Abstract
Quantitative measurements of serum hepatitis C virus (HCV) RNA are becoming increasingly important in the management of HCV-infected patients. Here we compared two quantitative assays, the COBAS AMPLICOR HCV Monitor 2.0 assay (Roche Diagnostics) and the branched DNA-based VERSANT HCV RNA 3.0 assay (Bayer Diagnostics) for HCV RNA measurement in 344 samples derived from 120 patients with chronic genotype 1 HCV infection. The overall concordance between the results of the two tests was 95%, and the HCV RNA titers within the dynamic ranges of the assays correlated very well (r2 = 0.86). Furthermore, both tests performed equally well in determining an early viral response at week 1 or 4 during antiviral therapy. We also compared two qualitative HCV RNA detection assays: the COBAS AMPLICOR HCV 2.0 assay versus the transcription-mediated amplification (TMA)-based VERSANT HCV RNA qualitative assay. Stored samples from sustained responders to interferon-ribavirin therapy were retested by the HCV TMA assay and were found to contain no detectable HCV RNA, demonstrating complete concordance between the results of PCR and TMA. However, HCV RNA was detected by the TMA assay in end-of-treatment (ETR) samples from 33% of patients with relapses who were HCV RNA negative according to the COBAS AMPLICOR assay. This observation suggests that a TMA assay can lead to a more correct definition of the ETR response.
Measurement of hepatitis C virus (HCV) RNA levels has become an integral and increasingly important part of the management of patients with chronic HCV infection: (i) demonstration of the presence of HCV RNA (qualitative test) in the serum or plasma of subjects who test positive for anti-HCV antibodies is essential to prove the existence of an ongoing HCV infection; (ii) definition of the genotype of the infecting HCV isolate is needed to select the appropriate treatment schedule and to estimate the treatment outcome; (iii) measurement of the viral load (quantitative test) at the start of therapy and after 12 weeks of treatment is needed to decide about the usefulness of further treatment (stopping rule); and finally, (iv) demonstration of the presence or absence of HCV RNA (qualitative test) at the end of treatment (ETR) or at the end of follow-up (EFU), 6 or more months later, discriminates sustained responders (SRs) from relapsers (RELs) (3, 7, 8, 12, 17, 19, 21-23, 25, 30, 42, 43).
Hepatitis C viral genomes generally appear in the body fluids of infected patients in numbers too low to be detected by simple and direct molecular hybridization-based techniques. Their detection and quantification require strong “amplification” methods that can be introduced as “target amplification,” as applied in PCR and transcription-mediated amplification (TMA), or as “signal amplification,” as applied in the branched DNA (bDNA) method. The assays most commonly used to quantify HCV RNA are based on the PCR or bDNA technique (3, 8, 23), methods that differ in analytical sensitivities and dynamic ranges. Whereas the former techniques are very sensitive and detect the smallest amounts of viral RNA, the latter techniques have a wide dynamic range and rarely need sample dilution. Apart from inherent technical shortcomings, the greatest limitations in managing patients with chronic HCV infection have been the lack of standardization of these assays. The recent introduction of a World Health Organization (WHO) international standard and the calibration of today's assays in international units (IU/ml) ensures consistency in the measurement and reporting of hepatitis C viral loads (28).
A semiautomated, PCR-based, quantitative assay for HCV RNA detection was produced by Roche Diagnostics (Mannheim, Germany) and marketed as the “complete bioanalytical system” (COBAS) AMPLICOR HCV Monitor assay. The performance characteristics of the second version of this assay (“HCV Monitor 2.0 assay”) have been analyzed in detail, and the dynamic range of quantification is 600 to 500,000 IU/ml (18, 27, 41). Roche also developed a new instrument for automated RNA extraction, the COBAS Ampliprep analyzer, which proved to be suitable for the routine molecular biology laboratory and equaled the manual extraction method in performance (37). Bayer Diagnostics (Tarrytown, N.Y.) introduced the semiautomated bDNA-based VERSANT HCV RNA 3.0 Assay (“HCV bDNA 3.0 assay”) for the quantification of HCV RNA in plasma. This direct assay has a broad dynamic range of 615 to 7,700,000 IU/ml. Because viral loads seldom exceed the upper limit of this assay, retesting of diluted samples is rarely needed (27, 39). Although the analytical sensitivity of the HCV bDNA 3.0 assay is now similar to the detection cutoff of the HCV Monitor 2.0 assay, these assays are still less sensitive than techniques for qualitative HCV RNA detection.
Qualitative assays for the detection of HCV RNA reveal the presence of HCV RNA and report a sample as “positive” or “negative” but do not measure the viral load. These assays remain clinically useful because of their superior analytical sensitivities in comparison with those of the current quantitative assays. The qualitative test from Roche Diagnostics is a PCR-based method called the HCV AMPLICOR 2.0 assay, which reliably detects HCV RNA concentrations down to 50 IU/ml. The qualitative test from Bayer Diagnostics, the TMA-based VERSANT HCV RNA Qualitative assay (“HCV TMA assay”), has a lower detection limit of 10 IU/ml.
In the present study we measured the HCV RNA levels in sera from 120 chronic HCV patients using two commercial quantitative assays: the HCV Monitor 2.0 assay with automated HCV RNA extraction (Ampliprep) and the HCV bDNA 3.0 assay, which is a direct technology that does not use HCV RNA extraction. The data obtained by both assays are compared, the correlations are analyzed, and the potential clinical impacts of discrepant HCV RNA quantifications by both assays on the tailoring of antiviral treatment are examined. We further analyzed the performances of two qualitative assays, the HCV AMPLICOR 2.0 assay and the HCV TMA assay, in a series of serum samples that scored “undetectable” by one or both of the quantitative assays. The analytical and diagnostic sensitivities of the TMA-based assay were further examined with a series of samples obtained from RELs at the ETR and from SRs during posttreatment follow-up (FU) and at the EFU. All these samples scored negative by the PCR-based qualitative test. The clinical implications of testing with highly sensitive HCV RNA detection systems are discussed.
MATERIALS AND METHODS
Patient selection and sample collection.
Serum samples derived from 120 patients chronically infected with HCV of genotype 1 were used here. All subjects participated in a therapeutic trial designed to study the influence of daily induction dose (5 MU/day) versus thrice-weekly dose (5 MU three times per week) of alpha interferon (IFN-α) monotherapy for 4 weeks, followed by a combination with ribavirin (1,000 or 1,200 mg/day) for the 4 next weeks and then maintenance treatment with the combination of IFN-α (3 MU/three times per week) and ribavirin (1,000 or 1,200 mg/day) in naïve chronic hepatitis C patients. The total treatment duration was 1 year (48 weeks) (40). Blood was taken before treatment and at weeks 1, 4, 8, 12, 24, and 48, which was the ETR. FU and EFU samples were obtained 6 and 12 months after ETR, respectively. Based on the qualitative HCV RNA results obtained by the HCV AMPLICOR 2.0 assay in samples collected at ETR, during FU, and at EFU, the patients could be classified as SRs, RELs, and nonresponders (NRs) to IFN-α-ribavirin treatment. Of the 454 chronic HCV patients (from the original study [40]), we first selected all genotype 1-positive patients. From these patients we selected 120 individuals (40 SRs, 40 RELs, and 40 NRs) from whom sufficient serum volumes were available at the different time points. The most important demographic and virological data for these 120 patients are shown in Table 1.
TABLE 1.
Demographic and virologic characteristics of patients
| Category (no. of patients)a | Ageb (yr) | Gender (no. of males/no. of females) | No. of patient infected with HCV genotypec:
|
|||
|---|---|---|---|---|---|---|
| 1a | 1b | 1b + 1a | 1b + non-1a | |||
| Virologic SRs (40) | 41.3 ± 12.4 | 17/7 | 8 | 28 | 4 | 0 |
| Virologic REL (40) | 43.8 ± 13.3 | 15/7 | 7 | 28 | 3 | 2 |
| Virologic NRs (40) | 51.5 ± 11.0 | 19/6 | 8 | 26 | 6 | 0 |
SRs, HCV RNA undetectable 24 weeks after discontinuation of therapy; RELs (ETR responders), HCV RNA undetectable at the end of treatment but with virologic relapse thereafter; NRs, HCV RNA positive at end of treatment and thereafter.
Values represent by averages ± standard deviations.
Genotyping was performed by a line probe assay (VERSANT HCV genotyping assay; Bayer Diagnostics).
For TMA testing, SR specimens were tested at week 72 (FU) or week 96 (EFU), depending on the serum availability at the given time point. If serum was available from both time points, the latest sample was analyzed. Furthermore, the size of the subset tested (16 of 40 SRs at ETR, 31 of 40 SRs at FU or EFU, and 21 of 40 RELs at ETR) was determined by the number of serum specimens available at each time point.
All sera were prepared, aliquoted, and stored at −80°C. Aliquots that had never been thawed before were used for these experiments.
All subjects gave written, informed consent to participate in these studies, which were approved by the Ethical Review Board of the Ghent University Hospital.
HCV genotyping.
Genotyping was performed by using a line probe assay (VERSANT HCV genotyping assay; Bayer Diagnostics), in accordance with the manufacturer's instructions.
COBAS Ampliprep automated sample preparation.
The COBAS Ampliprep, a new instrument developed to automate sample preparation (15), was used here to treat 250 μl of serum or plasma, according to the manufacturer's instructions. We (unpublished data) and others (37) demonstrated that the results obtained by the COBAS Ampliprep and COBAS AMPLICOR HCV Monitor test were comparable to those obtained by the COBAS AMPLICOR HCV Monitor test by use of a manual extraction protocol.
HCV RNA determinations.
Measurements of serum HCV RNA levels were performed by two qualitative and two quantitative detection methods in the context of a comparative evaluation of these methods. HCV RNA was detected qualitatively by the HCV AMPLICOR 2.0 assay (Roche Diagnostics) (24) and the HCV TMA assay (Bayer Diagnostics) (34). Quantitative HCV RNA measurements were performed by the HCV Monitor 2.0 assay (Roche Diagnostics) and the third-generation HCV bDNA 3.0 assay (Bayer Diagnostics). All procedures were performed according to the manufacturers' instructions.
(i) HCV Monitor 2.0 assay.
The HCV Monitor 2.0 assay has a reported lower limit of quantification of 600 IU/ml and an upper limit of 5 × 105 IU/ml (18, 27). Specimens yielding values above the upper limit were routinely diluted 100-fold and retested. Measured values were multiplied by this dilution factor to obtain the actual HCV RNA concentration in IU/ml.
(ii) HCV bDNA 3.0 assay.
The bDNA technology has a sandwich nucleic acid hybridization format. Briefly, the HCV bDNA 3.0 assay provides an improved means for singular measurements of plasma and serum samples. Greater sensitivity was achieved by making improvements in probe design and increasing the number of probes, and the assay background was reduced by the incorporation of synthetic nonnatural nucleosides. A second level of background reduction was achieved by redesigning the label extenders. The whole process is performed in a semiautomated Bayer system 340 bDNA analyzer, which automatically performs all incubations, washing steps, readings, and data processing. The instrument can process two 96-microwell plates per run, i.e., 168 patient samples and 24 calibrators/day. The dynamic range of quantification of the HCV bDNA 3.0 assay reaches from 615 to 7.7 × 106 IU/ml. The assay proved to be linear within its entire dynamic range of HCV RNA quantification (31). Therefore, samples never had to be diluted. The specificity, reproducibility, sensitivity, and linear range of quantification of the HCV bDNA 3.0 assay were described in detail by Ross et al. (31). The lowest concentration of HCV RNA that produces a quantitative result in 95% of replicate specimens (95% detection limit) was found to be 990 IU/ml (10).
(iii) HCV AMPLICOR 2.0 assay.
In the HCV AMPLICOR 2.0 assay, cDNA is made from HCV RNA by reverse transcription and is then amplified by PCR under a single set of conditions with the DNA polymerase of Thermus thermophilus (a single-tube, single-enzyme, and single-primer-set process). The assay uses 200 μl of plasma for RNA extraction and is independent of the HCV genotype (18). The lower detection limit of the HCV AMPLICOR 2.0 assay is 50 IU/ml (27).
(iv) HCV TMA assay.
The HCV TMA assay consists of three steps that are performed in a single tube: target capture, target amplification, and specific detection of target amplicons by a hybridization protection assay (33). An internal control is added to each sample before the extraction step and is processed through the assay. The appropriate signal generated from the internal control is used as an indication that the result is valid. The assay employs 500 μl of plasma or serum and has previously been described in detail (34). According to the WHO HCV RNA standard, the analytical sensitivities of the TMA assay are 96% at 5 IU/ml and 100% at 10 IU/ml. The sensitivity of the assay is not influenced by the HCV genotype (35). The clinical specificity is >99.5%. No effect on the assay performance was observed by potentially interfering endogenous substances or multiple freeze-thaw cycles (13, 14, 32).
Data analysis.
Linear regression analysis was done by using scatter plots for log-transformed HCV RNA levels. Comparison of the frequencies between different groups was analyzed by the χ2 test. Prediction of therapy outcome was analyzed by using negative predictive values (NPVs; %) and positive predictive values (PPVs; %) for SRs. NPV refers to the number of samples with true-negative results/(number of samples with true-negative results + number of samples with false-negative results), and PPV refers to the number of samples with true-positive results/(number of samples with true-positive results + number of samples with false-positive results).
RESULTS
Performances of two quantitative HCV RNA detection assays: correlation between the HCV bDNA 3.0 assay with manual RNA extraction and the HCV Monitor 2.0 assay with automated RNA extraction (Ampliprep).
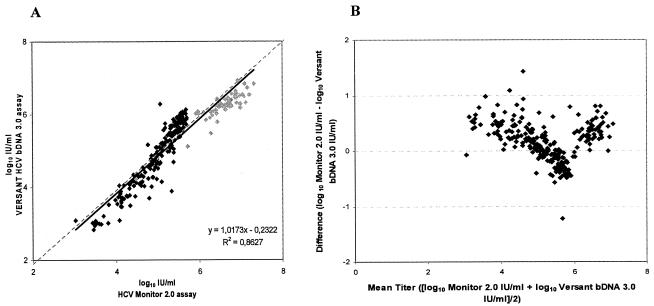
In a first series of experiments the HCV RNA contents of 344 blood samples were quantified by the HCV Monitor 2.0 assay and the HCV bDNA 3.0 assay. Two hundred eighty samples scored positive and 48 scored negative in both tests, while 16 scored positive in one or the other assay (Table 2). This results in an overall concordance of 95.3% (328 of 344) between both systems. A regression analysis of the 280 data sets (positive test results in both assays) revealed that both assays showed a good correlation (r2 = 0.86) (Fig. 1A). However, 14% of all paired results differed by more than 0.5 log10 IU/ml, which is a deviation that is regarded as clinically important in consecutive viral load determinations (1, 29). Figure 1B, in which the differences between the values reported by both formats is plotted against the average of the two values, further confirms the observation that the HCV Monitor 2.0 assay “overestimates” the HCV RNA content and that the HCV bDNA 3.0 assay “underestimates” the HCV RNA content. The absolute quantification results revealed that the mean HCV RNA concentration was 5.35 ± 0.84 log10 IU/ml when it was determined by the HCV Monitor 2.0 assay and 5.21 ± 0.93 log10 IU/ml when it was determined by the HCV bDNA 3.0 assay. The median difference between both assays was 0.16 log10 IU/ml. Overall, we can conclude that both quantification assays yielded comparable results.
TABLE 2.
Performances of two quantitative HCV-RNA detection assaysa
| HCV bDNA 3.0 assay result | No. of samples with the following HCV Monitor 2.0 assay result:
|
|
|---|---|---|
| + | − | |
| + | 280 | 3 |
| − | 13 | 48 |
A series of 344 samples were quantitatively measured by the HCV bDNA 3.0 assay and the HCV Monitor 2.0 Assay.
FIG. 1.
Comparison of VERSANT HCV bDNA 3.0 assay and HCV Monitor 2.0 assay. (A) Overall correlation of both tests within the dynamic ranges of the assays. The graph represents the 280 samples from Table 2 with positive test results by both quantitative assays. The results are shown after log10 transformation of the data. Samples were diluted 100-fold for determination of HCV RNA loads above the upper limit of the HCV Monitor 2.0 assay (500 000 IU/ml; grey shading). Linearity is shown as a dotted line, and the correlation curve is shown as a solid line. (B) For each specimen, the difference between the values reported by the VERSANT HCV bDNA 3.0 format and the HCV Monitor 2.0 format is plotted against the average of the two values.
Only 16 samples were classified differently by the two assays (Table 2): 13 samples were positive by the HCV Monitor 2.0 assay (geometric mean, 3.4 log10 IU/ml; range, 2.8 to 6.9 log10 IU/ml) and the results were below the 95% detection limit of the HCV bDNA 3.0 assay (<990 IU/ml) (10), whereas 3 samples were positive by the HCV bDNA 3.0 assay (geometric mean, 3.9 log10 IU/ml; range, 3.4 to 4.9 log10 IU/ml) and the results were below the detection limit of the HCV Monitor 2.0 assay kit (<600 IU/ml) (18).
Clinical relevance of discordant HCV RNA quantification by different assays.
The clinical relevance of discordant HCV RNA results by both assays was tested by examining the possible impact of the results on therapeutic decisions. According to the European (European Association for the Study of the Liver [EASL]) (11) and American (American Society for the Study of Liver Disease [AASLD]) (38) consensus recommendations, naïve patients infected by HCV of genotype 1 should receive medication (IFN plus ribavirin) for 48 weeks. However, therapy should be discontinued in patients who do not show an early virological response (EVR) after 3 months of therapy. This EVR has been defined as a drop in viral load of at least 2 log10 units or a complete absence of HCV RNA by week 12 of therapy. In the present study, early viral dynamics were monitored retrospectively at weeks 1 and 4 of therapy in a cohort of SRs, RELs, and NRs. In Table 3 the 2-log HCV RNA decline at weeks 1 and 4 was analyzed retrospectively in the three different patient groups. When SRs were compared to non-SRs, which are RELs and NRs combined, the PPVs for SR are comparable (<1% difference) for both assays (week 1, 70.8% and 70.0%, respectively; week 4, 56.6% and 57.4%, respectively). The NPVs are slightly higher (>3% difference) when the results were measured by the HCV bDNA 3.0 assay (week 1, 74.7% and 77.8%, respectively; week 4, 85.2% and 89.7%, respectively). Overall, both HCV RNA quantification assays yielded clinically comparable results with regard to the early viral dynamics.
TABLE 3.
Comparison of HCV RNA 2-log decline at weeks 1 and 4, as determined by the HCV Monitor 2.0 assay and the HCV bDNA 3.0 assay
| Wk | Assay | No. of patients with 2-log HCV RNA decrease/total no.a
|
PPV (%)b (SR↔NSR) | NPV (%)c (SR↔NSR) | P value (χ2 test) | ||
|---|---|---|---|---|---|---|---|
| SRs | NSRs
|
||||||
| RELs | NRs | ||||||
| 1 | HCV Monitor 2.0 | 17/39 | 7/35 | 0/37 | 70.8 | 74.7 | 3.5 × 10−5 |
| HCV bDNA 3.0 | 21/39 | 9/34 | 0/38 | 70.0 | 77.8 | 2.8 × 10−6 | |
| 4 | HCV Monitor 2.0 | 30/39 | 22/37 | 1/38 | 56.6 | 85.2 | 2.6 × 10−6 |
| HCV bDNA 3.0 | 31/37 | 22/37 | 1/38 | 57.4 | 89.7 | 1.2 × 10−7 | |
HCV RNA measurements that scored “undetectable” by the quantitative HCV RNA detection assays were retested by the TMA-based VERSANT HCV qualitative assay. Samples scoring “reactive” by this assay were given an HCV RNA value of 2.79 log10 IU/ml; samples scoring “nonreactive” were given an HCV RNA value of 1.00 log10 IU/ml.
PPV, % meeting the criteria for sustained viral response who were SRs.
NPV, % not meeting the criteria for a sustained viral response who were nonsustained responders (NSRs).
The absence of a 2-log drop at week 4 of therapy has an NPV for SRs of 85 to 90%, which means that 10 to 15% of these patients will still turn out to be SRs.
Performances of two qualitative HCV RNA detection assays.
Qualitative HCV RNA assays remain clinically useful because they are analytically more sensitive than the current quantitative assays. Here we have retested 64 samples with HCV RNA levels below the detection limit of both or one or the other of the quantitative assays (Table 2) by two qualitative HCV RNA assays: the COBAS HCV AMPLICOR 2.0 assay and the TMA-based HCV RNA qualitative assay. Table 4 demonstrates that all 16 (13 + 3) discordant samples shown in Table 2 were reactive by both qualitative assays. This suggests that these samples have viral loads between 50 and ±600 IU/ml and demonstrates that values close to the detection limits of quantitative assays should be interpreted with caution. Among the 48 serum samples that were concordantly “negative” (below detection limit) by both quantitative assays (Table 2), 26 samples (54%) were reactive in the HCV TMA assay and 20 samples (42%) scored positive by the AMPLICOR assay.
TABLE 4.
Performances of two qualitative HCV RNA detection assaysa
| Assay result | No. of samples with the indicated result by the following assay:
|
||||
|---|---|---|---|---|---|
| Total | HCV qualitative assayb (HCV TMA assay)
|
COBAS AMPLICOR HCV 2.0 assayc
|
|||
| R | NR | + | − | ||
| Negative by HCV bDNA 3.0 assay and positive by HCV Monitor 2.0 assay | 13 | 13 | 0 | 13 | 0 |
| Positive by HCV bDNA 3.0 assay and negative by HCV Monitor 2.0 assay | 3 | 3 | 0 | 3 | 0 |
| Negative by HCV bDNA 3.0 assay and negative by HCV Monitor 2.0 assay | 48 | 26 | 22 | 20 | 28 |
A series of 64 samples with HCV RNA loads below the detection limit of one or both of the quantitative assays were retested by two qualitative HCV RNA assays: the COBAS AMPLICOR HCV 2.0 assay and the TMA-based VERSANT HCV qualitative assay.
The analytical sensitivity of the HCV TMA assay is 10 IU/ml. R, reactive; NR, nonreactive.
The lower detection limit of the COBAS AMPLICOR assay is 50 IU/ml.
Clinical relevance of discordant qualitative HCV RNA measurements by different assays.
The clinical relevance of discordant qualitative HCV RNA measurements was evaluated by examining the effect on the classification of subjects as RELs and SRs. The initial classification of the patients was based on the results obtained by the HCV AMPLICOR 2.0 assay. Individuals with undetectable HCV RNA at ETR, during FU, and at EFU were considered SRs. RELs had undetectable HCV RNA at ETR but had a virological relapse (i.e., they were HCV RNA positive) after the discontinuation of therapy. To check whether these individuals were correctly classified, we retrospectively retested ETR samples from RELs and FU or EFU samples from SRs by the more sensitive TMA method. Detection of HCV RNA in ETR samples of RELs would require reclassification of these patients as NRs rather than RELs. Detection of residual HCV RNA in FU or EFU samples of SRs would reclassify these patients as RELs rather than SRs. For the group of SRs, 16 samples obtained at ETR (week 48) and 31 samples obtained during FU (week 72) or at EFU (week 96) were available. In the group of RELs, 21 samples obtained at ETR could be examined.
Among patients who were SRs to IFN-α plus ribavirin treatment, residual HCV RNA was not detected by the TMA assay in the 31 FU or EFU samples, nor was it detected in the 16 ETR samples (results not shown). Complete concordance between the AMPLICOR assay and the TMA assay-based results was observed for this group, which brings us to the conclusion that all SRs (at least the 31 SRs examined) were correctly classified by the HCV AMPLICOR 2.0 assay.
Table 5 shows a full comparison of the sensitivities of the HCV AMPLICOR 2.0 assay and the TMA assay for all the samples we had available from the relapse patients. We observed three different groups. In the first group, which consisted of 13 patients, HCV RNA could not be detected by either assay at ETR. These individuals can be considered as “true” relapsers. The treatment week in which the HCV RNA result turned negative by both assays differed from patient to patient: subject 8 cleared the virus after 1 week of treatment, and in subject 38 this occurred at the very end of treatment. In the second group, which consisted of one patient (subject 3), TMA revealed a breakthrough at week 16 that went undetected by the AMPLICOR assay. In the third group (7 of 21 relapsers), HCV RNA was not detected by the AMPLICOR assay at ETR but could be found by the TMA assay. Three of these relapsers (RELs 16, 37, and 39) were TMA reactive throughout the treatment period and can therefore be considered NRs. Three others (subjects 4, 12, and 40) were HCV RNA negative by both assays at some point during treatment and can therefore be considered patients with “breakthroughs.”
TABLE 5.
Performances of two qualitative HCV RNA detection assays in relapse patientsa
| Time of analysis | Assayb | Result for relapse patient:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 15 | 34 | 36 | 20 | 35 | 11 | 14 | 17 | 19 | 23 | 33 | 38 | 3 | 4 | 12 | 13 | 16 | 37 | 39 | 40 | ||
| Wk 1 | Ampl. 2.0 | −c | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| TMA | −c | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Wk 4 | Ampl. 2.0 | −d | −d | − | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | − |
| TMA | − | +d | +d | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | − | |
| Wk 8 | Ampl. 2.0 | −c | −c | −c | −d | + | + | −d | + | −d | −d | + | + | − | − | + | + | + | −d | − | ||
| TMA | −c | −c | −c | +d | + | + | +d | + | +d | +d | + | + | − | − | + | + | + | +d | − | |||
| Wk 12 | Ampl. 2.0 | − | + | + | + | −d | −d | −d | − | |||||||||||||
| TMA | − | + | + | + | +d | +d | +d | − | ||||||||||||||
| Wk 16 | Ampl. 2.0 | − | − | −c | −d | −d | −d | −d | −d | −d | − | − | −d | −d | −d | |||||||
| TMA | − | − | −c | +d | +d | +d | +d | +d | +d | − | − | +d | +d | +d | ||||||||
| Wk 20 | Ampl. 2.0 | − | − | −c | −c | −d | + | −d | −d | −d | − | |||||||||||
| TMA | − | − | −c | −c | +d | + | +d | +d | +d | − | ||||||||||||
| Wk 24 | Ampl. 2.0 | − | − | − | − | + | −d | −d | −d | − | ||||||||||||
| TMA | − | − | − | − | + | +d | +d | +d | − | |||||||||||||
| Wk 36 | Ampl. 2.0 | − | − | −d | −d | −d | ||||||||||||||||
| TMA | − | − | +d | +d | +d | |||||||||||||||||
| ETR | Ampl. 2.0 | − | − | − | − | − | − | − | −c | −c | −c | −c | −c | −c | − | −d | −d | −d | −d | −d | −d | −d |
| TMA | − | − | − | − | − | − | − | −c | −c | −c | −c | −c | −c | − | +d | +d | +d | +d | +d | +d | +d | |
| FU | Ampl. 2.0 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| EFU | Ampl. 2.0 | + | + | + | + | + | NDe | + | + | + | + | + | + | + | + | ND | ND | ND | ND | ND | ND | + |
A series of samples from 21 relapse patients taken at different time points during treatment were measured by both qualitative HCV RNA assays.
Ampl. 2.0, COBAS AMPLICOR HCV 2.0 assay; TMA, VERSANT HCV qualitative assay (HCV TMA assay).
Treatment-week wherein HCV RNA turns negative by both assays.
Discordant HCV-RNA results by both qualitative assays.
ND, not determined.
Furthermore, we observed that the treatment weeks in which the HCV RNA result turned negative by the AMPLICOR 2.0 assay and the TMA assay were identical for seven relapsers (RELs 3, 4, 12, and 40 with breakthroughs and RELs 8, 35, and 36). In 13 relapsers, the TMA assay was able to detect HCV RNA several weeks after the AMPLICOR 2.0 assay result had turned negative.
DISCUSSION
The routine diagnosis of HCV infection is currently based on the detection of antibodies to HCV. The crucial differentiation between ongoing and resolved infection, however, is achieved by a qualitative HCV RNA determination, such as that measured by the HCV AMPLICOR assay. This and other qualitative, nonquantitative assays can detect the presence of HCV RNA, but they cannot measure HCV viral loads. Advances in molecular biology, however, have made it possible to quantify HCV RNA levels in human body fluids. Subsequently, the quantification of HCV RNA has become an increasingly important tool in different clinical settings, for instance, (i) in the evaluation of patients infected chronically with HCV prior to the commencement of antiviral treatment, (ii) in response to this treatment, and (iii) in the design of stopping rules based on the definition of an EVR which is predictive of the response to continued treatment. To meet these clinical goals, quantification of serum HCV RNA needs to be specific, sensitive, reproducible, and standardized. Initially, “in-house” reverse transcription-PCR amplification-based techniques were used; but they lacked standardization, reproducibility, and accuracy (26). The subsequent development of commercial assays, namely, the noncompetitive PCR-based AMPLICOR HCV Monitor technique (Roche Diagnostics) and the “branched DNA”-based signal amplification technique (VERSANT HCV RNA 3.0 assay [bDNA]; Bayer Diagnostics) enabled HCV RNA to be quantified in any laboratory equipped for molecular biology-based methods. The performance of these assays has been extensively reviewed (26, 27). A WHO international standard has been used to calibrate HCV RNA concentration panels and to express HCV RNA loads in IU/ml. Nowadays, both companies have semiautomated HCV RNA quantification assays that give results in IU/ml: the COBAS AMPLICOR HCV Monitor assay version 2.0 and the VERSANT HCV bDNA 3.0 assay. In the present study we compared the quantitative results obtained by both assays. Since comparison of the HCV RNA data from 344 patient specimens resulted in only 5% discrepant results, we can conclude that there is an overall concordance between both systems. Comparison of the HCV RNA titers within the dynamic ranges of both assays revealed a good correlation (r2 = 0.86), and we can conclude that both assays perform equally well in determining HCV viral loads. The slightly stronger correlations reported by two other groups (2, 31) can probably be explained by the automated Ampliprep HCV RNA extraction device that was used in the present study. We further observed that 14% of all paired results differed by more than 0.5 log10 IU/ml, i.e., a deviation that is generally regarded as clinically important in consecutive viral load determinations (29). This agrees with the observation of Ross et al. (31), who reported that 11% of the results differed by this amount.
Discordant samples consisted of 3 samples (0.9%) in which HCV RNA was undetectable when it was measured by the HCV Monitor assay but that scored HCV RNA positive when it was measured by the HCV bDNA assay and 13 samples (3.8%) with opposite detection patterns. Recently, similar percentages were observed by Shiffman et al. (36). Retesting of these discrepant samples by the TMA assay revealed that all samples contained HCV RNA. We thus conclude that a greater number of false-negative results was obtained by the bDNA 3.0 assay. The higher analytical sensitivity of the Monitor platform is not surprising, since the 95% limit of detection of the bDNA 3.0 assay is estimated to be 990 IU/ml (10), whereas the detection limit of the HCV Monitor 2.0 assay is 600 IU/ml (18).
The clinical relevance of discordant HCV RNA quantifications obtained by both assays was evaluated for the possible impact on the discontinuation of antiviral therapy. It was demonstrated that testing for HCV RNA at critical time points during therapy might allow clinicians to identify NRs early in the treatment course (6, 16). According to the European (EASL) (11) and American (AASLD) (38) consensus guidelines, it was recommended that therapy be discontinued in patients who do not achieve an EVR (i.e., do not achieve a fall in HCV RNA levels to undetectable levels or a decrease of at least 2 log10 units) after 3 months of therapy (12, 20). However, because of considerable side effects and treatment costs, it is highly desirable to identify NRs as early as possible. In the present study we investigated the prognostic relevance for SRs of determining an EVR at week 1 or 4 of antiviral therapy. Simultaneously, we evaluated the performance characteristics of both quantitative methods in identifying the presence of an EVR. The results demonstrated similar EVR determinations when they were measured by both assays. However, for prospective purposes, the number of EVRs observed in SRs and not observed in RELs and NRs is important. Comparable PPVs for SRs were found when EVR was determined by both assays. NPVs were found to be slightly elevated when they were measured by the HCV bDNA 3.0 assay. We can conclude that both HCV RNA quantification systems yielded clinically comparable results with regard to the early viral dynamics. However, NPVs for SRs of 85 to 90% at week 4 is unacceptable as a decision threshold for treatment discontinuation because this would imply that, by using this test combination as a stopping rule, treatment would be discontinued in too many patients who might ultimately respond after completion of the full course of therapy. Recently, Davis (6) and Castro et al. (4) also described the NPVs of various time point definitions of EVR. Despite the differences in their experimental setups, similar NPVs for EVR (82%) at week 4 were observed. This finding justifies the EASL and AASLD recommendations to use the viral measurements obtained at week 12, because at that time the NPV for SRs of “the absence of a 2-log drop” is at least 97% (12).
In the second part of this study we evaluated two commercially available qualitative assays for HCV RNA determination. Qualitative assays remain clinically useful because they are more sensitive (they have lower detection cutoffs) for HCV RNA detection than the current quantitative assays. The performances of the two available qualitative HCV RNA detection platforms, TMA and AMPLICOR (v2.0) were compared by retesting 48 samples that scored below the detection cutoffs by both quantitative assays. Whereas 26 of the 48 samples scored “reactive” by the HCV TMA assay, only 20 samples scored positive by the Roche AMPLICOR assay. We can conclude that the higher frequency of TMA-reactive samples corresponds with the reported higher sensitivity of the TMA-based assay.
The clinical relevance of discordant HCV RNA qualitative measurements was evaluated by the possible “misclassification” of RELs and SRs. Routine classification of patients was based on HCV RNA testing by the COBAS AMPLICOR assay. Individuals with undetectable HCV RNA at ETR and FU-EFU were classified as SRs. In individuals classified as RELs, HCV RNA was not detectable at ETR, but a virological relapse occurred after the discontinuation of therapy. Since it was not inconceivable that the high sensitivity of the HCV TMA assay allowed the detection of residual HCV RNA in ETR specimens from RELs and in FU or EFU samples of virological SRs (33), we retrospectively tested stored ETR, FU, or EFU samples. None of the SR samples contained detectable HCV RNA by the TMA assay. A complete concordance between PCR- and TMA-based assay results was thus observed, and we can conclude that the sensitivities of the currently available PCR-based assays are sufficient for assessment of SRs 24 or 48 weeks after the termination of therapy. However, HCV RNA was detectable by TMA in ETR samples from 33% of the RELs who were HCV RNA negative, according to COBAS AMPLICOR HCV version 2.0. These observations are in accordance with previously reported results (5, 34), in which residual HCV RNA was detected by the HCV TMA assay in ETR samples in 34 to 36% of REL patients after standard therapy with IFN-α with or without ribavirin. Sarrazin et al. (34) further detected residual HCV RNA in 33% of ETR samples from REL patients after standard IFN-α-2a therapy and in 7% of REL patients following pegylated IFN-α-2a therapy. The lower rate of detection of residual HCV RNA in patients treated with pegylated IFN-α-2a may be due to the pharmacokinetics of this drug, which may lead to maintained antiviral pressure on HCV replication beyond the time of treatment discontinuation. Clinically, based on the TMA results, a considerable proportion (plus or minus one-third) of previous RELs has to be considered NRs or patients with a “breakthrough.” Future studies will show whether these patients benefit from prolonged antiviral therapy, as was suggested recently (9).
Acknowledgments
This work was supported by the Concerted Research Initiative of the University of Ghent (GOA, no. 12050203).
We are grateful to C. Laurent from Bayer Diagnostics for his scientific and technical support in performing the qualitative (TMA) and quantitative (bDNA) VERSANT HCV RNA assays. We thank N. Aelbrecht from Roche Diagnostics for technical support in performing automated RNA extractions with the Cobas Ampliprep analyzer.
Both companies are acknowledged for their kind gifts of reagents and equipment.
REFERENCES
- 1.Anastassopoulou, C. G., G. Touloumi, A. Katsoulidou, H. Hatzitheodorou, M. Pappa, D. Paraskevis, M. Lazanas, P. Gargalianos, and A. Hatzakis. 2001. Comparative evaluation of the QUANTIPLEX HIV-1 RNA 2.0 and 3.0 (bDNA) assays and the AMPLICOR HIV-1 MONITOR v1.5 test for the quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Virol. Methods 91:67-74. [DOI] [PubMed] [Google Scholar]
- 2.Beld, M., R. Sentjens, S. Rebers, C. Weegink, J. Weel, C. Sol, and R. Boom. 2002. Performance of the New Bayer VERSANT HCV RNA 3.0 assay for quantitation of hepatitis C virus RNA in plasma and serum: conversion to international units and comparison with the Roche COBAS AMPLICOR HCV Monitor, Version 2.0, assay. J. Clin. Microbiol. 40:788-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carithers, R. L., Jr., A. Marquardt, and D. R. Gretch. 2000. Diagnostic testing for hepatitis C. Semin. Liver Dis. 20:159-171. [DOI] [PubMed] [Google Scholar]
- 4.Castro, F. J., J. I. Esteban, A. Juarez, S. Sauleda, L. Viladomiu, M. Martell, F. Moreno, H. Allende, R. Esteban, and J. Guardia. 2002. Early detection of nonresponse to interferon plus ribavirin combination treatment of chronic hepatitis C. J. Viral Hepat. 9:202-207. [DOI] [PubMed] [Google Scholar]
- 5.Comanor, L., F. Anderson, M. Ghany, R. Perrillo, E. J. Heathcote, C. Sherlock, I. Zitron, D. Hendricks, and S. C. Gordon. 2001. Transcription-mediated amplification is more sensitive than conventional PCR-based assays for detecting residual serum HCV RNA at end of treatment. Am. J. Gastroenterol. 96:2968-2972. [DOI] [PubMed] [Google Scholar]
- 6.Davis, G. L. 2002. Monitoring of viral levels during therapy of hepatitis C. Hepatology 36:S145-S151. [DOI] [PubMed] [Google Scholar]
- 7.Davis, G. L., and J. Y. Lau. 1997. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology 26:122S-127S. [DOI] [PubMed] [Google Scholar]
- 8.de Medina, M., and E. R. Schiff. 1995. Hepatitis C: diagnostic assays. Semin. Liver Dis. 15:33-40. [DOI] [PubMed] [Google Scholar]
- 9.Drusano, G. L., and S. L. Preston. 2004. A 48-week duration of therapy with pegylated interferon alpha 2b plus ribavirin may be too short to maximize long-term response among patients infected with genotype-1 hepatitis C virus. J. Infect. Dis. 189:964-970. [DOI] [PubMed] [Google Scholar]
- 10.Elbeik, T., J. Surtihadi, M. Destree, J. Gorlin, M. Holodniy, S. A. Jortani, K. Kuramoto, V. Ng, R. Valdes, Jr., A. Valsamakis, and N. A. Terrault. 2004. Multicenter evaluation of the performance characteristics of the Bayer VERSANT HCV RNA 3.0 assay (bDNA). J. Clin. Microbiol. 42:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Association for the Study of the Liver. 1999. EASL International Consensus Conference on Hepatitis C. Paris, 26-28, February 1999, consensus statement. J. Hepatol. 30:956-961. [PubMed] [Google Scholar]
- 12.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 13.Gorrin, G., M. Friesenhahn, P. Lin, M. Sanders, R. Pollner, B. Eguchi, J. Pham, G. Roma, J. Spidle, S. Nicol, C. Wong, S. Bhade, and L. Comanor. 2003. Performance evaluation of the VERSANT HCV RNA qualitative assay by using transcription-mediated amplification. J. Clin. Microbiol. 41:310-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendricks, D. A., M. Friesenhahn, L. Tanimoto, B. Goergen, D. Dodge, and L. Comanor. 2003. Multicenter evaluation of the VERSANT HCV RNA qualitative assay for detection of hepatitis C virus RNA. J. Clin. Microbiol. 41:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jungkind, D. 2001. Automation of laboratory testing for infectious diseases using the polymerase chain reaction—our past, our present, our future. J. Clin. Virol. 20:1-6. [DOI] [PubMed] [Google Scholar]
- 16.Layden, J. E., and T. J. Layden. 2002. Viral kinetics of hepatitis C: new insights and remaining limitations. Hepatology 35:967-970. [DOI] [PubMed] [Google Scholar]
- 17.Layden, T. J., B. Mika, and T. E. Wiley. 2000. Hepatitis C kinetics: mathematical modeling of viral response to therapy. Semin. Liver Dis. 20:173-183. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsay, K. L. 1997. Therapy of hepatitis C: overview. Hepatology 26:71S-77S. [DOI] [PubMed] [Google Scholar]
- 20.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 21.Martinot-Peignoux, M., P. Marcellin, M. Pouteau, C. Castelnau, N. Boyer, M. Poliquin, C. Degott, I. Descombes, V. Le Breton, V. Milotova, et al. 1995. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology 22:1050-1056. [PubMed] [Google Scholar]
- 22.McHutchison, J. G., and T. Poynard. 1999. Combination therapy with interferon plus ribavirin for the initial treatment of chronic hepatitis C. Semin. Liver Dis. 19(Suppl. 1):57-65. [PubMed] [Google Scholar]
- 23.Morishima, C., and D. R. Gretch. 1999. Clinical use of hepatitis C virus tests for diagnosis and monitoring during therapy. Clin. Liver Dis. 3:717-740. [DOI] [PubMed] [Google Scholar]
- 24.Nolte, F. S., M. W. Fried, M. L. Shiffman, A. Ferreira-Gonzalez, C. T. Garrett, E. R. Schiff, S. J. Polyak, and D. R. Gretch. 2001. Prospective multicenter clinical evaluation of AMPLICOR and COBAS AMPLICOR hepatitis C virus tests. J. Clin. Microbiol. 39:4005-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orito, E., M. Mizokami, K. Suzuki, K. Ohba, T. Ohno, M. Mori, K. Hayashi, K. Kato, S. Iino, and J. Y. Lau. 1995. Loss of serum HCV RNA at week 4 of interferon-alpha therapy is associated with more favorable long-term response in patients with chronic hepatitis C. J. Med. Virol. 46:109-115. [DOI] [PubMed] [Google Scholar]
- 26.Pawlotsky, J. M. 1997. Measuring hepatitis C viremia in clinical samples: can we trust the assays? Hepatology 26:1-4. [DOI] [PubMed] [Google Scholar]
- 27.Pawlotsky, J. M. 2002. Molecular diagnosis of viral hepatitis. Gastroenterology 122:1554-1568. [DOI] [PubMed] [Google Scholar]
- 28.Pawlotsky, J. M., M. Bouvier-Alias, C. Hezode, F. Darthuy, J. Remire, and D. Dhumeaux. 2000. Standardization of hepatitis C virus RNA quantification. Hepatology 32:654-659. [DOI] [PubMed] [Google Scholar]
- 29.Pawlotsky, J. M., M. Martinot-Peignoux, J. D. Poveda, A. Bastie, V. Le Breton, F. Darthuy, J. Remire, S. Erlinger, D. Dhumeaux, and P. Marcellin. 1999. Quantification of hepatitis C virus RNA in serum by branched DNA-based signal amplification assays. J. Virol. Methods 79:227-235. [DOI] [PubMed] [Google Scholar]
- 30.Reichard, O., G. Norkrans, A. Fryden, J. H. Braconier, A. Sonnerborg, and O. Weiland. 1998. Comparison of 3 quantitative HCV RNA assays—accuracy of baseline viral load to predict treatment outcome in chronic hepatitis C. Scand J. Infect. Dis. 30:441-446. [DOI] [PubMed] [Google Scholar]
- 31.Ross, R. S., S. Viazov, S. Sarr, S. Hoffmann, A. Kramer, and M. Roggendorf. 2002. Quantitation of hepatitis C virus RNA by third generation branched DNA-based signal amplification assay. J. Virol. Methods 101:159-168. [DOI] [PubMed] [Google Scholar]
- 32.Ross, R. S., S. O. Viazov, S. Hoffmann, and M. Roggendorf. 2001. Performance characteristics of a transcription-mediated nucleic acid amplification assay for qualitative detection of hepatitis C virus RNA. J. Clin. Lab. Anal. 15:308-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarrazin, C. 2002. Highly sensitive hepatitis C virus RNA detection methods: molecular backgrounds and clinical significance. J. Clin. Virol. 25(Suppl. 3):S23-S29. [DOI] [PubMed] [Google Scholar]
- 34.Sarrazin, C., G. Teuber, R. Kokka, H. Rabenau, and S. Zeuzem. 2000. Detection of residual hepatitis C virus RNA by transcription-mediated amplification in patients with complete virologic response according to polymerase chain reaction-based assays. Hepatology 32:818-823. [DOI] [PubMed] [Google Scholar]
- 35.Sawyer, L. K., M. Friesenhahn, D. Duey, M. McMorrow, and B. Eguchi. 2000. Clinical laboratory evaluation of a new sensitive and specific assay for qualitative detection of hepatitis C virus RNA in clinical specimens. J. Hepatol. 32(Suppl. 2):116A. [Google Scholar]
- 36.Shiffman, M. L., A. Ferreira-Gonzalez, K. R. Reddy, R. K. Sterling, V. A. Luketic, R. T. Stravitz, A. J. Sanyal, C. T. Garrett, M. De Medina, and E. R. Schiff. 2003. Comparison of three commercially available assays for HCV RNA using the international unit standard: implications for management of patients with chronic hepatitis C virus infection in clinical practice. Am. J. Gastroenterol. 98:1159-1166. [DOI] [PubMed] [Google Scholar]
- 37.Stelzl, E., A. Kormann-Klement, J. Haas, E. Daghofer, B. I. Santner, E. Marth, and H. H. Kessler. 2002. Evaluation of an automated sample preparation protocol for quantitative detection of hepatitis C virus RNA. J. Clin. Microbiol. 40:1447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strader, D. B., T. Wright, D. L. Thomas, and L. B. Seeff. 2004. Diagnosis, management, and treatment of hepatitis C. Hepatology 39:1147-1171. [DOI] [PubMed] [Google Scholar]
- 39.Trimoulet, P., P. Halfon, E. Pohier, H. Khiri, G. Chene, and H. Fleury. 2002. Evaluation of the VERSANT HCV RNA 3.0 assay for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 40:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Vlierberghe, H., G. Leroux-Roels, M. Adler, N. Bourgeois, F. Nevens, Y. Horsmans, J. Brouwer, I. Colle, J. Delwaide, R. Brenard, B. Bastens, J. Henrion, R. A. de Vries, C. de Galocsy, P. Michielsen, G. Robaeys, and L. Bruckers. 2003. Daily induction combination treatment with alpha 2b interferon and ribavirin or standard combination treatment in naive chronic hepatitis C patients. A multicentre randomized controlled trial. J. Viral Hepat. 10:460-466. [DOI] [PubMed] [Google Scholar]
- 41.Yu, M. L., W. L. Chuang, C. Y. Dai, S. C. Chen, Z. Y. Lin, M. Y. Hsieh, L. Y. Wang, and W. Y. Chang. 2000. Clinical evaluation of the automated COBAS AMPLICOR HCV MONITOR test version 2.0 for quantifying serum hepatitis C virus RNA and comparison to the quantiplex HCV version 2.0 test. J. Clin. Microbiol. 38:2933-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeuzem, S. 1999. Clinical implications of hepatitis C viral kinetics. J. Hepatol. 31(Suppl. 1):61-64. [DOI] [PubMed] [Google Scholar]
- 43.Zeuzem, S., J. H. Lee, A. Franke, B. Ruster, O. Prummer, G. Herrmann, and W. K. Roth. 1998. Quantification of the initial decline of serum hepatitis C virus RNA and response to interferon alfa. Hepatology 27:1149-1156. [DOI] [PubMed] [Google Scholar]