 or LC3 induces targeted autophagic degradation of protein aggregates and mitochondria. Autophagy. 2023;19(11):2997–3013. doi: 10.1080/15548627.2023.2234797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Galluzzi L., Bravo-San Pedro J.M., Levine B., Green D.R., Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov. 2017;16(7):487–511. doi: 10.1038/nrd.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pant D.C., Nazarko T.Y. Selective autophagy: the rise of the zebrafish model. Autophagy. 2021;17(11):3297–3305. doi: 10.1080/15548627.2020.1853382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Li J., Yang D., Li Z., Zhao M., Wang D., Sun Z., et al. PINK1/Parkin-mediated mitophagy in neurodegenerative diseases. Ageing Res Rev. 2023:84101817. doi: 10.1016/j.arr.2022.101817. [DOI] [PubMed] [Google Scholar]
- 247.Koklesova L., Mazurakova A., Samec M., Kudela E., Biringer K., Kubatka P., et al. Mitochondrial health quality control: measurements and interpretation in the framework of predictive, preventive, and personalized medicine. Epma j. 2022;13(2):177–193. doi: 10.1007/s13167-022-00281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Fowler A.M., Strigel R.M. Clinical advances in PET-MRI for breast cancer. Lancet Oncol. 2022;23(1):e32–e43. doi: 10.1016/s1470-2045(21)00577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Golubnitschaja O., Polivka J., Jr., Potuznik P., Pesta M., Stetkarova I., Mazurakova A., et al. The paradigm change from reactive medical services to 3PM in ischemic stroke: a holistic approach utilising tear fluid multi-omics, mitochondria as a vital biosensor and AI-based multi-professional data interpretation. Epma j. 2024;15(1):1–23. doi: 10.1007/s13167-024-00356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Patton E.E., Zon L.I., Langenau D.M. Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials. Nat Rev Drug Discov. 2021;20(8):611–628. doi: 10.1038/s41573-021-00210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Arosa L., Camba-Gómez M., Golubnitschaja O., Conde-Aranda J. Predictive, preventive and personalised approach as a conceptual and technological innovation in primary and secondary care of inflammatory bowel disease benefiting affected individuals and populations. Epma j. 2024;15(1):111–123. doi: 10.1007/s13167-024-00351-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Zhang Y., Gao Z., Pan Z., Fu H., Jiang F., Yan H., et al. Crizotinib induces pulmonary toxicity by blocking autophagy flux in alveolar epithelial cells. Biochem Pharmacol. 2023;215115636 doi: 10.1016/j.bcp.2023.115636. [DOI] [PubMed] [Google Scholar]
- 253.You L., Shou J., Deng D., Jiang L., Jing Z., Yao J., et al. Crizotinib induces autophagy through inhibition of the STAT3 pathway in multiple lung cancer cell lines. Oncotarget. 2015;6(37):40268–40282. doi: 10.18632/oncotarget.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tavakol S., Ashrafizadeh M., Deng S., Azarian M., Abdoli A., Motavaf M., et al. Autophagy modulators: mechanistic aspects and drug delivery systems. Biomolecules. 2019;9(10) doi: 10.3390/biom9100530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Song S., Tan J., Miao Y., Zhang Q. Crosstalk of ER stress-mediated autophagy and ER-phagy: Involvement of UPR and the core autophagy machinery. J Cell Physiol. 2018;233(5):3867–3874. doi: 10.1002/jcp.26137. [DOI] [PubMed] [Google Scholar]
- 256.Georgakopoulos N.D., Wells G., Campanella M. The pharmacological regulation of cellular mitophagy. Nat Chem Biol. 2017;13(2):136–146. doi: 10.1038/nchembio.2287. [DOI] [PubMed] [Google Scholar]
- 257.Li J., Zhu R., Chen K., Zheng H., Zhao H., Yuan C., et al. Potent and specific Atg8-targeting autophagy inhibitory peptides from giant ankyrins. Nat Chem Biol. 2018;14(8):778–787. doi: 10.1038/s41589-018-0082-8. [DOI] [PubMed] [Google Scholar]
- 258.Yuan Y., Zhang X., Fan X., Peng Y., Jin Z. The emerging roles of circular RNA-mediated autophagy in tumorigenesis and cancer progression. Cell Death Discov. 2022;8(1):385. doi: 10.1038/s41420-022-01172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.He H., Zhou C., Chen X. ATNC: Versatile nanobody chimeras for autophagic degradation of intracellular unligandable and undruggable proteins. J Am Chem Soc. 2023;145(45):24785–24795. doi: 10.1021/jacs.3c08843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Humpton T.J., Alagesan B., DeNicola G.M., Lu D., Yordanov G.N., Leonhardt C.S., et al. Oncogenic KRAS induces NIX-mediated mitophagy to promote pancreatic cancer. Cancer Discov. 2019;9(9):1268–1287. doi: 10.1158/2159-8290.Cd-18-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lu J., Linares B., Xu Z., Rui Y.N. Mechanisms of FA-phagy, a new form of selective autophagy/organellophagy. Front Cell Dev Biol. 2021;9799123 doi: 10.3389/fcell.2021.799123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Macke A.J., Pachikov A.N., Divita T.E., Morris M.E., LaGrange C.A., Holzapfel M.S., et al. Targeting the ATF6-mediated ER stress response and autophagy blocks integrin-driven prostate cancer progression. Mol Cancer Res. 2023;21(9):958–974. doi: 10.1158/1541-7786.Mcr-23-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Shao D., Kolwicz S.C., Jr., Wang P., Roe N.D., Villet O., Nishi K., et al. Increasing fatty acid oxidation prevents high-fat diet-induced cardiomyopathy through regulating parkin-mediated mitophagy. Circulation. 2020;142(10):983–997. doi: 10.1161/circulationaha.119.043319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zheng W., Feng Z., You S., Zhang H., Tao Z., Wang Q., et al. Fisetin inhibits IL-1β-induced inflammatory response in human osteoarthritis chondrocytes through activating SIRT1 and attenuates the progression of osteoarthritis in mice. Int Immunopharmacol. 2017;45135–47 doi: 10.1016/j.intimp.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 265.Wu Q., Gao C., Wang H., Zhang X., Li Q., Gu Z., et al. Mdivi-1 alleviates blood-brain barrier disruption and cell death in experimental traumatic brain injury by mitigating autophagy dysfunction and mitophagy activation. Int J Biochem Cell Biol. 2018;9444–55 doi: 10.1016/j.biocel.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 266.Guo Y., Zhang H., Yan C., Shen B., Zhang Y., Guo X., et al. Small molecule agonist of mitochondrial fusion repairs mitochondrial dysfunction. Nat Chem Biol. 2023;19(4):468–477. doi: 10.1038/s41589-022-01224-y. [DOI] [PubMed] [Google Scholar]