Abstract
Lack of standardization in antibiogram (ABGM) preparation (the overall profile of antimicrobial susceptibility results of a microbial species to a battery of antimicrobial agents) has not been addressed until recently. The objective of this study was to analyze current antibiograms using the recently published NCCLS M39-A guidelines for preparation of antibiograms to identify areas for improvement in the reporting of antibiogram susceptibility data. Antibiograms from across the United States were obtained by various methods, including direct mailings, Internet searches, and professional contacts. Each ABGM collected was analyzed using prospectively defined elements from the M39-A guidelines. Additionally, seven quality indicators were also evaluated to look for the reporting of any atypical or inappropriate susceptibility data. The 209 antibiograms collected from 149 institutions showed at least 85% compliance to 5 of the 10 M39-A elements analyzed. Clinically relevant elements not met included annual analysis, duplicate isolate notation, and the exclusion of organisms with fewer than 10 isolates. As for the quality indicators evaluated, unexpected results included the 7% of antibiograms that reported <100% vancomycin susceptibility for Staphylococcus aureus, 24% that had inconsistent beta-lactam susceptibility for Staphylococcus aureus, 20% that reported <100% imipenem susceptibility for Escherichia coli, and 37% that reported >0% ampicillin susceptibility for Klebsiella pneumoniae. These findings suggest that antibiograms should be reviewed thoroughly by infectious disease specialists (physicians and pharmacists), clinical microbiologists, and infection control personnel for identification of abnormal findings prior to distribution.
The NCCLS (now known as the CLSI [Clinical and Laboratory Standards Institute]) defines an antibiogram (ABGM) as an overall profile of antimicrobial susceptibility results of a microbial species to a battery of antimicrobial agents (17), which should reflect patient care needs along with the institution's formulary (15). When properly prepared and interpreted, ABGMs are an important resource for healthcare providers. While patient-specific cultures and susceptibility reports are pending, the ABGM may guide empirical therapy decisions based on likely pathogens and their probable susceptibilities to anti-infectives available at the institution (6, 21, 24). Clinicians and local infection control personnel use ABGM data in monitoring resistance trends, identifying outbreaks, developing quality improvement initiatives, and forming infection control policies and procedures (1, 6, 7, 8, 25). Microbiology and laboratory personnel also use ABGMs as a quality assurance measure for the Joint Commission on Accreditation of Healthcare Organizations (6, 11, 19). Pharmacy and Therapeutics committees use ABGM data when making anti-infective formulary decisions and establishing drug use policies (6).
The lack of standardization in the preparation and data assimilation of ABGMs has not been addressed until recently, though susceptibility testing has been standardized for years (12, 18). Because of this lack of standardization, empirical therapy selection based on ABGM data may be compromised. Furthermore, the comparison of ABGM data between institutions may be less meaningful when data have been summarized using various methods. In an attempt to resolve these issues, the NCCLS has published the M39-A guidelines. This document provides recommendations on the collection, analysis, and presentation of cumulative antimicrobial susceptibility test data with the goal of guiding clinicians in the appropriate selection of empirical therapy (17).
The objective of this study was to identify areas of improvement in ABGM data presentation, which can ultimately improve empirical anti-infective selection. Recent ABGMs from different types of institutions from across the United States were analyzed using the NCCLS M39-A published guidelines on antibiogram development. Several quality indicators were also evaluated for the reporting of any atypical or inappropriate susceptibility data. It should be noted that analysis of specific antibiotic resistance patterns were not a focus of this study.
MATERIALS AND METHODS
Antibiogram collection process.
Direct mailings, professional contacts, and Internet searches were used to collect ABGMs for 2000, 2001, and 2002 from university-affiliated, state of Kansas, Kansas City metropolitan area, and various community hospitals. Hospitals were identified using a University HealthSystem Consortium database and an Internet search using http://www.yellowpages.com/. During the fall of 2002, letters requesting antibiograms and demographic survey completion were sent to 264 different institutions nationwide along with a preaddressed, postage-paid return envelope. Reminder cards were sent to nonrespondents. Recipient institutions were assured that their specific susceptibility data would remain completely confidential. Additional ABGMs were collected through professional contacts until March 2003.
An Internet search was conducted using the Google and Dogpile search engines during winter 2003. Search terms included “antibiograms,” “hospital,” “2001,” “2002,” “2003,” “university,” “susceptibility reports,” and “university hospitals” in various combinations. Inclusion criteria included ABGMs during the 2000, 2001, and 2002 calendar years reporting results from human isolates.
Demographic survey.
The demographic survey included institution type, number of licensed beds, departments involved in publishing ABGM data, publication frequency, and availability of any electronic distribution methods. Incomplete surveys were completed using the institution's Internet website or by telephone interview. The survey instruments were sequentially numbered prior to mailing to identify nonrespondents.
Antibiogram analysis.
Each ABGM was analyzed using prospectively defined elements from the M39-A guidelines. The following elements were evaluated: (i) methods of summarizing susceptibility data, (ii) reporting results as “percent susceptible,” (iii) organisms' morphological grouping, (iv) duplicate isolate notation, (v) description of exact collection period, (vi) number of isolates for each organism, (vii) reporting only organisms with greater than 10 isolates, (viii) antimicrobial description (generic name, trade name), (ix) utilization of NCCLS antimicrobial abbreviations, and (x) utilization of “dash” to describe susceptibility data not reported. For the purpose of analysis, frequency of publication was also considered a prospectively defined element, for a total of 11 elements. Other M39-A recommendations evaluated were based on institutional need. These included separation of urine susceptibility data from nonurine susceptibility data, separation of susceptibility data by hospital location (e.g., intensive care units), and resistance trends descriptions (17).
Quality data indicators.
Although data concerning antibiotic resistance patterns were not a focus of this study, ABGMs were evaluated for atypical or inappropriate susceptibility data to assess the quality of the data being reported. These indicators were selected from the recent NCCLS M-100 performance standards for susceptibility testing (17). These data included the following: (i) inconsistent beta-lactam susceptibility for Staphylococcus aureus, (ii) vancomycin susceptibility of less than 100% for S. aureus, (iii) Enterococcus spp. tested against cephalosporins or trimethoprim-sulfamethoxazole, (iv) imipenem susceptibility of less than 100% for Escherichia coli, (v) imipenem susceptibility for Stenotrophomonas maltophilia, (vi) less than 100% susceptibility to third-generation cephalosporins for Haemophilus influenzae, and (vii) ampicillin susceptibility for Klebsiella pneumoniae. In addition, we noted the frequency ABGMs report of Streptococcus pneumoniae susceptibility.
Statistical analysis.
Demographic information and preselected ABGM elements were descriptively compared and evaluated for statistical significance by the use of bivariate analyses. Fischer's exact or a χ2 test was used when appropriate. Data were analyzed using SPSS for Windows, release 9.0 (Chicago, IL).
RESULTS
One hundred seven hospitals responded to letter requests, yielding a 41% response rate. A total of 209 ABGMs were acquired using all three collection methods, which represented 149 hospitals. Seventeen additional hospitals indicated that ABGMs are not compiled at their institution. Fifty-seven percent of the ABGMs were from 2001, 33% were from 2000, and 11% were from 2002.
Table 1 represents cross-tabulations for institutional demographics based on hospital size. Antibiograms from large hospitals (>250 beds) represented 61% of the sample, while 59% of institutions were community hospitals. Over one-third of the institutions (36%) distribute ABGMs electronically (intranet, 22%; Internet, 14%).
TABLE 1.
Demographics by institution
| Parameter | No. of ABGMs from hospitals with bed size of:
|
P value (χ2 test) | |||
|---|---|---|---|---|---|
| 0-50 | 51-250 | >250 | Total (%) | ||
| No. of institutions | 23 | 51 | 92 | 166 | 0.072 |
| No. of institutions with ABGMsa | 9 | 49 | 91 | 149 | |
| Type of Institution | |||||
| Academic | 1 | 2 | 54 | 57 (34.3) | <0.001 |
| Community | 19 | 44 | 35 | 98 (59.0) | |
| Veterans Administration | 0 | 4 | 1 | 5 (3.0) | |
| Other | 3 | 1 | 2 | 6 (3.7) | |
| ABGM compiled by: | |||||
| Laboratory personnel | 8 | 41 | 65 | 114 (76.5) | 0.139 |
| Pharmacy personnel | 0 | 0 | 4 | 4 (2.7) | |
| Laboratory and pharmacy personnel | 0 | 8 | 16 | 24 (16.1) | |
| Other | 1 | 0 | 6 | 7 (4.7) | |
| Electronic reporting | |||||
| No | 5 | 26 | 35 | 66 (55.5) | 0.003 |
| Yes; intranet | 2 | 8 | 22 | 32 (26.9) | |
| Yes; Internet | 0 | 1 | 20 | 21 (17.6) | |
Evaluated in analysis.
Antibiogram evaluation.
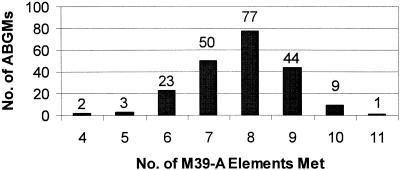
Fig. 1 represents the frequency of analyzed ABGMs meeting the 11 predefined M-39A elements. Over 85% of the ABGMs met at least seven of the elements evaluated. Only 1 ABGM of the 209 evaluated met all 11 elements. Table 2 represents cross-tabulations for the 11 preselected M39A ABGM elements by hospital size.
FIG. 1.
Frequency of analyzed ABGMs meeting the 11 predefined M-39A elements.
TABLE 2.
Summary of data by ABGM
| Parameter | No. of ABGMs from hospitals with bed size of:
|
|||
|---|---|---|---|---|
| 0-50 | 51-250 | >250 | Total (%) | |
| Summarized data presentationa | 9 | 68 | 122 | 199 (95.2) |
| Organisms separated by morphology | 10 | 63 | 114 | 187 (89.5) |
| Reported as % susceptible | 10 | 73 | 120 | 203 (97.1) |
| Duplicate isolate notation | 2 | 5 | 17 | 24 (11.5) |
| ABGM frequency | ||||
| More than annually (<12 mo) | 1 | 4 | 20 | 25 (16.8) |
| Annually | 6 | 44 | 70 | 120 (80.5) |
| Less than annually (>12 mo) | 2 | 1 | 1 | 4 (2.7) |
| Collection period description (i.e., mo and yr) | 5 | 52 | 94 | 151 (72.2) |
| No. of isolates reported | 11 | 73 | 119 | 203 (97.1) |
| Less than 10 isolatesb | 6 | 37 | 42 | 85 (40.7) |
| Name used in antimicrobial agent description | ||||
| All generic | 11 | 56 | 99 | 166 (79.4) |
| All generic and all brand | 0 | 3 | 6 | 9 (4.3) |
| All generic and some brand | 0 | 4 | 5 | 9 (4.3) |
| Some generic and some brand | 0 | 11 | 14 | 25 (12.0) |
| Antimicrobial abbreviations used | ||||
| NCCLS recommended | 1 | 14 | 7 | 22 (10.5) |
| Other | 1 | 6 | 30 | 37 (17.7) |
| Presentation of susceptibility data not reported | ||||
| Dash | 0 | 6 | 20 | 26 (12.4) |
| Blank | 6 | 48 | 75 | 129 (61.7) |
| Other method (e.g., shading, asterisk, etc.) | 5 | 20 | 29 | 54 (25.8) |
| Total no. of ABGMs | 11 | 74 | 124 | 209 |
Defined as data presented in tabular form.
Defined as the number of ABGMs with at least one organism reported with less than 10 isolates (n = 203).
Susceptibility data presentation.
The NCCLS recommends that final verified susceptibility results be reported on ABGMs (17). In this study, computer-generated, raw susceptibility summaries were not classified as “final, verified results” and represented only 5% of the entire sample.
Antibiogram methodology.
The NCCLS advocates the use of “percent susceptibility” for each data box, clarification of where the isolates came from (i.e., use of duplicates), and description of the collection period (17). Reporting the susceptibility data as “percent susceptible” for each organism-antimicrobial agent combination was the most commonly used method (97%). Additionally, seven quality indicators were also evaluated to look for the reporting of any atypical or inappropriate susceptibility data.
Isolates from the same patient should be excluded from ABGMs for a 1-year time period (17). This recommendation could not be evaluated; therefore, we evaluated ABGM documentation of inclusion or exclusion of duplicate isolates. Only 12% of ABGMs documented how duplicates were managed.
Most ABGMs are compiled on an annual basis, as NCCLS recommends. They generally included the dates (month and year) for the collection period (72%). Of those, 70% reflected calendar years (January through December). Others periods of time included July to June and May to April.
Morphological grouping.
Most ABGMs separated organisms by morphology (90%), as recommended, with less than 4% reporting fungal susceptibility information. The remainder (10%) listed organisms alphabetically or by prevalence of occurrence.
Number of isolates.
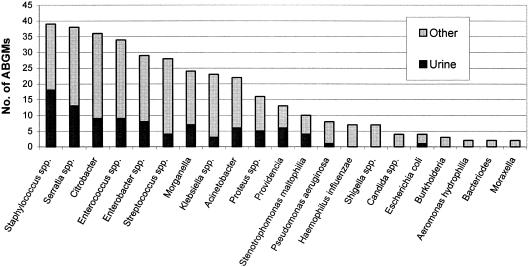
The inclusion of the total number of isolates collected for each organism is recommended along with the inclusion of data only for those organisms with 10 or more isolates (17). Ninety-seven percent of ABGMs reported the number of isolates tested per organism-antimicrobial combination. Interestingly, 42% of ABGMs contained susceptibility data for organisms with fewer than 10 isolates. As shown in Fig. 2, many of these were clinically uncommon species or urine isolates.
FIG. 2.
Organisms with fewer than 10 isolates reported.
Antimicrobial description.
The M39-A document vaguely recommends the use of complete antimicrobial names (17). Generic nomenclature was used most often (88%), followed by a mixture of some generic and some brand names (12%). Combination products (e.g., piperacillin-tazobactam) were most commonly implicated when ABGMs utilized brand names. The NCCLS recommends that when abbreviations are used for ABGMs, they should either agree with abbreviations used on patient susceptibility reports or utilize those listed in Appendix F of the M39-A document (17). Since it is not possible to ascertain abbreviation congruency between patient susceptibility reports and individual ABGMs, those contained in the M39-A document were the basis for this assessment. Twenty-eight percent of ABGMs contained abbreviations, and of those, only 37% used the NCCLS abbreviations.
Data not reported.
Use of a dash in each ABGM data box when a drug is either not tested at that institution or when it is known to be clinically ineffective is recommended in the M39-A guidelines (17). Our data show that only 12% of ABGMs utilized the dash method, 25% used another method, and the remainder simply left the data box blank.
Other M39-A recommendations based on institutional need.
The guidelines recommend that selectively tested antibiotics should be noted in some manner (e.g., footnotes) (17). Forty-seven percent of ABGMs separated urine isolates from nonurine isolates. Susceptibility information may be separated by specialty care areas, such as intensive care units or outpatient areas within the institution, to allow for more meaningful comparisons between like units and the overall institution ABGM (17). A few ABGMs had separated specific location susceptibility information into intensive care units (n = 20) and outpatient areas (n = 30). To assist clinicians with interpreting significant ABGM changes from year to year, institutions can provide a summary of major resistance trends in a variety of ways, such as with tables or graphs (1). Over 14% of ABGMs reported resistance trends. Various methods included the use of brief summaries, graphs, upward- and downward-pointing arrows in the data boxes, or numbers in boldface or nonboldface type. A couple of institutions distributed a brief letter with each new ABGM.
Quality data indicators.
Overall, 14.3% of ABGMs included unusual results, based upon the seven prospectively defined quality indicators assessing unusual results and indicating unverified ABGMs. Table 3 summarizes the results for these data quality indicators by institution size. The number of ABGMs assessed for each quality indicator varied widely, since not all ABGMs included results for each organism-antimicrobial combination evaluated.
TABLE 3.
Quality indicators
| Organism reported | Drug tested (% susceptibility) | No. of ABGMs from hospitals with bed size of:
|
|||
|---|---|---|---|---|---|
| 0-50 | 51-250 | >250 | No. reported/ total | ||
| S. aureus | Beta-lactam (inconsistent) | 3 | 19 | 27 | 49/201 |
| Vancomycin (<100) | 0 | 10 | 3 | 13/198 | |
| Enterococcus spp. | Cephalosporins | 1 | 2 | 0 | 3/198 |
| Trimethoprim-sulfa- methoxazole | 2 | 1 | 0 | 3/198 | |
| E. coli | Imipenem (<100) | 3 | 13 | 18 | 34/170 |
| S. maltophilia | Imipenem (>0) | 1 | 2 | 14 | 17/39 |
| H. influenzae | Broad-spectrum ceph- alosporins (<100) | 0 | 4 | 6 | 10/33 |
| K. pneumoniae | Ampicillin (>0) | 3 | 27 | 48 | 78/160 |
| Total number of ABGMs | 11 | 74 | 124 | 209 | |
Twenty-four percent of ABGMs reported inconsistent susceptibility rates for S. aureus across the beta-lactam class, specifically the penicillinase-resistant penicillins, cephalosporins, and carbapenems. Of further interest, 7% of the study ABGMs (13 of 198) reported less than 100% vancomycin susceptibility (range, 98 to 99% susceptible) for S. aureus. Few ABGMs (3 of 198) reported susceptibility data for Enterococcus spp. to cephalosporins and trimethoprim-sulfamethoxazole. Despite the widespread availability of national surveillance data which routinely characterize the susceptibility trends for pneumococcus, only 20% of ABGMs included susceptibility data for this pathogen.
Of ABGMs that reported E. coli, 20% showed less than 100% imipenem susceptibility (range, 93 to 99% susceptible), while 43% reported S. maltophilia as having susceptibility to imipenem (range, 2 to 11% susceptible). Thirty percent (10 of 33) of ABGMs reported less than 100% susceptibility (range, 75 to 97% susceptible) of H. influenzae isolates to broad-spectrum cephalosporins. Ampicillin showed susceptibility (range, 1 to 19% susceptible) to K. pneumoniae in 49% of ABGMs.
DISCUSSION
In an era of antimicrobial misuse, increasing anti-infective resistance, and reduced emphasis on antibiotic development by pharmaceutical manufacturers, the need for reliable, accurate ABGM data to guide appropriate antibiotic selection is critical (9, 20, 26). The recent publication of the first approved standards for cumulative susceptibility data presentation underscores the importance of these documents and their use in clinical practice. A couple of survey studies evaluate ABGM preparation but, to our knowledge, this study is the first of its size to evaluate actual ABGMs within the context of these standards (4, 8).
Perhaps the most interesting finding came from the evaluation of final, verified ABGM data, as done by evaluating eight separate quality indicators from the M-100 performance standards (18). These surrogate markers represent ABGM validity. With only eight documented cases of vancomycin-intermediate S. aureus (VISA) and three documented cases of vancomycin-resistant S. aureus (VRSA) in the United States (3, 5, 13), the 7% of ABGM analyzed with less than 100% vancomycin resistance for S. aureus lead us to believe either that the susceptibility data had not been verified at the institution level or that VISA or VRSA is more common than the published reports indicate. None of the institutions we evaluated were the institutions with published VISA/VRSA cases in the literature. Inconsistent beta-lactam susceptibilities for S. aureus may lead inexperienced clinicians to choose an inappropriate empirical regimen.
Although Halstead et al. reported that 90% of their respondents had a system in place to alert staff of atypical results (8), our findings indicate that many of these ABGMs may not have been thoroughly screened prior to distribution, since 14% of ABGMs had unusual susceptibility results. It is recommended that microbiology personnel or other clinicians review draft versions of ABGMs prior to distribution (10).
While the ABGM can be an important tool to increase awareness of hospital resistance patterns, study data revealed that a substantial number of small hospitals do not prepare ABGMs. This lack of ABGM preparation may be due to the fact that these small institutions have limited resources available or that their cultures are sent to outside laboratories, given that there is low demand for this service (23).
The NCCLS recommends that ABGMs be prepared on an annual basis to allow for proper trend interpretation without confounders of seasonal variations (17). Although others contend that more frequent analysis of susceptibility data can reveal early identification of resistance trends (15), the results of our study show that the majority of institutions report cumulative susceptibility data on an annual basis. Also, there was no consistency with dates used to label antibiogram documents. For example, a “2002” label on an antibiogram could refer to the actual collection date or the current year of release, which often leads to confusion by clinicians using these documents.
As expected, microbiology laboratory personnel compiled the majority of ABGMs; however, pharmacists were involved in the preparation process at 19% of institutions. Our findings underscore the value of using a multidisciplinary approach (physicians, infection control personnel, microbiologists, pharmacists) in reviewing antibiogram data prior to publication to avoid reporting misleading or suspicious susceptibility information. Also, with the increased use and accessibility of technology, it is not surprising that over 35% of institutions report their ABGMs on hospital Internet or intranet sites. It is advisable to provide an annual summary that highlights current susceptibility information, as several institutions in this study have done. This can assist clinicians with understanding institutional shifts in susceptibility from one reporting period to another and can be helpful for making empirical antibiotic decisions.
Many practitioners are completely unaware of how duplicate isolates from the same patient can skew susceptibility data on the antibiogram. Frequently, such duplicate isolates falsely elevate resistance rates. This has recently been addressed in both the literature and by the NCCLS (10, 14, 16, 22, 27), yet only 12% of ABGMs analyzed included some notation of how duplicate isolates from the same patient were managed during the compilation of susceptibility data. The proportion of institutions that report only nonduplicate isolates may be higher than is actually reported on ABGMs (7). Furthermore, it is apparent that to fully comply with the NCCLS duplicate isolates recommendation, laboratory information systems need to be modified to facilitate the removal of duplicate isolates, which is a tedious and time-consuming process when done manually (10, 16).
Nearly all ABGMs reported the number of isolates represented; however, 41% reported susceptibility information on less than 10 isolates. The NCCLS recognizes that this number of isolates is an arbitrary number; however, it was chosen to assure that a low sample size does not mislead practitioners. The question is whether small numbers will negatively influence susceptibility conclusions or would provide insight into the susceptibility data of less prevalent organisms (e.g., fungal pathogens). The small sample size of isolates reported also could influence many smaller hospitals in electing not to prepare ABGMs or to prepare them infrequently.
The use of complete antimicrobial names, NCCLS-defined abbreviations, or abbreviations used on patient reports is recommended. We interpreted this recommendation, regarding “complete antimicrobial names,” as meaning complete generic names, which follows the pattern exhibited throughout the M-39A document. The use of brand names for combination products could be due to spatial concerns on the ABGM or ease of recognition of brand names. Because the abbreviations for anti-infectives that were used on patient reports were unavailable, we analyzed this recommendation based solely upon the M-39A document. Therefore, the full implication of the recommendation is difficult to assess in the present study.
In our analysis, we discovered that institutions most frequently deviated from the guidelines when indicating that an antibiotic-pathogen combination was neither reported nor tested. With only 12% of respondents using the dash method, perhaps any method denoted on the ABGM, other than leaving blank data boxes, should be an acceptable alternative. Additionally, the use of reporting results from selectively tested isolates, specific locations, or resistance trends may be interesting, yet their use may not be feasible for smaller institutions based on need and quantity of isolates tested.
Our study contained several potential limitations. The most notable was that all conclusions were established based on ABGM evaluation without detailed knowledge of each institution's hospital policies and procedures. Furthermore, the M39-A guidelines contain approximately 40 recommendations, many of which could not be evaluated by inspecting only the antibiogram documents. For example, the recommendations regarding data analysis systems, patient demographic information, specimen identification and specifications, and antimicrobial susceptibility testing methods could not be assessed by the methods outlined. Additionally, readability may have been hampered in some cases by the access to photocopied versions of some of the ABGMs. The use of multiple ABGMs from the same institution could have influenced our study results. It is also possible that respondents who returned more than one ABGM were more interested in the process and/or had better access to published ABGMs data than other respondents. We included these multiple ABGMs in the analysis because ABGM preparation tends to evolve, and we believed that other institutions could learn from the changes that are made from year to year.
Despite these limitations, we and other organizations recognize the importance of ABGMs as a clinical tool. In 2002, the Centers for Disease Control and Prevention began promoting its latest campaign, “12 steps to prevent antimicrobial resistance among hospitalized adults” (2). The importance of ABGM data is featured in Step 6, “Use local data.” Specifically, the campaign asks that healthcare professionals know their antibiogram, formulary, and patient population. In addition, the Joint Commission on Accreditation of Healthcare Organizations recognizes the ABGM as a quality assurance measure for clinical laboratories and therefore as a fulfillment of Standard IM.8 (6, 11).
Given the emphasis that government agencies, accreditation organizations, and the medical literature have placed on antimicrobial susceptibility patterns and inappropriate use of antimicrobials, it is evident that ABGMs have undergone scrutiny. The lack of standardization limits the versatility of ABGMs across various areas of healthcare, including empirical antimicrobial selection, ABGM surveillance, and assistance in hospital and public health policy. Adoption of the M39-A guidelines for preparation of antibiogram data should improve the quality, standardization of reporting, and interpretation of results by infectious disease specialists along with microbiology and infection control personnel.
Acknowledgments
This work was not supported by outside funds.
We thank the institutions participating in this research. The authors also recognize the contribution of Kristine Brunton for providing additional ABGMs for institutions in Kansas and Kelly Smith for her Internet search efforts.
REFERENCES
- 1.Calfee, D. P., and B. M. Farr. 2002. Infection control and cost control in the era of managed care. Infect. Control Hosp. Epidemiol. 23:407-410. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2004. Campaign to prevent antimicrobial resistance in healthcare settings [Online.] http://www.cdc.gov/drugresistance/healthcare/default.htm.
- 3.Centers for Disease Control and Prevention. 2003. VISA/VRSA—vancomycin-intermediate/resistant Staphylococcus aureus. Fact sheet. [Online.] http://www.cdc.gov/ncidod/hip/ARESIST/visa.htm.
- 4.Ernst, E. J., D. J. Diekema, B. J. Bootsman, T. Vaughn, J. W. Yankey, S. D. Flach, M. M. Ward, C. L. J. Franciscus, E. Acosta, M. A. Pfaller, and B. N. Doebbeling. 2004. Are United States hospitals following national guidelines for the analysis and presentation of cumulative antimicrobial susceptibility data? Diagn. Microbiol. Infect. Dis. 49:141-145. [DOI] [PubMed] [Google Scholar]
- 5.Fridkin, S. K. 2001. Vancomycin-intermediate and -resistant Staphylococcus aureus: what the infectious disease specialist needs to know. Clin. Infect. Dis. 32:108-115. [DOI] [PubMed] [Google Scholar]
- 6.Ginocchio, C. 2002. Role of NCCLS in antimicrobial susceptibility testing and monitoring. Am. J. Health Syst. Pharm. 59(Suppl. 3):S7-S11. [DOI] [PubMed] [Google Scholar]
- 7.Gums, J. G. 2002. Assessing the impact of antimicrobial resistance. Am. J. Health Syst. Pharm. 59(Suppl. 3):S4-S6. [DOI] [PubMed] [Google Scholar]
- 8.Halstead, D. C., N. Gomez, and Y. S. McCarter. 2004. Reality of developing a community-wide antibiogram. J. Clin. Microbiol. 42:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hecker, M. T., D. C. Aron, N. P. Patel, M. K. Lehmann, and C. J. Donskey. 2003. Unnecessary use of antimicrobials in hospitalized patients. Arch. Intern. Med. 163:972-978. [DOI] [PubMed] [Google Scholar]
- 10.Horvat, R. T., N. E. Klutman, M. K. Lacy, D. Grauer, and M. Wilson. 2003. Effect of duplicate isolates of methicillin-susceptible and methicillin-resistant Staphylococcus aureus on antibiogram data. J. Clin. Microbiol. 41:4611-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joint Commission on Accreditation of Healthcare Organizations. 2000. Comprehensive accreditation manual for hospitals: the official handbook. Joint Commission on Accreditation of Healthcare Organizations, Oakbrook Terrace, Ill.
- 12.Jones, R. N. 2001. Method preferences and test accuracy of antimicrobial susceptibility testing. Arch. Pathol. Lab. Med. 125:1285-1289. [DOI] [PubMed] [Google Scholar]
- 13.Kacica, M., and L. C. McDonald. 2004. Brief report: vancomycin-resistant Staphylococcus aureus—New York, 2004. Morb. Mortal. Wkly. Rep. 53:322-323. [PubMed] [Google Scholar]
- 14.Lacy, M. K., N. E. Klutman, R. T. Horvat, and A. Zapantis. 2004. Antibiograms: new NCCLS guidelines, development, and clinical application. Hosp. Pharm. 39:542-553. [Google Scholar]
- 15.Lamp, K. 1996. Antibiograms. Pharm. Pract. Manag. Q. 16:52-56. [PubMed] [Google Scholar]
- 16.Lee, S., Y. K. Cho, S. Kim, E. S. Lee, S. Y. Park, and Y. Seo. 2004. Comparison of trends of resistance rates over 3 years calculated from results for all isolates and for the first isolate of a given species from a patient. J. Clin. Microbiol. 41:4776-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NCCLS. 2002. Analysis and presentation of cumulative susceptibility test data; approved guideline. NCCLS document M39-A. NCCLS, Wayne, Pa.
- 18.NCCLS. 2002. Performance standards for antimicrobial susceptibility testing: twelfth informational supplement. NCCLS document M100-S12. NCCLS, Wayne, Pa.
- 19.Patton, K. A. 2002. Role of JCAHO standards and clinical practice guidelines in promoting appropriate antimicrobial use. Am. J. Health Syst. Pharm. 59(Suppl. 3):S16-S18. [DOI] [PubMed] [Google Scholar]
- 20.Pflomm, J. 2002. Strategies for minimizing antimicrobial resistance. Am. J. Health Syst. Pharm. 59(Suppl. 3):S12-S15. [DOI] [PubMed] [Google Scholar]
- 21.Rush, D. R. 1991. Antimicrobial formulary management: meeting the challenge in the community hospital. Pharmacotherapy 11(1, Pt. 2):19S-26S. [PubMed] [Google Scholar]
- 22.Shannon, K. P., and G. L. French. 2002. Antibiotic resistance: effect of different criteria for classifying isolates as duplicates on apparent resistance frequencies. J. Antimicrob. Chemother. 49:201-204. [DOI] [PubMed] [Google Scholar]
- 23.Stein, C. R., D. J. Weber, and M. Kelley. 2003. Using hospital antibiogram data to assess regional pneumococcal resistance to antibiotics. Emerg. Infect. Dis. 9:211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ting, Y., and D. Miles. 2002. Developing a web-based tool using hospital-specific cumulative antibiogram data. Hosp. Pharm. 37:1190-1195. [Google Scholar]
- 25.Van Beneden, C. A., C. Lexau, W. Baughman, B. Barnes, N. Bennett, P. M. Cassidy, M. Pass, L. Gelling, N. L. Barrett, E. R. Zell, and C. G. Whitney. 2003. Aggregated antibiograms and monitoring of drug-resistant Streptococcus pneumoniae. Emerg. Infect. Dis. 9:1089-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wester, C. W., L. Durairaj, A. T. Evans, D. N. Schwartz, S. Husain, and E. Martinez. 2002. Antibiotic resistance, a survey of physician perceptions. Arch. Intern. Med. 162:2210-2216. [DOI] [PubMed] [Google Scholar]
- 27.White, R. L., L. V. Friedrich, D. S. Burgess, E. W. Brown, and L. E. Scott. 2001. Effect of removal of duplicate isolates on cumulative susceptibility reports. Diagn. Microbiol. Infect. Dis. 39:251-256. [DOI] [PubMed] [Google Scholar]