Abstract
Plasmodium falciparum msp-1 and msp-2 genes were quantified by fragment analysis in matched placental, peripheral, and cord blood samples. In the three compartments, the multiplicity of infection values were similar, and parasite populations only partially overlapped, as reported. However, identical alleles represented 80 to 95% of the overall parasite populations of each compartment, demonstrating much more homogenous parasite populations than previously thought.
In areas where malaria is endemic, it is a major cause of morbidity and mortality. During pregnancy, especially the first pregnancy, women are more susceptible and more frequently infected with high levels of placental parasitemia, causing complications for both mothers and newborn babies (5). The mature forms of Plasmodium falciparum sequester in deep microvessels in cerebral malaria and in placental intervillous spaces in pregnant women, while ring stages circulate in the peripheral blood. This phenomenon distorts the analysis of parasites in peripheral blood, revealing only parasites circulating at sampling time. With respect to pregnancy, few studies have investigated the homology of parasite populations from placental, peripheral (21), and cord (12, 13) blood from women at delivery. These studies mostly agree in showing parasite populations partially overlapping and marked differences in the various compartments from most women. While these studies (based on PCR genotyping of polymorphic markers) allow the identification of alleles, they don't permit their quantification. Alternatively, in this work, an attempt was made to obtain quantitative data on allele distribution. We quantified the differences in Plasmodium falciparum msp-1 and msp-2 polymorphisms in matched peripheral and placental samples and matched placental and cord blood samples by using a fragment analysis method.
This study was carried out in Thiadiaye, southeast of Dakar. A total of 281 pregnant women were sequentially enrolled and followed until delivery. Placental, peripheral, and cord blood samples were obtained after delivery from all women. Genomic DNA was extracted from the placenta blood samples, and a P. falciparum species-specific PCR (22) identified 60 positive samples. A similar PCR assay performed on the corresponding 60 peripheral and 60 cord blood samples identified 39 positive placental/peripheral blood pairs and 11 positive placental/cord blood pairs. The study was approved by the ethical committee, Ministry of Health, Senegal, and informed consent was obtained from all patients.
A fluorescent PCR analyzed block 3 and block 2 of the msp-2 and of msp-1 domains, as described (11). Primers were msp-1 f-5′-CACATGAAAGTTATCAAGAACTTGTC-3′ (sense, fluorescein labeled) and 5′-GTACGTCTAATTCCATTTGCACG-3′ (antisense) and msp-2 f-5′-GAAGGTAATTAAAACATTGTC-3′ (sense, fluorescein labeled) and 5′-GAGGGATGTTGCTGCTCCACA-3′ (antisense) (Genset SA Europe) (19). Amplification products were processed in an ABI Prism 310 genetic analyzer (Perkin Elmer Applied Biosystems) and analyzed using Genescan software (Applied Biosystems) (11). Each genotype is characterized by its size and the area under the curve (AUC) of the peak corresponding to msp-1 or msp-2 PCR products. Each peak AUC is proportional to the quantity of PCR products for the corresponding allele: this allows a precise relative quantification of this genotype.
Numbers and ratios of alleles were quantified for the msp-1 and msp-2 loci in all samples. Relative quantification of each allele derived from the ratio of the AUC corresponding to this allele/the sum of the AUC of all alleles present in this sample. Alleles representing less than 2% of the overall parasite population from a given sample were not considered unless they were present in both matched samples.
The alleles present in matched placental/peripheral and placental/cord blood samples were compared, and the proportion of matched samples with identical and partially concordant alleles (paired samples with both identical and distinct alleles) was calculated. The proportion of parasites sharing identical alleles in matched blood samples was quantified as the sum of the relative proportions of all shared alleles from each compartment.
Forty-three PCR-positive placentas were paired with PCR-positive samples from peripheral or cord blood (including 39 placental/peripheral blood pairs and 11 placental/cord blood pairs). Seven women were PCR positive for all three compartments.
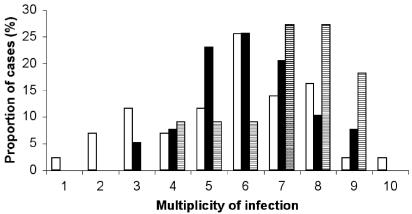
The fragment analysis method showed a large number of msp-1 and msp-2 alleles in all three compartments (Table 1). Among the 93 placental, peripheral, and cord blood samples analyzed, all but one corresponded to a polyclonal infection. The mean numbers of alleles were similar in peripheral (2.0 and 6.1 for msp-1 and msp-2, respectively), placental (2.1 and 5.6), and cord blood (1.4 and 7.1) samples (t test for paired samples; both P > 0.16 for placenta versus peripheral samples; P = 0.01 and 0.8 for msp-1 and msp-2, respectively, for placenta versus cord blood samples). The number of msp-2 alleles in placental blood was higher when the corresponding cord blood was infected (7.3 alleles) than when it was not (5.0 alleles) (Student t test; P = 0.001). The multiplicity of infection (mean number of alleles combining msp-1 and msp-2 genotyping), ranging over 1 to 10 alleles, in the peripheral, placental, and cord blood is shown in Fig. 1. Multiplicity of infection was not related to parasite density, being assessed on thick blood smears (Spearman correlation test; P = 0.3).
TABLE 1.
Allelic diversity, mean number of alleles, and the range of detected alleles in 43 women from Thiadiaye at delivery as deduced from msp-1 and msp-2 fragment-analysis method in placental, peripheral, and cord blood samples
| Gene and characteristic | Value for blood from:
|
||
|---|---|---|---|
| Periphery (n = 39) | Placenta (n = 43) | Cord (n = 11) | |
| msp-1 | |||
| Allelic diversity (no.) | 25 | 24 | 7 |
| Mean no. of alleles ± SD | 2.00 ± 1.12 | 2.14 ± 0.79 | 1.36 ± 0.81 |
| No. of alleles (min-max) | 1-5 | 1-4 | 1-3 |
| msp-2 | |||
| Allelic diversity (no.) | 26 | 21 | 17 |
| Mean no. of alleles ± SD | 6.10 ± 1.53 | 5.63 ± 2.12 | 7.09 ± 1.58 |
| No. of alleles (min-max) | 3-9 | 1-10 | 4-9 |
FIG. 1.
Results of combining the msp-1 and msp-2 genotyping data. The percentages of samples with different multiplicities of infection in the peripheral (black), placental (white), and cord (hatched) blood samples were calculated. Primers used were msp-1 f-5′-CACATGAAAGTTATCAAGAACTTGTC-3′ (sense, fluorescein labeled) and 5′-GTACGTCTAATTCCATTTGCACG-3′ (antisense) and msp-2 f-5′-GAAGGTAATTAAAACATTGTC-3′ (sense, fluorescein labeled) and 5′-GAGGGATGTTGCTGCTCCACA-3′ (antisense).
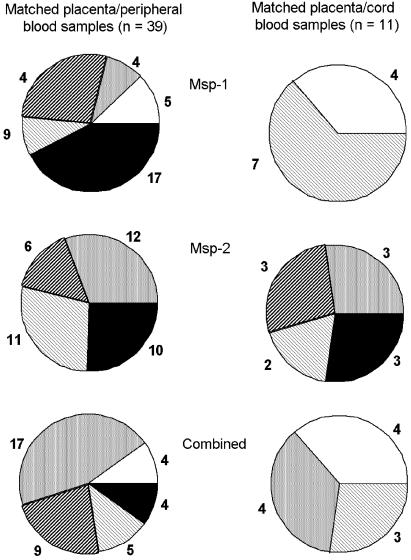
The proportion of matched placental/peripheral and placental/cord blood samples with identical and partially concordant msp-1 and msp-2 alleles (Fig. 2) shows three different situations: (i) concordant combined msp-1 and msp-2 profiles, representing identical parasite populations in both compartments; (ii) partial sharing of alleles corresponding to either one compartment parasite population being a subpopulation of the other or both compartments sharing alleles but also harboring nonshared alleles; and (iii) discordant allele profiles, suggesting completely separate populations in both compartments.
FIG. 2.
Msp-1 and msp-2 genotype composition in matched placental/peripheral samples and placental/cord blood samples with identical and partially concordant alleles, from infected women delivering in Thiadiaye, Senegal; the proportions of women with identical (black), completely discordant (white), and partially concordant alleles in matched samples are shown and the numbers of women are indicated. Three profiles are illustrated: msp-1 and msp-2 alleles taken separately and combined msp-1/msp-2 profiles. The group of matched samples with partially concordant alleles is subdivided into three subgroups with identical alleles in both compartments and additional alleles in (i) the placenta (light stripes), (ii) the peripheral or cord blood (heavy stripes), and (iii) both compartments (grey).
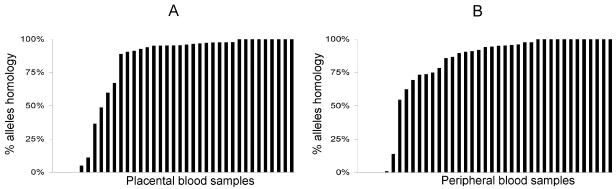
The percentage of identical alleles in matched pairs of samples was calculated from the quantitative data obtained by fragment analysis. For all matched samples, the sum of the relative quantification corresponding to alleles shared between the two compartments is shown in Fig. 3, representing the proportion of shared alleles in placental blood (A) and in peripheral blood (B) among the whole parasite population of each compartment. A median of 95.2% of the parasites from the placenta were also present in the peripheral blood, and 94.1% of parasites from peripheral blood were also in the placenta. A median of 79.9% of the parasites from the placenta were also present in the cord blood, and 95.9% of parasites from the cord blood were also in the placenta (data not shown).
FIG. 3.
Quantitative assessment of identical alleles in 39 matched placental/peripheral samples in placenta (A) and in peripheral blood (B) collected from infected women at delivery in Thiadiaye, Senegal. Each vertical bar represents a sample from one woman. Bars are ordered by increasing percentage of identical alleles between compartments.
Most studies on parasite population dynamics (6, 9, 11) and multiplicity of infection (1, 17) have investigated peripheral blood, where evidence is still lacking that detected parasites represent the overall infecting population. In women at delivery, there is a unique opportunity to analyze parasite genotypes in different compartments. A few studies addressed this issue, but only on a qualitative basis (2, 13, 21). A qualitative analysis of our data largely confirms those from previous studies, demonstrating a large allelic polymorphism and a high multiplicity of infection in the three compartments (4, 14, 20). Our data also confirm that in most matched placental/peripheral blood pairs, some alleles were shared, while others were detected only in one compartment (13, 21). One plausible explanation for the differences of allele composition in matched samples is the absence in one compartment of a highly synchronized genotype, from which all forms are either circulating ring forms or mature forms sequestering in the placenta (9). Conversely, asynchronous genotypes simultaneously present in both compartments lead to identical alleles in peripheral and placenta blood. In matched placental/cord blood samples, the alleles distribution differs more than in matched placental/peripheral samples, confirming the lower homology between cord and placenta blood (12, 13). Furthermore, full allelic discordance in some placental/cord blood pairs suggests the absence of contamination of cord blood by placental blood at delivery.
For the first time, we have performed the fragment analysis method (11) not only to enumerate alleles but also to quantify with a high sensitivity different parasite populations in different compartments. Specific allelic family PCR tends not to detect alleles present at proportions less than 10% (7, 8, 18), while we detected alleles at a ratio as low as 0.4% in paired samples from some individuals. Although several msp-1 and msp-2 alleles are not shared between the three compartments, the quantitative assessment demonstrates that almost all nonshared alleles constitute minor populations. Shared alleles in peripheral and placental represent 95.2% of the parasites from the placenta and 94.1% of those from peripheral blood. A high homology was also observed in matched placental/cord blood samples. Overall, these data strengthen the hypothesis that parasite populations in the peripheral, placental, and cord blood mostly derive from a single population. Moreover, parasites from peripheral and placental compartments share similar phenotypes of adherence to chondroitin sulfate A, the placental receptor of cytoadherence (23). The adhesion of infected erythrocytes to syncytiotrophoblasts in placenta is associated with the expression of distinct variant surface antigens on the membrane of infected erythrocytes. The major one is PfEMP-1 encoded by the var gene family (3, 10, 15). Further investigation should determine the expressed var genes responsible for parasite sequestration in placenta.
In conclusion, quantitative measurement of parasite alleles completes qualitative information and suggests a more homogenized view of the allelic distribution in different compartments. Identical alleles in placental, peripheral, and cord blood represent 80 to 95% of the parasite populations present in each of these compartments. The high degree of homology between placental and peripheral blood is consistent with the spontaneous clearance of parasites from peripheral blood within few hours after delivery (16), suggesting that the main, if not the only, organ of sequestration of P. falciparum in pregnant women is the placenta.
Acknowledgments
We acknowledge the support of the maternity staff of Thiadiaye hospital in sample collection, as well as the work of the field team of the IRD research unit in Senegal. We are grateful to the mothers who participated in the study. We are especially grateful to Nadine Fievet for her involvement in thick-blood-smear reading and Sandrine Houzé for help in P. falciparum PCR optimization.
This work was supported by grants from the Institut de Médecine et d'Epidémiologie Africaine (IMEA), the European Commission (QLK2-CT-2001-01302), the Fondation pour la Recherche Médicale (FRM), and the French Ministry of Health to National Malaria Reference Centre, Hôpital Bichat-Claude Bernard, APHP. Nicaise Tuikue Ndam was recipient of a grant from the French Ministry of Education and Research.
REFERENCES
- 1.Arnot, D. 1998. Unstable malaria in Sudan: the influence of the dry season. Clone multiplicity of Plasmodium falciparum infections in individuals exposed to variable levels of disease transmission. Trans. R. Soc. Trop. Med. Hyg. 92:580-585. [DOI] [PubMed] [Google Scholar]
- 2.Beck, S., F. P. Mockenhaupt, U. Bienzle, T. A. Eggelte, W. N. Thompson, and K. Stark. 2001. Multiplicity of Plasmodium falciparum infection in pregnancy. Am. J. Trop. Med. Hyg. 65:631-636. [DOI] [PubMed] [Google Scholar]
- 3.Biggs, B. A., L. Gooze, K. Wycherley, W. Wollish, B. Southwell, J. H. Leech, and G. V. Brown. 1991. Antigenic variation in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 88:9171-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Contamin, H., T. Fandeur, S. Bonnefoy, F. Skouri, F. Ntoumi, and O. Mercereau-Puijalon. 1995. PCR typing of field isolates of Plasmodium falciparum. J. Clin. Microbiol. 33:944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cot, M., and P. Deloron. 2003. Malaria prevention strategies. Brit. Med. Bull. 67:137-148. [DOI] [PubMed] [Google Scholar]
- 6.Daubersies, P., S. Sallenave-Sales, S. Magne, et al. 1996. Rapid turnover of Plasmodium falciparum populations in asymptomatic individuals living in a high transmission area. Am. J. Trop. Med. Hyg. 54:18-26. [DOI] [PubMed] [Google Scholar]
- 7.Durand, R., J. Eslahpazire, S. Jafari, et al. 2000. Use of molecular beacons to detect an antifolate resistance-associated mutation in Plasmodium falciparum. Antimicrob. Agents Chemother. 44:3461-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eldin de Pecoulas, P., L. K. Basco, B. Abdallah, M. K. Dje, J. Le Bras, and A. Mazabraud. 1995. Plasmodium falciparum: detection of antifolate resistance by mutation-specific restriction enzyme digestion. Exp. Parasitol. 80:483-487. [DOI] [PubMed] [Google Scholar]
- 9.Farnert, A., G. Snounou, I. Rooth, and A. Bjorkman. 1997. Daily dynamics of Plasmodium falciparum subpopulations in asymptomatic children in a holoendemic area. Am. J. Trop. Med. Hyg. 56:538-547. [DOI] [PubMed] [Google Scholar]
- 10.Forsyth, K. P., G. Philip, T. Smith, E. Kum, B. Southwell, and G. V. Brown. 1989. Diversity of antigens expressed on the surface of erythrocytes infected with mature Plasmodium falciparum parasites in Papua New Guinea. Am. J. Trop. Med. Hyg. 41:259-265. [PubMed] [Google Scholar]
- 11.Jafari, S., J. Le Bras, O. Bouchaud, and R. Durand. 2004. Plasmodium falciparum clonal population dynamics during malaria treatment. J. Infect. Dis. 189:195-203. [DOI] [PubMed] [Google Scholar]
- 12.Kamwendo, D. D., F. K. Dzinjalamala, G. Snounou, M. C. Kanjala, C. G. Mhango, M. E. Molyneux, and S. J. Rogerson. 2002. Plasmodium falciparum: PCR detection and genotyping of isolates from peripheral, placental, and cord blood of pregnant Malawian women and their infants. Trans. R. Soc. Trop. Med. Hyg. 96:145-149. [DOI] [PubMed] [Google Scholar]
- 13.Kassberger, F., A. Birkenmaier, A. Khattab, P. G. Kremsner, and M. Q. Klinkert. 2002. PCR typing of Plasmodium falciparum in matched peripheral, placental and umbilical cord blood. Parasitol. Res. 88:1073-1079. [DOI] [PubMed] [Google Scholar]
- 14.Mercereau-Puijalon, O. 1999. Molecular analysis of Plasmodium falciparum infections in man. Transfus. Clin. Biol. 6:44-56. [DOI] [PubMed] [Google Scholar]
- 15.Newbold, C. I., R. Pinches, D. J. Roberts, and K. Marsh. 1992. Plasmodium falciparum: the human agglutinating antibody response to the infected red cell surface is predominantly variant specific. Exp. Parasitol. 75:281-292. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen-Dinh, P., R. W. Steketee, A. E. Greenberg, J. J. Wirima, O. Mulenda, and S. B. Williams. 1988. Rapid spontaneous postpartum clearance of Plasmodium falciparum parasitaemia in African women. Lancet 2:751-752. [DOI] [PubMed] [Google Scholar]
- 17.Ntoumi, F., H. Contamin, C. Rogier, S. Bonnefoy, J. F. Trape, and O. Mercereau-Puijalon. 1995. Age-dependent carriage of multiple Plasmodium falciparum merozoite surface antigen-2 alleles in asymptomatic malaria infections. Am. J. Trop. Med. Hyg. 52:81-88. [DOI] [PubMed] [Google Scholar]
- 18.Parzy, D., C. Doerig, B. Pradines, A. Rico, T. Fusaï, and J. C. Doury. 1997. Proguanil resistance in Plasmodium falciparum African isolates: assessment by mutation-specific polymerase chain reaction and in vitro susceptibility testing. Am. J. Trop. Med. Hyg. 57:646-650. [DOI] [PubMed] [Google Scholar]
- 19.Ranford-Cartwright, L. C., P. Balfe, R. Carter, and D. Walliker. 1993. Frequency of cross-fertilization in the human malaria parasite Plasmodium falciparum. Parasitology 107:11-18. [DOI] [PubMed] [Google Scholar]
- 20.Robert, F., F. Ntoumi, G. Angel, D. Candito, C. Rogier, T. Fandeur, J. L. Sarthou, and O. Mercereau-Puijalon. 1996. Extensive genetic diversity of Plasmodium falciparum isolates collected from patients with severe malaria in Dakar, Senegal. Trans. R. Soc. Trop. Med. Hyg. 90:704-711. [DOI] [PubMed] [Google Scholar]
- 21.Schleiermacher, D., J. Y. Le Hesran, J. L. Ndiaye, R. Perraut, A. Gaye, and O. Mercereau-Puijalon. 2002. Hidden Plasmodium falciparum parasites in human infections: different genotype distribution in the peripheral circulation and in the placenta. Infect. Genet. Evol. 2:97-105. [DOI] [PubMed] [Google Scholar]
- 22.Singh, B., A. Bobogare, J. Cox-Singh, G. Snounou, M. S. Abdullah, and H. A. Rahman. 1999. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am. J. Trop. Med. Hyg. 60:687-692. [DOI] [PubMed] [Google Scholar]
- 23.Tuikue Ndam, N. G., N. Fievet, G. Bertin, G. Cottrell, A. Gaye, and P. Deloron. 2004. Variable adhesion abilities and overlapping antigenic properties in placental Plasmodium falciparum isolates. J. Infect. Dis. 190:2001-2009. [Online.]. [DOI] [PubMed] [Google Scholar]