Abstract
Animal foraging is an essential and evolutionarily conserved behavior that occurs in social and solitary contexts, but the underlying molecular pathways are not well defined. We discover that conserved autism-associated genes (NRXN1(nrx-1), NLGN3(nlg-1), GRIA1,2,3(glr-1), GRIA2(glr-2), and GLRA2,GABRA3(avr-15)) regulate aggregate feeding in C. elegans, a simple social behavior. NRX-1 functions in chemosensory neurons (ADL and ASH) independently of its postsynaptic partner NLG-1 to regulate social feeding. Glutamate from these neurons is also crucial for aggregate feeding, acting independently of NRX-1 and NLG-1. Compared to solitary counterparts, social animals show faster presynaptic release and more presynaptic release sites in ASH neurons, with only the latter requiring nrx-1. Disruption of these distinct signaling components additively converts behavior from social to solitary. Collectively, we find that aggregate feeding is tuned by conserved autism-associated genes through complementary synaptic mechanisms, revealing molecular principles driving social feeding.
Subject terms: Social behaviour, Molecular neuroscience, Neural circuits
C. elegans aggregate in large clumps during feeding. Here, the authors find conserved autism-associated genes mediate distinct molecular and circuit signaling components that tune C. elegans feeding behavior from solitary to social feeding.
Introduction
Social behaviors are broadly defined as interactions between individuals of the same species, which can range in complexity and include mating, kin selection, parental guidance, predation, and hierarchical dominance1,2. One highly conserved social behavior is the formation of groups to forage or feed. Social feeding behavior is exhibited by ant colonies3–5, shoaling fish6,7, large predator herds8–12, and hunter-gatherer societies13. Social feeding can confer advantages or disadvantages depending on context, such as access to resources, predator threat, disease risk, and competition over food or mates2,14,15. An animal’s propensity to join a group is the result of multiple, complex, and sometimes competing environmental factors that guide their behavior16–19. The neuronal mechanisms underlying social feeding are not well understood, in part due to the complexity of the behavioral decisions and the underlying neuronal circuits controlling them.
The nematode C. elegans exhibits a wide variety of foraging behaviors and strategies20. For example, on a bacterial food lawn, most wild isolate strains feed in large clumps of aggregating animals; however, other strains feed alone or display an intermediate level of aggregate feeding behavior20. Moreover, a gain-of-function polymorphism in the conserved neuropeptide receptor gene npr-1(ortholog to human NPY1R) was identified in the laboratory strain, N2 Bristol, which converts behavior from social to solitary feeding20. Social feeding behavior in the wild can, therefore, be genetically modeled in the solitary control strain through loss of function mutation in the npr-1 gene (npr-1(ad609))20. Aggregate feeding is controlled by a small sensory circuit that integrates environmental cues like oxygen levels, carbon dioxide levels, food, and aversive chemosensory stimuli, along with classical social cues like pheromones and touch21–29. npr-1 inhibits aggregation behavior acting in RMG interneurons21,22 downstream of multiple highly electrically connected sensory neurons, including URX, ADL, ASH, ASK, ADE, and AWB30–32. Moreover, the extent of social feeding is regulated by the binding affinity of flp-21 and flp-18 neuropeptide ligands33 that are released from several sensory neurons (ASE33, ASK34, ADL35, ASH36) and act on the NPR-1 receptor. In addition, aggregation behavior requires the gap junction innexin gene, unc-9, in select neurons30. However, less is known about the function of chemical signaling in social feeding and how neuronal circuit properties and synapses differ between solitary and social feeders.
Individuals diagnosed with neurodevelopmental conditions, including autism, can exhibit changes in social behavior and altered sensitization to sensory stimuli37–40. Genomic studies have associated hundreds of genetic loci with increased risk for autism41–44, including the synaptic adhesion molecules neurexins (NRXN1,2,3) and their canonical post-synaptic partners neuroligins (NLGN1,2,3,4)(Supplementary Table 1)45–50. The association of neurexins and neuroligins with autism strongly suggests roles for these genes in regulating social behaviors45–50. Neurexins are conserved synaptic adhesion molecules that organize chemical synaptic properties, including neuronal connectivity, synaptic plasticity, and excitatory/inhibitory balance28. Mammals have three neurexin genes that encode one long (α) and one/two short (β and γ (specific to NRXN1)) isoforms of the protein45. Mutations in these genes in rodents alter motor activity, anxiety-like behavior (avoidance), social approach, memory, performance of stereotyped behavior, and pre-pulse inhibition51–58. Neurexin mutations also impact chemical synapse function, structure, and signaling, including presynaptic density, release probability, calcium dynamics, and post-synaptic currents55–62.
C. elegans has a single ortholog of neurexins, nrx-1, which is 27% identical to human NRXN1 at the amino acid level based on DIOPT alignment63, with nearly identical domain structure (Supplementary Table 1)64. In C. elegans, nrx-1 contributes to retrograde inhibition of neurotransmitter release at neuromuscular junctions, regulation of GABA receptor diffusion and alignment of GABA, synaptic clustering, and synapse formation65–70. However, these synaptic functions of nrx-1 have rarely been linked to distinct behaviors, with the exception of male mating, where nrx-1 impacts male response to hermaphrodite contact71 and time to spicule protraction72. Despite these advances, we still have much to learn about the functions of nrx-1 in circuits and synapses outside of the neuromuscular junction and how nrx-1 mechanistically alters complex behaviors.
Using npr-1(ad609) mutant C. elegans to model social feeding behavior, we find three molecularly independent synaptic mechanisms (synaptic adhesion molecules NRX-1 and NLG-1 and the classical excitatory neurotransmitter, glutamate) that work together to tune foraging behavior from solitary to social. We also use genetic methods to identify the downstream glutamate receptors that regulate aggregation behavior, homologs of which are also associated with autism. Despite nrx-1 and eat-4 (the vesicular glutamate transporter) both functioning in ASH and ADL sensory neurons to modulate aggregation behavior, they do so through distinct mechanisms. NRX-1 regulates an increase in the number of ASH pre-synaptic release sites in aggregating animals, while faster glutamate release from ASH neurons in aggregating animals occurs independently of NRX-1. These additive neuronal mechanisms exemplify the complexity of C. elegans foraging strategies and, although not a model for autism, provide insights and a framework for how variation in social behavior is achieved at genetic, molecular, and circuit levels, which may be applied to more complex organisms.
Results
NRX-1(α) functions in ADL and ASH sensory neurons for aggregation behavior
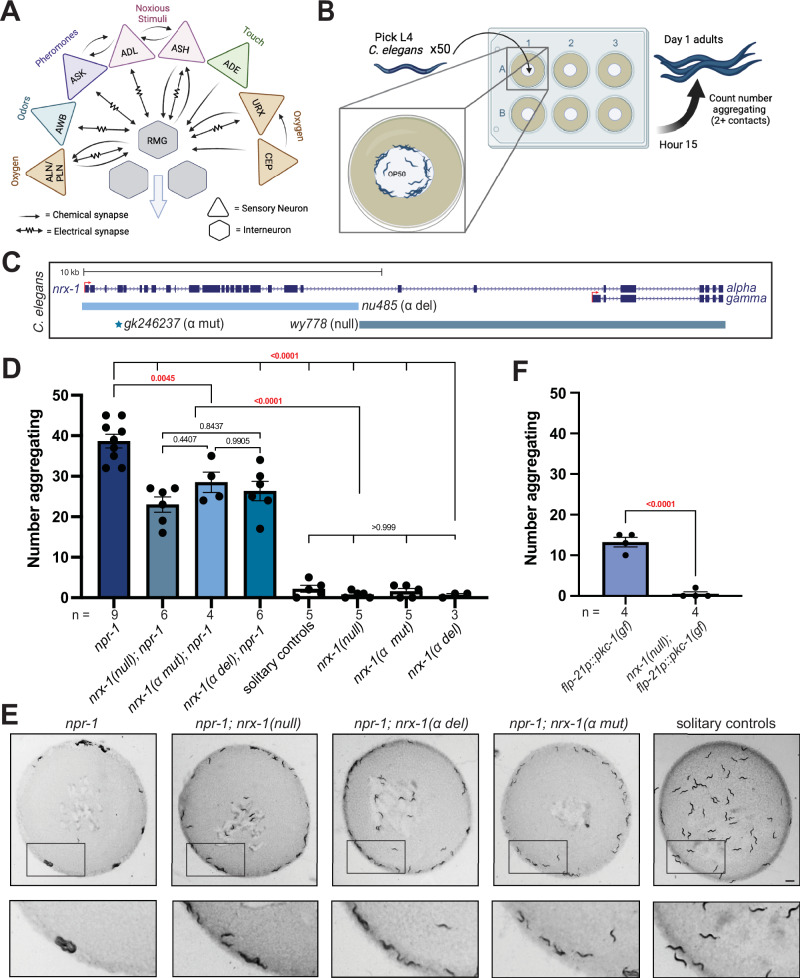
Neurexin genes, including nrx-1 in C. elegans, are broadly expressed in neurons in mammals and invertebrates. We used a database of neuronal gene expression profiles (CENGEN)73 to confirm that nrx-1 transcripts are present in the RMG interneurons and upstream sensory neurons implicated in aggregation behavior (Fig. 1A). Given the broad expression of nrx-1 in RMG interneurons and its synaptic partners, we asked if nrx-1 functions in aggregation behavior. We quantified aggregation behavior as the number of C. elegans in contact with two or more animals based on previous literature20, for day 1 adults using longitudinal, blinded image analysis (Fig. 1B). As expected, npr-1(ad609) mutants aggregate significantly more than solitary controls, with an average of 78% of npr-1 animals aggregating compared to 4% of solitary control animals aggregating (npr-1 average = 38.67, SEM = 1.675 vs. solitary control average = 2.2, SEM = 0.860, n = 50 animals)(Fig. 1D, E). Solitary controls consisted of the N2 Bristol strain and solitary animals from the N2 background with an integrated transgene (otIs525) and/or him-8 mutation used for genetic crosses. Aggregation behavior was not impacted by him-8; otIs525 in the solitary (N2) or aggregating background (npr-1(ad609))(Supplemental Fig. 1A). We tested three mutant alleles of nrx-1: a large deletion in nrx-1 that disrupts both the long (α) and short (γ) isoforms (wy778), an α-isoform specific deletion (nu485), and a nonsense mutation leading to a premature stop codon early in the α-isoform (gk246237)(Fig. 1C). In the npr-1(ad609) aggregating background, all three alleles of nrx-1 significantly decreased the number of aggregating C. elegans compared with npr-1(ad609) alone (Fig. 1D, E). Notably, npr-1(ad609) animals with any of the nrx-1 mutant alleles showed intermediate aggregation behavior compared to solitary controls or nrx-1 mutants alone, which showed almost no aggregation behavior. Thus, we found that nrx-1 is essential for aggregation behavior induced by npr-1 mutation, such that disruption of nrx-1 reduced aggregation behavior of npr-1 animals by ~ 40%, which is primarily mediated by the α isoform. Animals with the npr-1 variant of a wild social isolate strain (215 F in Hawaiian CB4856) in an otherwise N2 background (qgIR1) also displayed increased aggregation behavior (Supplemental Fig. 1B, C). Aggregate feeding in this strain was dependent on nrx-1, confirming that nrx-1 contributes to social feeding.
Fig. 1. NRX-1 is essential for aggregation behavior.
A Circuit diagram of sensory integration circuit. Connectome based on NemaNode and WormWiring data (Created in BioRender. Cowen, M. (2020) BioRender.com/g12s038). B Schematic of medium throughput aggregation behavior assay with 50 day 1 adult worms per well of a 6-well WormCamp imaged using WormWatcher platforms and scored for aggregation behavior defined as two or more animals in direct contact (Created in BioRender. Hart, M. (2023) BioRender.com/b48j541). C Schematic of C. elegans nrx-1 gene showing mutant alleles used and isoforms removed by functional null and α-isoform specific mutants. D Graph showing the number of aggregating animals in various genetic backgrounds. All mutant nrx-1 alleles (wy778 = nrx-1 null, gk246237 = nrx-1 α mut, nu485 = nrx-1 α del) show decreased aggregation behavior. E Representative images of aggregation behavior in npr-1(ad609), npr-1(ad609);nrx-1(wy778), npr-1(ad609);nrx-1(nu485), npr-1(ad609);nrx-1(gk246237) mutants, and solitary controls (Scale bar = 1 mm). F Graph showing the number of aggregating animals in flp-21p::pkc-1(gf) strain compared to flp-21p::pkc-1(gf);nrx-1(wy778). The number of biological replicates (n) are displayed in the figure, bars show the mean number of aggregating C. elegans, error bars indicate SEM. One-way ANOVA with Tukey’s post-hoc test was used for comparisons (t test for panel F), exact p-values are shown on graphs (red indicates significance, black indicates non-significance). Source data are provided in the Source Data file.
Previous work found that activating sensory neurons and the RMG interneurons through expression of a constitutively active Protein Kinase C (flp-21p::pkc-1(gf)) increases neurotransmission and induces aggregation behavior in solitary animals21. We used these animals to test whether nrx-1 was needed for aggregation behavior induced independently of npr-1 mutation. We found that, as previously reported, flp-21p::pkc-1(gf) induced social feeding, albeit at lower levels than npr-1(ad609) mutants21 (Fig. 1F). We next found that nrx-1(wy778); flp-21p::pkc-1(gf) animals aggregated less than flp-21p::pkc-1(gf) alone (Fig. 1F). Therefore, nrx-1 is also necessary for aggregation behavior induced by increased neuronal signaling within the sensory integration circuit that drives aggregation behavior.
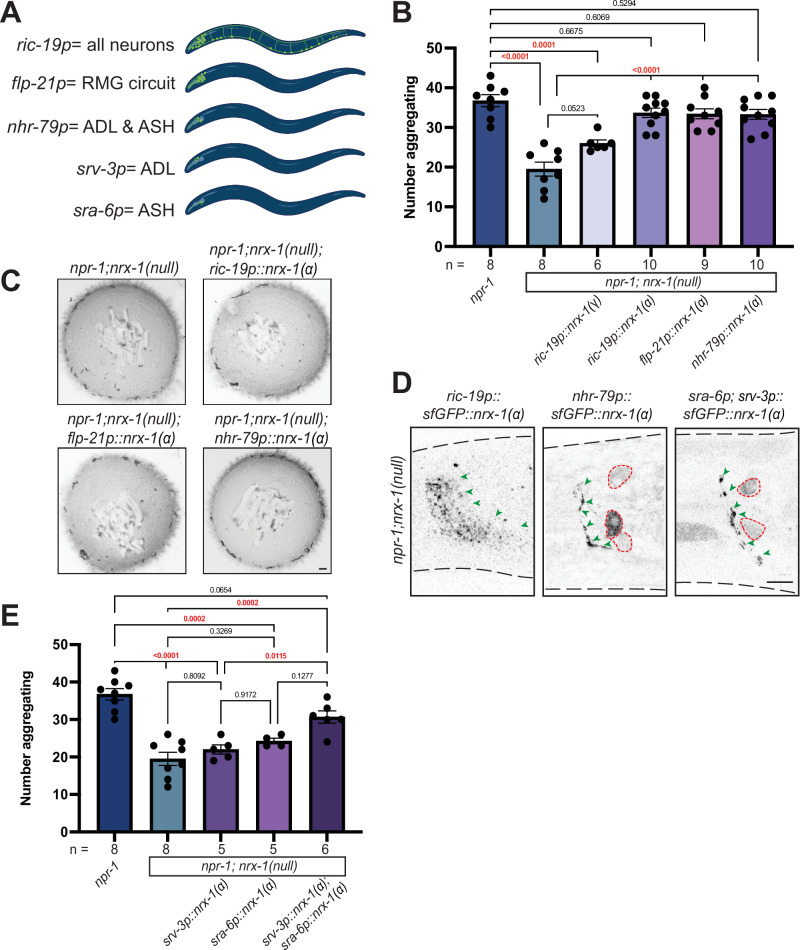
To localize the function of nrx-1 in aggregation behavior, we created animals expressing nrx-1 isoforms under various neuron-specific promoters. We tested a large panel of promoters and quantified the impact on aggregation behavior of nrx-1 null mutants in the npr-1 aggregating background (Supplemental Fig. 1D). Expression of NRX-1(α) in all neurons using the ric-19 promoter completely restored aggregation behavior in npr-1; nrx-1(wy778) mutants to the level of aggregating npr-1(ad609) animals (Fig. 2A–C). Expression of NRX-1(γ) in neurons under the same ric-19 promoter had little impact, confirming a specific role for the α-isoform in modifying aggregation behavior (Fig. 2A, B). Further, expression of the α-isoform of NRX-1 in the RMG interneurons and several sensory neurons, including ADL and ASH (flp-21p), or in both ADL and ASH sensory neurons (nhr-79p), restored aggregation behavior to levels comparable to pan-neuronal expression (Fig. 2A–C). Neuronal expression of NRX-1 in solitary controls, which should result in overexpression of the α-isoform, did not increase aggregation behavior (Supplemental Fig. 1E). Collectively, these data show that NRX-1(α) functions in two pairs of sensory neurons for aggregation behavior.
Fig. 2. NRX-1(α) acts in ADL and ASH sensory neurons for aggregate feeding.
A Schematics showing the neurons where each promoter is expressed. ric-19p expresses in all neurons, flp-21p expresses in several sensory neurons and RMG interneurons, nhr-79p expresses in ADL and ASH sensory neurons, srv-3p expresses in ADL neurons, and sra-6p expresses in ASH neurons (Created in BioRender. Hart, M. (2023) BioRender.com/w06b193). Graph showing number of aggregating animals (B) and representative images of aggregation behavior assay plates (C) in npr-1(ad609);nrx-1(null) mutants with NRX-1(α) driven by ric-19, flp-21, and nhr-79 promoters, and NRX-1(γ) driven by the ric-19 promoter, and controls (Scale bar = 1 mm). D Confocal image of NRX-1(α) expression in all neurons (ric-19p::sfGFP::nrx-1), ADL and ASH neurons (nhr-79p::sfGFP::nrx-1), and ADL and ASH neurons (sra-6p::sfGFP::nrx-1 & srv-3p::sfGFP::nrx-1). Green arrows indicate NRX-1 axonal expression. Red dashed lines show cell bodies. ric-19p::sfGFP::nrx-1(α) imaging performed in nrx-1(wy778) (Scale bar = 10 μm). E Graph showing the number of aggregating animals in various genetic backgrounds. Data for npr-1 and npr-1;nrx-1 are plotted in both 2B and 2E. The number of biological replicates (n) are displayed in the figure, bars show the mean number of aggregating C. elegans, and error bars indicate SEM. One-way ANOVA with Tukey’s post-hoc test was used for comparisons, exact p-values are shown on graphs (red indicates significance, black indicates non-significance). Source data are provided in the Source Data file.
We confirmed the expression and localization of the various nrx-1 transgenes by fusing a superfolder GFP to the nrx-1 coding sequence and monitoring fluorescence in the corresponding neurons (Fig. 2D and Supplemental Fig. 1F)74. In all transgenic animals, sfGFP::NRX-1(α) localized along the neurites and processes of the neurons in a punctate pattern; with some expression also observed within the cell body (Fig. 2D). To determine if nrx-1 functions in ADL and/or ASH neurons for aggregation behavior, we expressed sfGFP::NRX-1(α) specifically in ADL using the srv-3 promoter or specifically in ASH using the sra-6 promoter. Expression of sfGFP::NRX-1(α) in ADL or ASH individually did not restore aggregation behavior to npr-1 levels. However, a combination of these same two transgenes increased aggregation behavior, confirming the function of NRX-1(α) in both pairs of sensory neurons (Fig. 2D, E). These data are consistent with previous results showing that ablating both ADL and ASH disrupts aggregation behavior28.
NLG-1 is essential for aggregation behavior independently of NRX-1
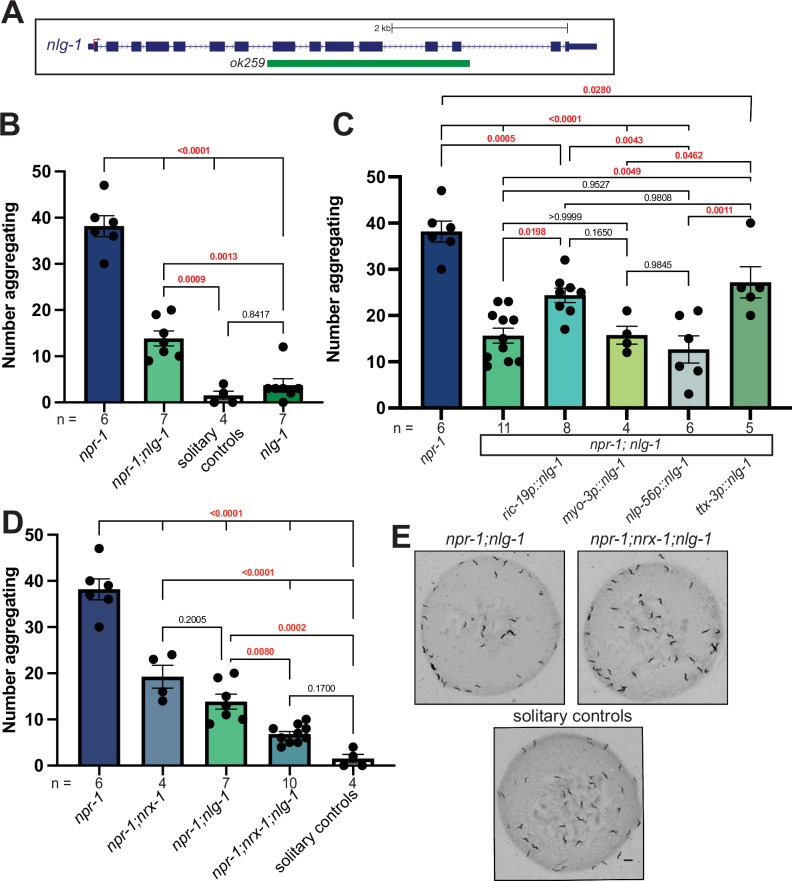
nlg-1 is the single C. elegans ortholog of the neuroligin synaptic adhesion genes NLGN1,2,3,475 and a well-characterized trans-synaptic partner of NRXN1(nrx-1)76. Using a large deletion in nlg-1(ok259)75, we asked if disruption of nlg-1 also altered aggregation behavior in npr-1(ad609) mutants. We found that loss of nlg-1 led to a significant decrease in aggregation behavior of npr-1(ad609) mutant animals but had no effect in solitary control animals (Fig. 3A, B, and E). To localize the function of nlg-1 in aggregation behavior we used a similar transgenic rescue approach as for nrx-1. Expression of sfGFP::NLG-1 in all neurons using the ric-19 promoter partially restored aggregation behavior (Fig. 3C). In contrast, expression of sfGFP::NLG-1 in ADL and ASH (nhr-79 promoter), ADL (srv-3 promoter), ASH (sra-6 promoter), AIA interneurons (ins-1 promoter), RMG interneurons (nlp-56 promoter), body wall muscles (myo-3 promoter), or via the nlg-1 promoter (integrated transgene), did not impact aggregation behavior (Fig. 3C and Supplemental Fig. 2A). However, expression of sfGFP::NLG-1 in AIY interneurons (ttx-3 promoter) increased the number of aggregating animals similar to levels with NLG-1 expression in all neurons (Fig. 3C). We also confirmed expression of all sfGFP::NLG-1 transgenes by analyzing expression of the sfGFP tag (Supplemental Fig. 2B). Together, these results indicate that NLG-1 functions in AIY interneurons and other neurons to partially modify aggregation behavior.
Fig. 3. NLG-1 contributes independent of NRX-1 in aggregation behavior.
A Schematic of C. elegans nlg-1 gene showing the deletion allele assessed. B Graph showing number of aggregating animals in npr-1(ad609), npr-1(ad609);nlg-1(ok259), nlg-1(ok529), and solitary controls. nlg-1 deletion decreased aggregation behavior in npr-1 animals. C Graph showing number of aggregating animals in npr-1(ad609);nlg-1(ok259) mutants with NLG-1 driven by ric-19, myo-3, nlp-56, and ttx-3 promoters and controls. ric-19p expresses in all neurons, myo-3p expresses in muscles, nlp-56p expresses in RMG neurons, and ttx-3p expresses in AIY neurons. D Graph showing number of aggregating animals in npr-1(ad609), npr-1(ad609);nrx-1(wy778), npr-1(ad609);nlg-1(ok259), npr-1(ad609);nrx-1(wy778);nlg-1(ok259), and solitary controls. E Representative images of aggregation behavior in npr-1(ad609);nlg-1(ok259), npr-1(ad609);nrx-1(wy778); nlg-1(ok259) and solitary controls (Scale bar = 1 mm). Data for npr-1 and npr-1;nlg-1 are plotted in 3B, 3C, and 3D. Data for solitary controls are plotted in 3B and 3C. The number of biological replicates (n) are displayed in the figure by order and color, bars show the mean number of aggregating C. elegans, and error bars indicate SEM. One-way ANOVA with Tukey’s post-hoc test was used for comparisons, exact p-values are shown on graphs (red indicates significance, black indicates non-significance). Source data are provided in the Source Data file.
To test whether nrx-1 and nlg-1 function together, we created an npr-1(ad609);nrx-1(wy778);nlg-1(ok259) triple mutant. We found a significant decrease in aggregation behavior in the triple mutant animals compared to either double mutant (Fig. 3D, E). These findings suggest that both nrx-1 and nlg-1 are critical for aggregation behavior but function in parallel, non-epistatic molecular pathways. These data are consistent with prior observations, which showed that nrx-1 and nlg-1 can function together70,77, independently67–69, or even antagonistically71,72.
Glutamate signaling from ADL and ASH neurons is necessary for aggregation behavior
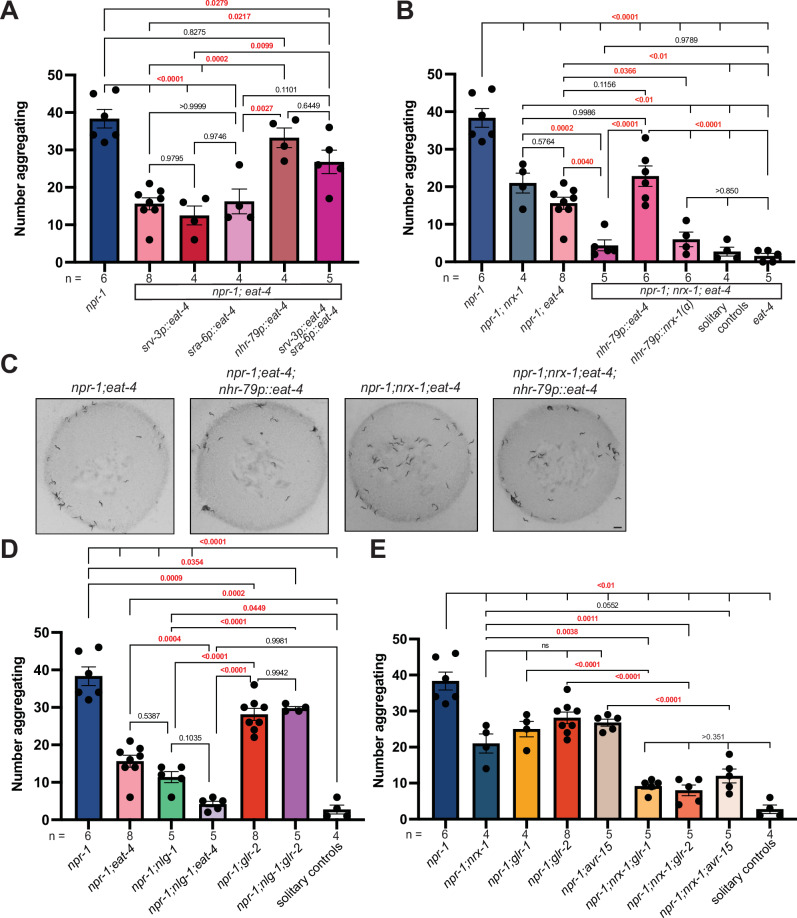
ADL and ASH sensory neurons signal via glutamate to modify animal behavior. Moreover, silencing the gap junctions in ADL and ASH has been shown to not impact social feeding behavior30, likely implicating glutamate signaling from these neurons. We hypothesized that mutations in the glutamate transporter EAT-4, a homolog of the human VGLUTs, might also affect aggregation behavior. Disruption of VGLUT(eat-4) in an aggregating npr-1 background significantly decreased aggregation behavior compared to npr-1 mutants (Fig. 4A, C). To test if glutamate functions specifically in ADL and ASH neurons for aggregation behavior, we expressed EAT-4 using the nhr-79 promoter, which restored aggregation behavior of npr-1(ad609); eat-4(ky5) double mutants to the same level as npr-1 mutants (Fig. 4A, C). Like NRX-1, we found that expression of EAT-4 is needed in both ADL and ASH neurons, while expression in either neuron alone was insufficient to restore aggregation behavior of npr-1(ad609); eat-4(ky5) mutants (Fig. 4A). We confirmed expression of all EAT-4 transgenes with visualization of GFP (an original sra-6p::eat-4 transgene with low levels of expression did not impact behavior on its own or in combination with srv-3p::eat-4)(Supplementary Fig. 3).
Fig. 4. Aggregation behavior depends on glutamate signaling from ADL and ASH neurons.
A Graph showing number of aggregating animals in npr-1(ad609) compared to npr-1;eat-4(ky5) mutants and number of aggregating animals in npr-1(ad609);eat-4(ky5) mutants with EAT-4 driven by srv-3, sra-6, nhr-79, and srv-3/sra-6 combined promoters. B Graph showing number of aggregating animals in npr-1(ad609), npr-1(ad609);nrx-1(wy778), npr-1(ad609);eat-4(ky5), npr-1(ad609);nrx-1(wy778);eat-4(ky5) mutants. Graph also includes npr-1(ad609);nrx-1(wy778);eat-4(ky5) mutants with EAT-4 driven under the nhr-79 promoter, npr-1(ad609);nrx-1(wy778);eat-4(ky5) mutants with NRX-1(α) driven under the nhr-79 promoter, and solitary controls. C Representative images of aggregation behavior in npr-1(ad609);eat-4(ky5), npr-1(ad609);eat-4(ky5); nhr-79p::eat-4, npr-1(ad609);nrx-1(wy778);eat-4(ky5), and npr-1(ad609);nrx-1(wy778);eat-4(ky5); nhr-79p::eat-4 animals (Scale bar = 1 mm). D Graph showing number of aggregating worms in npr-1(ad609), npr-1(ad609);eat-4(ky5), npr-1(ad609);nlg-1(ok259), npr-1(ad609);nlg-1(ok259);eat-4(ky5), npr-1(ad609);glr-2(ok2342), npr-1(ad609);nlg-1(ok259);glr-2(ok2342) mutants, and solitary controls. E Graph showing number of aggregating animals in npr-1(ad609), npr-1(ad609);nrx-1(wy778), npr-1(ad609);glr-1(n2461), npr-1(ad609);glr-2(ok2342), npr-1(ad609);avr-15(ad1051), npr-1(ad609);nrx-1(wy778);glr-1(n2461), npr-1(ad609);nrx-1(wy778); glr-2(ok2342), and npr-1(ad609);nrx-1(wy778);avr-15(ad1051) mutants. Data for npr-1 and npr-1;eat-4 are plotted in 4 A, 4B, and 4D. Data for npr-1;nrx-1 are plotted in 4B and 4E. Data for solitary controls are plotted in 4B, 4D, and 4E. The number of biological replicates (n) are displayed in the figure, bars show the mean number of aggregating C. elegans, and error bars indicate SEM. One-way ANOVA with Tukey’s post-hoc test was used for comparisons, exact p-values are shown on graphs (red indicates significance, black indicates non-significance). Source data are provided in the Source Data file.
The shared role of nrx-1 and eat-4 in ADL and ASH sensory neurons suggested that nrx-1 and eat-4 function together in these neurons to regulate aggregation behavior. However, we found that npr-1(ad609); nrx-1(wy778); eat-4(ky5) triple mutants further reduced aggregation behavior compared to either npr-1(ad609); nrx-1(wy778) or npr-1(ad609); eat-4(ky5) double mutants (Fig. 4B). This result indicates that glutamate and nrx-1 function in parallel, non-epistatic, pathways to affect aggregation behavior. Since we found that nlg-1 and nrx-1 also function independently, we asked if eat-4 and nlg-1 may function through the same molecular pathway. We found that when glutamate signaling and nlg-1 are perturbed in an npr-1(ad609);eat-4(ky5);nlg-1(ok259) triple mutant, aggregation behavior is lowered to a level similar to that of solitary controls and is significantly decreased compared to npr-1(ad609);eat-4(ky5) double mutants, but not npr-1(ad609);nlg-1(ok259)(Fig. 4D). Therefore, we conclude that (1) multiple molecular signaling components contribute to aggregation behavior, (2) nrx-1 functions in genetically distinct or parallel pathways to nlg-1 and eat-4, (3) while nlg-1 and eat-4 function in partially overlapping pathways. Remarkably, we found that the loss of each component individually reduced aggregation behavior significantly, but the combination of any two reduced aggregation behavior further towards solitary behavior. This demonstrates that aggregation behavior is regulated by heterogenous genetic pathways which together tune behavior between solitary and social feeding.
To further explore the interplay of nrx-1 and glutamate in ADL and ASH sensory neurons, we expressed EAT-4 or NRX-1(α) in these neurons in npr-1(ad609); nrx-1(wy778); eat-4(ky5) triple mutants using the nhr-79 promoter or combining the srv-3 and sra-6 promoters. Expression of EAT-4 in ADL and ASH with nhr-79p or combination of srv-3 and sra-6 in the npr-1(ad609); nrx-1(wy778); eat-4(ky5) triple mutants restored aggregation behavior to the level of npr-1(ad609);nrx-1(ad609) (Fig. 4B), providing further evidence that the role of glutamate in aggregation behavior is independent of nrx-1 despite functioning in the same sensory neurons. Expression of NRX-1(α) in ADL and ASH in npr-1(ad609); nrx-1(wy778); eat-4(ky5) triple mutants alone did not alter aggregation behavior (Fig. 4B), suggesting a dependence of NRX-1 signaling on functional glutamate neurotransmission. Together with the additive behavioral findings for nrx-1 and eat-4, this result implies a dual role for nrx-1 in aggregation behavior — one dependent on glutamate and one independent of glutamate that may occur in non-glutamate neurons.
Multiple glutamate receptors regulate aggregation behavior
Our results thus far have focused on the pre-synaptic mechanisms regulating aggregation behavior. To explore how aggregate feeding is controlled on the post-synaptic side, we next tested the role of glutamate receptors. We analyzed mutants in glutamate receptors including GRIA1,2,3(glr-1), GRIA2(glr-2), GRIN2B(nmr-2), GRM3(mgl-1), and GLRA2,GABRA3(avr-15). We found that glr-1(n2461), glr-2(ok2342), and avr-15(ad1051), but not mgl-1(tm1811) or nmr-2(ok3324), reduced aggregation behavior in the npr-1(ad609) background (Fig. 4E and Supplemental Fig. 3B). Notably, while glr-1 and glr-2 are excitatory AMPA-like receptors78, avr-15 is an inhibitory glutamate-gated chloride channel79 (whereas GLRA2,GABRA3 are gated by glycine and GABA, respectively) suggesting that a complex balance of glutamate signaling is involved in aggregation behavior.
We next wondered whether nrx-1 or nlg-1 function at the level of post-synaptic glutamate receptors similar to their role at other synapses80,81. To probe this, we generated triple mutant combinations for npr-1, nrx-1, and each glutamate receptor, as well as those with nlg-1 and glr-2. We found nrx-1(wy778) with each glutamate receptor mutation further reduced aggregation behavior compared with nrx-1 or each respective receptor mutant alone in an aggregating background (Fig. 4E). These data suggest that nrx-1 acts additively with the receptors, where loss of a single receptor reduces aggregation behavior, and loss of nrx-1 may lower functionality of the other two remaining receptors or act through independent mechanisms as indicated by the results with loss of glutamate itself. Surprisingly, we found that the reduced aggregation phenotype of nlg-1 is dependent on glr-2, as the nlg-1; glr-2 combination increased aggregation compared to nlg-1 alone, to levels similar to glr-2 alone (Fig. 4D). Therefore nlg-1 and nrx-1 have distinct genetic interactions with glr-2.
Glutamate release is higher in aggregating C. elegans
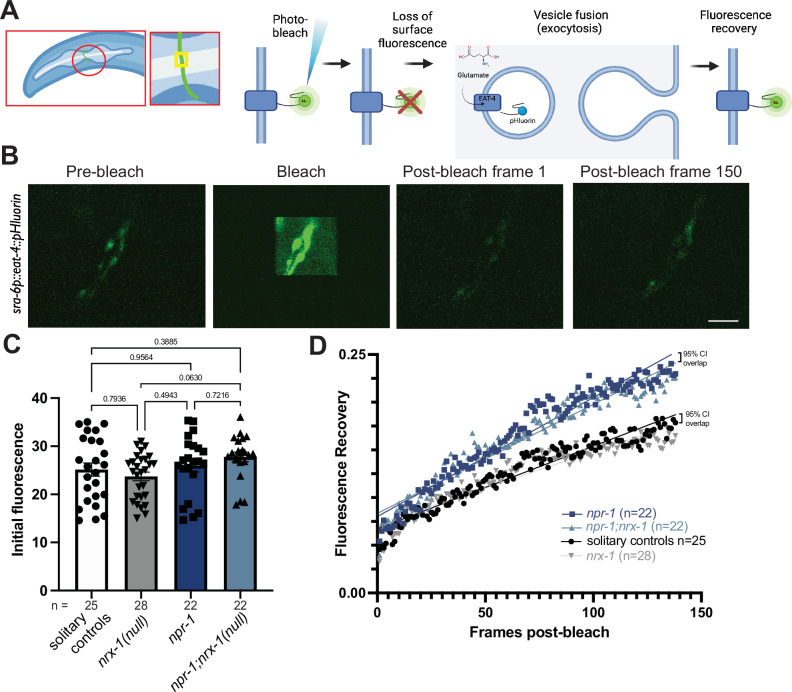
To determine how glutamate signaling contributes to solitary versus aggregate feeding behavior, we used fluorescence recovery after photobleaching (FRAP) of the pH-sensitive GFP-tagged vesicular glutamate transporter, EAT-4::pHluorin82, which measures exocytosis and the amount of membrane fusion as a proxy for glutamate vesicle release. To gain temporal information of synaptic release, we photobleached fluorescence at ASH pre-synaptic sites and recorded its recovery for two minutes post-bleach (Fig. 5A, B and Supplemental Fig. 4). Recovery was normalized to pre-bleach fluorescence as the maximum (1) and post-bleach fluorescence as the minimum (0)83. The slope of the recovery allowed us to compare rates of ASH glutamate release between genotypes. Initial EAT-4::pHluorin levels in ASH were not different between genotypes (Fig. 5C and Supplemental Fig. 4). We found that ASH neurons had faster spontaneous glutamate release in aggregating npr-1(ad609) animals compared to solitary controls as exemplified by greater overall and faster fluorescence recovery (Fig. 5D and Supplemental Fig. 4). We next tested whether NRX-1 had a role in the increased rate of glutamate release and found that ASH neurons in npr-1(ad609);nrx-1(wy778) mutants also had faster glutamate release dynamics relative to solitary controls (Fig. 5E and Supplemental Fig. 4). nrx-1(wy778) mutants in a solitary background had similar ASH glutamate release dynamics to that of solitary controls (Fig. 5D and Supplemental Fig. 4). Notably, we found that glutamate release was higher in strains generated in an aggregating background (npr-1(ad609) or npr-1(ad609);nrx-1(wy778)) strains compared to strains generated in the solitary background (N2 and nrx-1(wy778)). Therefore, while aggregation behavior is affected by nrx-1, changes in glutamate dynamics occur independent of nrx-1, providing further evidence that nrx-1 and glutamate signaling regulate aggregate feeding through distinct mechanisms.
Fig. 5. Glutamate release is faster in aggregating C. elegans, independent of NRX-1.
A Schematic of sra-6p::eat-4::pHluorin experiment, including a schematic of a small neuron section bleached and EAT-4::pHluorin photobleaching and recovery process (Created in BioRender. Hart, M. (2022) BioRender.com/q97a726). B Representative images of ASH neurons prior to bleaching (pre-bleach), during bleach, immediately following bleach, and after a recovery period of two minutes (Scale bar = 5 μm). C Graph showing initial fluorescence values taken from first 10 pre-bleach frames of FRAP experiments. Bars show the mean initial fluorescence, error bars indicate SEM. One-way ANOVA with Tukey’s post-hoc test was used for comparisons, exact p-values are shown on graphs (red indicates significance, black indicates non-significance). D Graph of post-bleach recovery as a fraction of initial fluorescence by post-bleach frame up to frame 138 (120 s, frame taken every 0.87 s). A straight line is fit with a non-linear regression. Points on the graph are the genotype mean of all replicates at each post-bleach frame. The null hypothesis (slope same for all data sets) was rejected with p < 0.0001. 95% confidence intervals of the slope are not significantly different between npr-1 vs. npr-1;nrx-1 ([0.001175 to 0.001322], [0.001082 to 0.001206]) or solitary control vs. nrx-1 ([0.0008085 to 0.000926], [0.0007136 to 0.0008126]), represented on the graph as 95% confidence intervals (CI) overlap. 95% confidence intervals of the slope are significantly different between solitary control vs. npr-1 ([0.0008085 to 0.000926], [0.001175 to 0.001322]), solitary control vs. npr-1;nrx-1 ([0.0008085 to 0.000926], [0.001082 to 0.001206]), nrx-1 vs. npr-1 ([0.0007136 to 0.0008126], [0.001175 to 0.001322]), and nrx-1 vs. npr-1;nrx-1 ([0.0007136 to 0.0008126], [0.001082 to 0.001206]). The number of biological replicates (n) are displayed in the figure and performed on at least 3 separate days. Source data are provided in the Source Data file.
ASH pre-synaptic puncta are increased in aggregating C. elegans dependent on NRX-1
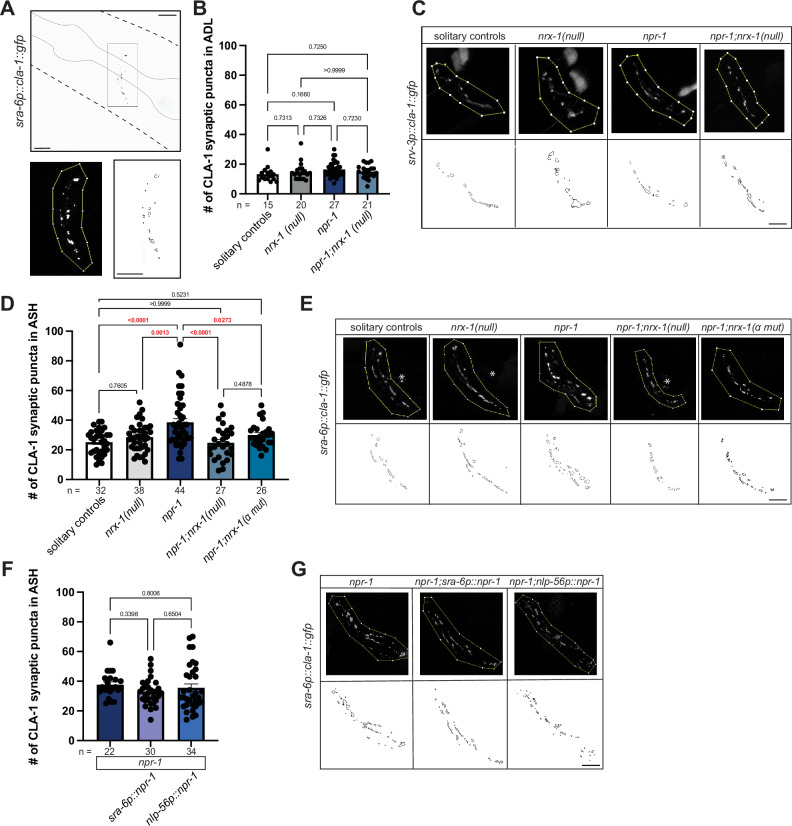
To investigate whether nrx-1 alters aggregation behavior through a role in synaptic structure or architecture, we analyzed the pre-synaptic morphology of the ADL and ASH neurons using enhanced resolution confocal microscopy (Leica Lightning Deconvolution analysis). Specifically, we used a GFP-tagged pre-synaptic marker clarinet CLA-1 (a bassoon ortholog) and quantified CLA-1::GFP puncta in the neurites of ADL or ASH sensory neurons using the srv-3 and sra-6 promoters via an unbiased particle analysis (see methods for details, Fig. 6A)84. We found no significant difference in ADL pre-synaptic puncta number between aggregating, solitary, or nrx-1 mutants (Fig. 6B, C). We next quantified pre-synaptic puncta in ASH neurons, and unlike ADL, we found that aggregating npr-1(ad609) mutants had a significant increase in the number of CLA-1::GFP puncta compared with solitary controls (Fig. 6D, E). Further, the number of ASH CLA-1::GFP puncta in a npr-1(ad609);nrx-1(wy778) double mutant was significantly lower than in npr-1 alone (Fig. 6D, E). These results indicate that aggregating animals have more ASH pre-synaptic puncta than solitary controls and that this increase is dependent on NRX-1. The impact of nrx-1(wy778) on CLA-1::GFP puncta in ASH was also context-dependent and only altered puncta number in the aggregating strain with no impact in the solitary control background.
Fig. 6. Higher number of ASH pre-synaptic puncta in aggregating C. elegans depends on nrx-1.
A Confocal micrograph of sra-6p::cla-1::gfp construct with pharynx outlined included as method schematic. Region of interests (ROI) in which counts were performed and puncta outlines generated by FIJI. Soma and projections outside of the nerve ring were not included in ROIs (Scale bar = 10 μm). Graph showing number (B) and representative images (Scale bars = 10 μm) (C) of srv-3p::cla-1::gfp puncta in ADL in solitary controls, nrx-1(wy778), npr-1(ad609), and npr-1(ad609);nrx-1(wy778) mutants. Graph showing number (D) and representative images (Scale bar = 10 μm) (E) of sra-6p::cla-1::gfp puncta in ASH in solitary controls, nrx-1(wy778), npr-1(ad609), npr-1(ad609);nrx-1(wy778) mutants, and npr-1(ad609);nrx-1(gk246237) mutants. White asterisks indicate ASH cell body GFP expression and not synaptic puncta. Graph showing number (F) and representative images (Scale bar = 10 μm) (G) of sra-6p::cla-1::gfp puncta in ASH in npr-1(ad609), npr-1(ad609); sra-6p::npr-1 (ASH), and npr-1(ad609); nlp-56p::npr-1 (RMG). The number of biological replicates (n) are displayed in the figure, bars show the mean number of puncta, and error bars indicate SEM. One-way ANOVA with Tukey’s post-hoc test was used for comparisons, exact p-values are shown on graphs (red indicates significance, black indicates non-significance). Source data are provided in the Source Data file.
To determine if a specific isoform of NRX-1 is responsible for regulating the higher number of pre-synaptic puncta number in aggregating strains, we tested an α-isoform specific mutant allele, nrx-1(gk24623). We found that npr-1(ad609);nrx-1(gk24623) mutants had fewer ASH CLA-1::GFP puncta relative to npr-1(ad609) aggregating animals, similar to what we observed in npr-1(ad609) animals carrying the allele of nrx-1 that knocks out both α and γ isoforms (Fig. 6D, E). This result suggests that pre-synaptic architecture, like aggregation behavior, is selectively mediated by NRX-1(α). The difference in ASH presynaptic architecture between solitary controls and aggregating npr-1(ad609) animals could be due to npr-1 functioning broadly to alter the activity of the circuit or in a cell-autonomous manner. Expression of NPR-1 in ASH (sra-6p::npr-1) or RMG (nlp-56p::npr-1) neurons in npr-1(ad609) animals did not impact ASH CLA-1::GFP puncta (Fig. 6F, G), which suggests that npr-1 is required in other neurons or more broadly in the circuit to impact ASH synaptic numbers. We did not find differences in the total area of ADL or ASH pre-synaptic puncta in any genotypes compared to solitary controls or aggregating animals (Supplemental Fig. 5). The nrx-1 dependent changes in ASH puncta suggest that nrx-1 mutations prevent the conversion of solitary to more social behavior through a reduction in glutamate input to other neurons (i.e., ADL, RMG), which lowers circuit activity and aggregation behavior (Fig. 1F).
Discussion
In this study, we identify the mechanisms by which neurexin isoforms regulate synapses, neuronal signaling, and social feeding behavior. In doing so, we identify multiple signaling pathways that modify the synaptic properties of sensory neurons and tune feeding behavior from social to solitary. We find that neurexin (nrx-1) and neuroligin (nlg-1) have an additive impact on behavior, which is surprising as neurexins and neuroligins are thought to be localized to pre- and post-synapses, respectively, and canonically bind each other. However, our results are consistent with those observed in Drosophila where dnl2;dnrx Δ83 double mutants show significant neuromuscular junction morphologic defects and lethality compared to either mutant alone85. Despite the ubiquitous expression of NRX-1 isoforms, we identify a specific role for the alpha (α) isoform of nrx-1 at ASH and ADL synapses within the well-studied sensory integration circuit controlling aggregation behavior. In contrast, our results with NLG-1 suggest a specific combinatorial expression of NLG-1 is needed for behavior with changes due to mis-expression of the nlg-1 transgene in all neurons, specific levels or timing of expression, or potential roles for nlg-1 in non-neuronal cells, aligning with known post-synaptic functions86. The requirement of NLG-1 in AIY neurons, which are not implicated in the aggregation behavioral circuit, suggests that additional neurons and circuits also influence aggregation, which has not been previously reported.
We find that glutamate signaling from ASH and ADL neurons also acts independently of NRX-1 to control aggregate feeding behavior. Further, nlg-1, but not nrx-1, interacts with with the glutamate receptor glr-2 to modify aggregate feeding, which is similar to another interaction with nrx-1 we recently observed87. In addition, we found that glr-1, glr-2, and avr-15 play a critical role in aggregation behavior. glr-1 and glr-2 are expressed in command interneurons (AVA, AVE, AVD) and AIB, which control backward locomotion and high-angle turning, while avr-15 is expressed in AIA, which inhibits turning73,88,89. We suggest that glutamate release from ADL and ASH neurons acts on these glutamate receptors to maintain animal position within the social aggregate. Moreover, mutations in the genes reported here (nrx-1, eat-4, glr-1, glr-2, and avr-15) result in intermediate reductions in aggregation behavior, whereas loss of sensory transduction channel subunits (tax-2, tax-4, osm-9, and ocr-2) and trafficking machinery (odr-4 and odr-8) abolish aggregate feeding. Together, this work expands the molecules and circuits regulating aggregation behavior and suggests that social feeding is tuned through modular usage of smaller distinct circuits and behaviors, which may explain the variation observed across wild isolates20.
Gap junctions and neuropeptide signaling are crucial for C. elegans aggregate feeding behavior, but chemical synaptic signaling has not been extensively characterized. We find that nrx-1 is needed for aggregation behavior induced by activation of neurons in the circuit and that aggregating animals have both increased numbers of pre-synaptic puncta and faster rates of glutamate release from ASH neurons compared to their solitary counterparts. We hypothesize that the ADL neurons act as an amplifier for ASH signaling, based on the bi-directional chemical synapses between ADL and ASH. Our finding that nrx-1 modifies pre-synaptic puncta number in ASH matches the general role of neurexins in the development and maintenance of pre-synaptic structures. While neurexins are broadly implicated in chemical synaptic properties and social behavior, rarely has a single isoform (NRX-1(α)), been simultaneously tied to both circuit mechanisms and behavior. Collectively, our studies identify a role for NRX-1(α) in the pre-synaptic architecture of specific synapses (from ASH), separately from glutamate release dynamics, in tuning aggregate feeding behavior. It will be important to analyze the contributions and interactions of the chemical, neuropeptide, and electrical synapses of ASH and ADL with RMG and other interneurons. In addition, it will be crucial to understand if the additional ASH pre-synaptic sites we observe in social feeding animals compared to solitary controls have the same or different post-synaptic partners, rates of release, and if nrx-1 impacts their formation, maintenance, or pruning.
The number of pre-synaptic release sites and the rate of release represent distinct, but related, mechanisms for regulating chemical synaptic signaling. We propose a tuning model in which glutamate signaling from ASH/ADL positively correlates with the level of aggregate feeding. High signaling via ASH in social animals can be lowered either via (1) a reduction of ASH synaptic puncta or (2) a decrease in the rate of glutamate release, which can be further reduced by these two mechanisms acting together. Loss of nrx-1, nlg-1, eat-4, glr-1, glr-2, or avr-15 alone led to intermediate levels of aggregation behavior, but disruption of more than one pathway produces more solitary-like behavior through distinct circuit functions. We suggest that ASH glutamate signaling acts as a dial for aggregation behavior, with the increased glutamate neurotransmission (via release rate or sites) driving aggregation behavior and vice-versa. An extension of this model is that it is not glutamate signaling specifically, but rather the overall activity level between sensory neurons and RMG interneurons that controls aggregation behavior. This model would explain how multiple sensory neurons (URX, ASK, ADL, ASH), modalities (oxygen, pheromones, aversive stimuli), and signaling components (NPR-1 inhibition, gap junctions, neuropeptides, release sites, exocytosis) function in the same behavior21–28,30,33. Experiments combining activation or silencing of specific neurons and manipulation of specific modalities, in parallel with calcium imaging of the associated neurons’ activity will be able to further build on this model.
Aggregate feeding involves the interaction of individual C. elegans with each other, matching a definition of social behavior2. However, since the first publication of aggregate feeding20, there has been a general skepticism about whether this behavior is social90,91. Studies have shown that oxygen is an important cue in maintaining these aggregates23–27, implying that this behavior might be driven by environmental cues. In contrast, other studies showed that pheromones and touch are also important for aggregation behavior21, suggesting a role for inter-individual interactions in this behavior. Moreover, C. elegans can participate in other behaviors that are canonically social. While C. elegans exist primarily as self-reproducing competent hermaphrodites, male C. elegans also exist. These males are attracted to hermaphrodites through pheromone and ascaroside signaling, prompting mate search and mating92,93—clear examples of social behaviors. In addition, adult hermaphrodites leave the bacterial food lawn in the presence of their larval progeny, likely to increase food availability to their developing offspring94. This potential parental response was shown to depend on nematocin, the C. elegans ortholog of the “social hormone” oxytocin94 and nlg-195. Despite these examples and the involvement of both environmental and social cues in aggregate feeding, the social drive to feed in groups remains controversial. Collectively, our study adds additional evidence of social behavior in nematodes.
Variants in human neurexins (NRXN1) and neuroligins (NLGN3) are associated with increased risk for autism (Supplementary Table 1), a neurodevelopmental condition characterized by altered social and communication behaviors, repetitive behaviors, and sensory processing/sensitivity48,49. Importantly, through our mechanistic exploration of the social feeding circuit and behavior, we uncovered novel roles for additional conserved autism-associated genes, including GRIA1,2,3(glr-1)96–98, GRIA2(glr-2)96–98, and GLRA2,GABRA3(avr-15)99–101 (Supplementary Table 1)102,103. Experiments in rodent models also implicate a role for many of these genes in social behaviors104–107. The involvement of these multiple conserved autism-associated genes, which affect social behaviors in mice, rats, and humans, may lend support for aggregate feeding as a simple form of social behavior. Variation in these genes in humans includes many genetic changes, often in the heterozygous state, whereas here, and in other model organisms, the genes are often studied in the homozygous loss of function context. Importantly, the functional study of conserved autism-associated genes we present does not imply a C. elegans model of autism or autism behaviors, which are human-specific. Rather, we leverage this pioneering genetic organism, its compact nervous system, and the evolutionarily important social feeding behavior to understand the circuit and molecular mechanisms by which behaviors are modified by conserved genes. These detailed mechanistic discoveries provide a framework to explore the molecular functions of autism-associated genes in social behaviors in more complex model systems and have implications for autism and neurodiverse communities.
Taken together, this work identifies multiple mechanisms that tune feeding behavior between social and solitary states. We define independent genetic pathways involving many conserved autism-associated genes and chemical signaling mechanisms, including glutamate release dynamics and pre-synaptic structural plasticity, that cooperate to determine foraging strategy. Our work suggests conserved roles for autism-associated genes in driving group interactions between animals across species and provides mechanistic insight into how these genes control neuronal and circuit signaling to modulate behavior. Lastly, our identification of conserved genes with known roles in social behavior suggests a social origin for aggregate feeding in C. elegans108.
Methods
C. elegans strain maintenance
All strains were maintained on Nematode Growth Medium (NGM) plates and seeded with Escherichia coli OP50 bacteria as a food source109. Strains were maintained on food by chunking and kept at ~ 22-23 °C. All strains and mutant alleles included are listed in Supplementary Table 2 in Figure order. Solitary controls consist of either N2 strain or transgenic strains expressing reporter constructs in the N2 background and/or him-8(e1489) mutation indicated in Supplementary Table 2, and aggregate feeding controls consist of DA609 with npr-1(ad609) or npr-1(ad609) with added reporters and/or him-8(e1489) mutation as indicated in Supplementary Table 2. The presence of the endogenous unc-119(ed3) mutant allele, which was used in the generation of TV13570(nrx-1(wy778)), was not confirmed in our strains. The presence of him-8(e1489) and otIs525[lim-6int4p::gfp], used in genetic crosses or as an anatomical landmark in indicated Figures, did not impact solitary or aggregate feeding behavior (Supplemental Fig. 1A). All experiments were performed on hermaphrodites, picked during larval stage 4 (L4), and confirmed as day 1 adults.
Cloning and constructs
All plasmids are listed in the Supplementary Table 3, along with primer sequences for each promoter. All plasmids were made by subcloning promoters or cDNA inserts into plasmids by Epoch Life Science Inc., as described below. Plasmids for nrx-1(α) transgenes were generated by subcloning each promoter to replace the ric-19 promoter in pMPH34 (ric-19::sfGFP::nrx-1(α)), which includes a Superfolder GFP tag fused to the N-terminus of the long α isoform of nrx-1. Plasmids for nlg-1 transgenes were generated by subcloning super folder GFP (primers: fwd—CTGCCCAGGATACGATCCATGAGCAAAGGAGAAGAAC; rev—AGATCCAGATCCGAGCTCTTTGTAGAGCTCATCC) to replace the N-terminal GFP11 fragment tag on nlg-1 in plasmid pMVC3110, then the ric-19 promoter was subcloned ahead of the artificial intron and start site of the resulting plasmid (primers: fwd—GCGCCTCTAGAGGATCCcattaaagagtgtgctcca; rev—TTTGGCCAATCCCGGgttcaaagtgaagagc). The plasmid pMPH45 includes the ric-19 promoter and a superfolder GFP tag fused to the N-terminus of nlg-1 (ric-19::sfGFP::nlg-1), which was subcloned with indicated promoters to replace the ric-19 promoter. Plasmids for eat-4 transgenes were generated by subcloning indicated promoters to replace the sre-1 promoter in pSM plasmid (sre-1p-eat-4::sl2::gfp). To generate plasmids for cla-1 transgenes, promoters indicated were subcloned to replace the lim-6 int4 promoter in pMPH21 (lim-6 int4::gfp::cla-1)77. To generate plasmids for npr-1 expression transgenes, npr-1 cDNA (primers: fwd—ACATAGGCCAAATGGAAGTTGAAAATTTTA; rev—AAAAAAAGATCATAAAAACTATTTCAGCAA) was subcloned to replace nlg-1 cDNA in plasmids sra-6p::nlg-1 cDNA::sl2::mCherry and nlp-56p::nlg-1 cDNA::sl2::mCherry to generate sra-6p::npr-1 cDNA::sl2::mCherry and nlp-56p::npr-1 cDNA::sl2::mCherry.
Transgenic animals
All plasmids and co-injection markers are indicated in Supplemental Table 2 and were injected to generate extrachromosomal arrays at 20 ng µl−1 unless otherwise indicated in Supplemental Table 2. For extrachromosomal transgenes, at least 2 independent transgenic lines were generated and analyzed to confirm expression levels and transmittance, after which a single line was selected for comprehensive analysis based on expression levels and moderate to high transmittance111.
Aggregate feeding behavior assay
Standard 6-well plates were filled with 6 mL of NGM. 75 μL of OP50 bacteria culture (OD600 = ~ 0.7) was added to the center of the well to form a circular food lawn. Plates were left at room temperature to dry. The day after seeding OP50, 60 L4 hermaphrodites of each genotype were moved to a clean plate then 50 animals were transferred to the aggregation behavior assay set-up. If transgenic strains were used, transgene-positive animals were identified by the presence of a fluorescent co-injection marker (listed in Supplemental Table 2). C. elegans were transferred to the center of the food lawn on each well. The experimenter was blinded to all genotypes at the time of loading. 10x Tween was put on the lid of the 6-well plate to prevent condensation from forming. Loaded 6-well plates were placed in the WormWatcher set up developed by Tau Scientific and the Fang-Yen Lab, and monitored for at least 15 h. Images were taken every 5 seconds for 1 min per hour.
To quantify aggregation behavior, the number of aggregating C. elegans was manually counted from blinded images, such that a C. elegans in contact with two or more other C. elegans was considered aggregating. In cases where the number of aggregating animals could not be clearly counted, the number of single animals was counted and subtracted from 50 to obtain a count of aggregating C. elegans. Data shown is from hour 15 after experimental set-up, therefore representing day 1 adult animals.
Confocal microscopy
Transgenic expression
For visualization of transgenic constructs, 5% agar was used to create a thin pad on a microscope slide. 5 μl of the paralytic sodium azide was pipetted on the agar pad. Adult animals expressing the co-injection markers were identified on the fluorescent microscope and moved to the agar pad and a coverslip was placed on top. C. elegans were imaged at 63X on a Leica SP8 point scanning Confocal Microscope, with z-stack images taken at 0.6 μm spanning expression. Images were processed in Adobe Photoshop (v21.1.1) to alter orientation and invert color. Figures were made in Adobe Illustrator (v24.1).
CLA-1 Puncta quantification
Relevant mutant strains were crossed with srv-3p::cla-1::sfGFP or sra-6p::cla-1::sfGFP in him-5 background. To visualize CLA-1::GFP puncta in ADL or ASH, microscope slides were prepared as described above. C. elegans were imaged at 63X, with an additional zoom of 2.5 X and a Z-stack size of 0.6μm. Following imaging, Lightning Deconvolution was applied to the images to reduce noise. The number of puncta was examined in FIJI using Particle Analysis (version 2.9.0/1.53t). Image z-stacks were combined to create a single image using a projection of max intensity. Images were auto-thresholded with a minimum of 50 and a maximum of 255. The region of interest for particle quantification was restricted to the expression of cla-1::gfp in the nerve ring and was drawn to exclude any background. If background fluorescence, resulting from the lin-44::gfp co-injection marker in these transgenic strains, was too high to distinguish puncta, images were not quantified. Particle analysis was performed with an area cut-off of 0.03 μm2 to remove small background particles, and bare outlines were generated.
Fluorescent recovery after photobleaching (FRAP)
For FRAP imaging, L4 C. elegans were picked 24 hours before imaging to appropriately stage the animals. The next day, no more than three C. elegans were placed on each microscope slide and paralyzed with 5 mM levamisole. Using the microlab FRAP module on the Leica SP8 Confocal Microscope at 63X with a zoom of 4.5, a 10 μm X 10 μm bleach area was defined, centered on the brightest part of the neurite. A recording session was set such that 10 frames were taken pre-bleach, 10 frames were taken with 50% laser power applied to the sample, and 138 frames were taken post-bleach with an interframe interval of 0.87 s for a total post-bleach recording of two minutes. During the two-minute recovery, animals were monitored to ensure they stayed in the frame. If drift was seen, minor manual adjustments to the z-plane were made to hold them in position. If drift was significant or if the animal moved, the recording was stopped and not included in our analysis.
To quantify the fluorescence recovery, all traces for each genotype were analyzed using the Stowers Institute Jay Plugins in FIJI (version 2.9.0/1.53t)112. The bleach region was set, and fluorescence at each frame was plotted. Graphs were then normalized with the maximum fluorescence set at 1 and the minimum set at 0.
Statistics and reproducibility
All statistical analyses were performed, and all data were plotted using GraphPad Prism 9.
For behavioral experiments, the hour 15 counts of aggregating animals were plotted for each genotype. Each data point represents an individual well of a 6-well plate. Within figures and corresponding supplemental figures in which the same genotype(s) were used in multiple plots, data were replotted (Fig. 2 and Supplemental Fig. 1, Fig. 3 and Supplementary Fig. 2 and Fig. 4 and Supplemental Fig. 3). Data was not replotted between unrelated figures and supplemental figures. At least three biological replicates were performed on at least three separate days per genotype, except where noted in figure legends. Plots include the standard error of the mean (SEM). To compare aggregation behavior levels across genotypes, a one-way ANOVA was performed with a Tukey’s Post-Hoc test applied. p-values are plotted on each graph. For graphs in which only two genotypes are shown (Fig. 1 and Supplemental Fig. 1), a t test was used.
For CLA-1::GFP puncta quantification, the number of puncta from each individual image was plotted with SEM and compared between genotypes using a one-way ANOVA with Tukey’s post-hoc test. Imaging sessions were performed on at least three separate days.
For FRAP experiments, the average normalized fluorescence value was plotted in GraphPad Prism 9 by frame post-bleach for each strain starting at frame 21 (frame 0 post-bleach) and ending at 158 (frame 138 post-bleach) with SEM. Fractional recovery data was fit to a straight line with non-linear regression. To determine whether the slopes of recovery plots differed between genotypes, the 95% confidence intervals of each slope were compared for overlap. Overlapping 95% confidence intervals were considered not significantly different. Non-overlapping 95% confidence intervals were considered to be significantly different. Experiments were performed on at least three separate days. Biorender.com was used to generate cartoons in Figs. 1A, 1B, 2A and 5A.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
The authors thank the Autism Spectrum Program of Excellence and the labs of Colin C. Conine, Chris Fang-Yen, David M. Raizen, John I. Murray, and Meera V. Sundaram for their feedback on this project, and specifically Anthony D. Fouad (Tau Scientific) and Vina Tikiyani for technical support. We also thank Theodore G. Drivas, Kevin C. Field, Brandon L. Bastien, Michael Rieger, Kathleen Quach, Marc V. Fuccillo, Aubrey Brumback, Jonathan T. Pierce, and members of the Hart and Chalasani labs for comments on the manuscript. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported in part by a Graduate research fellowship from NSF (M.H.C.), NIH 1R01MH096881, Nippert Foundation (S.H.C.), the Autism Spectrum Program of Excellence at the Perelman School of Medicine, and NIH 1R35GM146782 (M.P.H.).
Author contributions
M.H.C., S.H.C., and M.P.H. conceived and designed the study and experiments, and M.H.C., D.H., and K.Z. conducted all behavioral and microscopy experiments. M.P.H. designed and generated cloning and plasmids and transgenic strains. K.C.R. and D.H. generated transgenic animals and performed genetic studies. M.H.C. processed, analyzed, and interpreted all data. M.H.C. wrote the manuscript with assistance from M.P.H. and S.H.C., and all authors reviewed, revised, and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All data and materials are available upon request to the corresponding author. All data are available in the main text, or the supplementary materials, and source data are provided in the Source Data file. Source data are provided in this paper.
Competing interests
The authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53590-x.
References
- 1.Hamilton, W. D. The genetical evolution of social behaviour. I. J. Theor. Biol.7, 1–16 (1964). [DOI] [PubMed] [Google Scholar]
- 2.Chen, P. & Hong, W. Neural circuit mechanisms of social behavior. Neuron98, 16–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson, J. D., Arauco-Aliaga, R. P., Crow, S., Gordon, D. M. & Goldman, M. S. Effect of interactions between harvester ants on forager decisions. Front. Ecol. Evol. 4, 115 (2016). [DOI] [PMC free article] [PubMed]
- 4.Gordon D. M. Ant Encounters: Interaction Networks and Colony Behavior. In: Ant Encounters. Princeton (University Press, 2010).
- 5.Azorsa, F., Muscedere, M. L. & Traniello, J. F. A. Socioecology and evolutionary neurobiology of predatory ants. Front. Ecol. Evol. 9, 10.3389/fevo.2021.804200 (2022).
- 6.Peichel, C. L. Social behavior: How do fish find their shoal mate? Curr. Biol.14, R503–R504 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Pitcher, T. J., Magurran, A. E. & Winfield, I. J. Fish in larger shoals find food faster. Behav. Ecol. Sociobiol.10, 149–151 (1982). [Google Scholar]
- 8.Lührs, M. L., Dammhahn, M. & Kappeler, P. Strength in numbers: males in a carnivore grow bigger when they associate and hunt cooperatively. Behav. Ecol.24, 21–28 (2013). [Google Scholar]
- 9.Schaller G. B. The Serengeti Lion: A Study of Predator-Prey Relations. (University of Chicago Press, 1976).
- 10.Packer, C. & Ruttan, L. The evolution of cooperative hunting. Am. Nat.132, 159–198 (1988). [Google Scholar]
- 11.Lang, S. D. J. & Farine, D. R. A multidimensional framework for studying social predation strategies. Nat. Ecol. Evol.1, 1230–1239 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Creel, S. Cooperative hunting and group size: assumptions and currencies. Anim. Behav.54, 1319–1324 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Boyd, R. & Richerson, P. J. Large-scale cooperation in small-scale foraging societies. Evolut. Anthropol.: Issues, N., Rev.31, 175–198 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Kebede, A. & Internationals, O. Effect of social organization in wild animals on reproduction. J. Vet. Sci. Anim. Welf.3, 24–36 (2019). [Google Scholar]
- 15.Krause J., Ruxton G. Living in Groups. (Oxford University Press, 2002).
- 16.Blumstein, D. T., Hayes, L. D. & Pinter-Wollman, N. Social consequences of rapid environmental change. Trends Ecol. Evol.38, 337–345 (2023). [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues, A. M. M. Resource availability and adjustment of social behaviour influence patterns of inequality and productivity across societies. PeerJ6, e5488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman, T. & Candolin, U. Linking animal behavior to ecosystem change in disturbed environments. Front. Ecol. Evol. 10, 10.3389/fevo.2022.893453 (2022).
- 19.Evans J. C., Jones T. B., Morand-Ferron J. Dominance and the initiation of group feeding events: the modifying effect of sociality. Behav. Ecol. 29, 448–458 (2018).
- 20.de Bono, M. & Bargmann, C. I. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell94, 679–689 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Macosko, E. Z. et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature458, 1171–1175 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurent, P. et al. Decoding a neural circuit controlling global animal state in C. elegans. ELife4, e04241 (2015). [DOI] [PMC free article] [PubMed]
- 23.Cheung, B. H. H., Cohen, M., Rogers, C., Albayram, O. & de Bono, M. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr. Biol.15, 905–917 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Gray, J. M. et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature430, 317–322 (2004). [DOI] [PubMed] [Google Scholar]
- 25.McGrath, P. T. et al. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron61, 692–699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bretscher, A. J., Busch, K. E. & de Bono, M. A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA105, 8044–8049 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers, C., Persson, A., Cheung, B. & de Bono, M. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr. Biol.16, 649–659 (2006). [DOI] [PubMed] [Google Scholar]
- 28.de Bono, M., Tobin, D. M., Davis, M. W., Avery, L. & Bargmann, C. I. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature419, 899–903 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi, S., Chatzigeorgiou, M., Taylor, K. P., Schafer, W. R. & Kaplan, J. M. Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron78, 869–880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang, H. et al. Dissection of neuronal gap junction circuits that regulate social behavior in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA114, E1263–E1272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White, J. G., Southgate, E., Thomson, J. N. & Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci.314, 1–340 (1986). [DOI] [PubMed] [Google Scholar]
- 32.Witvliet, D. et al. Connectomes across development reveal principles of brain maturation. Nature596, 257–261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers, C. et al. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR−1. Nat. Neurosci.6, 1178–1185 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Individual Neurons—ASK. Accessed September 27, https://www.wormatlas.org/neurons/Individual%20Neurons/ASKframeset.html (2023).
- 35.Individual Neurons—ADL. Accessed September 27, https://www.wormatlas.org/neurons/Individual%20Neurons/ADLframeset.html (2023).
- 36.Individual Neurons—ASH. Accessed September 27, https://www.wormatlas.org/neurons/Individual%20Neurons/ASHframeset.html (2023).
- 37.Frye, R. E. Social skills deficits in autism spectrum disorder: Potential biological origins and progress in developing therapeutic agents. CNS Drugs32, 713–734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thye, M. D., Bednarz, H. M., Herringshaw, A. J., Sartin, E. B. & Kana, R. K. The impact of atypical sensory processing on social impairments in autism spectrum disorder. Dev. Cogn. Neurosci.29, 151–167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roley, S. S. et al. Sensory integration and praxis patterns in children with autism. Am. J. Occup. Ther. 69, 6901220010 (2015). [DOI] [PubMed]
- 40.Posar, A. & Visconti, P. Sensory abnormalities in children with autism spectrum disorder. J. de. Pediatr.94, 342–350 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Lang F. Encyclopedia of Molecular Mechanisms of Disease. (Springer Science & Business Media, 2009).
- 42.Bourgeron, T. Current knowledge on the genetics of autism and propositions for future research. C. R. Biol.339, 300–307 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet.51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satterstrom, F. K. et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of Autism. Cell180, 568–584 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Südhof, T. C. Synaptic neurexin complexes: A molecular code for the logic of neural circuits. Cell171, 745–769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.SFARI | SFARI Gene. SFARI. Published April 28, 2017. Accessed May 15, https://www.sfari.org/resource/sfari-gene/ (2023).
- 47.Wang, J. et al. Neurexin gene family variants as risk factors for autism spectrum disorder. Autism Res.11, 37–43 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Kim, H. G. et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet.82, 199–207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trobiani, L. et al. The neuroligins and the synaptic pathway in Autism Spectrum Disorder. Neurosci. Biobehav. Rev.119, 37–51 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Uchigashima, M., Cheung, A. & Futai, K. Neuroligin-3: A circuit-specific synapse organizer that shapes normal function and Autism Spectrum Disorder-associated dysfunction. Front. Mol. Neurosci.14, 749164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Etherton, M. R., Blaiss, C. A., Powell, C. M. & Südhof, T. C. Mouse neurexin-1α deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc. Natl. Acad. Sci. USA106, 17998–18003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tabuchi, K. & Südhof, T. C. Structure and evolution of neurexin genes: Insight into the mechanism of alternative splicing. Genomics79, 849–859 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Dachtler, J. et al. Deletion of α-neurexin II results in autism-related behaviors in mice. Transl. Psychiatry4, e484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Born, G. et al. Genetic targeting of NRXN2 in mice unveils role in excitatory cortical synapse function and social behaviors. Front. Synaptic Neurosci. 7, 3 (2015). [DOI] [PMC free article] [PubMed]
- 55.Chen, L. Y., Jiang, M., Zhang, B., Gokce, O. & Südhof, T. C. Conditional deletion of all neurexins defines diversity of essential synaptic organizer functions for neurexins. Neuron94, 611–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aoto, J., Földy, C., Ilcus, S. M. C., Tabuchi, K. & Südhof, T. C. Distinct circuit-dependent functions of presynaptic neurexin-3 at GABAergic and glutamatergic synapses. Nat. Neurosci.18, 997–1007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aoto, J., Martinelli, D. C., Malenka, R. C., Tabuchi, K. & Südhof, T. C. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell154, 75–88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Missler, M. et al. α-Neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature423, 939–948 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Anderson, G. R. et al. β-Neurexins control neural circuits by regulating synaptic endocannabinoid signaling. Cell162, 593–606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchigashima, M. et al. Specific Neuroligin3-αNeurexin1 signaling regulates GABAergic synaptic function in mouse hippocampus. Elife9, e59545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brockhaus, J. et al. α-Neurexins together with α2δ-1 auxiliary subunits regulate Ca2+ influx through Cav2.1 channels. J. Neurosci.38, 8277–8294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boxer, E. E. & Aoto, J. Neurexins and their ligands at inhibitory synapses. Front. Synaptic Neurosci. 14, 10.3389/fnsyn.2022.1087238 (2022). [DOI] [PMC free article] [PubMed]
- 63.Hu, Y. et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinforma.12, 357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Calahorro, F. Conserved and divergent processing of neuroligin and neurexin genes: from the nematode C. elegans to human. Invert. Neurosci.14, 79–90 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Tong, X. J. Hu, Z., Liu, Y., Anderson, D. & Kaplan, J. M. A network of autism linked genes stabilizes two pools of synaptic GABAA receptors. ELife4, e09648 (2015). [DOI] [PMC free article] [PubMed]
- 66.Tong, X. J. et al. Retrograde synaptic inhibition is mediated by α-Neurexin binding to the α2δ subunits of N-type calcium channels. Neuron95, 326–340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurshan, P. T. et al. γ-Neurexin and Frizzled mediate parallel synapse assembly pathways antagonized by receptor endocytosis. Neuron100, 150–166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Philbrook, A. et al. Neurexin directs partner-specific synaptic connectivity in C. elegans. Hobert O, ed. ELife7, e35692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maro, G. S. et al. MADD-4/Punctin and Neurexin Organize C. elegans GABAergic Postsynapses through Neuroligin. Neuron86, 1420–1432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu, Z. et al. Neurexin and neuroligin mediate retrograde synaptic inhibition in C. elegans. Science337, 980–984 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lázaro-Peña, M. I., Díaz-Balzac, C. A., Bülow, H. E. & Emmons, S. W. Synaptogenesis is Modulated by Heparan Sulfate in Caenorhabditis elegans. Genetics209, 195–208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hart, M. P. & Hobert, O. Neurexin controls plasticity of a mature, sexually dimorphic neuron. Nature553, 165–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor, S. R. et al. Molecular topography of an entire nervous system. Cell184, 4329–4347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pédelacq, J. D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol.24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Calahorro, F. & Ruiz-Rubio, M. Functional phenotypic rescue of caenorhabditis elegans neuroligin-deficient mutants by the human and rat NLGN1 genes. PLOS ONE7, e39277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Craig, A. M. & Kang, Y. Neurexin–neuroligin signaling in synapse development. Curr. Opin. Neurobiol.17, 43 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hart, M. P. Stress-induced neuron remodeling reveals differential interplay between neurexin and environmental factors in Caenorhabditis elegans. Genetics213, 1415–1430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rose, J. K., Kaun, K. R., Chen, S. H. & Rankin, C. H. GLR-1, a Non-NMDA glutamate receptor homolog, is critical for long-term memory in Caenorhabditis elegans. J. Neurosci.23, 9595–9599 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dent, J. A., Davis, M. W. & Avery, L. avr-15 encodes a chloride channel subunit that mediates inhibitory glutamatergic neurotransmission and ivermectin sensitivity in Caenorhabditis elegans. EMBO J.16, 5867–5879 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mondin, M. et al. Neurexin-neuroligin adhesions capture surface-diffusing AMPA receptors through PSD-95 scaffolds. J. Neurosci.31, 13500–13515 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Graf, E. R., Zhang, X., Jin, S. X., Linhoff, M. W. & Craig, A. M. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell119, 1013–1026 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ventimiglia, D. & Bargmann, C. I. Diverse modes of synaptic signaling, regulation, and plasticity distinguish two classes of C. elegans glutamatergic neurons. ELife6, e31234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hiroki, S. et al. Molecular encoding and synaptic decoding of context during salt chemotaxis in C. elegans. Nat. Commun.13, 2928 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xuan, Z. et al. Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. Davis GW, ed. ELife6, e29276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun, M. et al. Neuroligin 2 is required for synapse development and function at the drosophila neuromuscular junction. J. Neurosci.31, 687–699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hunter, J. W. et al. Neuroligin-deficient mutants of C. elegans have sensory processing deficits and are hypersensitive to oxidative stress and mercury toxicity. Dis. Model Mech.3, 366–376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bastien, B. L., Cowen, M. H. & Hart, M. P. Distinct neurexin isoforms cooperate to initiate and maintain foraging activity. Transl. Psychiatry13, 367 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wakabayashi, T., Kitagawa, I. & Shingai, R. Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neurosci. Res.50, 103–111 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Garrity, P. A., Goodman, M. B., Samuel, A. D. & Sengupta, P. Running hot and cold: behavioral strategies, neural circuits, and the molecular machinery for thermotaxis in C. elegans and Drosophila. Genes Dev.24, 2365–2382 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wade N. Can Social Behavior of Man Be Glimpsed in a Lowly Worm? The New York Times. https://www.nytimes.com/1998/09/08/science/can-social-behavior-of-man-be-glimpsed-in-a-lowly-worm.html. (2023).
- 91.Ardiel, E. L. & Rankin, C. H. C. elegans: social interactions in a “nonsocial” animal. Adv. Genet.68, 1–22 (2009). [DOI] [PubMed] [Google Scholar]
- 92.Liu, K. S. & Sternberg, P. W. Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron14, 79–89 (1995). [DOI] [PubMed] [Google Scholar]
- 93.Portman, D. S. Social and sexual behaviors in C. elegans: The first fifty years. J. Neurogenet.34, 389–394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scott, E. et al. An oxytocin-dependent social interaction between larvae and adult C. elegans. Sci. Rep.7, 10122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rawsthorne, H. et al. Neuroligin dependence of social behaviour in Caenorhabditis elegans provides a model to investigate an autism-associated gene. Hum. Mol. Genet. 29, 3546–3553 (2021). [DOI] [PMC free article] [PubMed]
- 96.Ismail, V. et al. Identification and functional evaluation of GRIA1 missense and truncation variants in individuals with ID: An emerging neurodevelopmental syndrome. Am. J. Hum. Genet.109, 1217–1241 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cai, Q. et al. Novel GRIA2 variant in a patient with atypical autism spectrum disorder and psychiatric symptoms: a case report. BMC Pediatr.22, 629 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moretto E., Passafaro M., Bassani S. in Neuronal and Synaptic Dysfunction in Autism Spectrum Disorder and Intellectual Disability. Sala, C., Verpelli, C., eds. 129–150 (Academic Press, 2016).
- 99.Fatemi, S. H., Reutiman, T. J., Folsom, T. D. & Thuras, P. D. GABAA receptor downregulation in brains of subjects with autism. J. Autism Dev. Disord.39, 223–230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niturad, C. E. et al. Rare GABRA3 variants are associated with epileptic seizures, encephalopathy and dysmorphic features. Brain140, 2879–2894 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Riley, J. D., Delahunty, C., Alsadah, A., Mazzola, S. & Astbury, C. Further evidence of GABRA4 and TOP3B as autism susceptibility genes. Eur. J. Med. Genet.63, 103876 (2020). [DOI] [PubMed] [Google Scholar]
- 102.McDiarmid, T. A. et al. Systematic phenomics analysis of autism-associated genes reveals parallel networks underlying reversible impairments in habituation. Proc. Natl. Acad. Sci. USA117, 656–667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wong, W. R. et al. Autism-associated missense genetic variants impact locomotion and neurodevelopment in Caenorhabditis elegans. Hum. Mol. Genet.28, 2271–2281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Adamczyk, A. et al. GluA3-deficiency in mice is associated with increased social and aggressive behavior and elevated dopamine in striatum. Behav. Brain Res.229, 265–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pilorge, M. et al. Genetic and functional analyses demonstrate a role for abnormal glycinergic signaling in autism. Mol. Psychiatry21, 936–945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Salpietro, V. et al. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat. Commun.10, 3094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.DeLorey, T. M., Sahbaie, P., Hashemi, E., Homanics, G. E. & Clark, J. D. Gabrb3 gene deficient mice exhibit impaired social and exploratory behaviors, deficits in non-selective attention and hypoplasia of cerebellar vermal lobules: a potential model of autism spectrum disorder. Behav. Brain Res.187, 207–220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crespi, B. J. The evolution of social behavior in microorganisms. Trends Ecol. Evol.16, 178–183 (2001). [DOI] [PubMed] [Google Scholar]
- 109.Brenner, S. The Genetics of CAENORHABDITIS ELEGANS. Genetics77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Feinberg, E. H. et al. GFP Reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron57, 353–363 (2008). [DOI] [PubMed] [Google Scholar]
- 111.Takeishi, A., Yeon, J., Harris, N., Yang, W. & Sengupta, P. Feeding state functionally reconfigures a sensory circuit to drive thermosensory behavioral plasticity. ELife9, e61167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stowers ImageJ Plugins. Accessed September 27, https://research.stowers.org/imagejplugins/. (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials are available upon request to the corresponding author. All data are available in the main text, or the supplementary materials, and source data are provided in the Source Data file. Source data are provided in this paper.