Abstract
G Protein-Coupled Receptors (GPCRs) are integral membrane proteins that have gained considerable attention as drug targets, particularly in cancer treatment. In this study, we explored the capacity of bioactive compounds derived from Gnetum gnemon (GG) for the development of of pharmaceuticals targeting GPCRs within the context of cancer therapy. Integrated approach combined network pharmacology and molecular docking to identify and validate the underlying pharmacological mechanisms. We retrieved targets for GG-derived compounds and GPCRs-related cancer from databases. Subsequently, we established a protein-protein interaction (PPI) network by mapping the shared targets. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were employed to predict the mechanism of action of these targets. Molecular docking was conducted to validate our findings. We identified a total of 265 targets associated with GG-derived bioactive compounds for the treatment of GPCRs-related cancer. Functional enrichment analysis revealed the promising therapeutic effects of these targets on GPCRs-related cancer pathways. The PPI network analysis identified hub targets, including MAPK3, SRC, EGFR, STAT3, ESR1, MTOR, CCND1, and PPARG, which demonstrate as treatment targets for GPCRs-related cancer using GG-derived compounds. Additionally, molecular docking experiments demonstrated the strong binding affinity of gnetin A, gnetin C, (-)-viniferin, and resveratrol dimer, thus inhibiting MAPK3, SRC, EGFR, and MTOR. Survival analysis established the clinical prognostic relevance of identified hub genes in cancer. This study presents a novel approach for comprehending the therapeutic mechanisms of GG-derived active compounds and thereby paving the way for their prospective clinical applications in the field of cancer treatment.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75240-4.
Keywords: GPCRs, Cancer, ADME, Network pharmacology, Molecular docking
Subject terms: Cancer, Drug discovery, Computational biology and bioinformatics, Gene ontology, Virtual drug screening
Introduction
G protein-coupled receptors (GPCRs) are transmembrane proteins that constitute the largest superfamily of receptors, responding to various ligand signals, such as neurotransmitters, hormones, and other small molecules1. GPCRs are essential for a broad spectrum of physiological processes, translating extracellular signals into intracellular responses and mediating signal transduction across the plasma membrane. With over 800 genes in this receptor family, GPCRs modulate numerous signaling processes involved in blood pressure regulation, behavior, cognition, immune response, mood, smell, and taste2. Classification of GPCRs is based on sequence and function, dividing them into six classes (A-F): rhodopsin-like receptors, secretin family, metabotropic glutamate receptors, fungal mating pheromone receptors, cAMP receptors, and frizzled (FZD) and smoothened (SMO) receptors3. Upon activation by external ligand signals, GPCRs interact with different heterotrimeric GTP-binding proteins (G proteins) or Arrestins, mediating signal flow through modulation of various downstream effectors or the GPCRs-mediated signal transduction pathway, resulting in the elicitation of cyclic adenosine 3,5-monophosphate (cAMP) response, calcium mobilization, or phosphorylation of extracellular regulated protein kinases 1/2 (pERK1/2)4,5. GPCRs have been associated with the pathogenesis of numerous diseases, including diabetes mellitus, obesity, depression, Alzheimer’s disease, and cancers1. Cancer, a complex disorder characterized by uncontrolled cell growth and proliferation, frequently exhibits overexpression of GPCRs in primary and metastatic tumor cells, associating them with tumor growth in various cancer types, including colon, ovarian, prostate, breast, lung, thyroid, pancreatic cancer, and melanoma5. Given their involvement in cancer hallmarks such as growth, proliferation, survival, metabolism, death and apoptosis, ion and nutrient transport, and migration, GPCRs have been emerged as potential targets for cancer therapy5. Activation of GPCRs can stimulate multiple mitogen-activated protein kinase (MAPK) cascades, involving a family of closely related serine/threonine kinases like ERK1/2, JNK1-3, p38MAPKs, and ERK5, playing a crucial role in linking membrane receptors to transcription factors6. Moreover, GPCRs also regulate the PI3K, AKT, and mTOR cascades, and the phosphorylation of multiple substrates has been demonstrated to play a significant role in cell metabolism, migration, growth, and survival7. Consequently, GPCR-related receptors and related signaling proteins expressed in cancer cells are promising as targets for the development of anticancer drugs.
Cancer treatment poses significant challenges due to the distinct pathophysiology of tumors and the development of resistance. Conventional approaches such as chemotherapy, radiotherapy, and immunotherapy have inherent limitations, including therapy resistance and adverse side effects. Currently prescribed cancer drugs often lead to severe side effects, such as neutropenia, chemotherapy-induced peripheral neuropathy, stomatitis, mucositis, diarrhea, nausea, and vomiting8. Complementary and alternative medicine is gaining recognition for its potential cost-effectiveness and might generally exhibit lower toxicity compared to conventional drugs, though the safety of plant-derived compounds can vary and should be evaluated on a case-by-case basis. Many phytochemicals function by regulating cancer-associated pathways, thus aiding in the prevention tumorigenesis and cancer progression9. Gnetum gnemon Linn (GG) is a plant that exhibits promising therapeutic potential for cancer treatment due to its antioxidant, anti-inflammatory, and anticancer properties. It has gained significant attention among alternative medicinal plants, and has been found effective against various types of cancer, including liver, skin, cervix uteri and vagina, lung, prostate, pancreatic, breast, and colon cancer10. The Melinjo seed extract (MSE) contains several resveratrol-containing compounds, including trans-resveratrol (resveratrol monomer), gnetin C (resveratrol dimer), and derivatives of gnetin C such as gnemonoside A and D (resveratrol dimer glycosides)11. The antitumor efficacy of MSE was validated in a widely used colon-26 tumor-bearing mouse model, where oral administration of the seed extract significantly inhibited tumor growth, intratumoral angiogenesis, and liver metastases in BALB/c mice bearing colon-26 tumors12. Furthermore, trans-resveratrol and gnetin C have been found to effectively suppress the growth of pancreatic, prostate, breast, and colon cancer cells. Gnetin C also induced both early and late stages apoptosis through caspase 3/7-dependent mechanisms12. Strong inhibitory effects of gnetin C on prostate cancer cells were observed, including specific downregulation of metastasis-associated protein 1 (MTA1) and the ETS-proto-oncogene 2 (ETS2), suggesting its potential as a targeted therapeutic agent for prostate cancer13. Therefore, GG contains bioactive compounds that exhibit anticancer properties against various types of cancer. However, the mechanisms through which these bioactive compounds from GG influence GPCRs-related cancers remain unclear.
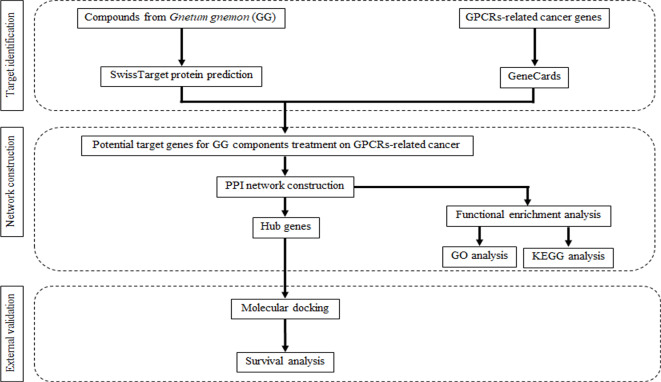
Network pharmacology is a pioneering research subfield that emerges from the analysis of network models and system biology14. It represents a cross-discipline that integrates fundamental theories and research methods from network science, bioinformatics, computer science, and mathematics. The primary objective of network pharmacology is to examine the intricate network of biological systems while identifying specific signal nodes for drug molecular design15. This field operates at the biological level, employing the mapping of the drug-target-disease network to investigate the interaction between the body and drugs. Additionally, network pharmacology explores the complexities of drug-protein interactions and protein-protein interactions (PPIs)16. Notably, it effectively accentuates important compounds found in traditional medicine and predicts their potential mechanisms by employing visual representations of the “drug-target-disease” network composition. Molecular docking, a technique used in drug design, focus on studying the interaction between receptors and drug molecules17. Over the past few years, this method has gained significant popularity and has been widely employed in drug research and exploration18. In this context of avoiding or minimizing the use of animals, the 3Rs—replacement, reduction and refinement – encompass alternative approach, inclusive of bioinformatic tools, serve as viable replacements for laboratory animals while expediting the process and reducing costs. The aim of this study is to explore the pharmacological mechanisms of bioactive compounds derived from GG for the treatment of GPCRs in cancer, utilizing network pharmacology and molecular docking experiments (Fig. 1).
Fig. 1.
General workflow of network pharmacology and molecular docking studies of current work.
Results
Chemoinformatics, drug likeness and ADME properties of compounds from GG
The primary objective of drug-likeness assessment is to predict potential therapeutic ligands. In this study, we examined 28 active components derived from GG and their chemoinformatics data by SwissADME19 are presented in Table 1. Promising ingredients were selected based on specific screening criteria, including a molecular weight (MW) below 500 g/mol, less than 10 rotatable bonds, no more than 10 hydrogen bond acceptors (HBA), no more than 5 hydrogen bond donors (HBD), a topological polar surface area (TPSA) of 140 Ų or less, and a lipophilicity below 5. Among the total compounds evaluated, 13 compounds exhibited compliance with Lipinski’s rules of five (RO5), indicating favorable drug-like properties. Conversely, 15 compounds were excluded from further consideration. Among the 13 compounds from GG that met RO5 criteria, their water solubility varied from poor to very soluble. Most compounds derived from GG exhibited excellent gastrointestinal absorption, with the exceptions being ursolic acid and gnetin D. However, caution should be exercised regarding the permeability of the blood-brain barrier for 3-methoxyresveratrol, pterostilbene, and tetramethoxystilbene. The majority of compounds from GG demonstrated low skin permeation, and it is also important to consider their potential as CYP enzyme inhibitors (Table 2). However, the ADME properties in this study are based on computational predictions, and further experimental studies are required.
Table 1.
Chemoinformatic properties and drug likeness of compounds from GG.
| Compounds from GG | Physicochemical Properties | Druglikeness by Lipinski (Pfizer) filter | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Formula | MW (g/mol) | No. rotatable bonds | HBA | HBD | Molar Refractivity | TPSA (Ų) | Lipophilicity | Water Solubility | ||
| Resveratrol dimer | C28H22O7 | 454.47 | 4 | 6 | 5 | 130.24 | 110.38 | 4.05 | Poorly soluble | Yes |
| 3-methoxyresveratrol | C15H14O3 | 242.27 | 3 | 3 | 2 | 72.35 | 49.69 | 2.88 | Soluble | Yes |
| Oxyresveratrol | C14H12O4 | 244.24 | 2 | 4 | 4 | 69.90 | 80.92 | 2.08 | Soluble | Yes |
| Gnemonol A | C42H32O10 | 696.70 | 4 | 10 | 8 | 191.57 | 180.30 | 4.94 | Poorly soluble | No |
| Gnemonol B | C56H42O12 | 906.93 | 8 | 12 | 9 | 254.97 | 209.76 | 7.31 | Insoluble | No |
| Gnemonol C | C56H42O13 | 922.92 | 8 | 13 | 10 | 257.00 | 229.99 | 6.92 | Insoluble | No |
| Gnemonol D | C42H32O10 | 696.70 | 6 | 10 | 9 | 194.63 | 180.30 | 5.31 | Poorly soluble | No |
| Gnemonol E | C42H32O10 | 696.70 | 6 | 10 | 8 | 194.63 | 180.30 | 5.30 | Poorly soluble | No |
| Gnemonol K | C42H32O9 | 680.70 | 6 | 9 | 7 | 192.61 | 160.07 | 5.64 | Insoluble | No |
| Gnemonol L | C42H32O9 | 680.70 | 6 | 9 | 7 | 192.61 | 160.07 | 5.64 | Insoluble | No |
| Gnemonol M | C30H26O8 | 514.52 | 4 | 8 | 6 | 140.5 | 139.84 | 3.69 | Poorly soluble | No |
| Gnemonoside A | C40H42O16 | 778.75 | 10 | 16 | 11 | 194.49 | 268.68 | 0.60 | Soluble | No |
| Gnemonoside B | C40H42O16 | 778.75 | 10 | 16 | 11 | 194.49 | 268.68 | 0.60 | Soluble | No |
| Gnemonoside C | C34H32O11 | 616.61 | 7 | 11 | 8 | 162.37 | 189.53 | 2.42 | Moderately soluble | No |
| Gnemonoside D | C34H32O11 | 616.61 | 7 | 11 | 8 | 162.37 | 189.53 | 2.46 | Moderately soluble | No |
| Gnemonoside E | C40H42O16 | 778.75 | 10 | 16 | 11 | 194.49 | 268.68 | 0.59 | Soluble | No |
| Gnemonoside F | C60H62O24 | 1167.12 | 15 | 24 | 16 | 288.98 | 397.52 | 0.37 | Moderately soluble | No |
| Pinostilbenoside | C21H24O8 | 404.41 | 6 | 8 | 5 | 104.47 | 128.84 | 1.06 | Soluble | Yes |
| Pterostilbene | C16H16O3 | 256.30 | 4 | 3 | 1 | 76.82 | 38.69 | 3.31 | Moderately soluble | Yes |
| Tetramethoxystilbene | C18H20O4 | 300.35 | 6 | 4 | 0 | 87.78 | 36.92 | 3.78 | Moderately soluble | Yes |
| 2,3-dihydroxypropyl icosanoate | C23H46O4 | 386.61 | 22 | 4 | 2 | 116.28 | 66.76 | 6.07 | Poorly soluble | Yes |
| Oleic acid | C18H34O2 | 282.46 | 15 | 2 | 1 | 89.94 | 37.30 | 5.71 | Moderately soluble | Yes |
| Ursolic acid | C30H48O3 | 456.70 | 1 | 3 | 2 | 136.91 | 57.53 | 5.88 | Moderately soluble | Yes |
| (-)-Viniferin | C28H22O6 | 454.47 | 4 | 6 | 5 | 130.24 | 110.38 | 4.05 | Poorly soluble | Yes |
| Gnetin A | C28H22O6 | 454.47 | 4 | 6 | 4 | 127.75 | 115.06 | 3.29 | Moderately soluble | Yes |
| Gnetin C | C28H22O6 | 454.47 | 4 | 6 | 5 | 130.24 | 110.38 | 4.12 | Poorly soluble | Yes |
| Gnetin D | C28H22O7 | 470.47 | 4 | 7 | 6 | 137.27 | 130.61 | 3.65 | Poorly soluble | Yes |
| Gnetin E | C42H32O9 | 680.70 | 6 | 9 | 7 | 192.61 | 160.07 | 5.73 | Poorly soluble | No |
MW Molecular weight; HBA Hydrogen bond acceptor; HBD Hydrogen bond doner; TPSA Topological polar surface area
Table 2.
ADME prediction of compounds from GG.
| Compounds from GG | Pharmacokinetics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GI absorption | BBB permeant | P-gp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | Log Kp (skin permeation) (cm/s) | ||
| Resveratrol dimer | High | No | No | No | No | Yes | No | No | -4.24 | |
| 3-methoxyresveratrol | High | Yes | No | Yes | No | Yes | No | Yes | -5.33 | |
| Oxyresveratrol | High | No | No | Yes | No | Yes | No | Yes | -5.82 | |
| Pinostilbenoside | High | No | Yes | No | No | No | No | No | -7.78 | |
| Pterostilbene | High | Yes | No | Yes | Yes | Yes | Yes | No | -5.18 | |
| Tetramethoxystilbene | High | Yes | No | Yes | Yes | Yes | Yes | No | -4.87 | |
| 2,3-dihydroxypropyl icosanoate | High | No | No | Yes | No | No | No | No | -2.62 | |
| Oleic acid | High | No | No | Yes | No | Yes | No | No | -2.6 | |
| Ursolic acid | Low | No | No | No | No | No | No | No | -3.87 | |
| (-)-Viniferin | High | No | No | No | No | Yes | No | No | -5.24 | |
| Gnetin A | High | No | No | No | No | Yes | No | No | -6.28 | |
| Gnetin C | High | No | No | No | No | Yes | No | No | -5.24 | |
| Gnetin D | Low | No | No | No | No | Yes | No | No | -5.58 | |
Target identification and analysis
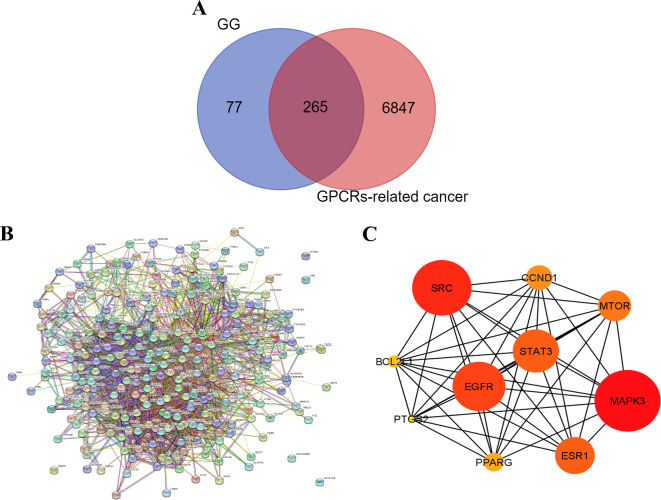
A screening process was performed using the Swiss target prediction databases20, leading to the identification of 342 target genes associated with the 13 active components derived from GG. Additionally, the GeneCards database supplied us with a comprehensive list of 7,112 targets linked to GPCRs in cancer21. Through a comparison between the targets of the 13 active components from GG and the GPCRs-related targets, a total of 265 intersecting genes were discovered. This intersection is visually depicted in Fig. 2A, utilizing a Venn diagram. These overlapping targets represent promising candidates for the therapeutic application of GG derived compounds in cancer treatment.
Fig. 2.
Protein-protein interaction (PPI) network and Hub gene analysis. (A) Venn diagram of the intersection relationship of target between bioactive compounds from GG and GPCRs-related cancer. (B) PPI network performed by STRING database, composed of 270 common target networks. (C) PPI network of the top 10 hub genes, analyzed by Cytoscape plugin cytoHubba.
GO enrichment analysis
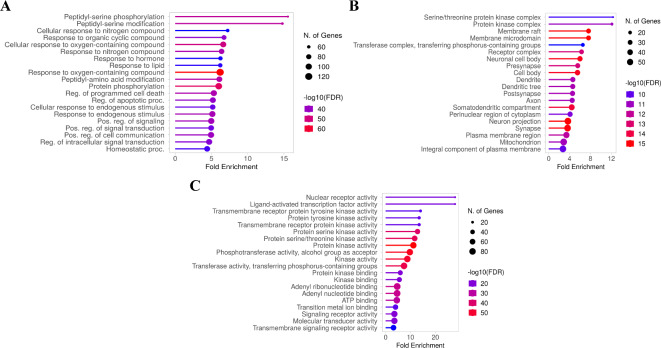
To explore the biological functions of GG compounds in relation to GPCRs-related cancer, a comprehensive GO enrichment analysis was performed on the 265 candidate targets using ShinyGO 0.7722. The analysis yielded a substantial number of significant GO terms (FDR, false discovery rate) < 0.05), amounting to 3,628 terms in total. These terms encompassed 1,743 biological process terms, 736 cellular component terms, and 1,149 molecular function terms (Fig. 3A-C). Figure 3C presents the top 20 outcomes of the GO enrichment analysis, with a specific focus on the molecular function category. Notably, the molecular function targets demonstrated a noteworthy enrichment false discovery rate (FDR), which was calculated based on the nominal P-value derived from the hypergeometric test. The identified targets predominantly exhibit association with key functions such as protein kinase activity, phosphotransferase activity (with alcohol group as acceptor), kinase activity, transferase activity (transferring phosphorus-containing groups), protein serine/threonine kinase activity, and protein serine kinase activity.
Fig. 3.
GO enrichment analysis for the targets in bioactive compounds from GG treating GCPRs-related cancer (p value < 0.05). GO analysis of biological processes (A), cellular components (B), and molecular functions (C) of potential target genes of GG in cancer.
KEGG enrichment analysis
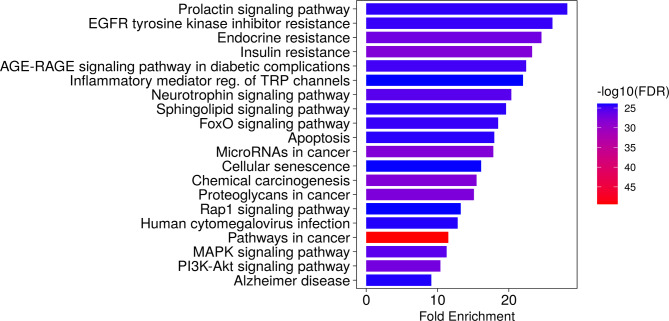
To elucidate the pathways relevant to GG compounds in relation to GPCRs-related cancer, an enrichment analysis of the candidate targets was conducted using the KEGG pathway database by ShinyGO 0.7722. The analysis revealed a total of 615 pathways (FDR < 0.05) that were identified as relevant. The targets exhibited robust associations with pathways known as pathways in cancer, insulin resistance, chemical carcinogenesis, PI3K-AKT signaling pathway, microRNAs in cancer, endocrine resistance, proteoglycans in cancer, neurotrophin signaling pathway, MAPK signaling pathway, and EGFR tyrosine kinase inhibitor resistance. Notably, these targets exhibited significant enrichment across various cancer-related pathways and signaling cascades. Among the identified pathways, the top 20 pathways displaying the highest enrichment of genes emerged as potential key routes for treating GPCRs-related cancer (Fig. 4). These findings emphasize the therapeutic potential of GG-derived compounds in modulating GPCRs-related cancer treatment by targeting these specific pathways.
Fig. 4.
The top 20 potential KEGG pathway enrichment of screened target genes in GPCRs-related cancer.
Network analysis of protein-protein interactions (PPI) and identification of key targets
The analysis of the PPI network involved the utilization of the STRING database23, and the resulting outcomes are presented in Fig. 2B. Within the PPI network, proteins are represented by circular nodes, with their respective 3D structures depicted within the nodes. The lines connecting the nodes illustrate the interactions between the proteins, with a greater number of lines denoting stronger interactions. Moreover, employing the cytoHubba plug-in in Cytoscape 3.9.124, topological analysis was conducted to identify the top 10 hub genes. These hub genes, identified as key regulator genes, hold significance for the treatment of GPCRs-related cancer, as illustrated in Fig. 2C. The color grading of the nodes corresponds to their score, with darker and redder nodes indicating higher scores. The top 10 hub key targets that emerged from this analysis include MAPK3, SRC, EGFR, STAT3, ESR1, MTOR, CCND1, PPARG, BCL2L1, and PTGS2 (Table 3).
Table 3.
Top 10 hub potential targets of GG for GPCRs-related cancer treatment.
| No. | UniProtKB | Gene names | Protein names |
|---|---|---|---|
| Directly associated with the GPCR family | |||
| 1 | P27361 | MAPK3 | Mitogen-activated protein kinase, ERK1 |
| 2 | P12931 | SRC | SRC proto-oncogene, non-receptor tyrosine kinase |
| 6 | P42345 | MTOR | Serine/threonine-protein kinase mTOR |
| 4 | P40763 | STAT3 | Signal transducer and activator of transcription 3 |
| Indirect GPCR-related targets | |||
| 5 | P03372 | ESR1 | Estrogen receptor 1 |
| 3 | P00533 | EGFR | Epidermal growth factor receptor |
| 7 | P24385 | CCND1 | Cyclin D1 |
| 8 | P37231 | PPARG | Peroxisome proliferator activated receptor gamma |
| Not involvement or related action through a GPCR mechanism | |||
| 9 | Q07817 | BCL2L1 | BCL2 like 1 |
| 10 | P35354 | PTGS2 | Prostaglandin-endoperoxide synthase 2 |
Through degree score analysis within the network pharmacology method, we have found that MAPK3 is the most valuable target, while PTGS2 is the most susceptible or least robust target among the top 10 targets for GG. To further investigate, we categorized the key targets into two groups: directly GPCR-related and indirect GPCR-related targets. Specifically, some of the key targets were directly associated with the GPCR family, including MAPK3, SRC, STAT3, and MTOR. On the other hand, EGFR, ESR1, CCND1, and PAPRG were considered indirect GPCR-related targets. Notably, BCL2L1 and PTGS2 were excluded from subsequent studies due to their lack of direct involvement or related action through a GPCR mechanism. Subsequently, molecular docking experiments were conducted to validate the interactions involving these targets.
Confirmation of hub target by molecular docking
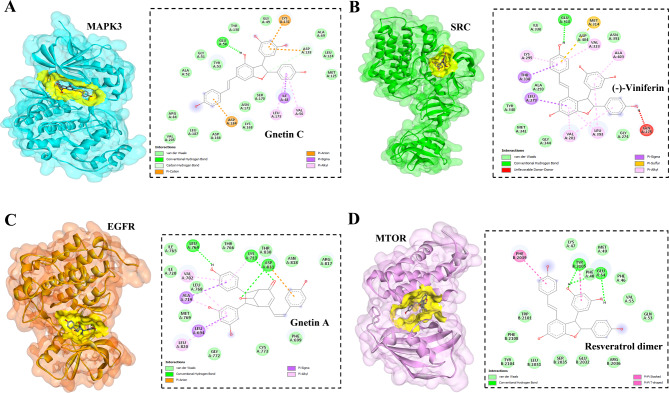
To ensure the validity of the drug-target interactions, the molecular docking analysis focused on the 10 hub proteins as selected targets. In this study, the stability or strong inhibition of the ligand-receptor binding was assessed based on the binding energies between the ligand and protein. A binding energy lower than − 9.5 kcal/mol served as the cut-off criterion, indicating a stable conformation of the ligand binding to the receptor. The results of molecular docking for the compounds derived from GG are summarized in Table 4. Among the evaluated proteins, compounds from GG did not exhibit significant inhibition of STAT3, ESR1, CCND1, and PPARG proteins, as indicated in Table 4. Notably, gnetin C demonstrated notable binding affinity to MAPK3, with a binding energy of -10.00 kcal/mol. Additionally, gnetin C formed hydrogen bonds with GLU50 and ASP128, as along with hydrophobic interactions involving ILE48, VAL56, LEU173, LYS131, ASP128, and ASP184, as depicted in Fig. 5A. Among the compounds, (-)-viniferin exhibited the strongest binding score with SRC, measuring − 9.80 kcal/mol. Furthermore, (-)-viniferin formed a hydrogen bond with GLU310 and engaged in several hydrophobic interactions, as shown in Fig. 5B. Various compounds derived from GG, including resveratrol dimer, ursolic acid, gnetin A, and gnetin D, displayed significant inhibition against EGFR. These compounds exhibited binding energies ranging from − 9.50 to -9.90 kcal/mol. Notably, gnetin A exhibited the highest binding energy and formed three hydrogen bonds with LYS721, ASP831, and LEU764 in its interaction with EGFR (Fig. 5C). Both resveratrol dimer and gnetin C demonstrated substantial binding affinity to MTOR with binding energies of -10.70 kcal/mol. Resveratrol established interactions with MTOR through the formation of three hydrogen bonds with PHE48, GLU54, and TYR105, along with engaging in a pi-pi stacking interaction (Fig. 5D). The bioactive compounds derived from GG exhibited notable binding energy and interactions with the top 4 key hub targets, indicating their potential significance in modulating these crucial molecular targets.
Table 4.
Results of molecular docking studies of 13 components and 8 hub targets.
| Potential effective ingredients | Binding energies (kcal/mol) | |||||||
|---|---|---|---|---|---|---|---|---|
| MAPK3 (PDB 4QTB) |
SRC (PDB 1Y57) |
EGFR (PDB 1M17) |
STAT3 (PDB 6NUQ) |
ESR1 (PDB 1ERR) |
MTOR (PDB 1FAP) |
CCND1 (PDB 2W96) |
PPARG (PDB 1FM6) |
|
| Resveratrol dimer | -8.70 | -9.10 | -9.50 | -7.20 | -7.40 | -10.70 | -7.60 | -7.30 |
| Oxyresveratrol | -8.60 | -7.80 | -7.60 | -5.80 | -7.10 | -7.70 | -6.20 | -7.00 |
| 3-methoxyresveratrol | -8.10 | -7.30 | -7.10 | -5.50 | -7.00 | -7.60 | -5.90 | -7.10 |
| Pinostilbenoside | -8.30 | -7.10 | -8.10 | -6.90 | -7.60 | -8.80 | -7.30 | -7.80 |
| Pterostilbene | -8.50 | -7.10 | -6.80 | -5.60 | -5.60 | -7.60 | -4.80 | -6.60 |
| Tetramethoxystilbene | -6.70 | -6.20 | -6.20 | -4.50 | -5.60 | -7.20 | -4.90 | -5.00 |
| 2,3-dihydroxypropyl icosanoate | -7.90 | -3.90 | -3.60 | -3.90 | -4.20 | -5.40 | -3.00 | -5.20 |
| Oleic acid | -4.40 | -4.00 | -3.70 | -3.60 | -3.90 | -3.90 | -3.40 | -3.80 |
| Ursolic acid | -8.50 | -8.10 | -9.50 | -7.60 | -7.40 | -9.80 | -7.10 | -8.60 |
| (-)-Viniferin | -8.50 | -9.80 | -9.30 | -6.70 | -7.90 | -10.20 | -8.10 | -7.40 |
| Gnetin A | -9.50 | -9.40 | -9.90 | -7.00 | -8.00 | -9.50 | -7.20 | -6.70 |
| Gnetin C | -10.00 | -8.40 | -9.30 | -7.50 | -8.10 | -10.40 | -8.00 | -8.40 |
| Gnetin D | -9.00 | -8.90 | -9.70 | -7.20 | -7.40 | -9.60 | -7.70 | -7.10 |
Fig. 5.
Molecular docking studies of active compounds from GG and against top hub targets. Molecular interactions of MAPK3 and gnetin C (A), SRC and (-)-viniferin (B), EGFR and gnetin A (C), and MTOR and resveratrol diner (D). The binding site of each protein was represented in yellow color.
Survival analysis
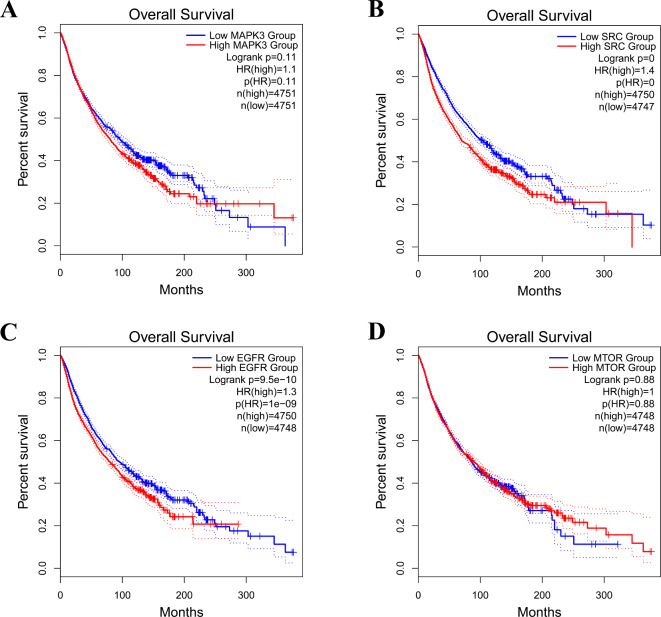
Survival analysis using Kaplan-Meier plots was performed for the top 4 hub genes, namely MAPK3, SRC, EGFR, and MTOR, which exhibited significant inhibition by GG-derived compounds. The analysis unveiled a remarkable association between increased expression of these hub genes and an unfavorable prognosis in cancer patients by the GEPIA database (p < 0.05)25, as illustrated in Fig. 6.
Fig. 6.
Kaplan-Meier overall survival analysis of cancer patients with GPCRs-related cancer based on expression of the top 4 hub genes.
Discussion
Cancer presents a substantial global health challenge and ranks among the leading causes of mortality worldwide. In 2020, GLOBOCAN reported approximately 19.3 million new cancer cases, resulting in nearly 10.0 million deaths26. The most frequently diagnosed cancers included breast cancer (11.7%), lung cancer (11.4%), colorectal cancer (10.0%), prostate cancer (7.3%), and stomach cancer (5.6%). Projections suggest a 47% increase in the global cancer burden, with an estimated 28.4 million cases expected by the year 204026. GPCRs, which constitute an extensive and diverse family of cell surface signaling receptors, have been implicated in numerous cancer types. Dysregulation of GPCR activity within cancer cells can manifest through various mechanisms, such as aberrant overexpression, activating mutations resulting in gain-of-function effects, and increased production and secretion of agonists. GPCRs are often overexpressed in several cancer types, including human chronic lymphocytic leukemia, breast cancer, colon cancer, pancreatic ductal adenocarcinoma, and cancer-associated fibroblasts (CAFs) cell lines27. For instance, protease-activated receptors (PARs), which constitute a distinct class of GPCRs associated with cancer, display notable expression and overexpression in solid tumors and cancer cells, including those of breast cancer, colon cancer, and melanoma28. The high expression of GPCRs in cancer cells suggest their potential involvement on malignant progression. Understanding the activation of GPCRs is paramount as it offers insights into the underlying mechanisms that can be targeted for the development of anticancer drugs. GPCRs play a pivotal role in regulating various pathways, such as Wnt, MAPK, and PI3K signaling, which are often disrupted in cancer due to mutations29. Formylpeptide receptor-2 (FPR2), a GPCR identified in colon cancer cells, activates the protein kinase B pathway (AKT pathway), thereby contributing to drug resistance30. However, the expression levels and molecular mechanisms of GPCRs may vary among different cancer types, owing to the distinct genetic and biochemical characteristics specific to each type of cancer. Currently, there are FDA-approved drugs available for cancer treatment that specifically target GPCRs. These include cabergoline, which targets the dopamine receptor D1 (DRD1) for neuroendocrine and pituitary cancer; lanreotide, targeting the somatostatin receptor (SSTR) for pancreatic cancer; Degarelix, aimed at the gonadotropin-releasing hormone receptor (GnRH) for prostate cancer; and Raloxifene, directed at the estrogen receptor (ER) for breast cancer31. Moreover, various other GPCRs, such as FPR2, Galphas-coupled beta-adrenergic receptor, Angiotensin II type 1 receptor (AT1R), GPR160, and ACKR3, have been proposed as potential targets for cancer treatment31. The increased expression of GPCRs and their associated signaling proteins in cancer cells presents promising opportunities for novel therapeutic targets, offering distinctive therapeutic possibilities.
Network pharmacology has emerged as a valuable approach for predictive analyses, particularly in complex diseases such as cancer32. In this study, we employed a comprehensive network pharmacology approach to elucidate the potential molecular mechanisms of GG compounds against cancer. Our approach encompassed drug-likeness evaluation, target identification, GO and KEGG pathway analysis, as well as PPI analysis. Through the integration of drug and disease databases, we identified a total of 265 common targets for constructing a PPI network. Among these targets, 260 nodes demonstrated protein-protein interactions (Fig. 2A and B). Subsequent KEGG analysis unveiled the specific pathways influenced by GG in GPCRs-related cancer. Notable pathways encompassed pathways in cancer, insulin resistance, chemical carcinogenesis, PI3K-AKT signaling pathway, microRNAs in cancer, endocrine resistance, proteoglycans in cancer, neurotrophin signaling pathway, MAPK signaling pathway, and EGFR tyrosine kinase inhibitor resistance. These pathways play pivotal roles in various facets of tumor development and progression, including tumor cell growth, apoptosis evasion, angiogenesis, invasion, and metastasis. This underscores their significant involvement in the pathological processes underpinning cancer33–35. After employing cytoHubba plug-ins in Cytoscape, we identified a set of 10 hub genes that demonstrate significant importance. These hub genes, namely MAPK3, SRC, EGFR, STAT3, ESR1, MTOR, CCND1, PPARG, BCL2L1, and PTGS2, play crucial roles in cellular signaling pathways. In particular, MAPK3, also recognized as ERK1, holds paramount significance as a key molecule in the ERK/MAPK pathway, a pivotal cell signaling pathway. The upregulation and heightened activity of MAPK3 are consistently linked to the initiation, progression, cancer cell migration, drug resistance in various carcinoma types, including liver, thyroid, lung, and gastric cancers36. It phosphorylates cytoplasmic proteins downstream, activating numerous nuclear transcription factors associated with apoptosis and cell proliferation29. Conversely, SRC shows abnormal overexpression and activation in various cancer types, including glioblastoma, liver, lung, colon, breast, bladder, and pancreatic cancers30,31. This heightened SRC expression significantly contributes to the progression of these malignancies. SRC can be activated by a diverse array of extracellular signals originating from integrins, G-protein-linked receptors, steroid receptors, and receptor tyrosine kinases (RTKs)37. Upon activation, SRC triggers the Ras/Raf/ERK signaling cascade and activates other kinases such as PI3K, MAPK, and AKT31. Functioning as an oncoprotein, SRC regulates transformed cells and plays a pivotal role in tumor progression and metastasis38. EGFR, a key hub gene in the growth factor receptor family with intrinsic tyrosine kinase activity, plays a central role in tumorigenesis, particularly in lung, breast, and glioblastoma cancers39. Elevated EGFR expression is associated with poor cancer patient prognoses. Ligand-induced EGFR activation triggers multiple signaling pathways, including Ras/MAPK, PI3K/AKT, and PLC/PKC cascades, making EGFR a target in current cancer therapies39. STAT3, an oncogene within the STAT family, undergoes activation via JAK, EGFR, and GPCR, resulting in its phosphorylation and subsequent nuclear translocation. Inside the nucleus, STAT3 upregulates target genes such as Bcl-xL, Cyclin D1, and VEGF40. STAT3 plays a crucial role in tumor cell proliferation, invasion, migration, resistance to therapy, and the prediction of poor prognoses41. Persistent STAT3 activation is consistently observed in esophageal squamous cell carcinoma, colorectal cancer, lung cancer, and gastric cancer, emphasizing its potential as a therapeutic target in cancer treatment42. Estrogen receptor (ESR1), a transcription factor, exerts significant influence on cell proliferation and differentiation in target tissues. ESR1, implicated in breast cancer, endometrial cancer, and osteoporosis, plays a central role in regulating gene expression associated with the cell cycle, proliferation, and apoptosis, involving key factors such as IGF1, Cyclin D1, c-Myc, FOXM1, GREB1, BCL2, and CXCL1243. MTOR, a serine/threonine kinase, plays a role in regulating a wide range of cellular processes, including cell survival, growth, metabolism, protein synthesis, autophagy, and homeostasis. Its importance spans across various cancer types, with regulatory interconnections with pathways such as PI3K/AKT, MAPK, VEGF, NF-κB, and p5344. Cyclin D1 (CCND1), a crucial regulator facilitating the transition from G1 to S phase in the cell cycle, has been found to be upregulated in a variety of solid and hematologic tumors45. Peroxisome proliferator-activated receptor gamma (PPARG), a ligand-inducible transcription factor and member of the nuclear receptor superfamily, regulates various physiological processes, including immunity, inflammation, vascular functions, cellular proliferation, differentiation, development, and apoptosis. Elevated PPARG expression has been noted in multiple cancer types, suggesting the potential for PPARG antagonists as therapeutic options for cancer treatment46. BCL2L1, also known as BCL-X, plays a pivotal role in enhancing cell survival and exerts anti-apoptotic effects, especially in cancer cells. The overexpression of BCL2L1 imparts resistance to chemotherapeutic agents, underscoring the potential for therapeutic interventions that target BCL2L1 in effective cancer treatment47. Prostaglandin-endoperoxide synthase 2 (PTGS2), an enzyme involved in prostaglandin synthesis, is induced by inflammation and expressed in tumor epithelial cells, especially in colorectal cancer and non-small cell lung cancer. Excessive prostaglandin production is linked to various facets of lung cancer progression, including angiogenesis, metastasis, and immunosuppression48. Significantly, BCL2L1 and PTGS2 were omitted from further investigation as they do not directly engage in or act via a GPCR mechanism.
Through molecular docking analyses, it was determined that (-)-viniferin, gnetin A, and the resveratrol dimer exhibited strong binding affinity toward SRC, EGFR, and MTOR, respectively. Particularly, gnetin C demonstrated a robust binding force with MAPK3 and PTGS2. A close examination of the protein-ligand interactions suggests that the GG extracts possess effective binding capabilities with these specific targets, achieving lower binding energies primarily through the formation of multiple hydrogen bonds. (-)-Viniferin, a stilbenoid compound, corresponds to cis-epsilon, whereas the resveratrol dimer corresponds to trans-epsilon-viniferin. (-)-Viniferin exhibited varying IC50 values in different cancer cell lines: 20.1 µM in C6 cells (Glioma), 76.2 µM in HepG2 cells (Hepatoblastoma), 21.5 µM in HeLa cells (Breast cancer), 47.2 µM in MCF-7 cells (Breast cancer), and 90.2 µM in HT-29 cells (Colorectal adenocarcinoma)49. Notably, this compound demonstrated activity in P-388 cells (Leukemia) with an IC50 value of 18.1 ± 0.7 µM50. Additionally, trans-epsilon-viniferin displayed variability in different cancer cell lines, with IC50 values as follows: 85.5 ± 8.1 µM in COLO205 cells (Colon carcinoma), 13.9 ± 0.1 µM in HT-29 cells (Colorectal adenocarcinoma), 7.7 ± 0.2 µM in HepG2 cells (Hepatoblastoma), 9.3 ± 0.3 µM in AGS cells (Gastric adenocarcinoma), and 5.6 ± 1.4 µM in HL-60 cells (Human leukemia)51. Previous studies have highlighted the antiproliferative and apoptosis-inducing effects of epsilon-viniferin on various cancer cell lines, including those of osteosarcoma, non-small cell lung cancer, human hepatoma, colon cancer, glioblastoma, and human melanoma52–54. Epsilon-viniferin has shown significant anticancer activity by modulating various pathways, including the induction of apoptosis, inhibition of cell cycle progression, and suppression of invasion and migration. Notably, in non-small cell lung cancer cells (A549 cells), epsilon-viniferin induces apoptosis by downregulating phospho-AKT expression and upregulating cleaved PARP and cleaved caspase-3 expression52. It also inhibited epithelial-mesenchymal transition (EMT), invasion, and migration in non-small cell lung cancer cells induced by TGF-β1 or IL-1β. These effects were achieved through the reduction of TGF-β1-induced reactive oxygen species (ROS) production and the downregulation of key factors associated with EMT and metastasis, including MMP2, vimentin, Zeb1, Snail, p-SMAD2, p-SMAD3, and ABCG2 in A549 cells. Furthermore, in xenograft metastatic mouse models of A549 cells, epsilon-viniferin significantly inhibited lung metastasis55. By modulating key regulators of the cell cycle, such as cyclins A, E, and D1, along with their associated CDK-1 and − 2, epsilon-viniferin compounds effectively disrupt melanoma cell cycle progression, particularly during the S phase54. Additionally, treatment with epsilon-viniferin remarkably suppressed lung cancer progression in nude mice bearing A549-cell xenografts52. In this study, we explored cis- and trans-epsilon-viniferin as inhibitors of SRC and MTOR. SRC, a protein involved in interactions with transmembrane receptors like integrin/FAK, RTKs, and GPCRs, activates critical downstream signaling pathways, including MAPK, ERK, PI3K/AKT/mTOR, IL-6/JAK/STAT3, and Rho/ROCK pathways. These pathways drive essential cancer processes, including survival, proliferation, angiogenesis, migration, invasion, and metastasis56. Therefore, the targeted inhibition of SRC and MTOR by cis- and trans-epsilon-viniferin highlights their potential to modulate and inhibit crucial signaling molecules in cancer cells, as discussed earlier. However, in vitro anti-cancer experiments should confirm the mechanism of action of cis- and trans-epsilon-viniferin, and variations in the mechanism might be observed depending on the type of cancer. Furthermore, epsilon-viniferin exhibits favorable drug-like properties and demonstrates promising ADME pharmacokinetic characteristics. These attributes position epsilon-viniferin as a promising and effective therapeutic agent with the potential to revolutionize current cancer treatment strategies.
Gnetin A and gnetin C are resveratrol derivatives that fall under the stilbene class, and they have been extracted from various species within the Gnetaceae family11. Gnetin A shares a similar chemical structure with gnetin C. In this study, gnetin A displayed a robust binding affinity to the tyrosine kinase of EGFR. However, there is currently limited information available regarding the anticancer properties of gnetin A, highlighting the need for further in vitro investigations in our future studies. Molecular docking of active compounds extracted from GG revealed the presence of chemical structures based on resveratrol. Resveratrol, a natural stilbenoid compound, has the ability to undergo oxidative coupling of two to eight resveratrol units, leading to the formation of oligostilbenoids in various plant families57. This natural phytoalexin compound, stilbenoid, is abundant in plants and notably prominent in red wine58. Researchers have shown considerable interest in its multiple health benefits, including cardiovascular protection, anti-inflammatory properties, anti-metastatic effects, and anti-cancer activities59. Gnetin C has demonstrated significant anticancer activity by inhibiting proliferation, migration, and angiogenesis in various cancer cell types and in in vivo models. Clinically achievable concentrations of gnetin C have been shown to significantly inhibit proliferation and induce apoptosis in pancreatic, prostate, breast, and colon cancer cells60. Gnetin C exhibited varying IC50 values in cancer cell proliferation, with the following results: 16.29 ± 1.11 µM in PANC-1 cells (pancreatic cancer), 13.83 ± 0.92 µM in AsPC-1 cells (pancreatic cancer), 12.22 ± 1.45 µM in Pan-02 cells (mouse pancreatic cancer), 10.28 ± 0.79 µM in PC-3 cells (prostate cancer), 9.85 ± 2.60 µM in DU-145 cells (prostate cancer), 8.95 ± 0.92 µM in LNCaP cells (prostate cancer), 9.01 ± 0.15 µM in PTEN-CaP8 cells (mouse prostate cancer), 13.13 ± 0.61 µM in MCF-7 cells (breast cancer), 11.78 ± 1.45 µM in HT-29 cells (breast cancer), and 11.3 ± 0.60 µM in Colon-26 (mouse colon cancer)60. MSE, which contains the active ingredient gnetin C, has been found to inhibit tumor growth, intratumoral angiogenesis, and the development of liver metastasis in mice with Colon-26 tumors52. The antitumor properties of gnetin C have been attributed to its ability to suppress the ERK1/2 pathway, leading to the inhibition of proliferation, migration, and tube formation in human umbilical vein endothelial cells61. Gnetin C has also demonstrated potent antitumor effects against patient-derived acute myeloid leukemia (AML) cells by targeting both the ERK1/2 and AKT/mTOR signaling pathways, which play critical roles in the survival and growth of AML cells61,62. The inhibition of the MAPK/ERK1/2 and AKT pathways by gnetin C in leukemia cells results in the inactivation of downstream members, including activated p90 ribosomal S6 kinase (RSK1) and mitogen- and stress-activated protein kinase (MSK1/2), leading to cell cycle arrest, apoptosis, and the inhibition of cell growth63. In our study, we propose that gnetin C exhibits a high binding affinity towards MAPK3 or ERK1, supporting previous findings and providing further evidence of its potential therapeutic efficacy and its ability to target crucial signaling pathways in cancer cells. Gnetin C has demonstrated inhibitory effects on PTGS2 (COX-2). Cyclooxygenase-2 (COX-2) is highly expressed in various human cancers, including colorectal, breast, ovarian, uterine cervix, lung, head, and neck cancers. The NF-κB/IκB pathway, which is regulated by the PI3K/AKT and ERK signaling pathways, plays a crucial role in inducing COX-2 expression. Elevated COX-2 expression can inhibit apoptosis, promote angiogenesis and invasiveness, contribute to inflammation and immunosuppression, and facilitate the conversion of procarcinogens into carcinogens, thereby promoting tumorigenesis64. Gnetin C, found in MSE, has demonstrated anti-inflammatory effects by downregulating IL-1β, a proinflammatory cytokine, in a mouse model. This effect may be associated with its inhibitory effects on COX-265. These findings emphasize the potential of gnetin C as a promising therapeutic agent for cancer treatment. It can inhibit not only COX-2 and its associated pro-tumorigenic effects but also exert anti-inflammatory actions by regulating IL-1β. Furthermore, gnetin C has been shown to downregulate the expression of MTA1 and exhibit significant inhibitory effects on cell viability, colony formation, cell death induction, and migration in DU145 and PC3M prostate cancer cells66. MTA1, a chromatin modifier protein, plays a significant role in promoting aggressiveness and metastasis in prostate cancer through its overexpression67. It activates downstream targets involved in inflammation, cell survival, and invasion, contributing to prostate cancer progression and metastasis. Alterations in MTA1 levels impact the PTEN/AKT pathway among other downstream pathways68. Treatment with gnetin C at a dose of 50 mg/kg results in the most potent suppression of tumor progression, as evidenced by reduced mitotic activity, angiogenesis, a significant increase in apoptosis, and confirmed downregulation of MTA1, Cyclin D1, and Notch 2 in xenograft prostate tumor tissues69. Recent studies have shown that gnetin C supplementation effectively reduces the progression of prostate cancer in a mouse model by inhibiting MTA1, which leads to decreased cell proliferation, inflammation, and angiogenesis. Notably, the administration of gnetin C did not result in any observable toxicity in mice13. In silico investigation of gnetin C through ADME analysis revealed favorable gastrointestinal absorption, the absence of blood-brain barrier permeation, and no inhibition of CYP enzymes, except for being a CYP2C9 inhibitor (Table 2). However, ADME properties are solely based on computational predictions in this study, and experimental studies will be necessary in subsequent work. In line with preclinical and clinical investigations, a 2-week daily consumption of pure gnetin C in healthy volunteers did not lead to any adverse events. Furthermore, gnetin C exhibited improved pharmacokinetic properties, including higher bioavailability, when compared to resveratrol70–72. Molecular docking models have offered valuable insights into the mechanism of action of these compounds on GPCRs-related targets, shedding light on their inhibitory effects on cancer. These insights find support in in vitro and in vivo experiments, providing preliminary confirmation of the therapeutic potential of various active components in GG. These components target key pathways that are implicated in cancer.
Moreover, there are several previous studies reported about in silico analysis of active compounds of GG. The natural compound epsilon-viniferin, an oligostilbene (a resveratrol dimer), binds to the hinge region between the α- and β-subunits of AMPK, interacting with its active site and contributing to improved hyperglycemia and hyperlipidemia. epsilon-viniferin’s hydroxyl groups form crucial hydrogen bonds with AMPK, involving residues ARG10 and LYS31 (α-subunit), as well as THR106, ARG107, and ASP108 (β-subunit)73. Gnetin C and trans-epsilon-viniferin bind to the active site of ACE (angiotensin-converting enzyme) through hydrogen bonds or hydrophobic interactions, with free energy binding values of -8.51 and − 8.13 kcal/mol, respectively74. HER2 proteins are pivotal in breast cancer cell growth and differentiation. epsilon-viniferin binds to HER2 receptors with a binding energy of -10.45 kcal/mol, forming six hydrogen bonds at MET766, CYS775, ASP855, LEU788, GLN791, and ASN84275. In an in silico study, Gnetin C and trans-resveratrol, active compounds from melinjo seeds, were assessed for binding affinity against ERα in breast cancer cells (MCF-7). The results revealed that both Gnetin C and trans-resveratrol can bind to the same amino acids (VAL54B, TYR55B, TYR216B, TRP227B, and LEU306B) with docking scores of -6.0 and − 7.9 kcal/mol, respectively76. In an in silico molecular docking study, gnetin C was evaluated for its interaction with the target protein VHR (Vaccinia H-1 related phosphatase), a receptor involved in multiple signaling pathways (MAPK, JNK, ERK1, p38, EGFR, and ErbB2/HER2) and crucial for the proliferation of HeLa cervical cancer cells. The docking results indicated that gnetin C binds to VHR with a binding energy of -8.3 kcal/mol, inhibiting receptor activity by interacting with specific amino acid residues: SER129, PRO162, ASN163, and GLU12677. VHR plays significant roles in cellular signaling, including cell-cycle regulation, DNA damage response, MAPK signaling, platelet activation, and angiogenesis77. Consequently, gnetin C and (-)-viniferin demonstrated specific binding to molecular targets in cancer, such as HER2, EGFR, ERα, and VHR, as revealed by in silico molecular docking studies. This study found that gnetin C and (-)-viniferin exhibit notable binding affinity to MAPK3 and SRC, with strong binding energy and hydrogen bond formation at the active sites of these targets. Therefore, gnetin C and (-)-viniferin inhibited MAPK3 and SRC, suggesting potential interactions with HER2, EGFR, ERα, and VHR. This could disrupt cancer cell signaling and growth. MAPK3 (ERK1) and SRC are signaling molecules associated with HER2, EGFR, and ERα in lung and breast cancer78,79. MAPK3 is also promoted by GPCRs via the Gαq/PLCβ/PKC pathway80, while SRC kinases form direct associations with GPCRs, resulting in complex interplay81. These kinases are also linked to cellular growth, cancer, and growth factor receptor tyrosine kinases. The SRC and STAT3 pathways serve as potential effectors for G proteins82. GPCRs activate STAT3 through JAKs, leading to cancer progression83. Additionally, our study reported that the resveratrol dimer and gnetin C exhibit substantial binding affinity to mTOR with strong binding energies. mTOR (mTORC2) activation is enhanced by β- and α-adrenergic signaling through GPCRs and the presence of growth factors via RTK receptors, and it also occurs in membrane subcellular compartments84. Previously, gnetin C has shown significant antitumor effects against patient-derived acute myeloid leukemia (AML) cells by targeting the ERK1/2 and AKT/mTOR signaling pathways, which are crucial for the survival and proliferation61,62. Therefore, mTOR, MAPK3, and SRC are key targets directly associated with the GPCR family and are targeted by gnetin C and (-)-viniferin, suggesting that the active compounds from GG may act as potent inhibitors of these targets, directly impacting GPCR-related pathways.
In conclusion, the consistent demonstration of GPCRs involvement in tumor progression and metastasis across various cancer types highlights their potential as promising targets for cancer treatment. Among the compounds found in the GG collection, a total of 13 exhibited drug-like properties, meeting the criteria of the Rule of Five (RO5). Through network pharmacology analysis, we identified 8 potential central targets related to GPCR for these GG compounds, including MAPK3, SRC, EGFR, STAT3, ESR1, MTOR, CCND1, and PPARG. By employing network pharmacology and molecular docking analysis, we predict that (-)-viniferin, gnetin A, gnetin C, and resveratrol dimer have the potential to exert an anti-cancer effect through interactions with multiple proteins, such as MAPK3, SRC and mTOR. These targets are closely associated with essential cellular processes in cancer, including proliferation, migration, apoptosis, and angiogenesis. However, it is imperative to conduct further in vitro and in vivo experimental studies to validate the effectiveness of these compounds and elucidate their mechanisms of action against specific types of cancer.
Methods
Chemoinformatics, drug likeness and ADME prediction
The chemoinformatics data and drug likeness of 28 compounds derived from GG were assessed using the SwissADME server (http://www.swissadme.ch), an online tool specifically designed for calculating pharmacokinetic properties, oral bioavailability, and drug-likeness19. To evaluate the drug-likeness of these compounds, Lipinski’s Rule of Five (RO5) was applied, which serves as a screening criterion for potential oral drugs in humans. The parameters taken into consideration included molecular weight (MW), lipophilicity(logP), topological polar surface area (TPSA), the number of rotatable bonds, hydrogen bond acceptor (HBA) and hydrogen bond donor (HBD) numbers, as well as water solubility. Only those compounds that met the drug-likeness criteria were selected for further analysis. A comprehensive flowchart illustrating the overall study process is provided in Fig. 1.
Target proteins prediction
The identification of targets for the bioactive compounds derived from GG was carried out using the Swiss Target Prediction databases (http://www.swisstargetprediction.ch/)20. To achieve this, the canonical SMILES representation of each compound sourced from GG was entered into the Swiss Target Prediction database. Subsequently, candidate targets with high probability scores were selected and then further standardized using the UniProt database (http://www.uniprot.org/).
GPCRs-related potential targets
To investigate and compile the targets associated with ‘GPCRs-related cancer,’ we employed the Human Gene Database (GeneCards, https://www.genecards.org/)21. Using the designated search term, we retrieved and consolidated the relevant targets. Afterward, we overlapped the predicted targets of the compounds derived from GG with those associated with GPCRs-related cancer, which resulted in the creation of a Venn diagram. This Venn diagram was generated using a web tool accessible at (https://bioinformatics.psb.ugent.be/webtools/Venn/) and visually illustrated the intersection of identified targets between the drug compounds and the disease. By extracting the common targets within this intersection, we obtained the target set of compounds from GG, which holds promise for potential treatment of GPCRs-related cancer.
GO and KEGG pathway enrichment analysis
To gain a deeper understanding of the importance of essential target genes, we conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. These analyses were performed using ShinyGO 0.77 (http://bioinformatics.sdstate.edu/go/), a bioinformatics tool specifically designed for gene function description and annotation22. We applied a significance threshold of p < 0.05 during the enrichment analysis. The results were effectively visualized using bubble and bar charts. GO serves as a comprehensive resource for functional genomics, offering detailed definitions of gene functions, including molecular functions. On the other hand, KEGG comprises graphical diagrams of biochemical pathways and potential signaling pathways. The outcomes of these analyses provide insights into the functional roles of essential target genes and highlight significant pathways associated with the compounds under investigation.
Construction of protein-protein interaction network
Protein-protein interactions (PPI) play a pivotal role in biological processes and are essential for a comprehensive understanding of the intricate workings of a living cell. In this study, we constructed the PPI network for the identified drug targets using the STRING database (https://string-db.org/)23, with a specific focus on the species ‘Homo sapiens.’ To ensure the reliability of the information, we set a confidence score threshold of > 0.9. Subsequently, we imported the results obtained from the PPI network into Cytoscape 3.9.1 (www.cytoscape.org/) for network generation and further analysis24. To pinpoint highly interconnected regions within the PPI network, we utilized the Cytoscape plugin cytoHubba (https://apps.cytoscape.org/apps/cytohubba; version 0.1). This plugin enabled us to identify clusters of proteins based on their degree level, with the top-ranked proteins designated as hub targets.
Molecular docking analysis between GG and hub genes
To gain a deeper understanding of the relationship, mode of interactions, and action mechanisms between the candidate proteins (or hub targets) and the bioactive compounds derived from GG, we employed molecular docking. 3D structures of the compounds were retrieved from the PubChem database (http://pubchem.ncbi.nlm.nih.gov/)85. Ligands were optimized by assigning bond orders, angles, and topology, while ensuring the addition of any missing and polar hydrogens at pH 7.4. To prepare the ligands for docking, we performed energy minimization using conjugate (steepest descent methods) and carried out charge addition for ionization correction. These optimization steps were conducted using UCSF Chimera 1.17.186. For the hub target proteins, we obtained their 3D crystal structures from the Protein Data Bank (PDB, https://www.rcsb.org/)87. Using BIOVIA Discovery Studio, we removed water molecules and small molecule ligands from the protein structures. Subsequently, we processed the hub targets with AutoDock tools, which involved steps such as hydrogenation, charge distribution, and atomic type addition. We performed molecular docking simulations using AutoDock Vina 1.1.2, enabling us to explore protein-ligand interactions and predict their binding modes88. The resulting docked structures were visualized and analyzed using the BIOVIA Discovery Studio visualizer89.
Overall survival analysis of hub genes
To investigate the impact of hub targets on overall survival (OS) in GPCRs-related cancers across all types, we utilized the Gene Expression Profiling Interactive Analysis (GEPIA2, http://gepia2.cancer-pku.cn/), an online server25. This server provides a cancer genomics dataset for analysis and offers the Kaplan-Meier plot, a widely used method for assessing survival outcomes. To determine the prognostic significance of the top 5 hub genes inhibited by compounds derived from GG, we employed the overall survival metric. The gene expression levels of cancer patients were categorized into two groups: high expression and low expression. Subsequently, we generated a Kaplan-Meier survival plot to compare the survival outcomes between these two groups. To assess the statistical significance of the observed differences, we calculated the hazard ratio (HR) with 95% confidence intervals and the log-rank p-value.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to express the sincere gratitude to Walailak University, Thailand for providing the necessary research and financially supported.
Author contributions
M.C., N.C.P., I.S., O.R., and A.T. conceived and designed the experiments. M.C., M.P., O.P., P.C., O.R., and A.T. performed the experiments. N.C.P., I.S., O.R., and A.T. analyzed the data and prepared all figures. N.C.P., I.S., and A.T. contributed materials/analysis tools. M.C., N.C.P., I.S., O.R., F.K., M.I., and A.T. wrote the paper. All authors reviewed the manuscript.
Data availability
All the data presented in this paper can be gathered from the open-source website platform, as mentioned in the article, and can be subjected to analysis using relevant software.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sriram, K. & Insel, P. A. GPCRs as targets for approved drugs: how many targets and how many drugs? Mol. Pharmacol. 93, 251–258. 10.1124/mol.117.111062 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomsen, W., Frazer, J. & Unett, D. Functional assays for screening GPCR targets. Curr. Opin. Biotechnol. 16, 655–665. 10.1016/j.copbio.2005.10.008 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Yang, D. et al. G protein-coupled receptors: structure- and function-based drug discovery. Sig Transduct. Target. Ther. 6, 7. 10.1038/s41392-020-00435-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wootten, D., Christopoulos, A., Marti-Solano, M., Babu, M. M. & Sexton, P. M. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell. Biol. 19, 638–653. 10.1038/s41580-018-0049-3 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Chaudhary, P. K. & Kim, S. An insight into GPCR and G-Proteins as Cancer drivers. Cells. 10, 3288. 10.3390/cells10123288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutkind, J. S. The pathways connecting G protein-coupled receptors to the nucleus through divergent mitogen-activated protein kinase cascades. J. Biol. Chem. 273, 1839–1842. 10.1074/jbc.273.4.1839 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Hemmings, B. A. & Restuccia, D. F. PI3K-PKB/AKT pathway. Cold Spring Harb Perspect. Biol. 4, a011189. 10.1101/cshperspect.a011189 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang, S. Y. et al. Potential of bioactive Food Components against Gastric Cancer: insights into molecular mechanism and therapeutic targets. Cancers. 13, 4502. 10.3390/cancers13184502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George, B. P., Chandran, R. & Abrahamse, H. Role of Phytochemicals in Cancer Chemoprevention: Insights. Antioxidants. 10, 1455. (2021). 10.3390/antiox10091455 [DOI] [PMC free article] [PubMed]
- 10.Anisong, N. et al. A comprehensive review on nutritional contents and functional properties of Gnetum gnemon Linn. Food Sci. Technol. 42, e100121. 10.1590/fst.100121 (2022). [Google Scholar]
- 11.Kato, E., Tokunaga, Y. & Sakan, F. Stilbenoids isolated from the seeds of Melinjo (Gnetum gnemon L.) and their biological activity. J. Agric. Food Chem. 57, 2544–2549. 10.1021/jf803077p (2009). [DOI] [PubMed] [Google Scholar]
- 12.Narayanan, N. K., Nargi, D., Randolph, C. & Narayanan, B. A. Liposome encapsulation of gnetin C, a novel resveratrol dimer from Gnetum gnemon, reduces cancer cell proliferation and induces apoptosis. PLoS ONE. 10, e0124807. 10.1371/journal.pone.0124807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parupathi, P. et al. Gnetin C intercepts MTA1-Associated neoplastic progression in prostate Cancer. Cancers. 14, 6038. 10.3390/cancers14246038 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang, M., Chen, J. L., Xu, L. W. & Ji, G. Navigating traditional Chinese medicine network pharmacology and computational tools. Evid Based Complement Alternat Med. 2013, 731969. 10.1155/2013/731969 (2013). [DOI] [PMC free article] [PubMed]
- 15.Hopkins, A. L. & Network Pharmacology Nat. Biotechnol. 25, 1110–1111. 10.1038/nbt1007-1110 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Chakraborty, C., Doss, C. G. P., Chen, L. & Zhu, H. Evaluating protein-protein interaction (PPI) networks for diseases pathway, target discovery, and drug-design using ‘in silico pharmacology’. Curr. Protein Pept. Sci. 15, 561–571. 10.2174/1389203715666140724090153 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Macalino, S. J., Gosu, V., Hong, S. & Choi, S. Role of computer-aided drug design in modern drug discovery. Arch. Pharm. Res. 38, 1686–1701. 10.1007/s12272-015-0640-5 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Pinzi, L. & Rastelli, G. Molecular Docking: shifting paradigms in Drug Discovery. Int. J. Mol. Sci. 20, 4331. 10.3390/ijms20184331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daina, A., Michielin, O., Zoete, V. & SwissADME A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717. 10.1038/srep42717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daina, A., Michielin, O. & Zoete, V. Swiss target prediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47, W357–W364. 10.1093/nar/gkz382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stelzer, G. et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 54 1.30.1–1.30.33 (2016). [DOI] [PubMed]
- 22.Ge, S. X., Jung, D. & Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 36, 2628–2629. 10.1093/bioinformatics/btz931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. 10.1093/nar/gkac1000 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560. 10.1093/nar/gkz430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. 10.3322/caac.21660 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Insel, P. A. et al. GPCRomics: GPCR expression in Cancer cells and tumors identifies New, potential biomarkers and therapeutic targets. Front. Pharmacol. 9, 431. 10.3389/fphar.2018.00431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi, X., Gangadharan, B., Brass, L. F., Ruf, W. & Mueller, B. M. Protease-activated receptors (PAR1 and PAR2) contribute to tumor cell motility and metastasis. Mol. Cancer Res. 2, 395–402 (2004). [PubMed] [Google Scholar]
- 29.Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature. 502, 333–339. 10.1038/nature12634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su, L. D., Peng, J. M. & Ge, Y. B. Formyl peptide receptor 2 mediated chemotherapeutics drug resistance in colon cancer cells. Eur. Rev. Med. Pharmacol. Sci. 22, 95–100. 10.26355/eurrev_201801_14105 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Usman, S., Khawer, M., Rafique, S., Naz, Z. & Saleem, K. The current status of anti-GPCR drugs against different cancers. J. Pharm. Anal. 10, 517–521. 10.1016/j.jpha.2020.01.001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Y. et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 48, 1031–1041. 10.1093/nar/gkz981 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derakhshani, A. et al. From Oncogenic Signaling pathways to single-cell sequencing of Immune cells: changing the Landscape of Cancer Immunotherapy. Molecules. 26, 2278. 10.3390/molecules26082278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rascio, F. et al. The pathogenic role of PI3K/AKT pathway in Cancer Onset and Drug Resistance: an updated review. Cancers. 13, 3949. 10.3390/cancers13163949 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smolarz, B., Durczyński, A., Romanowicz, H., Szyłło, K. & Hogendorf, P. miRNAs in Cancer. Int. J. Mol. Sci. 23, 2805. 10.3390/ijms23052805 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao, H. Y. et al. MiR-129 reduces CDDP resistance in gastric cancer cells by inhibiting MAPK3. Eur. Rev. Med. Pharmacol. 24, 11468. 10.26355/eurrev_202011_23759 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Pelaz, S. G. & Tabernero, A. Src: coordinating metabolism in cancer. Oncogene. 41, 4917–4928. 10.1038/s41388-022-02487-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang, T. & Qiu, Y. Interaction between Src and a C-terminal proline-rich motif of akt is required for akt activation. J. Biol. Chem. 278, 15789–15793. 10.1074/jbc.M212525200 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Uribe, M. L., Marrocco, I. & Yarden, Y. EGFR in Cancer: signaling mechanisms, drugs, and Acquired Resistance. Cancers. 13, 2748. 10.3390/cancers13112748 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stark, G. R. & &Darnell, J. J. The JAK-STAT pathway at twenty. Immunity. 36, 503–514. 10.1016/j.immuni.2012.03.013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groner, B. & von Manstein, V. Jak Stat signaling and cancer: opportunities, benefits and side effects of targeted inhibition. Mol. Cell. Endocrinol. 451, 1–14. 10.1016/j.mce.2017.05.033 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Wang, H. Q. et al. STAT3 pathway in cancers: past, present, and future. Med. Comm. 3, e124. 10.1002/mco2.124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hua, H. et al. Mechanisms for estrogen receptor expression in human cancer. Exp. Hematol. Oncol. 7, 24. 10.1186/s40164-018-0116-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian, T., Li, X. & Zhang, J. mTOR Signaling in Cancer and mTOR inhibitors in solid Tumor Targeting Therapy. Int. J. Mol. Sci. 20, 755. 10.3390/ijms20030755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, S. & Li, L. Degradation strategy of cyclin D1 in cancer cells and the potential clinical application. Front. Oncol. 12, 949688. 10.3389/fonc.2022.949688 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan, Y. et al. PPAR-α modulators as current and potential Cancer treatments. Front. Oncol. 11, 707. 10.3389/fonc.2021.599995 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Opferman, J. T., Kothari, A. & Anti-apoptotic BCL-2 family members in development. Cell. Death Differ. 25, 37–45. 10.1038/cdd.2017.170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benelli, R., Venè, R. & Ferrari, N. Prostaglandin-endoperoxide synthase 2 (cyclooxygenase-2), a complex target for colorectal cancer prevention and therapy. Transl Res. 196, 42–61. 10.1016/j.trsl.2018.01.003 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Kim, J. H. et al. Cytotoxic and Antimutagenic Stilbenes from seeds of Paeonia lactiflora. Arch. Pharm. Res. 25, 293–299. 10.1007/BF02976629 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Muhtadi Hakim, E. H. et al. Cytotoxic resveratrol oligomers from the Tree Bark of Dipterocarpus Hasseltii. Fitoterapia. 77, 550–555. 10.1016/j.fitote.2006.07.004 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Tian, X. et al. Chemical characterization of Main Bioactive constituents in Paeonia Ostii seed meal and GC-MS analysis of seed oil. J. Food Biochem. 44, e13088. 10.1111/jfbc.13088 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Huang, C. et al. ε-Viniferin and α-viniferin alone or in combination induced apoptosis and necrosis in osteosarcoma and non-small cell lung cancer cells. Food Chem. Toxicol. 158, 112617. 10.1016/j.fct.2021.112617 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Colin, D. et al. Antiproliferative activities of resveratrol and related compounds in human hepatocyte derived HepG2 cells are associated with biochemical cell disturbance revealed by fluorescence analyses. Biochimie. 90, 1674–1684. 10.1016/j.biochi.2008.06.006 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Nivelle, L. et al. Molecular analysis of differential antiproliferative activity of resveratrol, epsilon viniferin and labruscol on melanoma cells and normal dermal cells. Food Chem. Toxicol. 116, 323–334. 10.1016/j.fct.2018.04.043 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Chiou, W. C. et al. α-Viniferin and ε-Viniferin inhibited TGF-β1-Induced epithelial-mesenchymal transition, Migration and Invasion in Lung Cancer cells through downregulation of Vimentin expression. Nutrients. 14, 2294. 10.3390/nu14112294 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu, P. C., Yang, C. T., Jablons, D. M. & You, L. The crosstalk between Src and Hippo/YAP Signaling Pathways in Non-small Cell Lung Cancer (NSCLC). Cancers. 12 1361. 10.3390/cancers12061361 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen, J. et al. Update on Phytochemistry and Pharmacology of naturally occurring Resveratrol oligomers. Molecules. 22, 2050. 10.3390/molecules22122050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koushki, M. et al. A miraculous natural compound for diseases treatment. Food Sci. Nutr. 6, 2473–2490. 10.1002/fsn3.855 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xue, Y. Q. et al. Resveratrol oligomers for the prevention and treatment of cancers. Oxid. Med. Cell Longev. 2014, 765832. 10.1155/2014/765832 (2014). [DOI] [PMC free article] [PubMed]
- 60.Narayanan, N. K. et al. Antitumor activity of melinjo (Gnetum gnemon L.) seed extract in human and murine tumor models in vitro and in a colon26 tumor-bearing mouse model in vivo. Cancer Med. 4, 1767–1780. 10.1002/cam4.520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Espinoza, J. L. et al. The simultaneous inhibition of the mTOR and MAPK pathways with Gnetin-C induces apoptosis in acute myeloid leukemia. Cancer Lett. 400, 127–136. 10.1016/j.canlet.2017.04.027 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Breems, D. A. et al. Monosomal karyotype in acute myeloid leukemia: a better indicator of poor prognosis than a complex karyotype. J. Clin. Oncol. 26, 4791–4797. 10.1200/JCO.2008.16.0259 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Espinoza, J. L. & Inaoka, P. T. Gnetin-C and other resveratrol oligomers with cancer chemopreventive potential. Ann. N Y Acad. Sci. 1403, 5–14. 10.1111/nyas.13450 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Steelman, L. S. et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging. 3, 192–222. 10.18632/aging.100296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ikeda, E. et al. Healing effects of monomer and dimer resveratrol in a mouse periodontitis model. BMC Oral Health. 22, 460. 10.1186/s12903-022-02499-2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar, A., Dholakia, K., Sikorska, G., Martinez, L. A. & Levenson, A. S. MTA1-Dependent anticancer activity of gnetin C in prostate Cancer. Nutrients. 11, 2096. 10.3390/nu11092096 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dias, S. J. et al. Nuclear MTA1 overexpression is associated with aggressive prostate cancer, recurrence and metastasis in African americans. Sci. Rep. 3, 2331. 10.1038/srep02331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dhar, S. et al. Dietary pterostilbene is a novel MTA1-targeted chemopreventive and therapeutic agent in prostate cancer. Oncotarget. 7, 18469–18484. 10.18632/oncotarget.7841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gadkari, K. et al. Therapeutic potential of gnetin C in prostate Cancer: a pre-clinical study. Nutrients. 12, 3631. 10.3390/nu12123631 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakagami, Y. et al.. Immunomodulatory and metabolic changes after Gnetin-C supplementation in humans. Nutrients. 11, 1403. 10.3390/nu11061403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tani, H. et al. Pharmacokinetics and safety of resveratrol derivatives in humans after oral administration of melinjo (Gnetum gnemon L.) seed extract powder. J. Agric. Food Chem. 62, 1999–2007. 10.1021/jf4048435 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Ota, H. et al. Trans-resveratrol in Gnetum gnemon protects against oxidative-stress-induced endothelial senescence. J. Nat. Prod. 76, 1242–1247. 10.1021/np300841v (2013). [DOI] [PubMed] [Google Scholar]
- 73.Liu, R. et al. ε-Viniferin, a promising natural oligostilbene, ameliorates hyperglycemia and hyperlipidemia by activating AMPK in vivo. Food Funct. 11, 10084–10093. 10.1039/d0fo01932a (2020). [DOI] [PubMed] [Google Scholar]
- 74.Triputra, M. A. & Yanuar, A. Analysis of compounds isolated from Gnetum gnemon L. Seeds as potential ACE inhibitors through Molecular Docking and Molecular Dynamics simulations. J. Young Pharm. 10, 32–39. 10.5530/jyp.2018.2s.7 (2018). [Google Scholar]
- 75.Dewi, I. G. A. I. P., Yuda, P. E. S. K. & Rahadi, I. W. S. Silico Study and Pharmacokinetics Prediction of ɛ-Vinicompoundmpound as Anticancercandidatedidate. Jrki. 13, 121–130 (2023). [Google Scholar]
- 76.Romadhona, K. N., Shifa, N. A., Asmiyenti, D. & Djalil, A. D. Molecular Docking of Gnetin C and Trans-resveratrol of Melinjo Seeds (Gnetum Gnemon L.) Used as the Inhibitors of Breast Cancer Cells MCF-7. IJHMS. 4, 58–63.
- 77.Savitri, R. I., Arifin, N. H. & Febriansah, R. Antioxidant, cytotoxic activity and protein target inhibition of Ethyl acetate Fraction Melinjo seed (Gnetum gnemon L.) by in Vitro and in Silico studies on HeLa cervical Cancer cells. HAYATI J. Biosci. 30, 864–873. 10.4308/hjb.30.5.864-873 (2023). [Google Scholar]
- 78.Hsu, L. H., Chu, N. M., Kao, S. H. & Estrogen Estrogen receptor and Lung Cancer. Int. J. Mol. Sci. 18, 1713. 10.3390/ijms18081713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kay, C. et al. Current trends in the treatment of HR+/HER2 + breast cancer. Future Oncol. 17, 1665–1681. 10.2217/fon-2020-0504 (2021). [DOI] [PubMed] [Google Scholar]
- 80.New, D. C. & Wong, Y. H. Molecular mechanisms mediating the G protein-coupled receptor regulation of cell cycle progression. J. Mol. Signal. 26, 2. 10.1186/1750-2187-2-2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luttrell, D. K. & Luttrell, L. M. Not so strange bedfellows: G-protein-coupled receptors and Src family kinases. Oncogene. 23, 7969–7978. 10.1038/sj.onc.1208162 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Ram, P. & Iyengar, R. G protein coupled receptor signaling through the Src and Stat3 pathway: role in proliferation and transformation. Oncogene. 20, 1601–1606. 10.1038/sj.onc.1204186 (2001). [DOI] [PubMed] [Google Scholar]
- 83.Yu, H. et al. Revisiting STAT3 signaling in cancer: new and unexpected biological functions. Nat. Rev. Cancer. 14, 736–746. 10.1038/nrc3818 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Magaway, C., Kim, E. & Jacinto, E. Targeting mTOR and metabolism in Cancer: lessons and innovations. Cells. 8, 1584. 10.3390/cells8121584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim, S. et al. PubChem 2023 update. Nucleic Acids Res. 51, D1373–D1380. 10.1093/nar/gkac956 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 87.Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242. 10.1093/nar/28.1.235 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eberhardt, J., Santos-Martins, D., F Tillack, A. & Forli, S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python Bindings. J. Chem. Inf. Model. 61, 3891–3898. 10.1021/acs.jcim.1c00203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sama-Ae, I., Pattaranggoon, N. C. & Tedasen, A. In silico prediction of Antifungal compounds from Natural sources towards Lanosterol 14-alpha demethylase (CYP51) using Molecular docking and Molecular dynamic simulation. J. Mol. Graph. Model. 121, 108435. 10.1016/j.jmgm.2023.108435 (2023). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data presented in this paper can be gathered from the open-source website platform, as mentioned in the article, and can be subjected to analysis using relevant software.