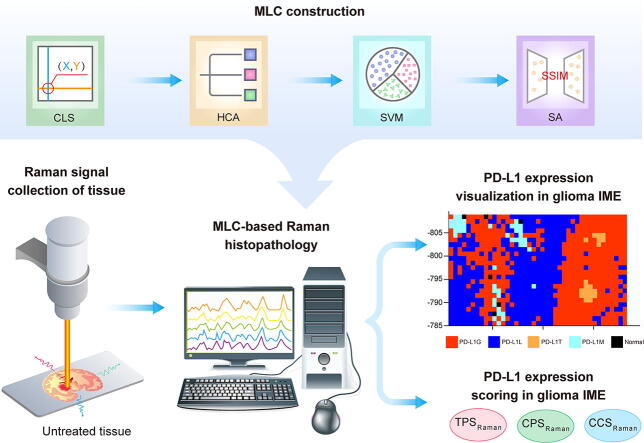
Graphical abstract
Keywords: Glioblastoma, PD-L1, Immune microenvironment, Raman spectroscopy, Machine learning
Highlights
-
•
This study achieved label-free visualization of PD-L1 expression at tissue level for the first time.
-
•
MLC-Raman histopathology achieved an accuracy of 0.99 in classifying distinct PD-L1 expression cells in GBM IME.
-
•
MLC-Raman histopathology could identify glioma, CD8+ T cells, macrophages and normal cells.
-
•
The imaging results exhibited strong concordance with MxIF and traditional pathologists' scoring.
-
•
The entire process from signal collection to visualization could be completed within 30 min.
Abstract
Introduction
Combination immunotherapy holds promise for improving survival in responsive glioblastoma (GBM) patients. Programmed death-ligand 1 (PD-L1) expression in immune microenvironment (IME) is the most important predictive biomarker for immunotherapy. Due to the heterogeneous distribution of PD-L1, post-operative histopathology fails to accurately capture its expression in residual tumors, making intra-operative diagnosis crucial for GBM treatment strategies. However, the current methods for evaluating the expression of PD-L1 are still time-consuming.
Objective
To overcome the PD-L1 heterogeneity and enable rapid, accurate, and label-free imaging of PD-L1 expression level in GBM IME at the tissue level.
Methods
We proposed a novel intra-operative diagnostic method, Machine Learning Cascade (MLC)-based Raman histopathology, which uses a coordinate localization system (CLS), hierarchical clustering analysis (HCA), support vector machine (SVM), and similarity analysis (SA). This method enables visualization of PD-L1 expression in glioma cells, CD8+ T cells, macrophages, and normal cells in addition to the tumor/normal boundary. The study quantified PD-L1 expression levels using the tumor proportion, combined positive, and cellular composition scores (TPS, CPS, and CCS, respectively) based on Raman data. Furthermore, the association between Raman spectral features and biomolecules was examined biochemically.
Results
The entire process from signal collection to visualization could be completed within 30 min. In an orthotopic glioma mouse model, the MLC-based Raman histopathology demonstrated a high average accuracy (0.990) for identifying different cells and exhibited strong concordance with multiplex immunofluorescence (84.31 %) and traditional pathologists' scoring (R2 ≥ 0.9). Moreover, the peak intensities at 837 and 874 cm−1 showed a positive linear correlation with PD-L1 expression level.
Conclusions
This study introduced a new and extendable diagnostic method to achieve rapid and accurate visualization of PD-L1 expression in GBM IMB at the tissular level, leading to great potential in GBM intraoperative diagnosis for guiding surgery and post-operative immunotherapy.
Introduction
Glioblastoma (GBM) is a highly invasive and location-specific malignant brain tumor that presents challenges in achieving complete surgical resection [1]. Additionally, it exhibits strong resistance to radiotherapy and chemotherapy [2], resulting in a mean overall survival of only 10 to 15 months for GBM patients [3]. In responsive GBM patients, combination immunotherapy rather than single pathway immunotherapy has shown promise in facilitating improvements in survival outcomes, indicating the potential clinical application of immunomodulation within the suppressive immune microenvironment (IME) [4], [5], [6], [7]. However, the expression level of programmed death-ligand 1 (PD-L1), which is a crucial predictive biomarker for immunotherapy, exhibits a highly heterogeneous distribution in the IME of GBM even within the same tissue block [8], [9]. This heterogeneity makes it challenging for post-operative histopathology to accurately capture the PD-L1 expression in the residual tumor’s IME, thereby limiting its capability to predict GBM patients’ responses to combination immunotherapy. Therefore, the intra-operative visualization of PD-L1 expression within critical functional regions of the residual GBM would greatly assist surgeons when comprehensively evaluating the balance between surgical tumor removal and post-operative immunotherapy.
Immunohistochemistry (IHC) is currently considered the gold standard for detecting immune biomarkers in tissues [10]. Other histopathological techniques, such as multiplex immunofluorescence (MxIF), multiplexed ion beam imaging (MIBI), and fluorescence in situ hybridization (FISH), have been employed to visualize PD-L1 expression level in GBM IME [11], [12], [13]. However, these methods are time-consuming and involve complex labeling steps through antigen–antibody or nucleic acid hybridization thus hindering their applications in intra-operative diagnosis. Furthermore, the currently available intra-operative navigation methods, such as fluorescence navigation, magnetic resonance imaging (MRI), and computed tomography (CT), present limitations in terms of specificity or spatial resolution, thereby restricting the visualization of PD-L1 expression level in GBM IME [14], [15], [16]. Consequently, an urgent need to develop a rapid, accurate, and label-free diagnostic method for intra-operative visualization of PD-L1 expression in GBM IME exists.
Raman spectroscopy, a member of molecular vibration spectroscopy, uses the inelastic light scattering phenomenon to analyze molecular components, such as proteins, nucleic acids, and lipids, in biological tissues without labeling [17]. Raman spectroscopy offers advantages, including non-destructiveness, low cost, and resistance to water and air disturbances, which makes it a promising tool for guiding intra-operative pathological diagnosis in brain tumors or molecular classification of GBM [18], [19], [20], [21]. However, the classification criteria used in the previous studies were based on patients or tissue blocks; therefore, these criteria overlooked the intra-tumoral heterogeneity. For tumor biomarkers, such as PD-L1, their distribution is highly heterogeneous and appears as scattered patterns, posing challenges in effectively capturing PD-L1 Raman signals for modeling and complex Raman signals for analysis. Therefore, to date, label-free Raman spectroscopy has not been capable of achieving the PD-L1 visualization at the tissue level.
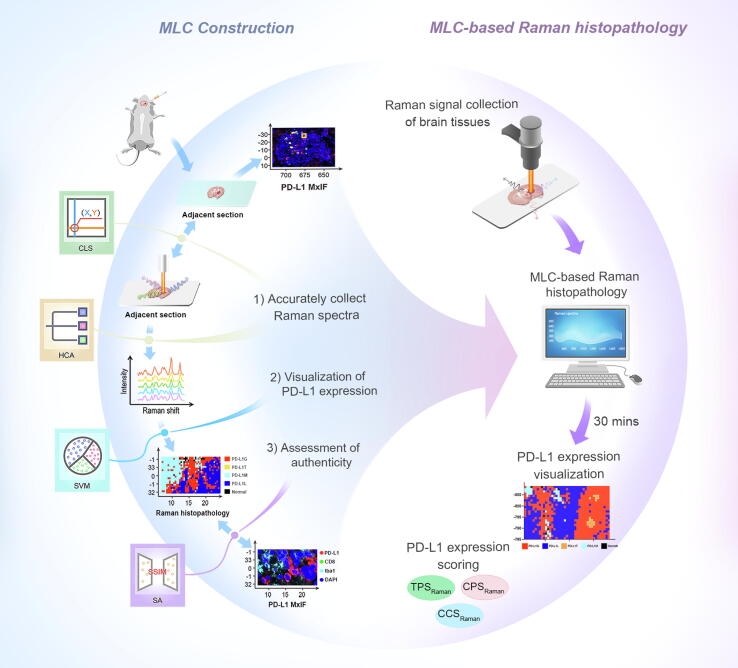
To address the above challenges, this study developed an innovative machine learning cascade (MLC)-based Raman histopathology. This method consisted of three steps: 1) the design of a coordinate localization system (CLS) and hierarchical clustering analysis (HCA) to overcome the heterogeneity of GBM IME and precisely collect Raman spectra training data on unprocessed tissue, 2) the use of a support vector machine (SVM) to extract useful signals from complex Raman spectra for visualizing PD-L1 expression level without labeling, and 3) application of a similarity analysis (SA) to validate the authenticity of MLC-based Raman histopathology using an external validation dataset (Scheme 1). In an orthotopic glioma mouse model, the resulting Raman histopathology method was successfully used to visualize the distribution of PD-L1 and differentiate glioma cells, CD8+ T cells, and macrophages with high PD-L1 expression from glioma cells with low PD-L1 expression and normal brain tissue with no PD-L1 expression. This visualization demonstrated a high degree of concordance with MxIF. Furthermore, the tumor proportion score (TPS), combined positive score (CPS), and cellular composition score (CCS) based on Raman were used to quantify the expression levels of PD-L1. Finally, the study examined the association between the extracted Raman spectral features and differentiating biomolecules within subgroups via the use of biochemical assays. This novel histopathological method offers potential for future intra-operative diagnosis of GBM, guidance for tumor excision, and facilitating tailor-made post-operative combination immunotherapy.
Scheme 1.
Schematic illustration of machine learning cascade-based Raman histopathology for rapid visualization of PD-L1 expression level in glioblastoma immune microenvironment.
Materials and methods
Ethics statement
All experiments involving animals were conducted according to the ethical policies and procedures approved by the Ethics Committee of the Nanjing Medical University (Approval no. 2021DZGKJDWLS-0029).
Orthotopic glioma mouse model construction
Ten C57BL/6 mice (6-week-old females, 18–20 g) underwent orthotopic implantation of GL261 cells, with eight used for model training and testing, and the other two mice from another batch used for external validation data. In vivo MRI was utilized to validate the successful establishment of the orthotopic glioma model. The details of cell culture, glioma orthotopic implantation, and mouse MRI results are described in the Supplementary Information.
Tissue preparation
To screen the most suitable Raman substrate slides, the signal-to-noise ratio (SNR) and background Raman signal intensity of four common Raman substrates, namely mirrored stainless steel (Lianxing hardware, #304), Raman grade calcium fluoride (CaF, Microphotons, #A80160131), quartz slides (Puiea Industry, #7360–0) and Raman grade monocrystalline silicon (Crystal Silicon Electronic & Technology Co., Ltd, #JX-S4N111200) were analyzed. The SNR was defined as the average peak intensities at 851, 1003, 1299, 1313, 1445, and 1659 cm−1 divided by their standard deviations. In addition, the optimal thickness of frozen sections was screened by comparing SNR based on mirrored stainless steel slides.
When the substantial tumors were grown for 25 days, the mice were anesthetized and perfused through the left ventricle with normal saline to eliminate the interference of hemoglobin (HB) to Raman spectra [22]. The HB-free brain was quickly removed and the middle part containing the tumor was snap-frozen in liquid nitrogen. Successive frozen sections were mounted on custom mirrored stainless steel slides and stored at − 80 °C for subsequent Raman signal acquisition. Each stainless steel slide was placed in a separate sealed section box to prevent changes in composition caused by air convection. The adjacent tissues were placed on a microscope glass slide used for MxIF. The central part of the tumor was used for histopathology study. Six to eight sections of each mouse brain tumor were used for Raman spectrum acquisition, and adjacent sections were used for MxIF imaging.
Histopathology imaging and PD-L1 scoring
The frozen MxIF sections were prepared using a four-color scheme. PD-L1 expression appeared as red fluorescence, CD8+ T cell as green, and ionized calcium-binding adapter molecule 1 (Iba1) as cyan. The central parts of the tumors were subject to hematoxylin and eosin (H&E) staining and subsequent immunohistochemistry (IHC). The specific experimental procedures of histopathology imaging including MxIF, H&E, and IHC are presented in Supplementary Information.
The expression level of PD-L1 in tumor or immune cells serves as a crucial quantitative indicator for tumor immunotherapy. Presently, the widely accepted quantitative indicators are the TPS and CPS [23]. PD-L1 TPS was defined as the percentage of viable tumor cells with partial or complete membrane staining of PD-L1 in at least 100 viable tumor cells. PD-L1 CPS was defined as the sum of PD-L1 stained tumor cells and tumor-associated immune cells per 100 tumor cells. In this study, the counting range of PD-L1 was matched to the Raman collection region in MxIF and not an absolute 100 cells. Therefore, these MXIF-based PD-L1 scoring indicators were labeled as TPSMxIF, CPSMxIF, and CCSMxIF. Two experienced immunohistochemistry pathologists performed TPSMxIF, CPSMxIF, and CCSMxIF while blinded to the predicted Raman results. The scoring results were averaged to obtain the final PD-L1 scores.
Raman signal collection
Raman spectroscopy was performed with a confocal Raman microscope (Renishaw plc #InVia Reflex). A 785 nm laser source with a round shape spot and power at about 100 mW was coupled with an N-Plan 100X (NA 0.85, WD 0.33) Leica objective. The detector was a charge-coupled device (CCD)-sensitive (1024 × 256 pixels) between 200 and 1060 nm and cooled at − 70 °C. A wavelength shift calibration was periodically performed using multiple standards (polystyrene, paracetamol, silica) and checked daily. The final laser power was filtered to reach around 50 mW.
The whole section was first viewed with a 5 × objective lens to locate the tumor. A 20 × objective lens was then used to find the glioma boundary corresponding to MxIF. After that, the magnification was switched to 50×, and the regions with different levels of PD-L1 expression were selected via a self-established CLS (see more details below). According to the MxIF results, the regions were divided into five subgroups: 1) glioma cells with high levels of PD-L1 expression (PD-L1G), 2) CD8+ T cells with high levels of PD-L1 expression (PD-L1T), 3) macrophages with high levels of PD-L1 expression (PD-L1M), 4) glioma cells with low levels of PD-L1 expression (PD-L1L), and 5) normal brain tissue. The Raman signals of each subgroup were collected in a rectangular grid with a size of approximately 10 μm × 10 μm and a step size of 1 µm. The Raman single collection time was 1 s with 10 accumulations for a biometric fingerprint region (674–1764 cm−1). For externally validated imaging data, the Raman signals were collected over an area of approximately 100 μm × 100 μm and step size from 1 μm to 5 μm. The acquisition time was optimized to six accumulations.
For the collection of the Raman spectra of a single cell, the living adherent cells were cleaned three times and then immersed in 1 × phosphate-buffered saline (PBS). The suspension cells were prepared with 1 × PBS and then seeded at a density of 5 × 105 on custom round mirrored stainless steel. The cells were kept on ice to avoid metabolic changes before Raman signal collection. Cells were located by 50 × light microscopy, and Raman spectra were collected from random areas on cells. The single collection time was 1 s with six accumulations. Six to eight Raman spectral data points were collected for each cell, with 4–5 cells acquired on each round mirrored stainless steel slide. To obtain the spectra for optimal cell activity, CD8+ T cells with favorable morphological characteristics such as small cell size and round shape were chose for Raman measurements. Raman signal collection for cells on each mirrored stainless steel slide was finished within 15 min. Approximately 40 spectra were collected for each cell to obtain their average spectra. The sources, activity, and purity of the cells used for single-cell Raman spectral acquisition are detailed in the Supplementary Information.
Raman spectra preprocessing
After Raman signal collection, the resulting spectra were preprocessed with WIRE 4.3 software (Renishaw plc) to reduce the interferences from the instrument or environment. The preprocessor procedure included quality control, spike removal, baseline correction, and normalization. Detailed procedures are described in Supplementary Information.
Construction and authenticity assessment of MLC-based Raman histopathology
Accurate collection of Raman spectra training data by CLS and HCA
The CLS based on computer vision technology was used to precisely position the laser at specific locations on the sample. The system used the original point, which serves as the reference point for the system, to establish coordinates for other points on the sample.
As the original point, ruler, and boxes were distinct, the system was designed to detect contours by setting the threshold. For instance, while detecting the box contour, the input red–green–blue (RGB) image was converted into a grayscale image, and pixels exceeding the threshold were removed from the threshold. A findContours function (OpenCV, Python 3.6.5., RRID: SCR_001658), which retrieves contours from the binary image, was then used to achieve the expected contour. Using the contour index, the locations of the rectangle’s vertices were obtained, and the location of the point could also be acquired in the same way. Finally, the coordinates were established by defining the location of the point as the origin. The coordinates of the bounding boxes could be calculated through the distance and ruler scale as shown below:
in which xv, yv and xp, yp represent the location of one vertex and the point in the pixel image, respectively. The len (ruler) is the length of the lower-right ruler. xd and yd represent the scaled coordinates of the vertex. Occasionally, when the angles of the MxIF and Raman optical image were inconsistent, the angles of the MxIF would be adjusted.
HCA is an effective method for identifying and excluding outlier Raman spectra in large datasets [24]. In this study, the HCA was used to differentiate the Raman spectra of undesired cell types from the region of interest (ROI) following specific steps:
-
1)
The preprocessed Raman spectra data of ROI were imported into R (version 4.2.2);
-
2)
The distance matrix among each Raman spectra data was calculated by the Euclidean method of distance function and reflected on the y-axis of a cluster dendrogram;
-
3)
Taking the distance matrix as input, the single clustering method in the hclust function was used for HCA;
-
4)
A plot function was used to draw a clustering tree, and Raman spectra data from other regions were screened according to the distance of a branch from the major cluster.
Visualization of PD-L1 expression level in glioma IME by SVM
Several classifiers, including SVM, linear discriminant analysis (LDA), Gradient Boosting Trees (GBT), and Random Forest (RF) were screened to extract useful signals from complex Raman spectra for classifying different cells with high or low expression of PD-L1 in glioma IME [25], [26].
First, the Raman spectra data after HCA screening were randomly divided into 80 % as the training dataset and 20 % as the testing dataset using the sample_n function from the dplyr R package. Next, we developed different classifiers with their respective tuning parameters optimized using a grid search: 1) SVM radial classifier with the radial basis kernel function and sequential minimal optimization for parameter optimization (e1071 R package); 2) LDA classifier with the tuning parameter, shrinkage (MASS R package); 3) GBM classifier with tuning parameters, including the number of trees, interaction depth, and shrinkage (gbm R package); and 4) RF classifier with tuning parameters, including the number of trees and the number of variables randomly sampled at each split (randomForest R package). For each classifier, the tuning parameters were optimized using 10-fold cross-validation (caret R package). The model performance was evaluated on testing datasets using metrics such as mean sensitivity, specificity, and accuracy. Additionally, multiclass receiver operating characteristic (ROC) curves were generated and the area under the curve (AUC) served as a measure of the model accuracy.
The model with the optimal performance was used to visualize and score PD-L1 expression. According to the immunofluorescence color results from different cells in glioma IME, corresponding pseudo colors were assigned to the predicted classification results of the SVM model to construct SVM-predicted imaging (ggplot2 R package).
Under the assumption of uniform cell sizes for each cell type, the number of cells is directly proportional to the number of Raman sampling points taken with a consistent step size. Consequently, SVM-predicted results were utilized to indirectly compute PD-L1 expression levels, including TPS and CPS. Additionally, the CCS was introduced to quantify the percentage of various tumor or immune cells that exhibit high PD-L1 expression within the overall cell population. This includes the glioma, CD8+ T cell, and macrophage composition scores (GCS, TCS, and MCS, respectively). These scores were specifically denoted as TPSRaman, CPSRaman, and CCSRaman. First, the table function (R 4.2.2) was used to calculate the frequency of each predicted value of SVM, and then the prop.table was used to obtain the proportion of different types of cells in the total number of cells, namely CCSRaman. The TPSRaman and CPSRaman were calculated according to the following formulas [23].
Authenticity assessment of SVM-predicted imaging by SA
Based on the SVM prediction results, visualization and scoring of PD-L1 were obtained. We compared the MxIF and SVM-predicted images of another batch of mice to evaluate the authenticity of the SVM algorithm.
The structural similarity (SSIM) method was adopted to evaluate the similarity between SVM-predicted imaging and MxIF. The SSIM was calculated as the ratio of similarities in luminance, contrast, and structure [27], defined as shown below:
in which x is the MxIF image, and y is the Raman map. L(x, y), c(x, y) and s(x, y) represent the luminance, contrast, and structure comparisons, respectively. μx, μy, σx, and σy represent the mean intensities and standard deviations of x and y, respectively. In this study, the constants of C1, C2, and C3 were included to prevent instability, and “α = β = γ = 1” was generally set. The SSIM was in the range of 0 to 1, and SSIM was equal to 1 if and only if x = y. Considering the calculation of SSIM (scikit-image, Python) was based on a single-color zone of MxIF or SVM-predicted imaging, k-means (OpenCV, Python) was used for segmenting color zones. The iteration details of SSIM color clustering by k-means are presented in Supplementary Information.
Exploration of the association between Raman spectral features and biomolecules
The relationship between the Raman spectra and the levels of PD-L1 expression or cell types was studied by analyzing the Raman spectra from reference biomolecules and single cells. For obtaining the typical Raman spectra of lipids, proteins, nucleic acids, and carbohydrates, 24 pure chemical compounds were selected due to their reported abundance in both normal brain tissue and glioma [28], [29]. These substances include cholesterol, cholesteryl stearate, sphingomyelin, phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), oleic acid, ganglioside, collagen, actin, histone, PD-L1, matrix metalloproteinases 2(MMP2), isocitrate dehydrogenase-1(IDH-1), deoxyribonucleic acid (DNA), ribonucleic acid (RNA), cytochrome c (Cyt c), β-carotene, glycogen, D-Glucosamine 6-phosphate (D-GlcN-6-P), hyaluronic acid (HA), arachidonic acid (ARA), and DL-lactic acid. Detailed information and the corresponding Raman spectra are presented in the Supplementary Information. For solid samples, 1 μg powder was placed on a mirrored stainless steel slide and pressed into a thin sheet. For liquid samples, 20 μL liquid was dropped onto a mirrored stainless steel slide after which their Raman spectra were collected with similar processes. Each chemical compound underwent Raman spectroscopy measurements in at least six different regions, and the resulting averaged spectra were used as the final reference spectra.
Principal component analysis (PCA) and RF importance plots were combined to extract Raman characteristic peaks among different subgroups. To apply PCA to Raman spectra data, the prcomp function from the stats R package was used to extract the difference spectra or PCA loadings from 1015 variables across different subgroups. A three-dimensional (3D) scatter plot using plot_ly (Plotly R package) was applied to display interactive 3D PCA plots. The rotation R package was used to obtain the score for all principal components (PCs), and the three PCs with the highest score were selected for further analysis. By fitting a linear combination of 24 preselected reference biomolecules to each PC (lm function in R), the biomolecules that contributed the most to the PC loadings were identified by examining the regression coefficient (coef function in R) [30]. The varImp function in the caret R package was used to calculate the importance of differential indicators among different groups from the RF model and the top 50 important indicators were selected.
Biochemical assays
Transfection, RNA extraction, real-time reverse transcriptase polymerase chain reaction (RT-qPCR), western blot analysis, and flow cytometry analysis were performed as described in detail in the Supplementary Information.
Statistics analyses
Statistical analyses can be found in the Supplementary Information.
Data availability
The data generated in this study are available upon request from the corresponding author.
Code availability
All code needed to replicate these results is available at https://github.com/Zhouqing96/Glioma_Raman_code.
Results
Construction of orthotopic glioma mouse model
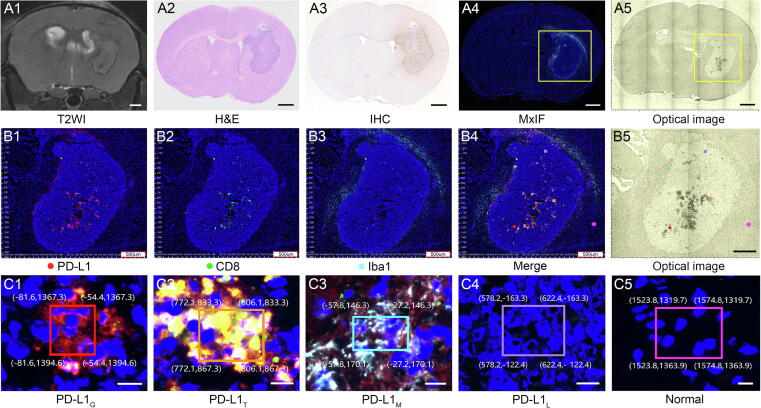
The C57BL/6 mice were implanted with GL261 cells for constructing an orthotopic glioma mouse model. In vivo T2-WI MRI showed that tumor allografts were mainly located around the corpus striatum and grew gradually after injection of GL261 cells (Supplementary Fig. S4). A representative mouse MRI, histological imaging (H&E, IHC, MxIF), and optical image are shown in Fig. 1A1-5. As a high throughput IHC method, MxIF can display different immunomarkers in tumor tissue via multiple fluorescence labeling, which was used as a reference for PD-L1 expression in glioma IME. The region with a high level of PD-L1 expression in tumor tissue is shown in red (Fig. 1B1), which corresponds to the IHC staining image (Fig. 1A3). In addition, CD8 shown in green, and IBa1 shown in cyan were used to label the major immune cells of CD8+ T cells and macrophages, respectively (Fig. 1B2 and B3). CD8+ T cells in tumor tissue and macrophages in the boundary zone between tumor and normal tissue showed high levels of PD-L1 expression, which are shown in orange and light blue in the merged image, respectively (Fig. 1B4).
Fig. 5.
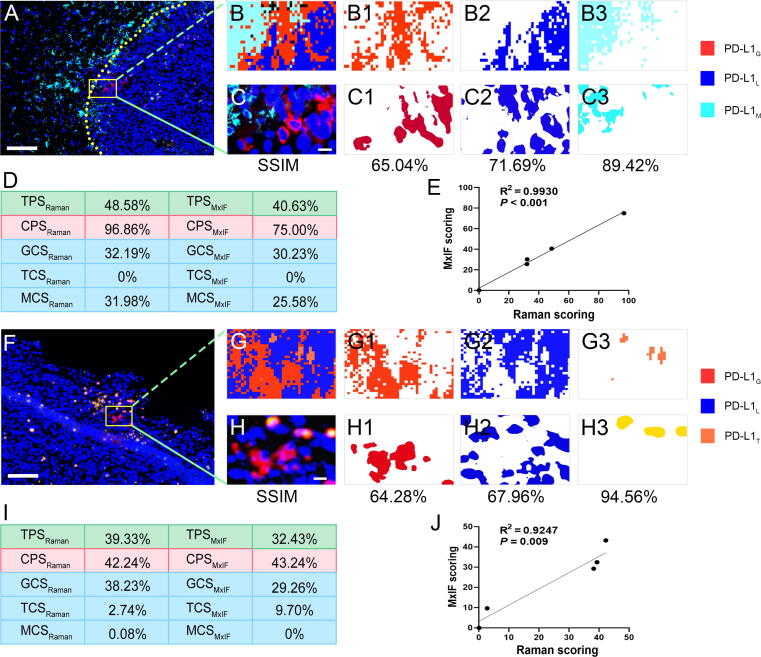
Representative images of PD-L1 expression in glioma IME and corresponding PD-L1 scoring. MxIF images of A, tumor boundary region containing PD-L1G, PD-L1L, and PD-L1M, and F, tumor peripheral region containing PD-L1G, PD-L1L, and PD-L1T. B, G, SVM-predicted imaging. C, H, Corresponding MxIF images. B1-3, C1-3, G1-3, H1-3, Different color blocks extracted by k-means according to the colors in B, C, G, and H, respectively. D, I, PD-L1 expression scoring based on Raman and MxIF. E, J, Correlation analysis of PD-L1 expression scoring between Raman and MxIF. Scale bar: A, F, 100 μm; C, H, 10 μm.
Fig. 1.
MRI, histology, and optical images of mouse orthotopic glioma. A1-A5, T2WI, H&E, IHC for PD-L1, MxIF for IME, and optical image for Raman measurements of mouse brain tissue bearing GL261 orthotopic allografts. B1-B4, Magnified different channels of MxIF images (B1, PD-L1; B2, CD8; B3, Iba1; B4, merge) with coordinate localization system from the yellow frame in A4. B5, Magnified image from the yellow frame of A5. C1-C5, Magnified MxIF images (C1, glioma cells; C2, CD8+ T cells; C3, microglial/macrophages with high PD-L1 expression; C4, glioma cells with low PD-L1 expression; C5, normal brain tissue) of the five frames in B4. The region of interest is selected by a boundary box with coordinate values given by the coordinate localization system. The positions of the five regions in B5 can be matched with B4 according to the coordinate values. Scale bar: A, 1 mm; B, 500 μm; C, 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Optimization of experimental conditions for Raman spectra collection
Before Raman spectra collection, several experimental conditions, such as Raman substrates used in measurements, storage time, and thickness of brain tissues were optimized. Firstly, stainless steel and CaF substrates exhibited minimal background Raman signals (Supplementary Fig. S5A). When preparing frozen tissue sections from white matter, gray matter, and tumors on these substrates, the stainless steel substrate yielded the highest SNR of sample Raman signals, which was used for the next set of experiments (Fig. S5B). The SNRs of Raman signals obtained from fresh tissue sections and frozen tissue sections stored for 1 h and 2 weeks were then compared, and no significant difference was observed (Fig. S5C). Furthermore, to keep consistent between frozen sections for Raman signal collection and MxIF, the thicknesses of tissue sections had been optimized to 5 μm (Fig. S5D).
Association between Raman spectral features and biomolecules
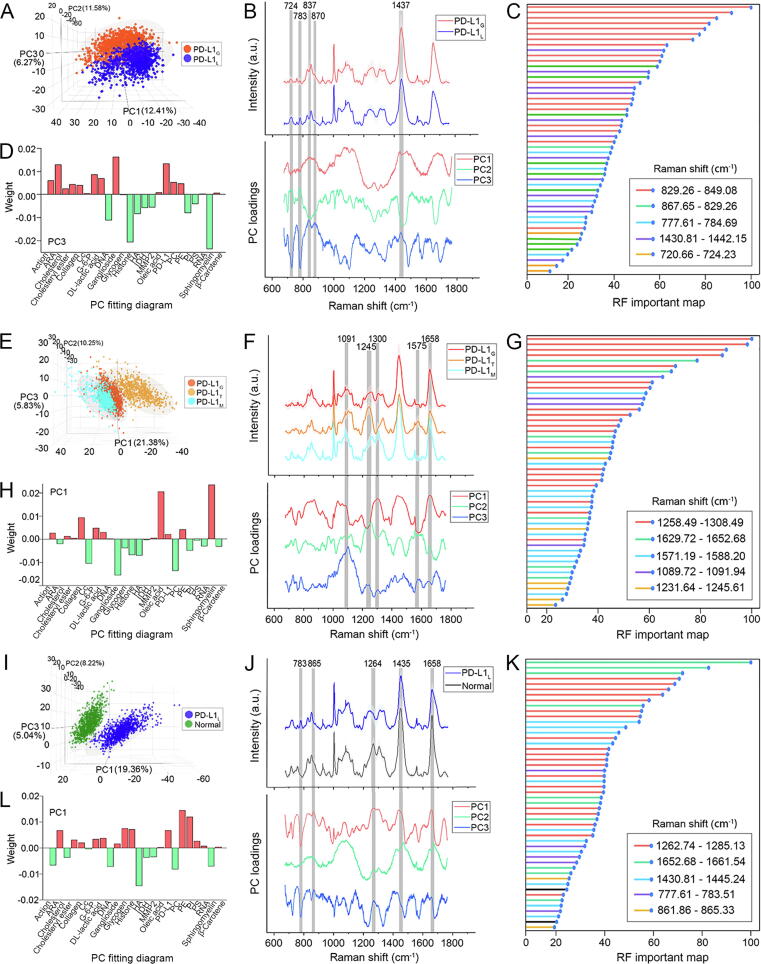
The association between Raman spectral features and biomolecules has previously been studied by qualitatively exploring the relationship between the Raman spectra and the levels of PD-L1 expression in different cells. First, PCA was employed to figure out the Raman spectra difference between PD-L1G and PD-L1L subgroups. The 3D PCA images indicated that the PC3 loading could best differentiate between these two subgroups, and the PC3 loading displayed many differential peaks existing in positive and negative directions (Fig. 6A and B). To screen the differential Raman peaks, RF was then used to process the Raman spectra and showed five extracted peaks at 723, 783, 837, 874, and 1437 cm−1 (Fig. 6C, the detailed RF map is presented in Supplementary Fig. S12A). Compared to PD-L1L, PD-L1G displayed stronger peak intensities at 837, 874, and 1437 cm−1 and weaker peak intensities at 724 and 783 cm−1. After that, the origin of the above Raman spectra difference was explored by applying a linear fit between the PC3 loading and the typical Raman spectra of 24 reference biomolecules in cells (Supplementary Fig. S2). The linear fit results could identify the biomolecules related to the spectral difference and rank their contributions (Fig. 6D) and demonstrated that ganglioside, PD-L1, and arachidonic acid (ARA) made notable contributions (shown in red) to high levels of PD-L1 expression, whereas sphingomyelin, histone, and DNA made notable contributions (shown in green) to low levels of PD-L1 expression.
Fig. 6.
Raman spectra differences among different subgroups and corresponding biological origins. A, E, I, 3D PCA image, B, F, J, Average Raman spectra and PC loadings, C, G, K, RF important maps (same color assigned to same Raman peaks) and D, H, L, PC fitting diagrams of PD-L1Gversus PD-L1L, PD-L1Gversus PD-L1Tversus PD-L1M and PD-L1Lversus normal brain tissue, respectively. The grey regions in B, F, and J indicate the differential Raman peaks extracted by RF. The red and green in D, H, and L mark the major contributive biomolecules in the positive and negative directions in PC loadings, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Based on a similar strategy, the Raman spectra difference among PD-L1G, PD-L1T, and PD-L1M subgroups and the spectra differences between PD-L1L and normal brain tissue were studied to reveal the biological association between cell types and Raman spectral features. Compared to PD-L1G, PD-L1T showed characteristic Raman peaks at 1091, 1245, and 1575 cm−1, and PD-L1M exhibited stronger peak intensities at 1300 and 1658 cm−1 (Fig. 6E–G, Fig. S12B), implying that ganglioside, phosphatidylcholine (PC), and cytochrome c (Cyt c) notably contributed to PD-L1T, while sphingomyelin and oleic acid notably contributed to PD-L1M (Fig. 6H). In addition, normal brain tissue displayed a characteristic Raman peak at 1264 cm−1 and stronger peak intensities at 865, 1435, and 1658 cm−1 when compared with PD-L1L (Fig. 6I–K, Fig. S12C). These spectra differences suggest that PE and PI are relatively abundant in normal brain tissue (Fig. 6L).
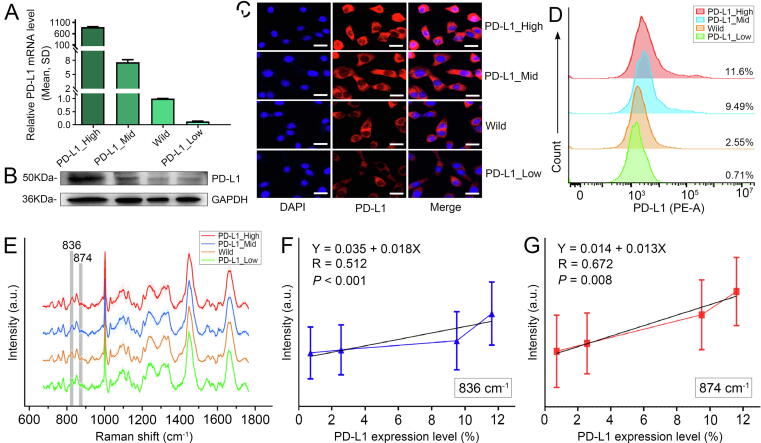
The PD-L1 protein is the most direct biomarker for evaluating PD-L1 expression and its relevance to Raman peaks has been quantitatively studied. By comparing the PC3 loading in Fig. 6B with typical Raman spectra of PD-L1 protein as shown in Supplementary Fig. S2, the characteristic Raman peaks of PD-L1 were determined to be at 837 and 874 cm−1. For quantitatively exploring the correlation between these Raman peaks and PD-L1 expression, GL261 cells with high, medium, and low levels of PD-L1 expression were established via plasmid and siRNA transfection. qPCR, western blot, and immunofluorescence imaging all confirmed the levels of PD-L1 expression in these cell lines (Fig. 7A–C, Supplementary Fig. S13). Flow cytometry showed that the expression levels of PD-L1 in PD-L1_High, PD-L1_Mid, wild type, and PD-L1_Low subgroups were 11.60 %, 9.49 %, 2.55 %, and 0.71 %, respectively (Fig. 7D). The average Raman spectra of different cells are shown in Fig. 7E. The peak intensities at 837 and 874 cm−1 were all positive linear correlation to the levels of PD-L1 expression (Fig. 7F and G). The intensity variations successfully showcased the increase in PD-L1 content in glioma cells with the increasing levels of PD-L1 expression.
Fig. 7.
Positive linear correlation between characteristic Raman peaks and levels of PD-L1 expression. A, qPCR, B, Western blot, C, immunofluorescence images, D, Flow cytometry and E, Average Raman spectra of GL261 cells with different levels of PD-L1 expression. The grey regions in E indicate the characteristic Raman peaks of PD-L1. F and G, Linear regression analysis between the levels of PD-L1 expression and the Raman peak intensities at 836 and 874 cm−1, respectively. Scale bar: 20 μm.
PD-L1 expression scoring and correlation with TPSMxIF, CPSMxIF, and CCSMxIF
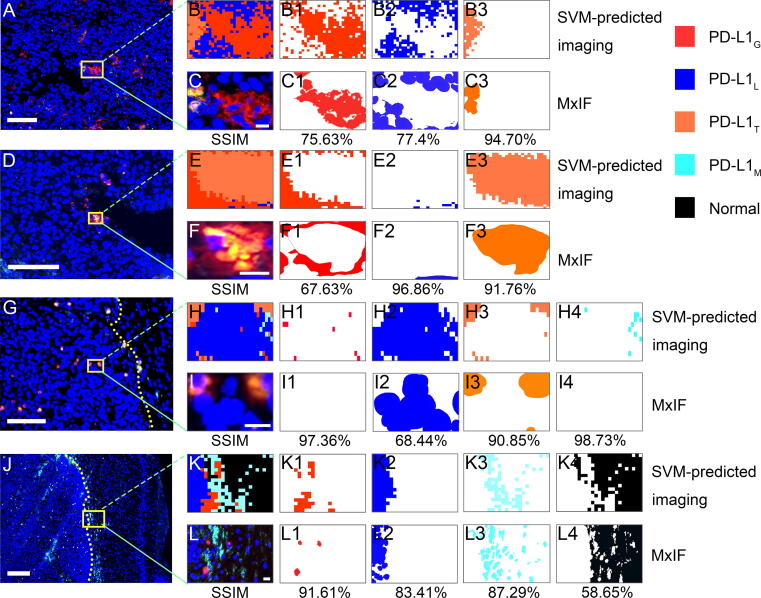
Constructed upon the accurate SVM-predicted imaging for visualizing PD-L1 expression in glioma IME, two representative SVM-predicted images were used to compare the PD-L1 scoring based on Raman and MxIF. These images encompassed PD-L1G, PD-L1L, PD-L1T, and PD-L1M. The predicted SVM imaging results are shown in Fig. 5B and Fig. 5G, while the corresponding adjacent sections of MxIF images are presented in Fig. 5C and Fig. 5H, respectively. SA shows the SVM-predicted images, and the adjacent MxIF shows consistency with average SSIM values of 75.38 % and 75.6 % for Fig. 5B and C and Fig. 5G and H, respectively. The computed TPSRaman, CPSRaman, CCSRaman, TPSMxIF, CPSMxIF, and CCSMxIF values are visually displayed in Fig. 5D and I respectively, and displayed no statistical difference based on paired-samples t-test (P = 0.118) or Wilcoxon matched pair sign rank test (P = 0.686). In addition, the PD-L1 expression scoring based on Raman demonstrated a strong correlation with the PD-L1 scoring conducted by two immunohistochemistry pathologists using MxIF (all R2 ≥ 0.9, P < 0.01) as shown in Fig. 5E and J. Collectively, this indicates that TPSRaman, CPSRaman, and CCSRaman could provide a quantitative assessment of PD-L1 expression level in glioma IME and exhibited a high level of correlation with MxIF. Additional PD-L1 expression scoring and correlation analysis between SVM-predicted imaging and the corresponding MxIF in Fig. 4 are presented in Supplementary Fig. S11.
Fig. 4.
Similarity analysis between the SVM-predicted imaging and MxIF from different regions. A, D, MxIF images of tumor core regions. G, MxIF images of tumor peripheral region. J, MxIF images of the boundary between the tumor and normal brain tissue. The yellow dashed lines in G and J indicate the boundaries. Scale bar: 100 μm. B, E, H, and K are SVM-predicted imaging. C, F, I, and L are the corresponding MxIF images derived from the yellow boxes in A, D, G, and J. B1-4, C1-4, E1-4, F1-4, H1-4, I1-4, K1-4, L1-4, Different color blocks extracted by k-means according to the colors in B, C, E, F, H, I, K, L, respectively. The percentages were the SSIM values of the two images above. Scale bar: A, D, G, J, 100 μm; C, F, I, L, 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Authenticity assessment of PD-L1 expression visualization via SA
An additional 4856 Raman spectra were collected from other batches of orthotopic glioma mouse models to perform SA between the SVM-predicted and MxIF images. In the core regions of the glioma, the SVM-predicted imaging provided a clear image of an increase in aggregation of PD-L1G and PD-L1T. The imaging results exhibited a high degree of similarity (mean SSIM of 84.00 %) with the corresponding MxIF images (Fig. 4A–F3). In the peritumoral region, scattered infiltration of PD-L1T was observed on the SVM-predicted imaging, a finding that showed a remarkable agreement with the MxIF (mean SSIM of 88.85 %) results (Fig. 4G–I4). The infiltration boundaries between the tumor and normal tissue showed a higher presence of PD-L1M (mean SSIM of 80.24 %) as shown in Fig. 4J–L4. The average SSIM value of each cell imaging was 84.31 % with PD-L1T exhibiting the highest SSIM of 92.44 %. The difference in SSIM values between the core and peripheral areas of the tumor in Fig. 4 may be attributed to the sparsely distributed PD-L1G cells, leading to higher mean values and lower variances. Overall, the SVM-predicted imaging could visualize the PD-L1 expression level in glioma IMT and delineate the infiltration boundaries distinctly. The imaging results displayed a high degree of similarity to the corresponding MxIF images, revealing significant heterogeneity in the spatial distribution of PD-L1 expression cells. The single-channel images of the MxIF results mentioned above are available in Supplementary Fig. S10.
PD-L1 expression visualization in glioma IME via SVM-predicted imaging
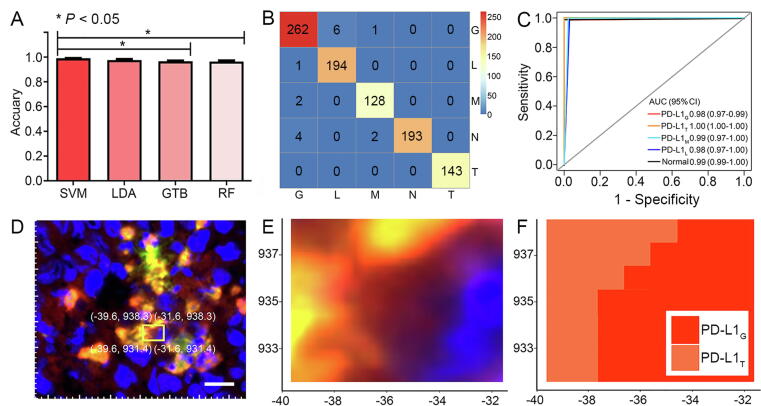
The visualization step of MLC was achieved using SVM-predicted imaging. After randomly selecting 80 % of Raman spectra as the training dataset and the rest as the testing dataset (Supplementary Table S2), the classified models aimed at distinguishing PD-L1 expression in different cells were established based on various supervised algorithms. In Fig. 3A, SVM exhibited an outstanding average accuracy of 0.990 for classifying PD-L1G, PD-L1T, PD-L1M, PD-L1L, and normal brain tissue, which was higher than other algorithms, such as LDA (0.976), GTB (0.966), and RF (0.965). Detailed statistical parameters are provided in Supplementary Table S3. Among all data, only three spectra of PD-L1M, seven spectra of PD-L1G, and six spectra of PD-L1L were incorrectly identified (Fig. 3B). Furthermore, the AUC based on the SVM showed that the classification accuracy for PD-L1T (1.000) was the highest followed by the normal brain tissue (0.996), PD-L1M (0.987), PD-L1L (0.984), and PD-L1G (0.982) as shown in Fig. 3C. Considering the highly classified accuracy of SVM, the visualization of PD-L1 expression in glioma IME could be achieved based on SVM-predicted imaging. As an example, when choosing a framed region containing PD-L1G and PD-L1T (Fig. 3D), the Raman spectra were collected by the coordinate localization system and then classified by SVM. The obtained SVM-predicted imaging clearly indicated the boundary between these two cells, resembling that observed in the MxIF image. (Fig. 3E and F).
Fig. 3.
Visualization of PD-L1 expression in glioma IME by SVM-predicted imaging. A, Average accuracy of different algorithms for classifying PD-L1G, PD-L1T, PD-L1M, PD-L1L and normal brain tissue. (One-way ANOVA, *P < 0.05). B, SVM confusion matrix for classification of five subgroups. C, ROC curve, and AUC (95CI) for different subgroups by SVM. D, MxIF images of glioma tissue containing PD-L1G and PD-L1T (the same region as Fig. 3C). E, Magnified image of the yellow frame in D. F, SVM-predicted imaging of the same region in E. Scale bar: 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Accurate collection of Raman spectra training data via CLS and HCA
After optimization of experimental conditions, Raman spectra were collected, analyzed, and assessed with the help of MLC for visualizing PD-L1 expression in glioma IME. CLS and HCA were designed to accurately collect the Raman spectra of specific regions for training classification models. Considering that MxIF could indicate different immune cells and their PD-L1 expression (Fig. 1B1–4), the CLS aimed at matching the ROI of Raman signal collection from optical images with adjacent MxIF images. After setting a point of origin (yellow dot) and framing the ROI as a rectangular box on MxIF images, this system could efficiently identify this box and yielded the coordinates (x, y) of box vertexes on section based on computer version technology (Fig. 1C1–5). Based on the resulting coordinates, it can precisely locate the same region on adjacent sections for Raman signal collection as the size and orientation of the two adjacent sections remain consistent. By virtue of the coordinate localization system, a total of 5029 Raman spectra were collected from the framed regions and divided into five subgroups consisting of PD-L1G, PD-L1T, PD-L1M, PD-L1L, and normal brain tissue.
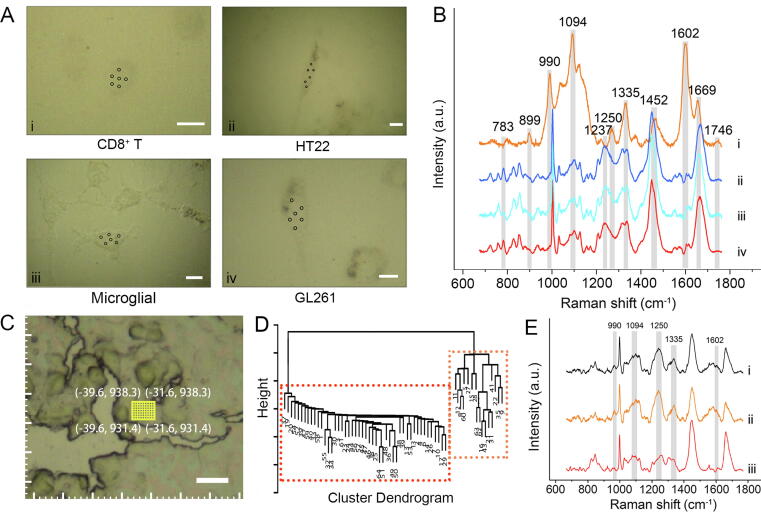
To potentially exclude the Raman signals from other cell types in framed regions, HCA was employed to further calibrate the spectra data. In this study, the Raman spectra of pure GL261 and HT22 cell lines, as well as primary CD8+ T from mouse peripheral blood and primary microglial cells from newborn mouse brain tissue were served as reference for obtaining the characteristic Raman peaks of glioma, normal brain tissue, CD8+ T cell, and macrophage in IME of glioma, respectively. The CD8+ T cells and microglial cells for Raman signal collection achieved activity levels of 93.63 ± 1.89 % and 96.5 ± 1.61 %, with purities of 87.9 ± 1.59 % and about 90.0 %, respectively (Fig. S6 and S7). The naive state of primary CD8+ T cells in peripheral blood was 32.56 ± 2.31 %, which was higher than that in the glioma IME with significant statistical difference (32.56 ± 2.31 % > 5.67 ± 2.82 %, Shapiro-Wilk, all P > 0.05, Levene's test, P = 0.81, two samples t test, P < 0.001, Fig. S8). It should be noted that these pure cells especially CD8+ T cells can not totally simulate the physiological states of cells in IME of glioma, so they only used for differentiate cell types. First, their Raman spectra at the single cell level were first collected (Fig. 2A and 2B). To highlight the Raman spectral differences, the Raman spectra of GL261, HT22, and primary microglial cells were overlapped (Supplementary Fig. S9). The main spectra difference among these four types of cells originated from DNA / RNA (783 and 1237 cm−1), saccharide (899 cm−1), ribose (990 cm−1), phospholipids (1094 and 1746 cm−1), proteins (1250 and 1669 cm−1), lipids (1335 and 1452 cm−1) and amino acids (1602 cm−1) [31]. The Raman vibrational mode involved in this study and the major assignment are shown in Supplementary Table S1. The Raman spectra collected from each ROI in tissue sections were then clustered based on unsupervised HCA, and undesired spectra dissimilar to the characteristic Raman signals of the corresponding cell line could be easily recognized and removed. As an example, a region of PD-L1G was located via the coordinate localization system (Fig. 2C), and the Raman spectra of this region were collected and classified into two categories (Fig. 2D). After removing the undesired category (orange part in Fig. 2D), which showed the characteristic Raman peaks at 990, 1094, 1250, 1335 and 1602 cm−1 of CD8+ T cells, the remaining spectra resembled the typical Raman spectra of GL261 cells (iv in Fig. 2B), indicating successful spectra calibration for obtaining pure PD-L1G signals (Fig. 2E). Using the above algorithm, a total of 352 Raman spectra were removed, and the remaining data included 1294 spectra of PD-L1G subgroup, 721 spectra of PD-L1T subgroup, 638 spectra of PD-L1M subgroup, 1058 spectra of PD-L1L subgroup, and 966 spectra of normal brain tissue subgroup.
Fig. 2.
Raman spectra classification and calibration by HCA. A, Optical images of each cell line. The black dots represent the collected regions of Raman spectra. B, Average Raman spectra of each cell line (i, CD8+ T; ii, HT22; iii, Microglial; iv, GL261 cells). The grey regions indicate the differentiated Raman peaks between cell lines. C, Optical image of PD-L1G region in brain tissue. D, HCA diagram of Raman spectra collected from the yellow frame in C. E, Average Raman spectra of whole data (i), orange part (ii), and red part (iii) in D. The grey regions indicate the characteristic Raman peaks of CD8+ T cells. Scale bar: 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
Understanding the PD-L1 expression level in IME of residual GBM within critical functional regions intraoperatively would be beneficial for predicting the response to combination immunotherapy in GBM patients and optimizing treatment strategies. In this study, an MLC-based Raman histopathology was developed to enable rapid, accurate, and label-free visualization of PD-L1 expression levels in the IME of residual GBM. Initially, an orthotopic glioma mouse model was established by implanting GL261 cells into C57BL/6 mice, and brain tissue sections were histologically imaged using MxIF as a reference for PD-L1 expression in glioma IME. Subsequently, experimental conditions such as the type of Raman substrates, storage time, and thickness of brain tissue sections were optimized before collecting Raman spectra.
As the core of this study, the MLC-based Raman histopathology consists of three steps (see Graphical Abstract). In the first step, the CLS and HCA were designed to accurately collect Raman spectra from specific regions in tissue sections. The CLS could provide the coordinates (x, y) of ROI on an MxIF image, and the same region on the adjacent tissue section could be found using the above coordinates for Raman signal collection. Thus, under the localization of MxIF, the Raman spectra of PD-L1G, PD-L1T, PD-L1M, PD-L1L, and normal brain tissue were collected and divided into the corresponding five subgroups (Fig. 1). HCA was subsequently used to calibrate the spectra data by excluding the Raman signals from undesired cells. This unsupervised algorithm classified the Raman spectra into several clusters and identified the undesired cell types based on the characteristic Raman spectra of pure cell lines (Fig. 2). This method effectively overcame the cellular and molecular heterogeneity of glioma tissue and accurately collected the Raman spectra of different cell types for training the model, which was the premise of accurate PD-L1 expression visualization in glioma IME. The next step involved using SVM to construct a classification model and generate SVM-predicted imaging and scoring. Compared to other supervised algorithms, such as LDA, GTB, and RF, SVM exhibited higher average accuracy, reaching up to 0.990 for classifying the five subgroups (Fig. 3). SVM has a radial basis function kernel and demonstrates excellent model generalization capability and proficiency in capturing complex and non-linear decision boundaries thus making it particularly suitable for Raman spectra data even with large numbers of variables (n = 1015) as previously described [32], [33]. Among the subgroups, PD-L1T showed the highest classification accuracy due to the prominent Raman peaks at 990, 1094, and 1237 cm−1 associated with DNA [34], which may originate from the dominant nucleus in T-cells. Owing to its high classification accuracy, SVM-predicted imaging clearly delineated the molecular boundaries of PD-L1 and the boundaries between tumor and normal brain tissue, to reveal the spatial heterogeneity of PD-L1 expression in cells. The imaging results successfully illustrated the aggregated PD-L1G and PD-L1T in the dense central region of the tumor. In contrast, scattered PD-L1T cells were observed in the peritumoral region, a finding that is consistent with the reported pattern of tumor-infiltrating lymphocyte infiltration in untreated human GBM [35]. This phenomenon may be attributed to the gradual infiltration of T-cells from peripheral blood or tertiary lymphoid structures into the tumor [36], [37]. In the third step, SA was performed to assess the authenticity of the SVM-predicted imaging by comparing it to MxIF. It is worth noting that a high degree of similarity between the SVM-predicted imaging and MxI with a mean SSIM of 84.31 % was found. The PD-L1T group showed the highest similarity (mean SSIM of 92.44 %), which was consistent with the classification results obtained by SVM (Fig. 4). Based on this result, PD-L1 expression scores were determined for different cell types. As a supplement to visualization, PD-L1 scoring provided a more objective and quantitative assessment of PD-L1 expression levels. In this study, TPSRaman, CPSRaman, and CCSRaman were found to be highly correlated with their corresponding TPSMxIF, CPSMxIF, and CCSMxIF (Fig. 5). Importantly, the entire process from signal collection to visualization and scoring of PD-L1 expression could be completed within 30 min, a finding that indicated the potential intra-operative application of MLC-based Raman histopathology.
To investigate the association between spectral features and biomolecules, the relationship between the Raman spectra and the levels of PD-L1 expression in different cell types was qualitatively explored (Fig. 6). Feature extraction methods, such as PCA and RF, along with the 24 reference biomolecules were used in this study to verify the biological sources of these Raman characteristic peaks. After extracting the differential Raman peaks, PD-L1G displayed stronger peak intensities at 837, 874, and 1437 cm−1 compared to PD-L1L. These spectral differences implied an increase in PD-L1 contents in PD-L1G. Similarly, the Raman spectra differences implied that PD-L1T contained higher levels of PC and Cyt c, and PD-L1M had more sphingomyelin and oleic acid when compared with PD-L1G. Among these biomolecules, Cyt c is abundant in activated T-cells and is involved in apoptosis induction [38], [39], while PC is present in T-cells and regulates cellular functions, such as proliferation inhibition [40], [41]. Sphingomyelin, a component of cell membranes, is relatively abundant in macrophages due to its higher proportion in cell membranes, while oleic acid has been demonstrated to be the main lipid component in macrophages [42]. The Raman spectra differences also indicated that normal brain tissue contained higher levels of PE and PI compared to PD-L1L, a finding that is consistent with the abundance of glycerophospholipids in brain tissue [43]. Furthermore, the correlation between characteristic Raman peaks of PD-L1 and the levels of PD-L1 expression was quantitatively studied (Fig. 7). The peak intensities at 837 and 874 cm−1 showed a positive linear correlation with the level of PD-L1 expression in GL261 cells. Thus, Raman spectra could provide information about the biomolecular compositions of cells, including PD-L1 and even their relative contents, enabling accurate visualization of PD-L1 in glioma IME.
This MLC-based Raman histopathology, due to its label-free, rapid, and accurate visualization of PD-L1 expression levels within gliomas, holds promise for clinical application, particularly in cases involving gliomas located in critical brain functional regions. By collecting Raman spectra from resected tissues after rinsing with normal saline and rapid freezing, intraoperative diagnosis can be obtained, assisting surgeons in making optimal resection decisions. Furthermore, it enables personalized combination immunotherapy targeting residual lesions. However, this preliminary study was conducted using an orthotopic glioma mouse model. Future work will focus on improving machine learning algorithms and extending the study to human GBM samples. Additionally, using MxIF from adjacent sections as a verification standard may lead to inconsistencies in cell morphology between the SVM-predicted imaging and the MxIF. Lastly, this method is currently limited to isolated tissues, and its signal intensity and penetration depth are unsatisfactory due to the inherent constraint of Raman spectroscopy. Subsequent studies may involve in vivo Raman imaging through biocompatible Raman probes and integrating combination immunotherapy to assess the effectiveness of the proposed method.
Conclusions
In summary, this study established an innovative diagnostic method called MLC-based Raman histopathology for label-free visualization of PD-L1 expression levels in brain tissues with gliomas. This technique is rapid, accurate, and convenient. Multiple machine learning algorithms, including CLS, HCA, SVM, and SA, were cascaded to accurately collect Raman spectra for training models, visualize PD-L1 expression levels in GBM IME, and finally assess the authenticity of imaging results. Furthermore, the study examined the association between Raman spectra features and biomolecules using biochemical assays. To the best of our knowledge, this study is the first to achieve label-free visualization of PD-L1 expression at a tissue level, and it could be extended to other tumor biomarkers or any target cells of interest. Considering the absence of labeling requirements and the miniaturization and low cost of Raman spectrometers, this study paves the way for intra-operative molecular pathological diagnosis of GBM, potentially improving GBM treatment strategies by considering functional localization and PD-L1 expression levels, and enabling post-operative tailor-made combination immunotherapy.
Authors’ disclosures
Lu reports grants from the National Key R&D Program of China and the National Natural Science Foundation of China during the conduct of the study. J. Guo reports grants from the Wuhan University of Technology during the conduct of the study. Q-Q. Zhou reports grants from the Nanjing Medical University during the conduct of the study. No disclosures were reported by the other authors.
CRediT authorship contribution statement
Qing-Qing Zhou: Conceptualization, Investigation, Data curation, Methodology, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing. Jingxing Guo: Investigation, Methodology, Formal analysis, Supervision, Funding acquisition, Project administration, Writing – review & editing. Ziyang Wang: Investigation, Formal analysis, Methodology. Jianrui Li: Investigation, Data curation, Resources. Meng Chen: Formal analysis, Methodology, Visualization. Qiang Xu: Formal analysis, Methodology, Writing – review & editing. Lijun Zhu: Investigation, Data curation. Qing Xu: Conceptualization, Investigation. Qiang Wang: Conceptualization, Investigation, Resources. Hao Pan: Conceptualization, Investigation, Resources. Jing Pan: Formal analysis, Methodology, Writing – review & editing. Yong Zhu: Conceptualization, Investigation, Writing – review & editing. Ming Song: Methodology, Formal analysis, Visualization. Xiaoxue Liu: Investigation, Data curation, Writing – review & editing. Jiandong Wang: Investigation, Resources. Zhiqiang Zhang: Conceptualization, Investigation, Formal analysis. Longjiang Zhang: Conceptualization, Formal analysis, Supervision. Yiqing Wang: Methodology, Formal analysis, Visualization. Huiming Cai: Methodology, Formal analysis, Project administration. Xiaoyuan Chen: Conceptualization, Supervision, Methodology, Formal analysis, Writing – review & editing. Guangming Lu: Conceptualization, Investigation, Methodology, Formal analysis, Supervision, Funding acquisition, Project administration, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
G. Lu acknowledges financial support from the National Key R&D Program of China (2020AAA0109505), the National Natural Science Foundation of China (82127806, 81790653). J. Guo acknowledges financial support from the Fundamental Research Funds for the Central Universities (WUT:3120622957, WUT:3120623054). Q-Q. Zhou acknowledges financial support from the Nanjing Medical University Imaging Elite Talent support program (JNYYHXKY202107). The authors thank H. Li (School of Science, China Pharmaceutical University) for guiding the Western blot experiment, N. You (Department of Radiology, Jinling Hospital, Affiliated Hospital of Medical School, Nanjing University,) for guiding the flow cytometry experiment, and thanks Y. Zhang (Department of Diagnostic Radiology, Jinling Hospital, Affiliated Hospital of Medical School) for scanning the mice MRI in vivo.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.12.002.
Contributor Information
Jingxing Guo, Email: jxguo@whut.edu.cn.
Xiaoyuan Chen, Email: chen.shawn@nus.edu.sg.
Guangming Lu, Email: cjr.luguangming@vip.163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Weller M., van den Bent M., Preusser M., Le Rhun E., Tonn J.C., Minniti G., et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol. 2021;18(3):170–186. doi: 10.1038/s41571-020-00447-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J., Li Y., Yu T.S., McKay R.M., Burns D.K., Kernie S.G., et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorland L.S., Fluge O., Gilje B., Mahesparan R., Farbu E. Treatment approach and survival from glioblastoma: results from a population-based retrospective cohort study from Western Norway. BMJ Open. 2021;11(3):e043208. doi: 10.1136/bmjopen-2020-043208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schalper K.A., Rodriguez-Ruiz M.E., Diez-Valle R., Lopez-Janeiro A., Porciuncula A., Idoate M.A., et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat Med. 2019;25(3):470–476. doi: 10.1038/s41591-018-0339-5. [DOI] [PubMed] [Google Scholar]
- 5.Ito H., Nakashima H., Chiocca E.A. Molecular responses to immune checkpoint blockade in glioblastoma. Nat Med. 2019;25(3):359–361. doi: 10.1038/s41591-019-0385-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh K., Batich K.A., Wen P.Y., Tan A.C., Bagley S.J., Lim M., et al. Designing Clinical Trials for Combination Immunotherapy: A Framework for Glioblastoma. Clin Cancer Res. 2022;28(4):585–593. doi: 10.1158/1078-0432.CCR-21-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bausart M., Preat V., Malfanti A. Immunotherapy for glioblastoma: the promise of combination strategies. J Exp Clin Cancer Res. 2022;41(1):35. doi: 10.1186/s13046-022-02251-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pombo Antunes A.R., Scheyltjens I., Duerinck J., Neyns B., Movahedi K., Van Ginderachter J.A. Understanding the glioblastoma immune microenvironment as basis for the development of new immunotherapeutic strategies. Elife. 2020:9. doi: 10.7554/eLife.52176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreatsoulas D., Bolyard C., Wu B.X., Cam H., Giglio P., Li Z. Translational landscape of glioblastoma immunotherapy for physicians: guiding clinical practice with basic scientific evidence. J Hematol Oncol. 2022;15(1):80. doi: 10.1186/s13045-022-01298-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuczkiewicz-Siemion O., Sokol K., Puton B., Borkowska A., Szumera-Cieckiewicz A. The Role of Pathology-Based Methods in Qualitative and Quantitative Approaches to Cancer Immunotherapy. Cancers (Basel) 2022;14(15) doi: 10.3390/cancers14153833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoranz A., Van Herck Y., Bolognesi M.M., Lynch S.M., Rahman A., Gallagher W.M., et al. Mapping the Immune Landscape in Metastatic Melanoma Reveals Localized Cell-Cell Interactions That Predict Immunotherapy Response. Cancer Res. 2022;82(18):3275–3290. doi: 10.1158/0008-5472.CAN-22-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCaffrey E.F., Donato M., Keren L., Chen Z., Delmastro A., Fitzpatrick M.B., et al. The immunoregulatory landscape of human tuberculosis granulomas. Nat Immunol. 2022;23(2):318–329. doi: 10.1038/s41590-021-01121-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M., Andreozzi M., Pockaj B., Barrett M.T., Ocal I.T., McCullough A.E., et al. Development and validation of a novel clinical fluorescence in situ hybridization assay to detect JAK2 and PD-L1 amplification: a fluorescence in situ hybridization assay for JAK2 and PD-L1 amplification. Mod Pathol. 2017;30(11):1516–1526. doi: 10.1038/modpathol.2017.86. [DOI] [PubMed] [Google Scholar]
- 14.Hollon T., Stummer W., Orringer D., Suero M.E. Surgical Adjuncts to Increase the Extent of Resection: Intraoperative MRI, Fluorescence, and Raman Histology. Neurosurg Clin N Am. 2019;30(1):65–74. doi: 10.1016/j.nec.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Tejada Solis S., de Quintana S.C., Gonzalez Sanchez J., Fernandez Portales I., Alamo D., de Pedro M., et al. Intraoperative imaging in the neurosurgery operating theatre: A review of the most commonly used techniques for brain tumour surgery. Neurocirugia (Astur : Engl Ed) 2020;31(4):184–194. doi: 10.1016/j.neucir.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Barbagallo G., Maione M., Peschillo S., Signorelli F., Visocchi M., Sortino G., et al. Intraoperative Computed Tomography, navigated ultrasound, 5-Amino-Levulinic Acid fluorescence and neuromonitoring in brain tumor surgery: overtreatment or useful tool combination? J Neurosurg Sci. 2019 doi: 10.23736/S0390-5616.19.04735-0. [DOI] [PubMed] [Google Scholar]
- 17.Zong C., Xu M., Xu L.J., Wei T., Ma X., Zheng X.S., et al. Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges. Chem Rev. 2018;118(10):4946–4980. doi: 10.1021/acs.chemrev.7b00668. [DOI] [PubMed] [Google Scholar]
- 18.Jermyn M, Mok K, Mercier J, Desroches J, Pichette J, Saint-Arnaud K, et al. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci Transl Med 2015:7(274):274ra19. 10.1126/scitranslmed.aaa2384. [DOI] [PubMed]
- 19.Uckermann O., Yao W., Juratli T.A., Galli R., Leipnitz E., Meinhardt M., et al. IDH1 mutation in human glioma induces chemical alterations that are amenable to optical Raman spectroscopy. J Neurooncol. 2018;139(2):261–268. doi: 10.1007/s11060-018-2883-8. [DOI] [PubMed] [Google Scholar]
- 20.Livermore L.J., Isabelle M., Bell I.M., Scott C., Walsby-Tickle J., Gannon J., et al. Rapid intraoperative molecular genetic classification of gliomas using Raman spectroscopy. Neurooncol Adv. 2019;1(1):vdz008. doi: 10.1093/noajnl/vdz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollon T., Jiang C., Chowdury A., Nasir-Moin M., Kondepudi A., Aabedi A., et al. Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging. Nat Med. 2023;29(4):828–832. doi: 10.1038/s41591-023-02252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo J., Liu Y., Ju H., Lu G. From lab to field: Surface-enhanced Raman scattering-based sensing strategies for on-site analysis. TrAC Trends in Analytical Chemistry. 2022;146 doi: 10.1016/j.trac.2021.116488. [DOI] [Google Scholar]
- 23.Wu H.X., Pan Y.Q., He Y., Wang Z.X., Guan W.L., Chen Y.X., et al. Clinical Benefit of First-Line Programmed Death-1 Antibody Plus Chemotherapy in Low Programmed Cell Death Ligand 1-Expressing Esophageal Squamous Cell Carcinoma: A Post Hoc Analysis of JUPITER-06 and Meta-Analysis. J Clin Oncol. 2023;41(9):1735–1746. doi: 10.1200/JCO.22.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langfelder P., Fast R. Functions for Robust Correlations and Hierarchical Clustering. Journal of Statistical Software. 2012;46(11):1–17. doi: 10.18637/jss.v046.i11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ralbovsky N.M., Lednev I.K. Towards development of a novel universal medical diagnostic method: Raman spectroscopy and machine learning. Chem Soc Rev. 2020;49(20):7428–7453. doi: 10.1039/d0cs01019g. [DOI] [PubMed] [Google Scholar]
- 26.Borek-Dorosz A., Nowakowska A.M., Leszczenko P., Adamczyk A., Pieczara A., Jakubowska J., et al. Raman-based spectrophenotyping of the most important cells of the immune system. J Adv Res. 2022;41:191–203. doi: 10.1016/j.jare.2021.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma K., Zhao T., Zeng K., Wang Z. Objective Quality Assessment for Color-to-Gray Image Conversion. IEEE Trans Image Process. 2015;24(12):4673–4685. doi: 10.1109/TIP.2015.2460015. [DOI] [PubMed] [Google Scholar]
- 28.Krafft C., Neudert L., Simat T., Salzer R. Near infrared Raman spectra of human brain lipids. Spectrochim Acta A Mol Biomol Spectrosc. 2005;61(7):1529–1535. doi: 10.1016/j.saa.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y., Liu C.H., Wu B., Yu X., Cheng G., Zhu K., et al. Optical biopsy identification and grading of gliomas using label-free visible resonance Raman spectroscopy. J Biomed Opt. 2019;24(9):1–12. doi: 10.1117/1.JBO.24.9.095001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaifulina R., Maher A.T., Kendall C., Nelson J., Rodriguez-Justo M., Lau K., et al. Label-free Raman spectroscopic imaging to extract morphological and chemical information from a formalin-fixed, paraffin-embedded rat colon tissue section. Int J Exp Pathol. 2016;97(4):337–350. doi: 10.1111/iep.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi H., Yanamisawa A., Kajimoto S., Nakabayashi T. Observation of the changes in the chemical composition of lipid droplets using Raman microscopy. Phys Chem Chem Phys. 2020;22(38):21646–21650. doi: 10.1039/d0cp03805a. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Wang Z., Dai H. Improved Parkinsonian tremor quantification based on automatic label modification and SVM with RBF kernel. Physiol Meas. 2023;44(2) doi: 10.1088/1361-6579/acb8fe. [DOI] [PubMed] [Google Scholar]
- 33.James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning with applications in R 2021; 2nd ed.
- 34.Robert C., Tsiampali J., Fraser-Miller S.J., Neumann S., Maciaczyk D., Young S.L., et al. Molecular monitoring of glioblastoma's immunogenicity using a combination of Raman spectroscopy and chemometrics. Spectrochim Acta A Mol Biomol Spectrosc. 2021;252 doi: 10.1016/j.saa.2021.119534. [DOI] [PubMed] [Google Scholar]
- 35.Mitsdoerffer M., Aly L., Barz M., Engleitner T., Sie C., Delbridge C., et al. The glioblastoma multiforme tumor site promotes the commitment of tumor-infiltrating lymphocytes to the T(H)17 lineage in humans. Proc Natl Acad Sci U S A. 2022;119(34) doi: 10.1073/pnas.2206208119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yost K.E., Chang H.Y., Satpathy A.T. Recruiting T cells in cancer immunotherapy. Science. 2021;372(6538):130–131. doi: 10.1126/science.abd1329. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher T.N., Thommen D.S. Tertiary lymphoid structures in cancer. Science. 2022;375(6576) doi: 10.1126/science.abf9419. [DOI] [PubMed] [Google Scholar]
- 38.MacIver N.J., Michalek R.D., Rathmell J.C. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi D., Ahmed S., Ishida M., Kasai S., Kikuchi H. Calcium/calmodulin signaling elicits release of cytochrome c during 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced apoptosis in the human lymphoblastic T-cell line. L-MAT Toxicology. 2009;258(1):25–32. doi: 10.1016/j.tox.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Azarcoya-Barrera J., Wollin B., Veida-Silva H., Makarowski A., Goruk S., Field C.J., et al. Egg-Phosphatidylcholine Attenuates T-Cell Dysfunction in High-Fat Diet Fed Male Wistar Rats. Front Nutr. 2022;9 doi: 10.3389/fnut.2022.811469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishiyama-Naruke A., Curi R. Phosphatidylcholine participates in the interaction between macrophages and lymphocytes. Am J Physiol Cell Physiol. 2000;278(3):C554–C559. doi: 10.1152/ajpcell.2000.278.3.C554. [DOI] [PubMed] [Google Scholar]
- 42.Feuerer N., Marzi J., Brauchle E.M., Carvajal Berrio D.A., Billing F., Weiss M., et al. Lipidome profiling with Raman microspectroscopy identifies macrophage response to surface topographies of implant materials. Proc Natl Acad Sci U S A. 2021;118(52) doi: 10.1073/pnas.2113694118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson S.N., Wang H.Y., Woods A.S. Direct profiling of lipid distribution in brain tissue using MALDI-TOFMS. Anal Chem. 2005;77(14):4523–4527. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.