To the Editor:
The PAX5 gene is one of the cancer predisposing genes that confer high penetrance susceptibility selectively to childhood BCP-ALL [1, 2]. Currently, 4 different PAX5 germline variants (p.Gly183Ser; p.Gly183Arg; p.Arg38His; c.1013-2A>G, exon 6 deletion) have been described in 26 patients of 13 families, with incomplete penetrance of the phenotype [3–9]. All family members who developed BCP-ALL had a second somatic hit affecting the wild-type PAX5 allele, either through somatic uniparental disomy or as a second structural variant [3–9].
Here we report a family with a new combination of PAX5 germline and somatic variants. For the first time, we describe a novel damaging frameshift PAX5 germline variant in two Italian siblings with BCP- ALL, associated with the same somatic PAX5 P80R variant in both cases, suggesting a possible novel mechanism for promoting leukemogenesis.
Case A was a 13-year-old boy with BCP-ALL who was enrolled in the AIEOP-BFM ALL2009 protocol (EudraCT 2007-004270-43) and classified as intermediate-risk based on minimal residual disease. Eight years later, his sister (14 years, case B) was also diagnosed with BCP- ALL. She was enrolled in the intermediate-risk group of the AIEOP-BFM ALL2017 protocol (EudraCT 2016-001935-12). Both patients were negative for CNS involvement, for recurrent fusion genes and for cytogenetic abnormalities. they were otherwise healthy and descended from healthy, unrelated parents; there was no history of cancer nor significant hematologic disorders in the pedigree.
After local IRB approval and family informed consent, a custom Next Generation Sequencing ALL predisposition panel (details in Supplementary) was applied and we identified a novel heterozygous germline variant in PAX5 shared by the two siblings; namely, a single nucleotide deletion resulting in a reading frameshift (NM_016734.2 PAX5 c.548delG, p.Gly183AlafsTer84, MAF 0), classified as likely pathogenic according to ACMG guidelines. No other known common pathogenic or likely pathogenic variants were identified through WES.
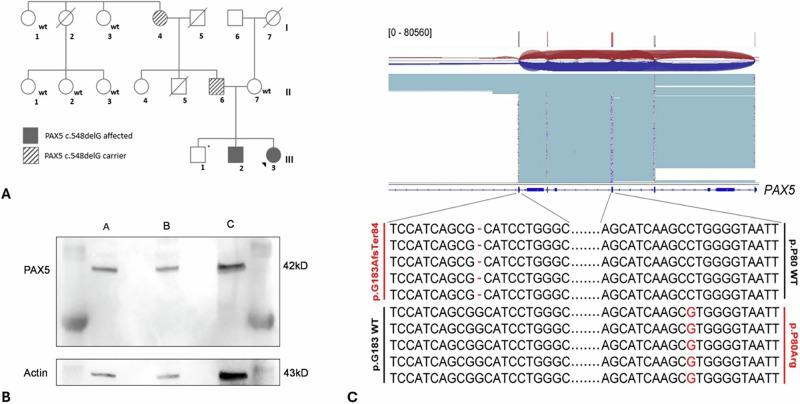
A selective familiar segregation analysis revealed a paternal origin of the PAX5 germline variant (father and grandmother of the patients) and demonstrated an incomplete penetrance of the phenotype (Fig. 1A). A healthy brother of the subjects (24 years old) was not tested for the variant because of a family decision.
Fig. 1. Characterization of germline and somatic variants in the family.
A Familial segregation of PAX5 p.Gly183AlafsTer84. The germline variant in the two siblings was demonstrated to be inherited from the father (II.6) and the grandmother (I,4) both asymptomatic carriers with no history of cancer. *: non tested. B Western blot analysis of Pax-5 expression. PAX5 expression was evaluated in CASE B (A), in the wild-type mother (B), control lymphoblastoid cell line (C); only the wild-type PAX5 protein (42 kD) was detected. C PAX5 variant allele analysis. Long read sequencing proved that the two identified variants are in trans, as shown in the image. The sequence variants are highlighted in red. Cumulative read depth is shown on top of the figure; sequence reads were aligned to the human GRCh38/hg38 reference.
In contrast to previously described PAX5 germline families [3–9], our patients exhibit a germline frameshift variant that is predicted to cause a premature arrest of the protein synthesis (predicted MW 30KDa). However, western blotting analysis in case B (protein material from patient A was not available) showed the presence of the wild-type Pax5 protein only (Fig. 1B), indicating a possible degradation of mutant mRNA by the nonsense-mediated mRNA decay pathway rather than the production of a truncated protein. This mechanism would therefore cause a PAX5 germline haploinsufficiency instead of a protein variant as in other families.
Strikingly, the disease samples from both siblings carried an identical additional somatic pathogenic PAX5 p.Pro80Arg variant (c.239C>G, p.Pro80Arg; variant fraction: 29% in Case A and 38% in Case B), which has been functionally characterized as a pathogenic variant promoting leukemogenesis [10–13]. Differently from what reported for other families with PAX5 germline variants and ALL, that shared a 9p deletion as a second hit [3–8], no structural chromosomal aberrations were identified, nor the digital MLPA analysis identified CNV alterations affecting the PAX5 locus. On the other hand, case A showed a homozygous deletion in CDKN2A (exons 1–4) and CDKN2B (exons 1–2), whereas case B showed a heterozygous deletion on IKZF1 (exons 1–8).
A long-read sequencing of the disease sample formally proved for the first time that the germline and somatic PAX5 variants were in trans (reads carrying p.Pro80Arg mutation in cis with allele p.Gly183 wild-type: 96%) (Fig. 1C), meaning that in the leukemic cells, both PAX5 alleles are impaired.
Whole-Transcriptome RNA-SEQ was negative to fusion genes in both patients. Furthermore, the gene expression classifier ALLCatchR was applied on RNA-SEQ data and both samples were assigned to “PAX5 P80R” sub-category with high-confidence score.
The combination of a germline PAX5 frameshift variant associated with a somatic in trans-gain-of-function PAX5 variant in the alternate allele recapitulates the published Pax5+/– mice model in which mice heterozygous for a loss-of-function Pax5 allele, after mutagenesis-induced secondary somatic variants resulted in the progression to B cell leukemia with moderate penetrance [14]. This is opposite to the heterozygous Pax5 P80R+ knock-in mice, which model the background of a gain-of-function variant with the disruption of the remaining PAX5 wild-type allele by deletion or frameshift mutations and almost complete penetrance [10–13]. The older age of both our patients with PAX5 germline haploinsufficiency (13 and 14 years old) compared to patients with PAX5 missense variants at the same position (mean age 3.9 years) might reflect the different underlying mechanisms, but more cases need to be accumulated to confirm this hypothesis. This discrepancy in age would be consistent with the fact that pre-leukemic PAX5 haploinsufficiency requires a longer time to acquire and select a specific somatic gain-of-function PAX5 variant (e.g., PAX5 P80R) compared to the inactivation of the wild-type allele by any means available (e.g., PAX5 structural variants) in cases of germline missense PAX5, as in previously described PAX5 families.
Two are the main questions arising from these evidences. First, we cannot exclude that in other families and individuals with PAX5 germline variants, alternative secondary genomic mutations or deletions targeting B-lymphoid development can be observed, including additional genes and pathways mutated in ALL (i.e. tumor suppressors, RAS and JAK-pathway genes, others), differentially contributing to leukemia progression, like in the Pax5+/− mice model [14, 15]. On the other hand, the extraordinary coincidence of the same somatic PAX5 P80R variant in both siblings with ALL indicates a strong selection in favor of this variant, with the consequence of a double hit on the same PAX5 gene as a leukemia driver.
Second, the evidence of somatic structural variants of 9p as a biomarker of possible germline PAX5 variants might miss cases with subtle but equally damaging somatic alterations as a second hit for progression to leukemia. This might further support the need for a larger screening of all children with ALL diagnosis in search of possible germline variants in cancer predisposing genes [1].
Still, in the herewith described family, the penetrance was not complete, since 2/4 known carriers (50%) did not develop leukemia since their adult age. To investigate a possible impairment of PAX5 function in the development of the B cell lineage, we examined the B-cell repertoire on fresh peripheral blood mononuclear cells by FACS analysis in both healthy carriers (n = 2), compared to healthy controls (n = 4); interestingly, both healthy carriers had a moderate reduction in memory B cells IgD− CD27+ (healthy carriers 9% vs 27% healthy donor) (Fig. 2), while no difference was observed in the amount of B cells (CD19+ B cells) and naive B cells (IgD + CD27−). Despite an impaired B cells differentiation, nonetheless, the two PAX5mut healthy carriers did not have a history of increased susceptibility to infections. These findings recapitulate the impairment in B cell maturation observed by Escudero and colleagues in PAX5 p.Gly183Arg germline families [8], which might be linked to the recent observation that the octapeptide sequence (as well as the partial homeodomain) is required for optimal B cell development in a mice model [15]. This observation on the medium/low penetrance and missing phenotype of the carriers, further support the possibility that families and individuals with PAX5 germline variants might be underestimated and that a larger screening of children diagnosed with ALL could identify more cases with cancer genes predisposition.
Fig. 2. Cytofluorimetric analysis of B cell repertoire in healthy PAX5 variant carriers.
a B cell repertoire on fresh peripheral blood mononuclear analysis showed a reduction in memory B cells IgD− CD27+ (mean ± SD of PAX5 mutated carriers vs wild-type healthy donors: 9 ± 6% vs 27% ± 11%; p = 0.07). b Dot plot of a PAX5 wild-type healthy donor compared with a PAX5 mutated carrier.
This report sheds light on diverse mechanisms contributing to leukemogenesis in the context of PAX5 germline mutation. Either missense or frameshift germline PAX5 variant can predispose to leukemia, associated with structural or missense somatic variants of the reciprocal PAX5 allele. Either an hypomorphic PAX5 missense variant or a PAX5 haploinsufficiency, as a consequence of frameshift variant, might be tolerated in the germline, possibly with different latency, but a complete deregulation of PAX5 is required for full-blown leukemogenesis.
Supplementary information
Acknowledgements
The authors deeply thank the “Comitato Maria Letizia Verga” for its support and the members of the family for their contribution.
Author contributions
LRB, GF, and GC contributed to study conception and design; LRB, CS and SR performed experiments; RP contributed to long-read sequencing; CB contributed to cytofluorimetric analysis; LRB and NS collected clinical data; LRB, CS, SR, and GF contributed to data analysis and interpretation; LRB, GF, and GC wrote the manuscript. AB and GC supervised the study; all authors revised the paper and approved the submitted version. SR is a fellow of the University of Milano-Bicocca, Milan, Doctoral Program in Molecular and Translational Medicine (DIMET).
Funding
This work was partly supported by the EU-COST ActionCA16223 “LEukaemiaGENe Discovery by data sharing, mining, and collaboration” (LEGEND); by grants from the Italian Association for Cancer Research (AIRC) (IG-2018 n. 21999 and IG-2023 n. 29175 to GC; IG-2023 n. 29191 to GF).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Laura Rachele Bettini, Grazia Fazio.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-024-02399-0.
References
- 1.Wagener R, Elitzur S, Brozou T, Kubaczkova V, Bibi S, Hof J, et al. Functional damaging germline variants in ETV6, IKZF1, PAX5 and RUNX1 predisposing to B-cell precursor acute lymphoblastic leukemia. Eur J Med Genet. 2023;66:104725. [DOI] [PubMed] [Google Scholar]
- 2.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukemia. Nature. 2007;446:758–64. [DOI] [PubMed] [Google Scholar]
- 3.Stasevich I, Inglott S, Austin N, Bown N, Hall G, Harrison CJ, et al. PAX5 alterations in genetically unclassified childhood precursor B-cell acute lymphoblastic leukemia. Br J Haematol. 2015;171:263–72. [DOI] [PubMed] [Google Scholar]
- 4.Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auer F, Ruschendorf F, Gombert M, Husemann P, Ginzel S, Izraeli S, et al. Inherited susceptibility to pre B-ALL caused by germline transmission of PAX5 c.547G>A. Leukemia. 2014;28:1136–8. [DOI] [PubMed] [Google Scholar]
- 6.Yazdanparast S, Khatami SR, Galehdari H, Aryan Y, Sohrabi F, Javadi GR, et al. One missense mutation in exon 2 of the PAX5 gene in Iran. Genet Mol Res. 2015;14:17768–75. [DOI] [PubMed] [Google Scholar]
- 7.Duployez N, Jamrog LA, Fregona V, Ducourneau B, Ferster A, Attal D, et al. Germline PAX5 mutation predisposes to familial B-cell precursor acute lymphoblastic leukemia. Blood. 2021;137:1424–8. [DOI] [PubMed] [Google Scholar]
- 8.Escudero A, Takagi M, Auer F, Eckert C, Scholl V, Moorman AV, et al. Clinical and immunophenotypic characteristics of familial leukemia predisposition caused by PAX5 germline variants. Leukemia. 2022;36:2338–42. [DOI] [PubMed] [Google Scholar]
- 9.Van Engelen N, Roest M, van Dijk F, Kroeze LI, Wegman-Ostrosky T, Koudijs MJ, et al. A novel germline PAX5 single exon deletion in a pediatric patient with precursor B-cell leukemia. Leukemia. 2023;37:1908–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Healy J, van Doorn-Khosrovani SB, van Wering ER, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–66. [DOI] [PubMed] [Google Scholar]
- 11.Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung M, Schieck M, Hofmann W, Attarbaschi A, Mann G, Neth O, et al. Frequency and prognostic impact of PAX5 p.P80R in pediatric acute lymphoblastic leukemia patients treated on an AIEOP-BFM acute lymphoblastic leukemia protocol. Genes Chromosomes Cancer. 2020;59:667–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passet M, Boissel N, Sigaux F, Saillour V, Thomas X, Chalandon Y, et al. PAX5 P80R mutation identifies a novel subtype of B-cell precursor acute lymphoblastic leukemia with favorable outcome. Blood. 2019;133:280–4. [DOI] [PubMed] [Google Scholar]
- 14.Dang J, Wei L, de Ridder J, Su X, Rust AG, Roberts KG, et al. PAX5 is a tumor suppressor in mouse mutagenesis models of acute lymphoblastic leukemia. Blood. 2015;126:2933–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia Z, Gu Z, Dai Y, Wu W, Zhu Y, Sun J, et al. PAX5 alterations in B-cell acute lymphoblastic leukemia. Front Oncol. 2022;12:1023606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.