Abstract
There is an urgent need for low-cost human immunodeficiency virus type 1 (HIV-1) viral load (VL) monitoring technologies in resource-limited settings. An automated TaqMan real-time reverse transcription-PCR (RT-PCR) assay was transferred to the laboratory of the Centre de Diagnostic et de Recherches sur le SIDA, Abidjan, Côte d'Ivoire, and assessed for HIV-1 RNA VL testing in 806 plasma samples collected within four ANRS research programs. The detection threshold and reproducibility of the assay were first determined. The quantitative results obtained with this assay were compared with two commercial HIV-1 RNA kits (the Versant version 3.0 and Monitor version 1.5 assays) in specimens harboring mainly the circulating recombinant form 02 strain (CRF02). The clinical evaluation of this test was done in different situations including the early diagnosis of pediatric infection and the monitoring of antiretroviral-treated patients. The quantification limit of our method was 300 copies/ml. The HIV-1 RNA values obtained by real-time PCR assay were highly correlated with those obtained by the Versant kit (r = 0.901; P < 0.001) and the Monitor test (r = 0.856; P < 0.001) and homogeneously distributed according to HIV-1 genotypes. For the early diagnosis of pediatric HIV-1 infection, the sensitivity and specificity of the real-time PCR assay were both 100% (95% confidence intervals of 93.7 to 100.0 and 98.3 to 100.0, respectively), compared to the Versant results. Following initiation of antiretroviral treatment, the kinetics of HIV-1 RNA levels were very comparable, with a similar proportion of adults and children below the detection limit during follow-up with our technique and the Versant assay. The TaqMan real-time PCR ($12 per test) is now routinely used to monitor HIV-1 infection in our laboratory. This technology should be further evaluated in limited-resource countries where strains other than CRF02 are prevalent.
In sub-Saharan Africa, the increased access to antiretroviral (ARV) drugs (due to significant price reductions) allows the large-scale implementation of programs focused on the prevention of mother-to-child transmission of human immunodeficiency virus type 1 (HIV-1) (6) and the scaling up of highly active antiretroviral therapy (HAART) (19, 21, 36, 37). In parallel, how to transfer and use accurate, simple, and low-cost HIV biological tools for monitoring the increasing number of HIV-positive individuals included in these programs is now a crucial issue in resource-limited settings (5).
In industrialized countries, commercial HIV-1 RNA viral load (VL) assays are used for the early diagnosis of HIV-1 infection in children (9, 20) and for monitoring patients receiving HAART (11). Unfortunately, in resource-constrained settings, these commercial assays are available only in reference laboratories because they are sophisticated and very expensive (for instance, between $50 and $100 per test in Côte d'Ivoire), largely exceeding the monthly cost of a standard HAART regimen (19). Two cheaper alternatives have been recently evaluated: the heat-dissociated HIV-1 p24 antigen enzyme-linked immunosorbent assay (2, 32) and the reverse transcriptase activity assay kit (Cavidi ExaVir) (3, 24). These commercial assays may be useful in provincial laboratories but have not been widely implemented because they are still labor intensive, relatively expensive ($21 and $15, respectively), and need more clinical validations. Besides dipsticks and chip technologies which are not yet ready to be used in routine practice (42), several real-time PCR tests could represent one other low-cost alternative HIV VL monitoring technology (18, 28). In developed countries, these assays have been mainly evaluated for viruses that are not of high commercial interest, such as human herpesvirus 8 (4), human T-cell leukemia type 1 (8), cytomegalovirus (23), and adenovirus (22), as well as for hepatitis B virus (17), and hepatitis C virus (41). For HIV infection, the assays have been used for HIV-2 (7), HIV-1 group O (16), and HIV-1 DNA proviral load (10, 12, 44). Moreover, plasma HIV-1 RNA has been assessed by real-time PCR for multiplex nucleic acid amplification testing in blood donations (25, 26, 40) and for HAART monitoring in HIV-1-seropositive patients (15, 30).
The main goals of the present study conducted in Abidjan (the capital city of Côte d'Ivoire, West Africa) were the following: (i) to transfer in a reference laboratory an automated TaqMan 5′ nuclease real-time PCR test applied to plasma HIV-1 RNA quantitation in order to evaluate its analytical performance characteristics in the context of the predominance of the circulating recombinant form 02 (CRF02) HIV-1 strain (38) and (ii) to perform a clinical evaluation of the assay against Bayer and Roche commercial assays, among patients (children and adults) enrolled within four distinct research programs conducted on site by the French Agence Nationale de Recherches sur le SIDA (ANRS).
(Part of this research was reported at the 2nd International AIDS Society Conference on HIV Pathogenesis and Treatment in Paris, France, 13 to 16 July, 2003, abstr. 277, 682, 683, 1032, and 1093; and at the 13th International Conference on AIDS and Sexually Transmitted Infections in Africa, Nairobi, Kenya, 21 to 26 September, 2003, abstr. 879186).
MATERIALS AND METHODS
Study populations and samples.
A total of 806 plasma samples (stored at −80°C) from 618 subjects included in four distinct ANRS research studies conducted in Abidjan were tested for HIV-1 RNA by using the TaqMan real-time RT-PCR assay (Table 1). All studies were approved by the Ethics Committee of the National AIDS Control program in Côte d'Ivoire.
TABLE 1.
Selected populations and samples tested by TaqMan HIV-1 RNA real-time RT-PCR assaya
| ANRS Program | Tested subjects (n) | Tested samples
|
Test(s) for comparison | |
|---|---|---|---|---|
| n | Timing | |||
| ANRS 1201/1202 Ditrame Plus Program | ||||
| HIV-1-positive mothers | 177 | 177 | Pre-inclusion | Versant v3.0 |
| HIV-1-infected neonates | 28 | 57b | D2, W4, W6, M3, M6 | Versant v3.0 |
| Uninfected neonates | 230 | 230c | W4 | Versant v3.0 |
| ANRS 1220 Primo-ci Cohort, HIV-1 seroconverters | 101 | 101 | D0 | Monitor v1.5 and Versant v3.0 |
| ANRS 1244/1278 Children Program, HIV-1-infected children with HAART | 36 | 149d | D0, M6, M12, M18, M24 | Versant v3.0 |
| ANRS 1269 Trivacan Trial, HIV-1-infected adults with HAART | 46 | 92 | D0, M3 | Versant v3.0 |
| Total | 618 | 806 | ||
Samples were collected from 2001 to 2003 in Abidjan, Côte d'Ivoire (West Africa). D, day; W, week; M, month; v, version.
Specimens positive with the Versant assay v3.0, and drawn at day 2 (n = 9), weeks 4 to 6 (n = 39) and months 3 to 6 (n = 9) of life in 28 HIV-1-infected children (see Materials and Methods).
Including 210 samples negative and 20 specimens with false-positive results by using the Versant assay v3.0 and taken at week 4 of life in 230 HIV-1-uninfected children (see Materials and Methods).
31 samples missing due to insufficient volume (n = 11), deaths (n = 3), or loss or incomplete follow-up (n = 17).
The first population consisted of 402 woman-infant pairs included in a program (ANRS 1201/1202 Ditrame Plus) for the prevention of mother-to-child transmission carried out from March 2001 to August 2002 (F. Dabis, D. K. Ekouevi, L. Bequet, F. Rouet, A. Horo, P. Fassinou, L. Dequae-Merchadoux, and V. Leroy, Abstr. 10th Conf. Retrovir. Opportunistic Infect., abstr. 854, 2003). The Versant branched DNA (bDNA) assay version 3.0 was used to detect early HIV-1 RNA in plasma from infants. HIV-1 infection in infants was diagnosed when plasma HIV-1 RNA levels were ≥5,000 copies/ml at 4 and 6 weeks of life. Children were considered to be HIV-1 negative when HIV-1 RNA levels were below the threshold (250 copies/ml) at 4 weeks. If the load was between 250 and 5,000 copies/ml at 4 weeks, it could indicate a false-positive result, meaning that a sample taken at 6 weeks was retested (34). Finally, HIV serology was assessed on available samples collected from 18 months of follow-up in order to validate HIV infection status. We evaluated the real-time reverse transcription-PCR (RT-PCR) by the following testing: (i) 177 maternal specimens taken at preinclusion and selected from four HIV-1 RNA classes according to bDNA results of 2.4 to 3.20 (n = 32), 3.21 to 3.91 (n = 37), 3.92 to 4.62 (n = 49), and >4.62 (n = 59) log10 copies/ml; (ii) 287 samples from children, consisting of all available specimens (n = 57) positive with the Versant assay and drawn between day 2 and month 6 of life among all 28 HIV-1-infected children, 210 randomly selected samples negative with the Versant test and taken at week 4 from 210 uninfected children, and all samples (n = 20) from 20 children identified as false-positive results at week 4 by the Versant assay but who were confirmed uninfected (negative HIV-1 RNA viral load at week 6 of life with the Versant assay and HIV serology negative at month 18).
The second group consisted of 180 HIV-1 seroconverter adults enrolled in the ANRS 1220 Primo-ci Cohort since June 1997 (35). The Roche RT-PCR assay version 1.5 was used to quantify plasma HIV-1 RNA at inclusion and every 6 months during follow-up. Real-time RT-PCR was used to determine baseline levels in samples from the first 101 enrolled subjects. Of these, 40 randomly selected specimens were also tested by the bDNA assay.
The third population was composed of 282 HIV-1-seropositive children (2 to 15 years old) enrolled in the ANRS 1244/1278 Pediatric Program since October 2000 (13). Among these, 150 received HAART because of a CD4+ cell percentage under 15% per mm3. The real-time RT-PCR test was compared with the Versant technique in samples from the first 36 ARV-treated children during a 24-month follow-up period.
The last study group included 840 adults enrolled in a structured treatment interruption trial (ANRS 1269 Trivacan) initiated in December 2002 (C. Danel, R. Moh, S. Sorho, A. Anzian, Y. Abo, H. Chenal, C. Kanga, S. Eholie, D. Sauvageot, D. Gabillard, F. Rouet, E. Bissagnene, X. Anglaret, and R. Salamon, Abstr. XV Int. AIDS Conf., abstr. WeOrB1284, 2004). Before randomization, patients with a CD4+ cell count ranging between 150 and 350 cells/mm3 received continuous HAART. The real-time RT-PCR test was compared with the Versant assay in samples taken from the first 46 ARV-treated individuals at baseline and 3 months after HAART initiation.
Plasma HIV-1 RNA quantification. (i) Real-time TaqMan RT-PCR test.
All tests were performed at the Centre de Diagnostic et de Recherches sur le SIDA (CeDReS, CHU Treichville, Abidjan, Côte d'Ivoire), which is the main reference laboratory for ANRS research studies conducted in Abidjan. The protocol was established by the Working Group for Viral Quantification of the ANRS (coordinated action AC11) (C. Rouzioux, F. Rouet, E. Nerrienet, C. Toure-Cane, O. Manigart, J. M. Reynes, T. Hung, M. Burgard, P. Colson, D. Descamps, M. Gueudin, O. Guisthau, P. Merel, M. Peeters, K. Saune, A. Vabret, and J. M. Seigneurin, Abstr. 2nd Conf. HIV Path. Treat., abstr. 682, 2003). Briefly, RNA was extracted from 200 μl of stored (−80°C) plasma by using the QIAGEN procedure (QIAGEN, Courtaboeuf, France). The TaqMan PCR targeted a conserved consensus region in the long terminal repeat (LTR) region of the HIV-1 major group. The sequences of the forward and reverse primers were 5′-GCCTCAATAAAGCTTGCCTTGA-3′ and 5′-GGCGCCACTGCTAGAGATTTT-3′, respectively. The internal HIV-1 TaqMan probe LTR was 5′-AAGTAGTGTGTGCCCGTCTGTTRTKTGACT-3′. This probe carried the 5′ reporter 6-carboxyfluorescein and the 3′ quencher 6-carboxytetramethylrhodamine (Applied Biosystems, Foster City, CA). All runs were performed in a 50-μl volume containing RNA extract (20 μl), primers (a 500 nM concentration of each), probe (200 nM), 1× PCR buffer, and 1× RT multiscribe/RNase Inhibitor Mix (Taqman One-Step RT-PCR Master Mix; Applied Biosystems). The external standard was a culture supernatant of an HIV-1 subtype B strain quantified by the Versant bDNA assay version 3.0. For each experiment, one aliquot of this standard was extracted together with the clinical samples and serially diluted (fivefold dilutions) to concentrations from 24,900,000 (7.4 log10) to 60 (1.8 log10) copies/ml.
Thermocycling conditions were 30 min at 48°C and 10 min at 95°C, followed by 50 cycles of 95°C for 15 sec and 60°C for 1 min. Amplification and data acquisition were carried out using the ABI Prism 7000 Sequence Detection System (Applied Biosystems). The percent amplification efficiency was calculated as [−1 + 10(1/slope)] × 100. The log10 of the number of targets initially present was proportional to the cycle threshold (CT) and was determined using the standard curve. The CeDReS participated in a north-south interlaboratory quality assurance and quality control (QA/QC) protocol (Rouzioux et al., Abstr. 2nd Conf. HIV Path. Treat., abstr. 682, 2003); results were highly concordant.
(ii) Commercial HIV-1 RNA assays.
The Versant bDNA HIV RNA kit version 3.0 (Bayer Diagnostics, Emeryville, CA) was used at the CeDReS, strictly according to the manufacturer's instructions (29). Quantitative results were recorded by the Quantiplex bDNA system 340 analyzer and determined from a standard curve by using DNA plasmid at four concentrations. In all cases, 200 μl of stored (−80°C) plasma was used. This approach was shown not to affect the quantitative results of the bDNA assay (43). A threshold of 250 copies/ml was used in all contexts (quantification in adults and HAART monitoring) except for the diagnosis of HIV-1 in children (see above). The Amplicor HIV-1 Monitor standard RT-PCR assay (version 1.5; Roche Molecular Systems, Pleasanton, CA) was performed at the Centre Intégré de Recherche Bioclinique d'Abidjan, Abidjan, Côte d'Ivoire) on stored (−80°C) plasmas with a threshold of 400 copies/ml (2). In both cases, specimens that gave results exceeding the upper limit of the test were retested after dilution.
HIV-1 genotyping assay.
Phylogenetic analyses were performed by using the consensus protocol of the ANRS Resistance study group (31). The relationships between reverse transcriptase sequences were estimated by using reference sequences of HIV-1 genetic subtypes and CRFs obtained from the Los Alamos Database (http://hiv-web.lanl.gov). Reverse transcriptase nucleotide sequences were aligned by using the CLUSTAL W program, version 1.7. Phylogenetic trees were constructed by using a Kimura two-parameter model and the neighbor-joining method with 100 bootstrapped data sets (14). Overall, 177 HIV-1 isolates were sequenced in the context of ANRS studies (71 from women enrolled in the Ditrame Plus study, 92 from adults recruited in the Primo-ci Cohort, and 14 from children in the ANRS 1244/1278 program).
Statistical analysis.
The standard was tested in 25 separate runs to determine the threshold, reproducibility, and linearity of the real-time RT-PCR technique. A low-positive control (quantified at 3.72 log10 copies/ml by the bDNA assay and at 3.80 log10 copies/ml by the Monitor RT-PCR kit) was also used to determine between-run variability. To assess within-run variability, the low-positive control was tested in 10 replicates, and clinical samples were initially tested in duplicate. Spearman correlation coefficients were calculated to determine the relationship between the HIV-1 RNA levels obtained with the Versant kit (or with the Monitor assay) and with the real-time PCR technique. The Bland-Altman method (1) was used to assess the agreement between values obtained with the three assays. For the early diagnosis of infection in children, the sensitivity of the real-time PCR was calculated as the number of positive results divided by the total number of positive samples obtained with the Versant assay on specimens from infected children. The specificity was calculated as the number of negative samples divided by the total number of negative specimens obtained with the Versant assay on specimens from uninfected children. The proportions of subjects with undetectable HIV-1 RNA levels after HAART initiation according to the Versant assay and the real-time PCR were compared using the McNemar matched chi-square test. The mean and median plasma HIV-1 RNA levels were compared using a paired Student's t test and the Mann-Whitney test, respectively.
RESULTS
Analytical evaluation. (i) Interassay reproducibility of the standard curve and determination of the detection threshold.
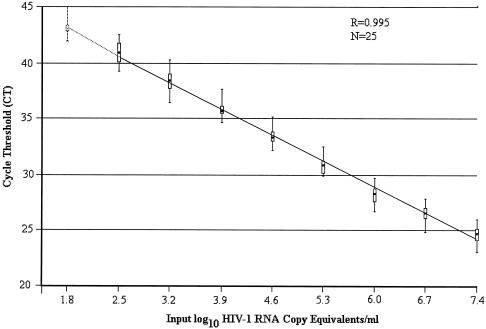
The interassay reproducibility of the standard curve was determined (Fig. 1). In all cases, there was a strong linear relationship between the CT values and the log10 of the number of HIV-1 RNA copies/ml. The median correlation coefficient was 0.995 (range, 0.991 to 0.998). The median slope was −3.38 (range, −3.02 to −3.76), corresponding to a median PCR efficiency of 98%. As expected, every fivefold decrease in the copy number yielded an increase in the CT value of approximately 2. The amplification/detection of the lower standard concentrations was 100% at 300 copies/ml and 33% at 60 copies/ml (Fig. 1, dotted section of the line). The quantification cutoff for the assay was thus set at 300 (2.48 log10) copies/ml, with a linear dynamic range of five orders of magnitude.
FIG. 1.
Standard curve of the TaqMan HIV-1 RNA real-time PCR test, Abidjan, Côte d'Ivoire (West Africa). The cycle threshold (CT) is the number of cycles before the fluorescence passes a fixed limit (time to positivity). The CT values were obtained from 25 different runs, with three distinct operators. The solid line connects the median values and 25% and 75% interquartile ranges (box plot) of the CT. The vertical lines show the ranges of CT.
(ii) Within- and between-run variations.
When testing the low positive control, mean values of 3.83 (standard deviation [SD], ±0.12) and 3.93 (SD, ±0.18) log10 copies/ml were obtained for within- and between-run assays, respectively, with coefficients of variation of 3.1 and 4.6%, respectively. When the first 185 clinical samples were tested in duplicate, an excellent within-run variation was also obtained (r = 0.989; P < 0.001). Thus, all subsequent clinical specimens were quantified just once.
(iii) Comparison with commercial plasma HIV-1 RNA assays.
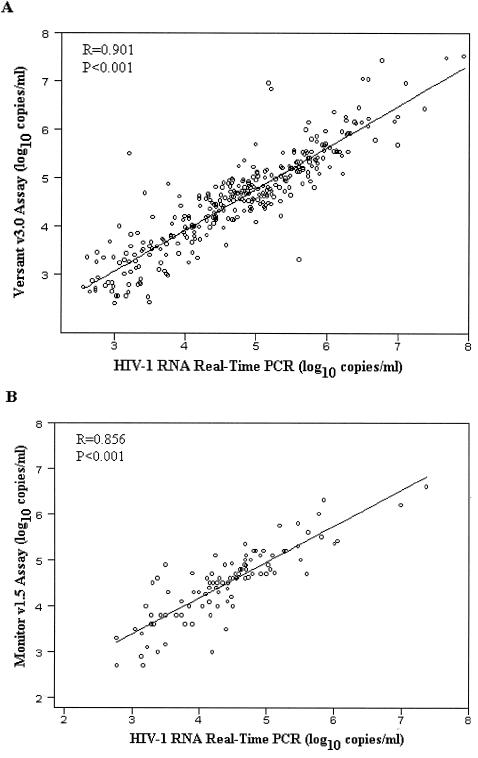
Of the 327 independent plasma specimens that were positive with the Versant bDNA assay, 320 (97.9%) also tested positive by real-time PCR. Seven were Versant positive but real-time PCR negative, with a Versant value of <3.0 log10 copies/ml for six samples and <4.0 log10 copies/ml for one sample. For the 320 specimens that tested positive with both techniques, plasma HIV-1 RNA levels were highly correlated (Fig. 2A) (r = 0.901; P < 0.001). The mean difference in the HIV-1 RNA values obtained with real-time PCR and the Versant assay was +0.20 log10 copies/ml (mean, 4.75 versus 4.55 log10 copies/ml, respectively) (P < 0.001).
FIG. 2.
Correlation between HIV-1 RNA viral load as measured with the TaqMan real-time PCR test and with two commercial RNA kits. (A) Comparison between the real-time PCR test and the Versant assay, version 3.0 (320 comparative measurements including 28 samples from children and 171 samples from mothers in the ANRS 1201/1202 Ditrame plus program, 39 adult samples from the Primo-ci Cohort, 36 samples from children in the ANRS 1244/1278 program, and 46 adult samples from the ANRS 1269 trial). (B) Comparison between the real-time PCR test and the Monitor assay version 1.5 (97 comparative measurements in the ANRS 1220 Primo-ci Cohort). The fitted regressions are represented by solid lines.
Among the 101 samples from the Primo-ci Cohort, 99 tested positive with the Monitor RT-PCR assay and 2 were negative. The real-time PCR test gave concordant positive results for 97 of the 99 (98.0%) specimens. Two samples were Roche positive but real-time PCR negative (Roche values, 4.80 and 2.80 log10 copies/ml, respectively). Real-time PCR failed to detect HIV-1 RNA in one of the two Roche-negative samples, whereas the other one was found to be positive (3.80 log10 copies/ml). For the 97 specimens that tested positive with both techniques, plasma HIV-1 RNA levels were highly correlated (Fig. 2B) (r = 0.856; P < 0.001). The mean difference in the plasma HIV-1 RNA levels obtained with real-time PCR and the Roche assay was −0.09 log10 copies/ml (mean, 4.43 versus 4.52 log10 copies/ml, respectively) (P = 0.058).
(iv) Impact of HIV-1 genotypic diversity.
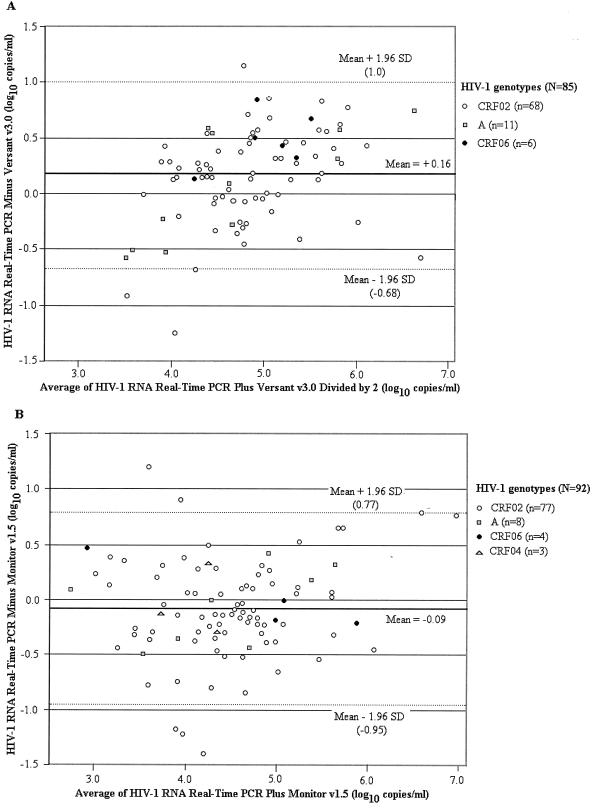
The phylogenetic analysis revealed that most of the isolates (145/177 or 81.9%) clustered within CRF02. As shown in Fig. 3A and B (Versant and Monitor assays compared to real-time PCR, respectively), the differences in log10 assay results versus the mean assay results were homogeneously distributed within the 95% confidence interval (95% CI) of the mean difference. In Fig. 3A, 96.5% (82/85) of the HIV-1 RNA values fell between these ranges. Differences of more than 0.5 log10 copies/ml were observed in 23.5% of specimens, including 9 (6 CRF02, 2 CRF06, and 1 subtype A) that were better quantified with the real-time PCR test versus 11 (8 CRF02 and 3 subtypes A) that were better quantified with the Versant assay. In Fig. 3B, 87 of 92 (94.6%) specimens fell between the 95% CI of the mean difference. Differences of more than 0.5 log10 copies/ml were obtained in 19.6% of specimens, including 10 that were better quantified with the real-time PCR test versus 8 that were better quantified with the Monitor assay.
FIG. 3.
Impact of HIV-1 genotype on plasma HIV-1 RNA levels measured with the TaqMan real-time PCR test and two commercial RNA kits. (A) Difference between the mean viral loads measured with the real-time PCR test and the Versant assay version 3.0 (85 comparisons). (B) Difference between the mean viral loads measured with real-time PCR test and Monitor assay, version 1.5 (92 comparisons). The mean differences are represented by solid lines; the 95% CIs are shown by dashed lines.
Clinical evaluation. (i) Early diagnosis of HIV-1 infection in children.
Fifty-seven samples from 28 infected children in the Ditrame Plus Program tested positive by the Versant assay. The sensitivity of the real-time PCR for these 57 samples was 100% (95% CI, 93.7 to 100.0) at day 2 (n = 9), 4 to 6 weeks (n = 39), and 3 to 6 months (n = 9). All children who had a positive test result by HIV enzyme-linked immunosorbent assays at month 18 (n = 14) had previously tested positive by real-time PCR. Of 210 specimens from 210 uninfected children that were negative with the Versant technique at 4 weeks of life, the real-time PCR technique also failed to detect HIV-1 RNA in any of these samples, yielding a specificity of 100% (95% CI, 98.3 to 100.0). None of the 138 children who tested HIV-antibody negative at ≥18 months had ever previously tested positive by real-time PCR. All the 20 samples previously identified as false-positive results by the Versant test were negative by real-time PCR. Among these, 14 children who could be followed up at month 18 were found to be HIV-antibody negative.
(ii) Kinetics of plasma HIV-1 RNA levels during HAART.
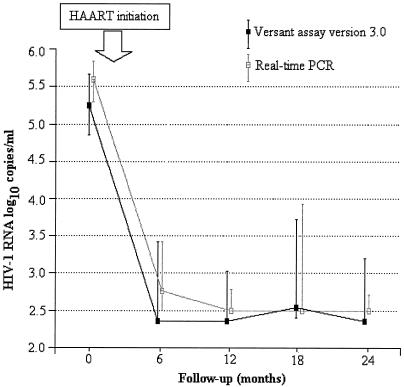
We monitored the kinetics of HIV-1 RNA following initiation of HAART in 36 children from the ANRS 1244/1278 program (Fig. 4). The median HIV-1 RNA levels at baseline were 5.23 and 5.57 log10 copies/ml with Versant and real-time PCR, respectively (P < 0.001). The proportions of children with undetectable HIV-1 RNA with these two techniques were not significantly different at months 6, 12, 18, and 24 (P values of 0.68, 0.75, 0.29, and 0.37, respectively, by the McNemar test).
FIG. 4.
Kinetics of plasma HIV-1 RNA decrease on HAART. The graph shows a comparison between the TaqMan real-time PCR test and the Versant assay based on the analysis of 149 samples taken from 36 ARV-treated children enrolled in the ANRS 1244/1278 cohort, Abidjan, Côte d'Ivoire. The solid line connects the median values of the individual data points. The vertical bars represent the 25% and 75% interquartile ranges.
In samples from 46 adults from the ANRS 1269 trial, the median HIV-1 RNA levels at baseline were 4.87 log10 copies/ml with the Versant assay and 5.22 log10 copies/ml with the real-time PCR (P < 0.001). The proportion of treated patients with undetectable levels at month 3 was not significantly different with the Versant assay (36/46 or 78.3%) and real-time PCR (30/46 or 65.2%) (McNemar test, P = 0.11).
DISCUSSION
This is the first report describing the transfer and validation of an automated real-time RT-PCR test (targeting the LTR gene) used as an HIV-1 diagnostic and monitoring technology in a resource-limited country such as Côte d'Ivoire (West Africa).
The test offered four advantages. First, the test had good analytical performance characteristics, similar to those obtained with commercial HIV-RNA assays. Indeed, this technique appeared to be sensitive in our setting where the CRF02 is the most prevalent strain. The cutoff of 300 copies/ml was similar to cutoffs obtained with the commercial assays, and there was no need for repeat testing in the case of high VLs. This test was precise and reproducible with good intra- and interrun SDs. Excellent correlations were obtained between real-time PCR test results and those obtained from commercially available HIV-1 RNA VL test kits. Its specificity was also excellent. Indeed, real-time PCR amplified and quantified HIV-1 RNA simultaneously in sealed plates that were never opened following amplification, minimizing the risk of PCR product carry-over contamination. Second, the test demonstrated its clinical usefulness for HIV-1 RNA VL monitoring in HIV-1-infected patients. It required just 0.2 ml of plasma, making it suitable for use on adults and children as well. Our test has given excellent results for both infant diagnosis and HAART efficacy monitoring. Third, the test was low cost; indeed, this test was performed in Côte d'Ivoire for $12.4 per sample, including reagents for extraction ($4.4), “mix” ($4.3), probe and primers ($0.2), consumables ($3.4 for microtubes, microplates, and filtered tips), and other materials ($0.1 for ethanol, distilled water, and free-powder gloves). In 2002 to 2004, it was 5- to 10-fold less costly than commercial HIV-RNA kits. Fourth, the test has good feasibility and practicality. It was easy to obtain reagents in Côte d'Ivoire, with delivery time not exceeding 1 month. The probe and primers could be purchased once per year, given the long half-life of these freeze-dried reagents. In contrast, the bDNA assay had to be ordered several times per year and required dry ice. Moreover, with two trained technicians, 70 HIV-1 RNA clinical results could be obtained in 4 h 30 min. Thus, this high-throughput technique required significantly less hands-on time than the commercial assays and did not need the dedicated laboratory space necessary for standard PCR techniques. By comparison, in our African setting, with two technicians, 84 clinical results were obtained in 36 h by using the Versant assay, and 44 were obtained in 7 h with the Amplicor kit.
The disadvantages of this test were as follows: (i) those common to all HIV-1 RNA VL assays such as complex technology, expensive equipment cost (the start-up costs of the instrumentation are currently between $30,000 and $50,000, depending on the material used), and equipment maintenance; and (ii) those more specifically observed with all “home-brew” real-time PCR assays such as the variability in reagents used (notably for the “mix” and standard curve), with no manufacturer's quality assurance. This highlights the importance of both internal controls which must be used during each run and interlaboratory QA/QC programs. The first training QA/QC exercise has shown that all laboratories (10 in France and 6 in Africa, including CeDReS) were able to produce consistent and reproducible results (Rouzioux et al., Abstr. 2nd Conf. HIV Path. Treat., abstr. 682, 2003).
In conclusion, our TaqMan real-time PCR test provided a significant virologic contribution to the early diagnosis of HIV infection in children, the reliable quantification of non-B HIV-1 subtypes circulating in Côte d'Ivoire, and the monitoring of HAART effectiveness, compared to commercial assays. Since April 2003, we have been using it routinely in our laboratory for the analysis of all specimens from ANRS programs. In many resource-limited countries, a CD4 count is considered essential and HIV-1 RNA VL optional for guiding HAART prescription and follow-up. However, given the performance characteristics and low cost of real-time PCR assays, we believe that they will become part of HAART monitoring, even in resource-limited settings, in order to better promote adherence and prevent ARV drug resistance. The future challenges for these promising real-time PCR techniques include, first, validating their performance characteristics in large cohorts of ARV-treated patients from other resource-limited countries where other clades of HIV-1 are circulating. For countries with multiple coexisting HIV-1 subtypes, such as Cameroon (39), the flexibility of real-time PCR with respect to primers and probe provides an advantage in dealing the HIV-1 diversity challenge. For this purpose, a newly designed minor groove binder HIV-1 probe has been developed, and its evaluation is in progress in the laboratory of virology in Necker Hospital (Paris). Moreover, real-time PCR can be adapted for multiple virus detection (HIV-2, hepatitis B virus, and hepatitis C virus) (33). The second challenge is to ensure reliable specimen transfer from rural regions to appropriate reference laboratories. As shown previously for commercial HIV-1 RNA kits (27), the usefulness of real-time PCR assays using dried blood spots has recently been demonstrated for the early detection of HIV-1 infection in children born to HIV-positive mothers in Thailand (W. Khamduang, N. Kanthawong, P. Leechanachai, P. Ananpatharachai, P. Attavijtrakarn, P. Wannarit, S. Hanpinitsak, S. Hotrawarikarn, S. Sirinontakan, S. Srirojana, S. Tanupattarachai, T. Hinjiranandana, W. Ardong, and N. Ngo-Giang-Huong Abstr. XV Int. AIDS Conf., abstr. MoPeB3162).
Acknowledgments
This work was supported by the French Agence Nationale de Recherches sur le SIDA (ANRS, Paris, France) and the French Ministry of Foreign Affairs as part of Coordinated Action AC12.
The authors wish to acknowledge the support of the Developing Country Unit of the ANRS, particularly Brigitte Bazin, Séverine Blesson, and Chantal Canon.
We thank C. Montcho, N. Coulibaly, and J. B. Kottan for their excellent technical assistance. We thank Serge Maurin (Applied Biosystems), Julien Gaha, Bertin Kouadio, Ezéchiel Akédier, and Philippe Lepère for supplying the reagents. We are grateful to the Working Group for Viral Quantification of the ANRS (AC11), coordinated by J. M. Seigneurin and C. Rouzioux.
Finally, we thank all HIV-positive subjects enrolled in the ANRS programs in Abidjan.
The ANRS Côte d'Ivoire PAC-CI Program, coordinated by T. N′Dri-Yoman and R. Salamon, is organized as described below.
For the the ANRS 1201/1202 Ditrame Plus Program Study Group, F. Dabis and V. Leroy (INSERM U 593, Université Victor Segalen, Bordeaux, France) were principal investigators; other investigators were C. Welffens-Ekra and M. Timité-Konan (Centre Hospitalier Universitaire de Yopougon, Abidjan, Côte d'Ivoire). L. Bequet, D. K. Ekouevi, I. Viho, and B. Tonwe-Gold provided coordination in Abidjan. Members of the clinical team were C. Amani-Bosse, I. Ayekoe, G. Bédikou, N. Coulibaly, P. Fassinou, A. Horo, R. Likikouët, and H. Touré and staff of the Health Centers of Anonkouakouté, Sagbé, Avocatier, Abobo-sud, Niangon-sud, Toit Rouge, and Wassakara. The laboratory team included staff of the Centre de Diagnostic et de Recherches sur le SIDA; A. Inwoley, C. Montcho, and F. Rouet (Centre Hospitalier Universitaire de Treichville); (P. Fian and O. Fofana (Abobo-Avocatier); and G. Kouaho (Yopougon). R. Becquet, L. Dequae-Merchadou, C. Sakarovitch, and D. Touchard (INSERM U 593, Bordeaux, France) and G. Allou and A. Gerard (PAC-CI, Abidjan, Côte d'Ivoire) were responsible for biostatistics and data management. H. Aka-Dago, A. Desgrées du Lou, S. Kouadio, A. Sihé, and B. Zanou were members of the psycho-social team. S. Blanche, J.-F. Delfraissy, P. Lepage, L. Mandelbrot, C. Rouzioux, and R. Salamon were members of the scientific committee.
For the ANRS 1220 Primo-ci Cohort Study Group, R. Salamon (INSERM U 593, Université Victor Segalen, Bordeaux, France) was the principal investigator. Coordination in Abidjan was provided by C. Huet and A. Minga. Members of the clinical team were Y. Abo, C. Danel, L. Dohoun, and A. Minga. Members of the laboratory team were N. Coulibaly and T. A. Tony (Centre Intégré de Recherches Biocliniques).). Biostatistics and data management were handled by F. Dabis (INSERM U 593, Bordeaux) and by A. Coulibaly, C. Huët, and E. Moro (PAC-CI Abidjan, Côte d'Ivoire). M. Moussa was a member of the psycho-social team.
For the ANRS 1244/1278 Pediatric Program Study Group, P. Msellati (UR 036, IRD, Abidjan, Côte d'Ivoire) was the principal investigator. (Coordination was provided by M. F. Anaky and R. Laguidé. R. Dossou, N. Elenga, P. Fassinou, A. Kouakoussui, and M. L. Wénin were members of the clinical team. H. Ménan, T. Ouassa, and F. Rouet (CeDReS, Centre Hospitalier Universitaire de Treichville) were members of the laboratory team. H. Atta (PAC-CI Abidjan) was responsible for biostatistics and data management. H. Aka-Dago and M. C. Cacou were members of the psychological team.
For the ANRS 1269 Trivacan Trial Study Group, R. Salamon (INSERM U 593, Université Victor Segalen, Bordeaux, France) and E. Bissagnéné (Centre Hospitalier Universitaire de Treichville, Abidjan, Côte d'Ivoire) were principal investigators. Coordination was provided by X. Anglaret (INSERM U 593, Bordeaux, France) and C. Danel (PAC-CI, Abidjan, Côte d'Ivoire). H. Chenal, S. Eholié, C. Kanga, A. Minga, R. Moh, C. Seyler, and S. Touré were members of the clinical team. A. Inwoley, F. Rouet, and R. Touré (CeDReS, Centre Hospitalier Universitaire de Treichville) were members of the laboratory team. Biostatistics and data management were handled by D. Gabillard (INSERM U 593, Bordeaux, France) and S. Sorho (PAC-CI, Abidjan, Côte d'Ivoire). F. Barré-Sinoussi, F. Boué, G. Chêne, F. Dabis, Y. A. Flori, P.-M. Girard, A. Kadio, C. Leport, T. N′Dri-Yoman, and Y. Souteyrand were members of the scientific committee.
REFERENCES
- 1.Bland, J. M., and D. J. Altman. 1995. Comparing methods of measurement: why plotting difference against standard method is misleading. Lancet 346:1085-1087. [DOI] [PubMed] [Google Scholar]
- 2.Bonard, D., F. Rouet, T. A. Toni, A. Minga, C. Huet, D. K. Ekouevi, F. Dabis, R. Salamon, and C. Rouzioux. 2003. Field evaluation of an improved assay using a heat-dissociated p24 antigen for adults mainly infected with HIV-1CRF02_AG strains in Cote d'Ivoire, West Africa. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 34:267-273. [DOI] [PubMed] [Google Scholar]
- 3.Braun, J., J. C. Plantier, M. F. Hellot, E. Tuaillon, M. Gueudin, F. Damond, A. Malmsten, G. E. Corrigan, and F. Simon. 2003. A new quantitative HIV load assay based on plasma virion reverse transcriptase activity for the different types, groups and subtypes. AIDS 17:331-336. [DOI] [PubMed] [Google Scholar]
- 4.Broccolo, F., G. Locatelli, L. Sarmati, S. Piergiovanni, F. Veglia, M. Andreoni, S. Butto, B. Ensoli, P. Lusso, and M. S. Malnati. 2002. Calibrated real-time PCR assay for quantitation of human herpesvirus 8 DNA in biological fluids. J. Clin. Microbiol. 40:4652-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowe, S., S. Turnbull, R. Oelrichs, and A. Dunne. 2003. Monitoring of human immunodeficiency virus infection in resource-constrained countries. Clin. Infect. Dis. 37:S25-S35. [DOI] [PubMed] [Google Scholar]
- 6.Dabis, F., and E. R. Ekpini. 2002. HIV-1/AIDS and maternal and child health in Africa. Lancet 359:2097-2104. [DOI] [PubMed] [Google Scholar]
- 7.Damond, F., M. Gueudin, S. Pueyo, I. Farfara, D. L. Robertson, D. Descamps, G. Chene, S. Matheron, P. Campa, F. Brun-Vezinet, and F. Simon. 2002. Plasma RNA viral load in human immunodeficiency virus type 2 subtype A and subtype B infections. J. Clin. Microbiol. 40:3654-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dehee, A., R. Cesaire, N. Desire, A. Lezin, O. Bourdonne, O. Bera, Y. Plumelle, D. Smadja, and J. C. Nicolas. 2002. Quantitation of HTLV-I proviral load by a TaqMan real-time PCR assay. J. Virol. Methods 102:37-51. [DOI] [PubMed] [Google Scholar]
- 9.Delamare, C., M. Burgard, M. J. Mayaux, S. Blanche, A. Doussin, S. Ivanoff, M.-L. Chaix, C. Khan, C. Rouzioux. 1997. HIV-1 RNA detection in plasma for the diagnosis of infection in neonates. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 15:121-125. [DOI] [PubMed] [Google Scholar]
- 10.Desire, N., A. Dehee, V. Schneider, C. Jacomet, C. Goujon, P. M. Girard, W. Rozenbaum, and J. C. Nicolas. 2001. Quantification of human immunodeficiency virus type 1 proviral load by a TaqMan real-time PCR assay. J. Clin. Microbiol. 39:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dybul, M., A. S. Fauci, J. G. Bartlett, J. E. Kaplan, A. K. Pau, et al. 2002. Guidelines for using antiretroviral agents among HIV-infected adults and adolescents Ann. Intern. Med. 137:381-433. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson, L. E., T. Leitner, B. Wahren, A. C. Bostrom, and K. I. Falk. 2003. A multiplex real-time PCR for quantification of HIV-1 DNA and the human albumin gene in CD4(+) cells. APMIS 111:625-633. [DOI] [PubMed] [Google Scholar]
- 13.Fassinou, P., N. Elenga, F. Rouet, R. Laguide, K. A. Kouakoussui, M. Timite, S. Blanche, and P. Msellati. 2004. Highly active antiretroviral therapies among HIV-1 infected children in Abidjan, Cote d'Ivoire. AIDS 18:1905-1913. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 2001. PHYLIP, phylogeny inference package, version 3.6 (alpha). University of Washington, Seattle, Washington.
- 15.Gibellini, D., F. Vitone, E. Gori, M. La Place, and M. C. Re. 2004. Quantitative detection of human immunodeficiency virus type 1 (HIV-1) viral load by SYBR green real-time RT-PCR technique in HIV-1 seropositive patients. J. Virol. Methods 115:183-189. [DOI] [PubMed] [Google Scholar]
- 16.Gueudin, M., J. C. Plantier, F. Damond, P. Roques, P. Mauclere, and F. Simon. 2003. Plasma viral RNA assay in HIV-1 group O infection by real-time PCR. J. Virol. Methods 113:43-49. [DOI] [PubMed] [Google Scholar]
- 17.Ho, S. K. N., W. C. Yam, E. T. K. Leung, L. P. Wong, J. K. H. Leung, K. N. Lai, and T. M. Chan. 2003. Rapid quantification of hepatitis B virus DNA by real-time PCR using fluorescent hybridization probes. J. Med. Microbiol. 52:397-402. [DOI] [PubMed] [Google Scholar]
- 18.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katzenstein, D., M. Laga, and J. P. Moatti.2003. The evaluation of the HIV/AIDS drug access initiatives in Côte d'Ivoire, Senegal and Uganda: how access to antiretroviral treatment can become feasible in Africa. AIDS 17:S1-S4. [PubMed] [Google Scholar]
- 20.Lambert, J. S., D. R. Harris, E. R. Stiehm, J. Moye, M. G. Fowler, W. A. Meyer III, J. Bethel, and L. M. Mofenson. 2003. Performance characteristics of HIV-1 culture and HIV-1 DNA and RNA amplification assays for early diagnosis of perinatal HIV-1 infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 34:512-519. [DOI] [PubMed] [Google Scholar]
- 21.Landman, R., R. Schiemann, S. Thiam, M. Vray, A. Canestri, S. Mboup, C. T. Kane, E. Delaporte, P. S. Sow, M. A. Faye, M. Gueye, G. Peytavin, C. Dalban, P. M. Girard, and I. Ndoye. 2003. Once-a-day highly active antiretroviral therapy in treatment-naive HIV-1-infected adults in Senegal. AIDS 17:1017-1022. [DOI] [PubMed] [Google Scholar]
- 22.Leruez-Ville, M., V. Minard, F. Lacaille, A. Buzyn, E. Abachin, S. Blanche, F. Freymuth, and C. Rouzioux. 2004. Real-time blood plasma polymerase chain reaction for management of disseminated adenovirus infection. Clin. Infect. Dis. 38:45-52. [DOI] [PubMed] [Google Scholar]
- 23.Leruez-Ville, M., M. Ouachee, R. Delarue, A. S. Sauget, S. Blanche, A. Buzyn, and C. Rouzioux. 2003. Monitoring cytomegalovirus infection in adult and pediatric bone marrow transplant recipients by a real-time PCR assay performed with blood plasma. J. Clin. Microbiol. 41:2040-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malmsten, A., X. W. Shao, K. Aperia, G. E. Corrigan, E. Sandstrom, C. F. R. Kallander, T. Leitner, and J. S. Gronowitz. 2003. HIV-1 viral load determination based on reverse transcriptase activity recovered from human plasma. J. Med. Virol. 71:347-359. [DOI] [PubMed] [Google Scholar]
- 25.Meng, Q., C. Wong, A. Rangachari, S. Tamatsukuri, M. Sasaki, E. Fiss, L. Cheng, T. Ramankutty, D. Clarke, H. Yawata, Y. Sakakura, T. Hirose, and C. Impraim. 2001. Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 39:2937-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mine, H., H. Emura, M. Miyamoto, T. Tomono, K. Minegishi, H. Murokawa, R. Yamanaka, A. Yoshikawa, and K. Nishioka. 2003. High throughput screening of 16 million serologically negative blood donations for hepatitis B virus, hepatitis C virus and human immunodeficiency virus type-1 by nucleic acid amplification testing with specific and sensitive multiplex reagent in Japan. J. Virol. Methods 112:145-151. [DOI] [PubMed] [Google Scholar]
- 27.Mwaba, P., S. Cassol, A. Nunn, R. Pilon, C. Chintu, M. Janes, and A. Zumla. 2003. Whole blood versus plasma spots for measurement of HIV-1 viral load in HIV-infected African patients. Lancet 362:2067-2068. [DOI] [PubMed] [Google Scholar]
- 28.Niesters, H. G. M. 2001. Quantitation of viral load using real-time amplification techniques. Methods 25:419-429. [DOI] [PubMed] [Google Scholar]
- 29.Pachl, C., J. A. Todd, D. G. Kern, P. J. Sheridan, S. F. Fong, M. Stempien, B. Hoo, D. Besemer, T. Yeghiazarian, B. Irvine, J. Kolberg, R. Kokka, P. Neuwald, and M. S. Urdea. 1995. Rapid and precise quantification of HIV-1 RNA in plasma using a branched DNA (bDNA) signal amplification assay. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 8:446-454. [DOI] [PubMed] [Google Scholar]
- 30.Palmer, S., A. P. Wiegand, F. Maldarelli, H. Bazmi, J. M. Mican, M. Polis, R. L. Dewar, A. Planta, S. Y. Liu, J. A. Metcalf, J. W. Mellors, and J. M. Coffin. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 41:4531-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasquier, C., N. Millot, R. Njouom, K. Sandres, M. Cazabat, J. Puel, and J. Izopet. 2001. HIV-1 subtyping using phylogenetic analysis of pol gene sequences. J. Virol. Methods 94:45-54. [DOI] [PubMed] [Google Scholar]
- 32.Ribas, S. G., P. Ondoa, J. Schupbach, G. van der Groen, and K. Fransen. 2003. Performance of a quantitative human immunodeficiency virus type 1 p24 antigen assay on various HIV-1 subtypes for the follow-up of human immunodeficiency type 1 seropositive individuals. J. Virol. Methods 113:29-34. [DOI] [PubMed] [Google Scholar]
- 33.Rouet, F., D. K. Ekouevi, A. Inwoley, M.-L. Chaix, M. Burgard, L. Bequet, I. Viho, V. Leroy, F. Simon, F. Dabis, and C. Rouzioux. 2004. Field evaluation of a rapid human immunodeficiency virus (HIV) serial serologic testing algorithm for diagnosis and differentiation of HIV type 1 (HIV-1), HIV-2, and dual HIV-1-HIV-2 infections in West African pregnant women. J. Clin. Microbiol. 42:4147-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouet, F., C. Montcho, C. Rouzioux, V. Leroy, P. Msellati, J. B. Kottan, B. You, I. Viho, and F. Dabis. 2001. Early diagnosis of paediatric HIV-1 infection among African breast-fed children using a quantitative plasma HIV RNA assay. AIDS 15:1849-1856. [DOI] [PubMed] [Google Scholar]
- 35.Salamon, R., C. Marimoutou, D. Ekra, A. Minga, E. Nerrienet, C. Huet, G. Gourvellec, D. Bonard, I. Coulibaly, P. Combe, F. Dabis, A. Bondurand, and L. Montagnier. 2002. Clinical and biological evolution of HIV-1 seroconverters in Abidjan, Cote d'Ivoire, 1997-2000. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 29:149-157. [DOI] [PubMed] [Google Scholar]
- 36.Seyler, C., X. Anglaret, N. Dakoury-Dogbo, E. Messou, S. Toure, C. Danel, N. Diakite, A. Daudie, A. Inwoley, C. Maurice, B. Tonwe-Gold, F. Rouet, T. N′Dri-Yoman, and R. Salamon. 2003. Medium-term survival, morbidity, and immuno-virological evolution in HIV-infected adults receiving antiretroviral therapy, Abidjan, Côte d'Ivoire. Antivir. Ther. 8:385-393. [PubMed] [Google Scholar]
- 37.Taylor, K., and P. De Young. 2003. WHO's 3-by-5 target. Lancet 362:918. [DOI] [PubMed] [Google Scholar]
- 38.Toni, T. D., P. Recordon-Pinson, A. Minga, D. Ekouevi, D. Bonard, L. Bequet, C. Huet, H. Chenal, F. Rouet, F. Dabis, M. E. Lafon, R. Salamon, B. Masquelier, and H. J. Fleury. 2003. Presence of key drug resistance mutations in isolates from untreated patients of Abidjan, Cote d'Ivoire: ANRS 1257 study. AIDS Res. Hum. Retrovir. 19:713-717. [DOI] [PubMed] [Google Scholar]
- 39.Vergne, L., A. Bourgeois, E. MpoudiNgole, R. Mougnutou, J. Mbuagbaw, F. Liegeois, C. Laurent, C. Butel, L. Zekeng, E. Delaporte, and M. Peeters. 2003. Biological and genetic characteristics of HIV infections in Cameroon reveals dual group M and O infections and a correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology 310:254-266. [DOI] [PubMed] [Google Scholar]
- 40.Vet, J. A. M., A. R. Majithia, S. A. E. Marras, S. Tyagi, S. Dube, B. Poiesz, and F. R. Kramer. 1999. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl. Acad. Sci. USA 96:6394-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White, P. A., Y. Pan, A. J. Freeman, G. Marinos, R. A. Ffrench, A. R. Lloyd, and W. D. Rawlinson. 2002. Quantification of hepatitis C virus in human liver and serum samples by using LightCycler reverse transcriptase PCR. J. Clin. Microbiol. 40:4346-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, W. J., C. L. Strout, T. Z. De Santis, J. L. Stilwell, A. V. Carrano, and G. L. Andersen. 2002. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes 16:119-127. [DOI] [PubMed] [Google Scholar]
- 43.Yeghiazarian, T., Y. Zhao, S. E. Read, W. Kabat, X. B. Li, S. J. Hamren, P. J. Sheridan, J. C. Wilber, D. N. Chernoff, and R. Yogev. 1998. Quantification of human immunodeficiency virus type 1 RNA levels in plasma by using small-volume-format branched-DNA assays. J. Clin. Microbiol. 36:2096-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, Y. Q., M. Yu, J. W. Miller, M. Z. Chen, E. G. Bremer, W. Kabat, and R. Yogev. 2002. Quantification of human immunodeficiency virus type I proviral DNA by using TaqMan technology. J. Clin. Microbiol. 40:675-678. [DOI] [PMC free article] [PubMed] [Google Scholar]