Abstract
To understand the transmission of Cryptosporidium infection in children, fecal specimens from 62 Kuwaiti children with gastrointestinal symptoms found to be positive by microscopy were genotyped and subtyped with a small subunit rRNA-based PCR-restriction fragment length polymorphism analysis and a 60-kDa glycoprotein-based DNA sequencing tool. The median age of infected children was 4.5 years, and 77% of infections occurred during the cool season of November to April. Fifty-eight of the children (94%) had Cryptosporidium parvum, three (5%) had Cryptosporidium hominis, and one (1%) had both C. parvum and C. hominis. Altogether, 13 subtypes of C. parvum (belonging to four subtype allele families) and C. hominis (belonging to three subtype allele families) were observed, with 92% of specimens belonging to the common allele family IIa and the unusual allele family IId. Thus, the transmission of cryptosporidiosis in Kuwaiti children differed significantly from other tropical countries.
Cryptosporidiosis is a significant cause of diarrheal diseases in both developing and industrialized nations. Recent molecular epidemiologic studies of cryptosporidiosis have helped researchers to better understand the transmission of cryptosporidiosis in humans and the public health significance of Cryptosporidium spp. in animals and the environment. Using genotyping tools, five species of Cryptosporidium (C. hominis, C. parvum, C. meleagridis, C. felis, and C. canis) have been shown to be responsible for most human infections. Of these five species, C. hominis and C. parvum are the two most common species (34). Because these five human pathogenic Cryptosporidium species have different spectrums of host specificity, the characterization of Cryptosporidium at the species level is useful in investigating infection and contamination sources. Recently, a number of subtyping tools have been developed and used to characterize the population structure and transmission dynamics of C. parvum and C. hominis (2, 8, 17, 18, 24-26, 29, 30).
Although cryptosporidiosis is prevalent in tropical regions, limited studies have been conducted to characterize Cryptosporidium spp. from humans at the molecular level. Several studies have examined the transmission of human cryptosporidiosis in South Africa, Malawi, Kenya, Uganda, Peru, and Thailand, all of which have shown a predominance of C. hominis in humans, indicating anthroponotic transmission plays a major role in the epidemiology of cryptosporidiosis in most tropical countries (7, 17, 25, 31-33). Only two of the studies subtyped small numbers of Cryptosporidium spp. (17, 25). In the present study, 62 Cryptosporidium-positive specimens were collected from children in Kuwait City between 1997 and 2004 and examined by a small subunit (SSU) rRNA-based PCR-restriction fragment length polymorphism (PCR-RFLP) analysis and a 60-kDa glycoprotein (GP60)-based PCR sequencing tool (2, 33). Results of the study have shown a predominance in children of C. parvum, which traditionally is associated with farm animals. Thus, anthroponotic transmission was possibly less important in human cryptosporidiosis in Kuwait City than in other tropical regions.
MATERIALS AND METHODS
Specimens.
Stool specimens from 62 Kuwaiti children submitted for Cryptosporidium diagnosis were used in this study. They were collected between January 2001 and February 2004, with the exception of four specimens, which were collected in 1997 and 1998. Minimum clinical data accompanied each stool specimen, such as age and sex of the patient, major clinical symptoms, and date of specimen collection. These children visited government hospitals for treatment because of gastrointestinal symptoms. They were diagnosed as positive for Cryptosporidium by microscopy of acid-fast stained fecal smears (11). Aliquots of the Cryptosporidium-positive specimens were preserved in 2.5% potassium dichromate and shipped to the laboratory at the Centers for Disease Control and Prevention in Atlanta, Georgia, for molecular analysis.
DNA isolation.
DNA was extracted from all 62 specimens after initial treatment with 1 M KOH for 15 min at 65°C followed by neutralization with 25% HCl. The DNA lysate was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) solution and purified using a QIAamp DNA stool kit (QIAGEN Inc, Valencia, CA) following the manufacturer's suggested protocols (35).
Cryptosporidium genotyping.
Initially, all specimens were genotyped by a PCR-RFLP technique (33). This technique amplifies an ∼830-bp fragment of the SSU rRNA gene by nested PCR and differentiates Cryptosporidium species or genotypes by banding patterns in restriction analysis of the secondary PCR products with the enzymes SspI and VspI.
C. parvum and C. hominis subtyping.
C. parvum and C. hominis in these specimens were further subtyped by a GP60-based tool (2), which amplifies an ∼850-bp fragment of the GP60 gene by nested PCR. For the specimens that failed to be amplified, a smaller fragment (∼400 bp) of the gene was amplified using primers AL3531 (5′-ATAGTCTCCGCTGTATTC-3′) and AL3533 (5′-GAGATATATCTTGGTGCG-3′) in primary PCR and AL3532 (5′-TCCGCTGTATTCTCAGCC-3′) and LX0029 (5′-CGAACCACATTACAAATGAAGT-3′). The nucleotide sequences obtained categorized C. parvum and C. hominis to many families of subtypes. This was done by alignment of GP60 sequences obtained in this study and reference sequences retrieved from GenBank using the program ClustalX (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/). To further support the grouping of subtypes, a neighbor-joining tree was generated from the aligned sequences using the program TreeCon (http://www.psb.rug.ac.be/bioinformatics/psb/Userman/treeconw.html) based on the genetic distances calculated by the Kimura two-parameter model. The GP60 tree was rooted with a GP60 sequence of C. meleagridis (AF401499). The reliability of branches was assessed by bootstrap analysis using 1,000 replicates.
Nucleotide sequence accession numbers.
The unique partial GP60 sequences generated in this study have been deposited in GenBank under accession numbers AY738184 to AY738196.
RESULTS
Cryptosporidiosis in children.
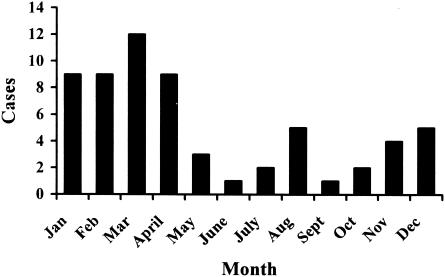
The 62 Kuwaiti children studied attended the hospitals because of acute gastroenteritis. Most of the children had diarrhea, but some of them also had accompanied abdominal pain (five children), vomiting (two children), or upper respiratory tract infection (two children). These children were between 1 and 19 years of age, with a median of 4.5 (5.4 ± 2.9) years. Thirty-six of the children were girls and 25 were boys (one child with unknown status). Seventy-seven percent of cases occurred during the months November to April (Fig. 1).
FIG. 1.
The seasonal distribution of cryptosporidiosis cases in Kuwaiti children.
Cryptosporidium genotypes.
DNA preparations of all 62 specimens yielded products of the expected size (∼830 bp) in the nested PCR analysis of the SSU rRNA gene. RFLP analyses of the secondary SSU rRNA PCR products with the restriction enzymes SspI and VspI showed that 58 of the children had C. parvum, three had C. hominis, and one had both C. parvum and C. hominis (Table 1). DNA sequencing of three C. parvum PCR products and one C. hominis product yielded SSU rRNA sequences identical to those previously reported for C. parvum and C. hominis, respectively (36).
TABLE 1.
Distribution of Cryptosporidium genotypes and subtype families in Kuwaiti children by age
| Age group | No. of cases | No. of patients infected witha:
|
||||
|---|---|---|---|---|---|---|
| C. parvum | C. hominis | IIa allele | IId allele | Other alleles | ||
| <2 years | 1 | 1 | 0 | 0 | 1 | 0 |
| 2-3 years | 20 | 19 | 2 | 11 | 8 | 3 |
| 4-6 years | 14 | 13 | 1 | 6 | 6 | 2 |
| 7-10 years | 15 | 15 | 0 | 2 | 12 | 1 |
| 11-19 years | 7 | 6 | 1 | 4 | 2 | 1 |
| Unknown | 5 | 5 | 0 | 5 | 0 | 0 |
| Total | 62 | 59 | 4 | 28 | 29 | 7 |
One child had mixed infections of C. parvum and C. hominis, and two additional children had mixed infections of two C. parvum subtypes.
C. parvum and C. hominis subtype alleles.
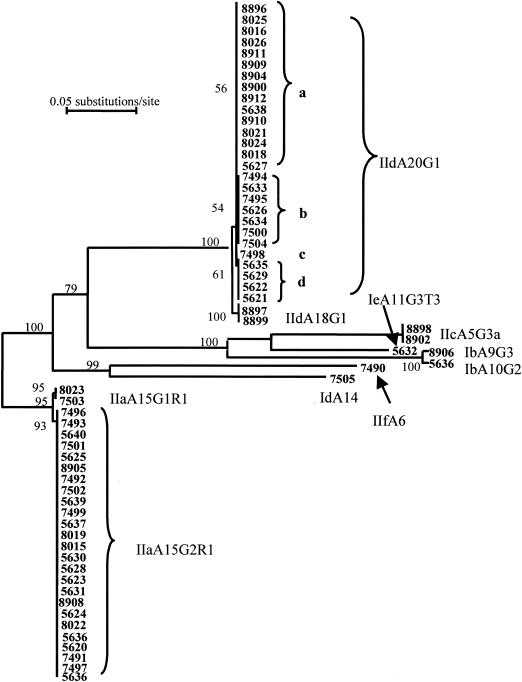
DNA of 53 specimens was also amplified using the regular GP60 primers. The DNA from the nine specimens (5620, 5624, 5627, 5636, 7491, 7497, 8021, 8022, and 8898) that failed to produce the expected PCR band (∼850 bp) was amplified by the new primer set, producing PCR products of about 400 bp. Both the long and short PCR products were sequenced, and the obtained sequences were aligned with sequences obtained from previous studies. Phylogenetic analysis of the sequences showed that these C. parvum and C. hominis isolates belonged to seven subtype families (Fig. 2). Most of the C. parvum specimens belonged to the previously described subtype families IIa and IId (Table 1). However, two specimens (8898 and 8902) had subtype family IIc (previously known as Ic), whereas another specimen (7490) belonged to a new C. parvum subtype family. The latter was named subtype family IIf. Generally, there was no significant difference between different age groups in the distribution of allele families IIa and IId. However, in the 7- to 10-year-old age group, 12 of the children were infected with allele IId but only 2 were infected with allele IIa (Table 1).
FIG. 2.
The presence of multiple C. parvum subtypes in Kuwaiti children as revealed by a neighbor-joining analysis of the sequences of GP60. IIa, IIc, IId, and IIf are C. parvum allele families; Ib, Id, and Ie are C. hominis allele families. The numbers on branches are bootstrap values greater than 50%.
The four specimens that were diagnosed to contain C. hominis were also subtyped. One specimen (7505) belonged to the subtype allele family Id and one (5632) to the allele family Ie, whereas two (5636 and 8906) belonged to the allele family Ib.
Nomenclature for the C. parvum and C. hominis subtypes.
Multiple subtypes were present in the C. parvum allele families IIa and IId in this study. Within each subtype family, subtypes differed from each other mostly in the number of trinucleotide repeats (TCA or TCG) coding for the amino acid serine. For example, in the allele family IIa, two subtypes were seen in Kuwaiti children; one subtype had 15 copies of the TCA repeat and 1 copy of the TCG repeat, whereas the other subtype had 15 copies of the TCA repeat and 2 copies of the TCG repeat. Therefore, the two subtypes were designated IIaA15G1R1 and IIaA15G2R1, respectively. In the subtype name IIaA15G1R1, IIa indicates that the subtype belongs to allele family IIa, A15 indicates that the subtype has 15 copies of the TCA repeat, and G1 indicates that the subtype has one copy of the TCG repeat. Because some subtypes have one copy of the sequence ACATCA immediately after the trinucleotide repeats whereas others have two copies of the sequence, R1 and R2 are used to differentiate these two types of sequences. Therefore, the only difference between subtypes IIaA15G1R1 and IIaA15G2R1 is the number of TCG repeats; one has one copy of the repeat, while the other has two copies.
Likewise, a similar system was used to name the subtypes in allele family IId. Most of the specimens had 20 copies of the TCA repeat and 1 copy of the TCG repeat. Two specimens, however, had 18 copies of the TCA repeat and 1 copy of the TCG repeat. The former was named IIdA20G1 and the latter IIdA18G1 to differentiate the two types of IId sequences. Unlike most other subtype allele families, subtypes within IId may also differ from each other slightly in the nonrepeat regions. For example, there were four types of sequences within IIdA20G1 seen in Kuwaiti children, which differ from each other in single nucleotide polymorphisms at three locations: a change of G to A (resulting in a change of amino acid from arginine to lysine) upstream of the trinucleotide repeats, a change of C to A (resulting in a change of amino acid from asparagine to glutamine) shortly after the repeats, and a change of G to A (a synonymous change) in the 3′ region (representative sequences for each subtype were submitted to the GenBank database). These four types of sequences were designated IIdA20G1a, IIdA20G1b, IIdA20G1c, and IIdA20G1d.
The same system was used to describe subtypes of other allele families. The only subtype (specimen 7505) in the C. hominis allele family Id was named IdA14 because of the presence of 14 copies of the TCA repeat, whereas the only subtype in the new C. parvum allele family IIf was named IIfA6 because of the presence of six copies of the TCA repeat. These two subtypes had no TCG repeat seen in many allele families. In contrast, the C. hominis subtype allele Ie seen in the study had 3 copies of the TCT repeat in addition to 11 copies of the TCA repeat and 3 copies of the TCG repeat and was named IeA11G3T3. Two subtypes of the C. hominis allele Ib, which were named IbA9G3 and IbA10G2 because of the differences in the number of TCA and TCG repeats, were seen in this study. The subtype in the C. parvum allele family IIc (previously known as Ic) was named IIcA5G3a because of the presence of five copies of the TCA repeat and three copies of the TCG repeat. Allele IIc differs significantly from other C. parvum and C. hominis allele families in the GP60 sequences. First, IIc has a short repeat region. Second, there is no difference in the number and type of the trinucleotide repeats; all subtypes in the family have five copies of the TCA repeat and three copies of the TCG repeat. Third, there are three major types of IIc sequences, which differ from each other significantly in the 3′ region of the gene. Thus, the subtype seen in this study was designated IIcA5G3a.
Distribution of C. parvum and C. hominis subtypes in Kuwaiti children.
Altogether, 13 subtypes belonging to seven C. parvum and C. hominis allele families were seen in Kuwaiti children. Most of the children (56 of 62) had subtypes in the C. parvum allele family IIa or IId, with almost equal numbers of children (28 and 29, respectively) infected with each allele. Most of the children infected with allele IIa parasites had subtype IIaA15G2R1; only two (specimens 7503 and 8023) had IIaA15G1R1. Likewise, most of the children with allele IId had GP60 sequences containing 20 copies of the TCA repeat and one copy of the TCG repeat (IIdA20G1); only two (specimens 8897 and 8899) had the subtype IIdA18G1. There were four subtypes within the sequence type IIdA20G1, with all recent specimens belonging to IIdA20G1a and most earlier specimens belonging to IIdA20G1b, IIdA20G1c, and IIdA20G1d (Fig. 2).
The other five allele families (C. parvum allele families IIc and IIf and C. hominis allele families Ib, Id, and Ie) were represented by only one or two children (Fig. 2). Three children were infected with mixed subtypes; specimen 5627 had IIdA20G1a and IIaA15G2R1 and specimen 5636 had IIaA15G2R1 and IbA10G2, whereas specimen 8896 had IIdA20G1a and an undecided subtype (poor sequence resolution because of the presence of mixed subtypes).
DISCUSSION
Results of the study confirm the presence of a unique endemicity of cryptosporidiosis revealed in an earlier study in Kuwait (11). Previous studies in various tropical countries have shown the highest prevalence of cryptosporidiosis in children younger than 2 years (3-5, 12-14, 21, 23, 27, 32). In contrast, Kuwaiti children infected with Cryptosporidium are significantly older than those in other areas of endemicity, a finding in agreement with an earlier study conducted in the same population (11). In a neighboring country, Saudi Arabia, children are also seemingly infected with Cryptosporidium later than those in other tropical countries (1). The delayed Cryptosporidium infection in Kuwaiti children was previously attributed to better hygiene in Kuwait than in other tropical countries (11).
In tropical countries, Cryptosporidium transmission in children is usually associated with the rainy season, and waterborne transmission is considered a major route in the epidemiology of cryptosporidiosis in these areas (3, 5, 13, 14, 21-23, 25, 27, 32). However, in Kuwait, there is very little rainfall, and Cryptosporidium transmission occurs mostly during the cool season of November to April. Waterborne transmission likely plays a less important role also because desalinated water is the major source of drinking water in Kuwait. In another study conducted in a tropical area with little rainfall, Lima, Peru, cryptosporidiosis in children was more frequent in the warm season (4). Summer in Lima, however, is significantly milder than in Kuwait City, where the daily high temperature frequently reaches over 40°C.
Differences in infection sources could also be an important factor responsible for the differences in Cryptosporidium transmission between Kuwait and other tropical countries. The distribution of Cryptosporidium genotypes in people is very different between Kuwait and other countries. Thus far, studies conducted in Peru, Thailand, Malawi, Uganda, Kenya, and South Africa showed a dominance of C. hominis in children or human immunodeficiency virus-positive adults (7, 17, 25, 31-33). In contrast, Kuwaiti children were almost exclusively infected with C. parvum. The only region where C. parvum is more often seen in humans than C. hominis is Europe, where several studies have shown a slightly higher prevalence of C. parvum than C. hominis in both immunocompetent and immunocompromised persons (2, 6, 9, 20). The differences in the distribution of Cryptosporidium genotypes in humans are considered an indication of differences in infection sources (15, 16, 20). Thus, the predominance of C. parvum in a population has been considered the result of zoonotic transmission. Indeed, even in areas with a high percentage of infections due to C. parvum, massive slaughter of farm animals during foot-and-mouth disease outbreaks can result in a reduction of the proportion of human infections due to C. parvum (10, 28).
Recent subtyping studies have shown that not all C. parvum infections in humans are results of zoonotic transmission (2, 19, 34). Among the C. parvum GP60 subtype families identified, alleles IIa and IIc (previously known as Ic) are the two most common ones. The former has been identified in both humans and ruminants, whereas the latter has been seen only in humans (2, 26, 34). In this study, two of the Kuwaiti children were infected with an allele IIc subtype, indicating that anthroponotic transmission of C. parvum occurs in Kuwait. Nevertheless, the low proportion of infections due to C. hominis suggests that anthroponotic transmission of cryptosporidiosis in Kuwait is probably not as important as in other countries.
Despite the high number of subtypes and allele families identified in Kuwaiti children, most of the parasites identified belong to two subtype families, IIa and IId. This is very different from results of studies conducted in South Africa, Malawi, Portugal, Northern Ireland, and the United States, in which positive samples were more or less evenly distributed among the common allele families Ia, Ib, Id, Ie, IIa, and IIc (2, 8, 17, 25, 29, 34). Even within each of the two common allele families in Kuwaiti children, IIa and IId, there were only minor genetic differences between the limited number of subtypes presented. The significance of this finding is not fully clear. Subtypes in the allele family IIa are commonly seen in both humans and ruminants in many areas, and in most areas studied, allele IIa is almost exclusively the only allele family found in cattle (2, 26). In contrast, subtypes of the allele family IId have so far only been found in six AIDS patients and three cattle in Portugal (2), none of which are the same subtypes seen in Kuwaiti children.
Not enough epidemiological data were collected to elucidate the source of C. parvum infection and reasons responsible for the unique endemicity of cryptosporidiosis in Kuwaiti children. A well-designed case control study, with detailed collection of data on water and food sources and animal contact, would be helpful in identifying risk factors involved in the acquisition of Cryptosporidium infection and in answering some of the above questions. Sampling of water and domestic animals would also be useful. In particular, subtyping C. parvum from domestic animals in Kuwait is needed in the determination of the extent of zoonotic transmission of cryptosporidiosis in Kuwaiti children.
Acknowledgments
We thank the Kuwait University (grant MI 03/03) for financial support.
REFERENCES
- 1.Al-Braiken, F. A., A. Amin, N. J. Beeching, M. Hommel, and C. A. Hart. 2003. Detection of Cryptosporidium amongst diarrhoeic and asymptomatic children in Jeddah, Saudi Arabia. Ann. Trop. Med. Parasitol. 97:505-510. [DOI] [PubMed] [Google Scholar]
- 2.Alves, M., L. Xiao, I. Sulaiman, A. A. Lal, O. Matos, and F. Antunes. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41:2744-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bern, C., B. Hernandez, M. B. Lopez, M. J. Arrowood, A. M. De Merida, and R. E. Klein. 2000. The contrasting epidemiology of Cyclospora and Cryptosporidium among outpatients in Guatemala. Am. J. Trop. Med. Hyg. 63:231-235. [PubMed] [Google Scholar]
- 4.Bern, C., Y. Ortega, W. Checkley, J. M. Roberts, A. G. Lescano, L. Cabrera, M. Verastegui, R. E. Black, C. Sterling, and R. H. Gilman. 2002. Epidemiologic differences between cyclosporiasis and cryptosporidiosis in Peruvian children. Emerg. Infect. Dis. 8:581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya, M. K., T. Teka, A. S. Faruque, and G. J. Fuchs. 1997. Cryptosporidium infection in children in urban Bangladesh. J. Trop. Pediatr. 43:282-286. [DOI] [PubMed] [Google Scholar]
- 6.Chalmers, R. M., K. Elwin, A. L. Thomas, and D. H. Joynson. 2002. Infection with unusual types of Cryptosporidium is not restricted to immunocompromised patients. J. Infect. Dis. 185:270-271. [DOI] [PubMed] [Google Scholar]
- 7.Gatei, W., J. Greensill, R. W. Ashford, L. E. Cuevas, C. M. Parry, N. A. Cunliffe, N. J. Beeching, and C. A. Hart. 2003. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. J. Clin. Microbiol. 41:1458-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guyot, K., A. Follet-Dumoulin, E. Lelievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter, P. R., R. M. Chalmers, Q. Syed, L. S. Hughes, S. Woodhouse, and L. Swift. 2003. Foot and mouth disease and cryptosporidiosis: possible interaction between two emerging infectious diseases. Emerg. Infect. Dis. 9:109-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iqbal, J., P. R. Hira, F. Al-Ali, and R. Philip. 2001. Cryptosporidiosis in Kuwaiti children: seasonality and endemicity. Clin. Microbiol. Infect. 7:261-266. [DOI] [PubMed] [Google Scholar]
- 12.Iqbal, J., M. A. Munir, and M. A. Khan. 1999. Cryptosporidium infection in young children with diarrhea in Rawalpindi, Pakistan. Am. J. Trop. Med. Hyg. 60:868-870. [DOI] [PubMed] [Google Scholar]
- 13.Javier Enriquez, F., C. R. Avila, J. Ignacio Santos, J. Tanaka-Kido, O. Vallejo, and C. R. Sterling. 1997. Cryptosporidium infections in Mexican children: clinical, nutritional, enteropathogenic, and diagnostic evaluations. Am. J. Trop. Med. Hyg. 56:254-257. [DOI] [PubMed] [Google Scholar]
- 14.Katsumata, T., D. Hosea, E. B. Wasito, S. Kohno, K. Hara, P. Soeparto, and I. G. Ranuh. 1998. Cryptosporidiosis in Indonesia: a hospital-based study and a community-based survey. Am. J. Trop. Med. Hyg. 59:628-632. [DOI] [PubMed] [Google Scholar]
- 15.Learmonth, J., G. Ionas, A. Pita, and R. Cowie. 2001. Seasonal shift in Cryptosporidium parvum transmission cycles in New Zealand. J. Eukaryot. Microbiol. 2001(Suppl.):34S-35S. [DOI] [PubMed] [Google Scholar]
- 16.Learmonth, J. J., G. Ionas, K. A. Ebbett, and E. S. Kwan. 2004. Genetic characterization and transmission cycles of Cryptosporidium species isolated from humans in New Zealand. Appl. Environ. Microbiol. 70:3973-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leav, B. A., M. R. Mackay, A. Anyanwu, R. M. O'Connor, A. M. Cevallos, G. Kindra, N. C. Rollins, M. L. Bennish, R. G. Nelson, and H. D. Ward. 2002. Analysis of sequence diversity at the highly polymorphic Cpgp40/15 locus among Cryptosporidium isolates from human immunodeficiency virus-infected children in South Africa. Infect. Immun. 70:3881-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallon, M., A. MacLeod, J. Wastling, H. Smith, B. Reilly, and A. Tait. 2003. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. J. Mol. Evol. 56:407-417. [DOI] [PubMed] [Google Scholar]
- 19.Mallon, M. E., A. MacLeod, J. M. Wastling, H. Smith, and A. Tait. 2003. Multilocus genotyping of Cryptosporidium parvum type 2: population genetics and sub-structuring. Infect. Genet. Evol. 3:207-218. [DOI] [PubMed] [Google Scholar]
- 20.McLauchlin, J., C. Amar, S. Pedraza-Diaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moodley, D., T. F. Jackson, V. Gathiram, and J. van den Ende. 1991. Cryptosporidium infections in children in Durban. Seasonal variation, age distribution and disease status. S. Afr. Med. J. 79:295-297. [PubMed] [Google Scholar]
- 22.Nath, G., A. Choudhury, B. N. Shukla, T. B. Singh, and D. C. S. Reddy. 1999. Significance of Cryptosporidium in acute diarrhoea in North-Eastern India. J. Med. Microbiol. 48:523-526. [DOI] [PubMed] [Google Scholar]
- 23.Newman, R. D., C. L. Sears, S. R. Moore, J. P. Nataro, T. Wuhib, D. A. Agnew, R. L. Guerrant, and A. A. M. Lima. 1999. Longitudinal study of Cryptosporidium infection in children in northeastern Brazil. J. Infect. Dis. 180:167-175. [DOI] [PubMed] [Google Scholar]
- 24.Peng, M. M., O. Matos, W. Gatei, P. Das, M. Stantic-Pavlinic, C. Bern, I. M. Sulaiman, S. Glaberman, A. A. Lal, and L. Xiao. 2001. A comparison of Cryptosporidium subgenotypes from several geographic regions. J. Eukaryot. Microbiol. 2001(Suppl.):28S-31S. [DOI] [PubMed] [Google Scholar]
- 25.Peng, M. M., S. R. Meshnick, N. A. Cunliffe, B. D. Thindwa, C. A. Hart, R. L. Broadhead, and L. Xiao. 2003. Molecular epidemiology of cryptosporidiosis in children in Malawi. J. Eukaryot. Microbiol. 50:557-559. [DOI] [PubMed] [Google Scholar]
- 26.Peng, M. M., M. L. Wilson, R. E. Holland, S. R. Meshnick, A. A. Lal, and L. Xiao. 2003. Genetic diversity of Cryptosporidium spp. in cattle in Michigan: implications for understanding the transmission dynamics. Parasitol. Res. 90:175-180. [DOI] [PubMed] [Google Scholar]
- 27.Perch, M., M. Sodemann, M. S. Jakobsen, P. Valentiner-Branth, H. Steinsland, T. K. Fischer, D. D. Lopes, P. Aaby, and K. Molbak. 2001. Seven years' experience with Cryptosporidium parvum in Guinea-Bissau, West Africa. Ann. Trop. Paediatr. 21:313-318. [DOI] [PubMed] [Google Scholar]
- 28.Smerdon, W. J., T. Nichols, R. M. Chalmers, H. Heine, and M. H. Reacher. 2003. Foot and mouth disease in livestock and reduced cryptosporidiosis in humans, England and Wales. Emerg. Infect. Dis. 9:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sulaiman, I. M., A. A. Lal, and L. Xiao. 2001. A population genetic study of the Cryptosporidium parvum human genotype parasites. J. Eukaryot. Microbiol. 2001(Suppl.):24S-27S. [DOI] [PubMed] [Google Scholar]
- 31.Tiangtip, R., and S. Jongwutiwes. 2002. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop. Med. Int. Health 7:357-364. [DOI] [PubMed] [Google Scholar]
- 32.Tumwine, J. K., A. Kekitiinwa, N. Nabukeera, D. E. Akiyoshi, S. M. Rich, G. Widmer, X. Feng, and S. Tzipori. 2003. Cryptosporidium parvum in children with diarrhea in Mulago Hospital, Kampala, Uganda. Am. J. Trop. Med. Hyg. 68:710-715. [PubMed] [Google Scholar]
- 33.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 34.Xiao, L., R. Fayer, U. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao, L., I. M. Sulaiman, U. M. Ryan, L. Zhou, E. R. Atwill, M. L. Tischler, X. Zhang, R. Fayer, and A. A. Lal. 2002. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int. J. Parasitol. 32:1773-1785. [DOI] [PubMed] [Google Scholar]
- 36.Xiao, L., L. Escalante, C. F. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]