Abstract
We analyzed Burkholderia cepacia complex isolates recovered from 1,218 cystic fibrosis (CF) patients and 90 patients without CF. Although all B. cepacia complex species were found, some were rarely identified. The distribution of species differed between the CF and non-CF populations and appears to be changing over time among CF patients.
During the past several years, a series of comprehensive taxonomic studies have made it clear that bacteria previously identified as “Burkholderia cepacia” actually comprise a group of closely related species referred to collectively as the B. cepacia complex (4, 5, 17-22). These species are phenotypically nearly indistinguishable and are characterized by unusually high interspecies DNA-DNA hybridization values (6). Initially referred to as genomovars, each species has now received a formal binomial designation. The group includes B. cepacia (formerly genomovar I), B. multivorans (genomovar II), B. cenocepacia (genomovar III), B. stabilis (genomovar IV), B. vietnamiensis (genomovar V), B. dolosa (genomovar VI), B. ambifaria (genomovar VII), B. anthina (genomovar VIII), and B. pyrrocinia (genomovar IX).
The taxonomic studies that defined the B. cepacia complex typically included analyses of strains recovered from both clinical specimens and the natural environment. Among the clinical specimens were isolates recovered from respiratory secretions of persons with cystic fibrosis (CF), a common inherited disorder wherein “B. cepacia” has been long recognized as an important opportunistic pathogen. Although each species of the B. cepacia complex was thus identified among CF sputum isolates, these studies were not designed to address the relative frequency of each species in this or other patient populations.
Large-scale surveys intended to more specifically examine the distribution of B. cepacia complex species in CF patients have revealed a disproportionate representation of species, with B. cenocepacia and B. multivorans being significantly more common than the remaining species. We previously found that B. cenocepacia was recovered from half of 606 B. cepacia complex-infected CF patients in the United States; B. multivorans accounted for 38% of infected patients (11). A study of 447 Canadian CF patients found an even greater proportion (83%) infected with B. cenocepacia (16). Although patients infected with other B. cepacia complex species were identified in these surveys, both studies were performed prior to the identification of B. anthina and B. pyrrocinia as members of the B. cepacia complex. More recent studies from several additional countries have identified various proportions of CF patients infected with species other than B. cenocepacia and B. multivorans, but these studies have involved relatively small numbers of patients (1, 7-10, 13). Thus, the relative proportion of B. cepacia complex species causing human infection, particularly in CF, requires further elucidation.
We have now identified in excess of 1,200 CF patients and 90 patients without CF infected with B. cepacia complex species in the United States. In this report we describe the distribution of B. cepacia complex species in these patient groups and demonstrate changes in the relative proportions of species recovered from clinical specimens during the past several years.
Bacterial isolates and study population.
Since Spring 1997, the Burkholderia cepacia Research Laboratory and Repository (BcRLR) has served as a referral laboratory for identification of members of the B. cepacia complex and related species. During this time, we have confirmed the recovery of members of the B. cepacia complex (see below) from 1,218 persons with CF and 90 patients identified by the referring physician as not having CF. The CF patients received care in 133 cities in the United States, and all were registered in the Cystic Fibrosis Foundation Patient Registry (Bethesda, MD). This group includes the 606 B. cepacia complex-infected patients described previously (11). The 90 patients without CF received care in 44 medical centers in the United States. No single center accounted for more than six patients, indicating that the non-CF isolates were not derived from large nosocomial outbreaks. The non-CF isolates were recovered between 1996 and 2004 from a variety of clinical specimens, including tracheal aspirate (n = 21), sputum (n = 15), blood (n = 14), urine (n = 6), bronchial lavage (n = 4), sinus (n = 4), bone marrow (n = 2), and wound (n = 2), and one each from stool, pancreatic aspirate, cornea, pleural biopsy, and spinal fluid; for 17 isolates, the clinical specimen type was not specified. For patients (both CF and non-CF) from whom multiple isolates were received, only the first confirmed as B. cepacia complex was included in the study. All bacterial isolates were stored in skim milk at −80°C.
Species identification and phylogenetic analysis.
Isolates were confirmed as B. cepacia complex by polyphasic analyses employing biochemical tests and genus- and species-specific16S rRNA-directed PCR assays as previously described (15). Differentiation of species within the B. cepacia complex was also accomplished by using species-specific recA-directed PCR and restriction fragment length polymorphism analyses as previously described (12). Isolates for which species identification remained equivocal after PCR and restriction fragment length polymorphism testing were further analyzed by determining the complete recA nucleotide sequence. DNA sequencing was performed with an Applied Biosystems ABI model 3700 sequencer using the protocols of the manufacturer (PE Applied Biosystems, Foster City, CA) and the BigDye Terminator Cycle Sequencing Ready Reaction Kit. Resultant sequences were visualized as chromatograms and manually edited using Chromas v.2.22 (Technelysium Pty. Ltd., Helensvale, Australia). Edited sequences were assembled using EditSeq (DNAStar Inc.) and identified by using BLASTN and comparison to sequences currently available in the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/BLAST).
Species distribution.
The distribution of B. cepacia complex species among the 1,218 infected CF patients, as well as among the 90 non-CF patients, was quite disproportionate (Table 1). Forty-five percent of CF patients were infected with B. cenocepacia, and approximately 39% with B. multivorans. Less than 1% of patients were infected with B. stabilis, B. ambifaria, B. anthina, or B. pyrrocinia. In 16 patients, the species of the infecting isolate was indeterminate based on B. cepacia complex species-specific rRNA and recA gene PCR assays. Phylogenetic analysis using recA gene sequences clearly placed these isolates within the B. cepacia complex, although assignment to one of the defined species of the complex was not possible. As previously described (12), recA analysis also allows separation of B. cenocepacia isolates into subgroups designated A, B, C, and D. Among the 556 B. cenocepacia CF isolates, 151 (27.2%) were subgroup A and 405 (72.8%) were subgroup B; no recA subgroup C or D isolates were identified.
TABLE 1.
Distribution of B. cepacia complex species among 1,218 CF and 90 non-CF patients
| Species | No. of patients infected (%)
|
|
|---|---|---|
| CF patients | Non-CF patients | |
| B. cepacia | 38 (3.1) | 17 (18.9)a |
| B. multivorans | 471 (38.7) | 14 (15.6)a |
| B. cenocepacia | 556 (45.6)b | 23 (25.6)ac |
| B. stabilis | 4 (0.3) | 3 (3.3)d |
| B. vietnamiensis | 72 (5.9) | 5 (5.6) |
| B. dolosa | 46 (3.8) | 0 (0) |
| B. ambifaria | 10 (0.8) | 4 (4.4)d |
| B. anthina | 2 (0.2) | 4 (4.4)d |
| B. pyrrocinia | 3 (0.3) | 0 (0) |
| Indeterminate | 16 (1.3) | 20 (22.2)a |
| Total | 1,218 (100) | 90 (100) |
Proportions significantly different between CF and non-CF patients (P < 0.001, chi-square test).
One hundred fifty-one (27.2%) are recA subgroup A, and 405 (72.8%) are recA subgroup B.
Eight (34.8%) are recA subgroup A, and 15 (65.2%) are recA subgroup B.
Proportions significantly different between CF and non-CF patients (P < 0.05, Fisher's exact test).
The distribution of B. cepacia complex species among the 90 non-CF patients differed from the distribution found among the CF patients (Table 1). In this patient group, B. cenocepacia and B. multivorans together accounted for only 41% of patients. B. cepacia and “species indeterminate” isolates infected approximately 19% and 22% of the patients, respectively. Again, very few patients were found to be infected with B. stabilis, B. ambifaria, or B. anthina, and no patients were infected with B. dolosa or B. pyrrocinia.
The reasons for the predominance of B. cenocepacia and B. multivorans among infected CF patients, despite the very close phylogenetic and phenotypic relatedness of the species within the B. cepacia complex, are unknown. Recent observations suggest that B. cenocepacia and B. multivorans are not more abundant in the natural environment (14), although the preferred niches of each species of the B. cepacia complex remain to be defined. Rather, the disproportionate representation of these two species suggests an enhanced capacity for human infection. We found that these species were also relatively common among patients without CF. The reasons for the relatively greater proportions of non-CF patients infected with B. cepacia (genomovar I) and “species indeterminate” strains are unknown.
The remaining B. cepacia complex species, in contrast, accounted for relatively little infection. B. stabilis, B. anthina, and B. pyrrocinia in particular were found in very few patients, although chronic infection of CF patients for periods of up to 5 years was noted with each of these species (data not shown). Also of note was the observation that 31 (67%) of the 46 CF patients we identified with B. dolosa infection received care at the same treatment center and were infected with the same strain (2).
In some countries, the proportions of CF patients infected with species other than B. cenocepacia and B. multivorans are greater than what we observed here. However, these studies have involved much smaller numbers of patients. For example, Drevinek and colleagues (9) found that 13 (54%) of 24 Slovak patients were infected with B. stabilis. Cunha and colleagues (7) identified B. cepacia and B. stabilis in 8 (36%) and 4 (18%) of 22 Portuguese CF patients, respectively. A more recent survey of 153 infected French CF patients revealed a distribution of species comparable to that found in our study, with 45% and 52% of patients infected with B. cenocepacia and B. multivorans, respectively (3).
Change in species distribution.
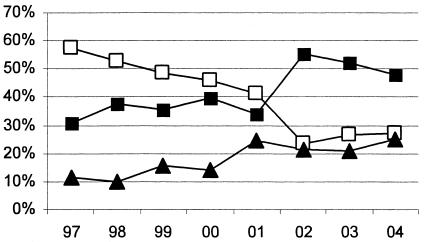
The analysis described in Table 1 provides a static view of the distribution of B. cepacia complex species among CF patients in the United States. We also sought to determine whether changes in this distribution may be occurring. Therefore, for each of the past several years we assessed the distribution of species among patients newly infected during that year. The results indicate that while the proportion of patients infected with B. cenocepacia between 1997 and 2002 decreased, the proportion of patients becoming infected with B. multivorans has increased (Fig. 1). The reasons for this shift are unclear. It is possible that more stringent infection control measures have limited the interpatient spread of so-called “epidemic” B. cenocepacia strains in CF treatment centers. It is also possible that this apparent change in relative frequency of infecting species actually reflects a bias in the strains being referred to the BcRLR for analysis. The unusual phenotype of B. multivorans often presents a greater diagnostic challenge, and it is possible that this species is preferentially referred for more advanced analysis.
FIG. 1.
Proportions of CF patients' first-time B. cepacia complex sputum culture isolates received by the BcRLR each year that were identified as B. cenocepacia (open squares), B. multivorans (closed squares), or any of the remaining seven B. cepacia complex species (solid triangles).
In summary, our analysis of B. cepacia complex isolates recovered from several hundred patients provides a robust assessment of the relative frequencies with which these species cause human infection. We confirm the dominance of B. cenocepacia and B. multivorans, particularly among CF isolates. As important, we demonstrate that despite their unusually close genetic relatedness, several species within the B. cepacia complex are rarely associated with human infection. The biologic determinants of this dramatic disparity have yet to be elucidated.
Acknowledgments
This work was supported by a grant (to J.J.L.) from the Cystic Fibrosis Foundation.
We acknowledge the generosity and cooperation of participating CF centers and microbiology laboratories for submission of clinical isolates.
REFERENCES
- 1.Agodi, A., E. Mahenthiralingam, M. Barchitta, V. Giannino, A. Sciacca, and S. Stefani. 2001. Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J. Clin. Microbiol. 39:2891-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biddick, R., T. Spilker, A. Martin, and J. J. LiPuma. 2003. Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiol. Lett. 228:57-62. [DOI] [PubMed] [Google Scholar]
- 3.Brisse, S., C. Cordevant, P. Vandamme, P. Bidet, C. Loukil, G. Chabanon, M. Lange, and E. Bingen. 2004. Species distribution and ribotype diversity of Burkholderia cepacia complex isolates from French patients with cystic fibrosis. J. Clin. Microbiol. 42:4824-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroecke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 5.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51:1481-1490. [DOI] [PubMed] [Google Scholar]
- 6.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunha, M. V., J. H. Leitao, E. Mahenthiralingam, P. Vandamme, L. Lito, C. Barreto, M. J. Salgado, and I. Sa-Correia. 2003. Molecular analysis of Burkholderia cepacia complex isolates from a Portuguese cystic fibrosis center: a 7-year study. J. Clin. Microbiol. 41:4113-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Detsika, M. G., J. E. Corkill, M. Magalhaes, K. J. Glendinning, C. A. Hart, and C. Winstanley. 2003. Molecular typing of, and distribution of genetic markers among, Burkholderia cepacia complex isolates from Brazil. J. Clin. Microbiol. 41:4148-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drevinek, P., O. Cinek, J. Melter, L. Langsadl, Y. Navesnakova, and V. Vavrova. 2003. Genomovar distribution of the Burkholderia cepacia complex differs significantly between Czech and Slovak patients with cystic fibrosis. J. Med. Microbiol. 52:603-604. [DOI] [PubMed] [Google Scholar]
- 10.Kidd, T. J., S. C. Bell, and C. Coulter. 2003. Genomovar diversity amongst Burkholderia cepacia complex isolates from an Australian adult cystic fibrosis unit. Eur. J. Clin. Microbiol. Infect. Dis. 22:434-437. [DOI] [PubMed] [Google Scholar]
- 11.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 12.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrucca, A., P. Cipriani, P. Valenti, D. Santapaola, C. Cimmino, G. L. Scoarughi, I. Santino, S. Stefani, R. Sessa, and M. Nicoletti. 2003. Molecular characterization of Burkholderia cepacia isolates from cystic fibrosis (CF) patients in an Italian CF center. Res. Microbiol. 154:491-498. [DOI] [PubMed] [Google Scholar]
- 14.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shelly, D. B., T. Spilker, E. J. Gracely, T. Coenye, P. Vandamme, and J. J. LiPuma. 2000. Utility of commercial systems for identification of Burkholderia cepacia complex from cystic fibrosis sputum culture. J. Clin. Microbiol. 38:3112-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storms, V., N. Van Den Vreken, T. Coenye, E. Mahenthiralingam, J. J. LiPuma, M. Gillis, and P. Vandamme. 2004. Polyphasic characterisation of Burkholderia cepacia-like isolates leading to the emended description of Burkholderia pyrrocinia. Syst. Appl. Microbiol. 27:517-526. [DOI] [PubMed] [Google Scholar]
- 18.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 19.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 20.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 21.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. DeVos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermis, K., T. Coenye, J. J. LiPuma, E. Mahenthiralingam, H. J. Nelis, and P. Vandamme. 2004. Proposal to accommodate Burkholderia cepacia genomovar VI as Burkholderia dolosa sp. nov. Int. J. Syst. Evol. Microbiol. 54:689-691. [DOI] [PubMed] [Google Scholar]