Abstract
A nested PCR assay was developed for the detection of spotted fever group (SFG) rickettsiae in serum samples. The assay was based on specific primers derived from the rickettsial outer membrane protein B gene (rompB) of Rickettsia conorii. An SFG rickettsia-specific signal is obtained from R. akari, R. japonica, R. sibirica, and R. conorii. Other bacterial species tested did not generate any signal, attesting to the specificity of the assay. As few as seven copies of the rompB gene of R. conorii could be detected in 200 μl of serum sample. The assay was evaluated with a panel of sera obtained from patients with acute-phase febrile disease tested by immunofluorescent antibody assay (IFA). The SFG rickettsia-specific DNA fragment was detected in 71 out of 100 sera, which were proven to have immunoglobulin M antibodies against SFG rickettsial antigen by IFA. The results were further confirmed by restriction fragment length polymorphism and sequencing analysis of the DNA fragments. The results indicated that this PCR assay is suitable for the diagnosis of spotted fever group rickettsiosis in Korea.
Rickettsioses are some of the oldest known arthropod-borne diseases caused by obligate intracellular bacteria belonging to the genus Rickettsia. The genus comprises the spotted fever group (SFG) rickettsiae and the typhus group (TG) rickettsiae (15). The TG includes Rickettsia prowazekii, the agent of epidemic typhus, and R. typhi, the agent of murine typhus (15). SFG rickettsioses are associated with arthropods, mainly ticks, mites, and fleas (15). SFG rickettsioses are widely distributed throughout the world in foci of endemicity, causing sporadic outbreaks in areas such as Japan, southern China, and eastern Russia—countries that surround Korea (6, 14, 17). No clinical human case of SFG rickettsiosis, however, has been reported in Korea until recently, when the evidence for the existence of SFG rickettsiosis surfaced in reliable reports. The presence of the nucleic acids of R. japonica and R. rickettsii in Haemaphysalis longicornis was demonstrated through the PCR analysis for the gltA and 16S rRNA genes (10). Serologic study of patients with acute febrile episodes showed the presence of immunoglobulin G (IgG) and IgM antibodies in the patients' sera against SFG rickettsiae, R. japonica, R. akari, R. conorii, and R. sibirica (7). Recently, the nested PCR analysis with primers targeting the partial rickettsial outer membrane protein B gene (rompB) of SFG rickettsiae demonstrated the existence of the SFG rickettsiae R. conorii, R. akari, R. japonica, and R. felis in human sera (3). In this study, a nested PCR assay was developed for the detection of SFG rickettsiae in serum samples. The assay was compared with the immunofluorescent antibody assay (IFA), retrospectively, on a panel of sera obtained from patients with acute-phase febrile disease, to evaluate its usefulness for the diagnosis of SFG rickettsiosis in Korea.
(This work was a part of the doctoral thesis of Y.-J. Choi.)
MATERIALS AND METHODS
Rickettsial strains.
The rickettsial strains used in this study were obtained from the American Type Culture Collection (Manassas, VA): R. typhi Wilmington (VR-144), R. prowazekii Breinl (VR-142), R. akari MK (VR-148), R. japonica YH (VR-1363), R. conorii Indian Tick Typhus (VR-597), and R. sibirica 246 (VR-151). Orientia tsutsugamushi Boryong was isolated and maintained at the Department of Microbiology, School of Medicine, Seoul National University. The rickettsia strains were cultivated in L929 (ATCC CCL-1) or Vero (ATCC CRL-1586) cell monolayers supplemented with Eagle's minimum essential medium (Gibco BRL, Grand Island, NY), 2% fetal bovine serum, 100 μg of streptomycin per ml, 100 U of penicillin per ml, and 2 mM l-glutamine in a humidified 5% CO2 atmosphere at 32°C.
Counting of rickettsial particles.
Rickettsial antigens were purified through an altered version of the Percoll density gradient centrifugation described by Tamura et al. (18). Purified rickettsial antigens (R. conorii) were serially diluted to 10−15 for the counting of rickettsial particles by IFA in a microcentrifuge tube (the final volume was 50 μl). The mouse polyserum against R. conorii at a 1:500 dilution ratio in phosphate-buffered saline (PBS; 138 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4 · 2H2O, 1.2 mM KH2PO4 [pH 7.4]) was used as a primary antibody. The reaction mixture was incubated at 37°C for 30 min and centrifuged at 12,000 × g for 15 min. The supernatant was removed, supplemented with 500 μl of PBS, and centrifuged at 12,000 × g for 15 min. The fluorescein isothiocyanate-conjugated donkey anti-mouse IgG (heavy-plus-light-chain-specific) (715-095-151; Jackson ImmunoResearch Lab, Inc., West Grove, PA) antibody at a 1:100 dilution ratio in PBS was used as a secondary antibody. Incubated at 37°C for 30 min and washed, the pellet was resuspended in 50 μl of PBS. The suspension was spotted on a spot slide. The slides were dried and examined at ×1,000 magnification using a fluorescence microscope (BX51; Olympus, Japan) for the counting of the rickettsial particles.
Serum samples and serologic testing.
A total of 200 human serum samples were used in this study (100 IFA-positive serum samples and 100 IFA-negative serum samples) and were selected from among 3,400 serum samples. The sera were obtained from South Korean patients with acute febrile illness from 1993 to 1999. The sera were submitted to the Institute of Endemic Disease at Seoul National University's Medical Research Center for laboratory diagnosis for scrub typhus, leptospirosis, and hemorrhagic fever with renal syndrome caused by hantavirus. Some of the sera were used for the nucleic acid detection study of SFG rickettsial agents. The rationale for selecting the 100 positive samples for the PCR analysis included the presence of IgM antibodies with titers from 1:40 to 1:160 against any of the tested SFG rickettsial antigens in the samples. The 100 negative serum samples for the PCR analysis have no IgG and IgM antibodies at the 1:40 dilution ratio of the samples to any of the antigens.
Design of PCR primers.
PCR primers were derived from conserved regions based on a multiple-sequence alignment of rompB sequences obtained from GenBank. Primer sequences are listed in Table 1, along with their positions relative to the nucleotide sequence of the rompB gene of the R. conorii strain Seven (GenBank accession number AF123721). Primers Rc.rompB.4,362p and Rc.rompB.4,836n were used in the initial attempts to amplify part of the rompB gene from the various SFG rickettsial species. Primers Rc.rompB.4,496p and Rc.rompB.4,762n were used in the second round of attempts to amplify the inner part of the first-round PCR amplicons. They were designed for the specific detection of SFG rickettsial DNA in clinical samples.
TABLE 1.
Oligonucleotide primers used for PCR of SFG rompB gene
| Primer | Nucleotide sequence (5′ to 3′) | Positiona |
|---|---|---|
| Rc.rompB.4362p | GTCAGCGTTACTTCTTCGATGC | 4362-4383 |
| Rc.rompB.4,836n | CCGTACTCCATCTTAGCATCAG | 4836-4815b |
| Rc.rompB.4,496p | CCAATGGCAGGACTTAGCTACT | 4496-4517 |
| Rc.rompB.4,762n | AGGCTGGCTGATACACGGAGTAA | 4762-4740b |
Corresponding position in R. conorii strain Seven rompB sequence (GenBank accession number AF123721).
Reverse orientation.
DNA extraction from patient serum.
The DNA extraction procedures used were altered versions of previous methods using Chelex-100 (12, 13, 16). Chelex-100 is an ion-exchange resin that scavenges multivalent metal ions. Two hundred microliters of serum sample was filled out to 1 ml with PBS and then centrifuged at 12,500 × g for 30 min. The pellet was dissolved in equal volumes of 2× lysis buffer (100 mM Tris-HCl [pH 9.0], 2 mM EDTA, 2% Triton X-100, 800-μg/ml proteinase K [Boehringer Mannheim GmbH, Germany]) and incubated at 60°C for 2 hours. After heat inactivation of the proteinase K (for 10 min in boiling water), 1/2 volume of 15% (wt/vol) Chelex-100 (Bio-Rad, Hercules, CA) was added, and the sample was incubated at 56°C for 30 min. The sample was vortexed at high speed for 10 seconds and was boiled for 10 min. After centrifugation for 2 minutes at 15,000 × g, the supernatant was used for DNA amplification. Chelex-100 particles were carefully separated from the sample by a brief centrifugation before the PCR.
Detection of rompB gene in human sera.
Detection of the SFG rickettsial rompB sequence in human sera was carried out using nested PCR. The primary amplification of the specimen was done in a final reaction volume of 50 μl. The reaction mixture contained 5 μl of prepared DNA sample, 20 pmol of Rc.rompB.4362p and Rc.rompB.4,836n, 200 μM of deoxynucleoside triphosphate mixture (Takara, Japan), 1× PCR buffer, 1.25 units of Taq polymerase (Takara EX Taq; Takara), and distilled water. First, PCR mixtures were incubated at 95°C for 5 minutes and subjected to 35 cycles of 95°C for 15 s, 54°C for 15 s, and 72°C for 30 s and then to a final extension at 72°C for 3 minutes in a GeneAmp PCR system 9600 (Perkin-Elmer Applied Biosystems, Foster City, CA). After this, 2 μl of the amplified product was again amplified in a nested fashion with inner primer sets (Rc.rompB.4,496p and Rc.rompB.4,762n). The nested PCR mixture contained 10 pmol of each primer in a PCR premixture tube (AccuPower PCR PreMix; Bioneer Corp., Korea) that contained 1 U of Taq DNA polymerase, 250 μM each of deoxynucleoside triphosphates, 50 mM of Tris-HCl (pH 8.3), 40 mM of KCl, 1.5 mM of MgCl2, and gel loading dye. The volume was then adjusted to 20 μl with distilled water. Nested PCR mixtures were incubated at 95°C for 5 minutes and subjected to 35 cycles of 95°C for 15 s, 56°C for 15 s, and 72°C for 30 s and then to a final extension at 72°C for 3 minutes. To avoid cross-contamination, three separate rooms with entirely separate equipment and solutions were used. Aerosol-resistant tips (Axigen Scientific, Inc., California) were used for the handling of all reagents in the PCR study. The amplification products were visualized by electrophoresis on a 1.5% agarose gel stained with ethidium bromide (0.5 μg/ml) and using a 1× TAE migration buffer (pH 8.0; 40 mM Tris-acetate, 1 mM EDTA).
Cloning, sequencing, and analysis of nucleotide.
All positive PCR products were cloned using pGEM-T Easy Vector System I (Promega, WI) according to the manufacturer's instructions. Verifying whether the clones contained inserts was accomplished by digestion of plasmid DNA with EcoRI (New England Biolabs) and separation in 1.5% agarose gels. Plasmids containing DNA inserts were sequenced for both strands using the Big Dye Terminator sequence kit and the ABI Prism 377 automated DNA sequencer (Perkin-Elmer Applied Biosystems) according to the manufacturer's protocol. The obtained sequences, except for the primer regions, were aligned with the corresponding sequences of other rickettsiae deposited in the GenBank database to identify known sequences with a high degree of similarity using multisequence alignment programs, the Phydit software (4) and the MegAlign software package (Windows version 3.12e; DNAStar, Wisconsin).
Nucleotide sequence accession numbers.
The sequences for the nine B clones determined in this study have been deposited in GenBank under accession numbers DQ019320 to DQ019328.
RESULTS
Sensitivity of the nested PCR.
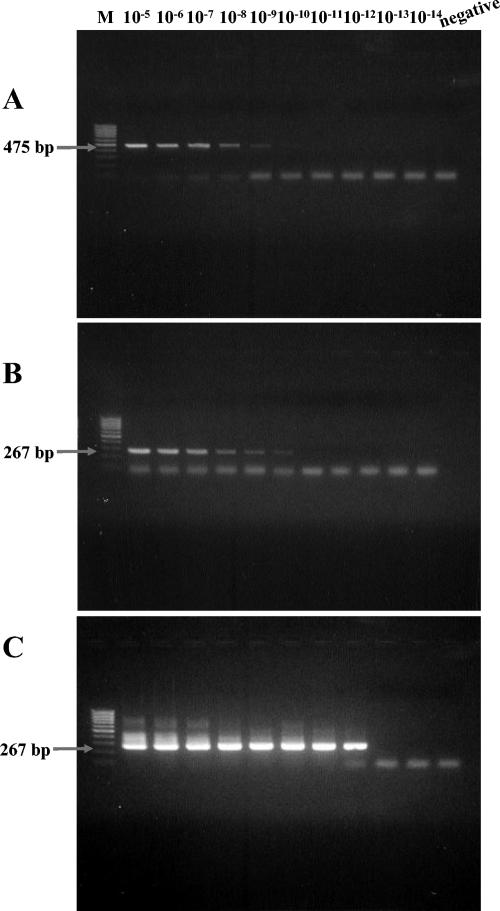
The primers amplified expected amplicons of 475 bp and 267 bp, respectively, in the first and second PCRs from R. conorii serially diluted in normal mouse serum (Fig. 1A and B). The sensitivity of the nested PCR assay for detection was evaluated with an R. conorii suspension with known concentrations of rickettsial particles. Rickettsial DNA was extracted from 5 μl of serially diluted suspension (from 10−5 to 10−14), and its final volume was 100 μl. Using 5 μl of the template DNA, PCR assays were performed with the primer pairs Rc.rompB.4,362p-Rc.rompB.4,836n and Rc.rompB.4,496p-Rc.rompB.4,762n. The sensitivity based on Rc.rompB.4,362p-Rc.rompB.4,836n primers was approximately 7,000 particles (positive result at a dilution rate of 10−9) per assay (starting with 200 μl of sample). The sensitivity based on the primer pair Rc.rompB.4,496p-Rc.rompB.4,762n was approximately 700 particles (positive result at a dilution rate of 10−10). Using a nested PCR assay with the two primer pairs increased the detection sensitivity for R. conorii to fewer than seven rickettsial particles per assay (positive result at a dilution rate of 10−12 [Fig. 1C]).
FIG. 1.
Sensitivity of the nested PCR assay with nested primer pairs Rc.rompB.4362p-Rc.rompB.4,836n in the primary reactions and Rc.rompB.4,496p-Rc.rompB.4,762n in the secondary reactions for the detection of SFG rickettsial rompB genes. Shown are results of agarose gel electrophoresis analysis on a 1.5% agarose gel of the PCR-amplified DNAs primed with primer pairs Rc.rompB.4362p-Rc.rompB.4,836n (A) and Rc.rompB.4,496p-Rc.rompB.4,762n (B) and the nested primer pairs Rc.rompB.4362p-Rc.rompB.4,836n and Rc.rompB.4,496p-Rc.rompB.4,762n (C) from serially diluted R. conorii suspensions (from 10−5 to 10−14). Lanes: M, size marker DNA (100-bp DNA ladder); 10−5 to 10−14, each dilution rate of R. conorii suspension; negative, no-template control. The numbers on the left indicate the molecular sizes (in base pairs) of the amplified PCR products.
Specificity of the nested PCR.
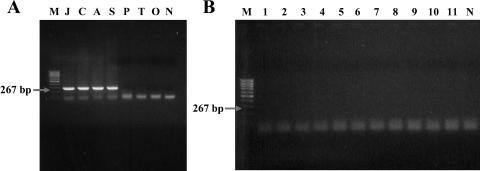
The specificity of nested primer pairs Rc.rompB.4,362p-Rc.rompB.4,836n and Rc.rompB.4,496p-Rc.rompB.4,762n in detecting SFG rickettsial agents was confirmed with DNA extracted from various rickettsiae: R. akari, R. conorii, R. sibirica, R. japonica, R. prowazekii, R typhi, and O. tsutsugamushi. Electrophoretic analysis of the nested PCR products revealed that the primer pairs amplified only the SFG rickettsial DNA templates and did not amplify DNA templates from TG rickettsial species and O. tsutsugamushi (Fig. 2A). Other infectious agents and/or ubiquitous bacteria, such as Escherichia coli, Salmonella enterica serovar Typhi, Shigella flexneri, Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus pyogenes, Proteus vulgaris, Neisseria sicca, Leptospira interrogans, Borrelia burgdorferi, and Candida albicans, did not produce any specific DNA fragment with the nested PCR system with the primer pairs (Fig. 2B). These results suggest that this PCR detection system has high specificity for SFG rickettsial species.
FIG. 2.
Specificity of the nested PCR assay with primer pairs Rc.rompB.4362p-Rc.rompB.4,836n and Rc.rompB.4,496p-Rc.rompB.4,762n for the detection of SFG rickettsial rompB genes. Shown are results of agarose gel electrophoresis analysis on a 1.5% agarose gel of the PCR-amplified DNAs primed with the nested primer pairs from various SFG and TG species (A) and other ubiquitous and/or infectious agents (B). Lanes: M, size marker DNA (100-bp DNA ladder); J, R. japonica; C, R. conorii; A, R. akari; S, R. sibirica; P, R. prowazekii; T, R typhi; O, O. tsutsugamushi; N, no-template control; 1, Escherichia coli; 2, Salmonella enterica serovar Typhi; 3, Shigella flexneri; 4, Staphylococcus aureus; 5, Staphylococcus epidermidis; 6, Streptococcus pyogenes; 7, Proteus vulgaris; 8, Neisseria sicca; 9, Leptospira interrogans; 10, Borrelia burgdorferi; 11, Candida albicans DNA as a template.
Detection of SFG rickettsial DNA sequences in human serum samples.
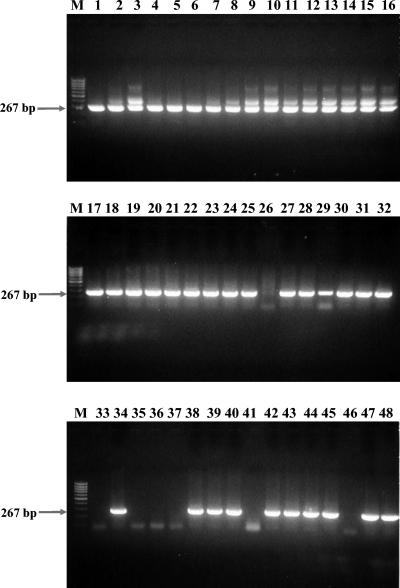
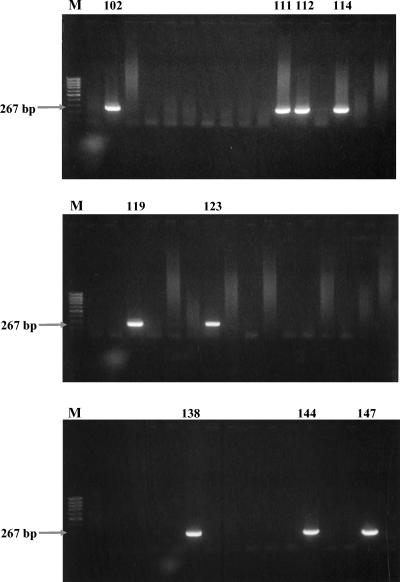
The nested PCR assay amplified the predicted products with DNA templates from 71 out of 100 seropositive sera and 19 out of 100 seronegative sera. Figure 3 shows PCR amplicon profiles from 48 samples of 100 seropositive samples. Figure 4 shows PCR amplicon profiles from 48 samples of 100 seronegative samples. The PCR products were also confirmed by sequencing analysis. Table 2 compares the similarity between partial rompB sequences of various rickettsial strains and the representative nested PCR clones. R. japonica, R. akari, R. conorii, and R. sibirica were detected by comparison of nucleotide sequences between amplified products and partial rompB sequences of reference strains. However, the nucleotide sequences of the amplified rompB gene region from R. conorii and R. sibirica could not be differentiated.
FIG. 3.
Agarose gel electrophoresis analysis on a 1.5% agarose gel of DNAs amplified by the PCR assay with the nested primer pairs with template DNAs from 48 seropositive samples. Lanes M, size marker DNA (100-bp DNA ladder); each numbered lane represents a separate seropositive sample. The numbers on the left indicate the molecular sizes (in base pairs) of the amplified PCR products.
FIG. 4.
Agarose gel electrophoresis analysis on a 1.5% agarose gel of DNAs amplified by the PCR assay with the nested primer pairs with template DNAs from 48 seronegative samples. Lanes M, size marker DNA (100-bp DNA ladder); each of the numbered lanes, 101 to 148, represents a separate seronegative sample. The numbers of the first lanes on the gels and those of lanes containing positive bands are shown. The numbers on the left indicate the molecular sizes (in base pairs) of the amplified PCR products.
TABLE 2.
Similarity matrix between partial rompB sequences of various rickettsial strains and representative nested PCR productsa
| Sequence | % Similarity of partial rompB sequence
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | B3 | B10 | B19 | B38 | B42 | B53 | B84 | B96 | B195 | |
| 1 | ||||||||||||||||
| 2 | 92.79 | |||||||||||||||
| 3 | 92.34 | 97.75 | ||||||||||||||
| 4 | 92.79 | 100 | 97.75 | |||||||||||||
| 5 | 82.43 | 84.68 | 84.68 | 84.68 | ||||||||||||
| 6 | 82.43 | 84.68 | 84.68 | 84.68 | 100 | |||||||||||
| 7 | 81.53 | 82.43 | 82.43 | 82.43 | 92.34 | 92.34 | ||||||||||
| B3 | 92.79 | 100 | 97.75 | 100 | 84.68 | 84.68 | 82.43 | |||||||||
| B10 | 98.65 | 94.14 | 92.79 | 94.14 | 82.88 | 82.88 | 81.98 | 94.14 | ||||||||
| B19 | 92.34 | 97.75 | 100 | 97.75 | 84.68 | 84.68 | 82.43 | 97.75 | 92.79 | |||||||
| B38 | 92.34 | 99.55 | 97.3 | 99.55 | 84.23 | 84.23 | 81.98 | 99.55 | 93.69 | 97.3 | ||||||
| B42 | 91.89 | 99.1 | 97.75 | 99.1 | 84.68 | 84.68 | 82.43 | 99.1 | 93.24 | 97.75 | 98.65 | |||||
| B53 | 91.89 | 96.85 | 99.1 | 96.85 | 83.78 | 83.78 | 81.53 | 96.85 | 91.89 | 99.1 | 96.4 | 96.85 | ||||
| B84 | 91.89 | 97.3 | 98.65 | 97.3 | 83.33 | 83.33 | 81.08 | 97.3 | 92.34 | 98.65 | 96.85 | 96.4 | 97.75 | |||
| B96 | 99.55 | 92.34 | 91.89 | 92.34 | 81.98 | 81.98 | 81.08 | 92.34 | 98.2 | 91.89 | 91.89 | 91.44 | 91.44 | 91.44 | ||
| B195 | 96.4 | 92.79 | 92.34 | 92.79 | 83.33 | 83.33 | 81.98 | 92.79 | 95.95 | 92.34 | 92.34 | 91.89 | 91.44 | 91.89 | 95.95 | |
DISCUSSION
A nested PCR assay with newly designed primer pairs Rc.rompB.4,362p-Rc.rompB.4,836n and Rc.rompB.4,496p-Rc.rompB.4,762n targeting the rompB gene was developed for the detection of SFG rickettsiae in clinical samples. This nested PCR is highly specific for SFG rickettsiae and is a reliable method for the detection of the pathogens in serum samples.
The PCR-based amplification method is a useful diagnostic tool in the early phase of illness. Rickettsiae may be detected by PCR methods of specific rickettsial DNA from several clinical samples that include serum, blood clot or whole blood, skin biopsy samples, paraffin-embedded tissues, cerebrospinal fluid, and arthropod tissues (9, 15). The combination of PCR-based amplification of rickettsial DNA and methods for the analysis of PCR products, such as restriction fragment length polymorphism (RFLP) or DNA sequencing, is the key to determining the true identity of infectious rickettsiae (9). This study aimed to detect the rompB sequence through the PCR amplification method because the complete sequence of the gene has been reported for many SFG and TG rickettsiae (5, 19), and the gene proved to be a good candidate for the detection and phylogenetic study of SFG and TG rickettsiae (2, 5, 8, 19).
The use of the nested PCR designed in this study could detect approximately seven particles of SFG rickettsial antigens in 200 μl of the serum sample. The sensitivity of the nested PCR test is 2 orders of magnitude greater than that of the normal PCR test, which increases the possibility of detecting rickettsial DNA in clinical samples. It did not generate nonspecific amplification in other infectious agents. The negative controls consistently failed to yield detectable PCR products, while the positive controls always gave the expected PCR products in repeated studies.
Since rickettsiae are intracellular, whole blood or buffy coat samples are considered the preferred type for the PCR tests (11). Most of the specimens provided by medical centers, however, were serum samples. This study also used serum samples from patients with febrile episodes. The nested PCR detected SFG rickettsial DNA in 71 out of 100 seropositive samples which have IgM antibodies against SFG rickettsiae. The positive results in this study may be due to the higher sensitivity of the nested PCR test. As shown in the results of the nested PCR, the amplified partial rompB gene products were also obtained from seronegative sera (19% of samples tested). This could be regarded as detection of the early stage of infection in which IgM antibodies had not yet been produced in patients with acute febrile illness. The differentiation of febrile exanthematous diseases from SFG rickettsioses is important in high-risk areas of endemicity. The nested PCR detection method could also be useful for establishing a definitive diagnosis of SFG rickettsioses during the critical stage.
The PCR amplicons were confirmed through RFLP and nucleotide sequencing analysis (RFLP analysis data not shown). Generally, PCR-RFLP analysis is the simplest method to determine whether a rickettsia isolate is identical to or different from reference strains (1). Several SFG rickettsiae, R. conorii, R. sibirica, R. akari, and R. japonica, were detected by comparing nucleotide sequences and RFLP patterns between amplified products and the partial rompB gene region of the reference strains. The PCR amplicons of R. conorii and R. sibirica, however, could not be differentiated from the nucleotide sequence. In addition, R. conorii and R. akari showed identical RFLP patterns, but R. japonica did not. Therefore, although the nested PCR assay using primer pairs Rc.rompB.4362p-Rc.rompB.4,836n and Rc.rompB.4,496p-Rc.rompB.4,762n effectively detected SFG rickettsial agents in the serum samples, it was not useful for precise differentiation of strains among the various SFG rickettsiae.
In conclusion, this study indicated that the nested PCR assay is a highly sensitive and specific method for the detection of SFG rickettsial agents in sera. Furthermore, SFG rickettsial agents were detected from 71% of seropositive sera and 19% of seronegative sera from patients with febrile episodes. These results suggest the existence of various SFG rickettsial infections in Korea. Therefore, this nested PCR assay may be useful for the diagnosis of SFG rickettsiosis in Korean patients with febrile episodes.
Acknowledgments
This paper was supported by Konkuk University in 2001.
REFERENCES
- 1.Beati, L., P. J. Kelly, P. R. Mason, and D. Raoult. 1994. Species-specific BALB/c mouse antibodies to rickettsiae studied by Western blotting. FEMS Microbiol. Lett. 119:339-344. [DOI] [PubMed] [Google Scholar]
- 2.Carl, M., C. W. Tibbs, M. E. Dobson, S. Paparello, and G. A. Dasch. 1990. Diagnosis of acute typhus infection using the polymerase chain reaction. J. Infect. Dis. 161:791-793. [DOI] [PubMed] [Google Scholar]
- 3.Choi, Y. J., W. J. Jang, J. H. Kim, J. S. Ryu, S. H. Lee, K. H. Park, H. S. Paik, Y. S. Koh, M. S. Choi, and I. S. Kim. 2005. Spotted fever group and typhus group rickettsioses in humans, South Korea. Emerg. Infect. Dis. 11:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun, J. 1995. Computer-assisted classification and identification of actinomycetes. Ph.D. thesis. University of Newcastle upon Tyne, Newcastle upon Tyne, United Kingdom.
- 5.Eremeeva, M., X. Yu, and D. Raoult. 1994. Differentiation among spotted fever group rickettsiae species by analysis of restriction fragment length polymorphism of PCR-amplified DNA. J. Clin. Microbiol. 32:803-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng, H. M., T. S. Chen, B. H. Lin, Y. Z. Lin, P. F. Wang, Q. H. Su, H. B. Xia, K. Kumano, and T. Uchida. 1991. Serologic survey of spotted fever group rickettsiosis in Hainan Island of China. Microbiol. Immunol. 35:687-694. [DOI] [PubMed] [Google Scholar]
- 7.Jang, W. J., J. H. Kim, Y. J. Choi, K. D. Jung, Y. G. Kim, S. H. Lee, M. S. Choi, I. S. Kim, D. H. Walker, and K. H. Park. 2004. First serologic evidence of human spotted fever group rickettsiosis in Korea. J. Clin. Microbiol. 42:2310-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labruna, M. B., J. W. McBride, D. H. Bouyer, L. M. Camargo, E. P. Camargo, and D. H. Walker. 2004. Molecular evidence for a spotted fever group rickettsia species in the tick Amblyomma longirostre in Brazil. J. Med. Entomol. 41:533-537. [DOI] [PubMed] [Google Scholar]
- 9.La Scola, B., and D. Raoult. 1997. Laboratory diagnosis of rickettsioses: current approaches to diagnosis of old and new rickettsial disease. J. Clin. Microbiol. 35:2715-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, J. H., H. S. Park, K. D. Jung, W. J. Jang, S. E. Koh, S. S. Kang, I. Y. Lee, Y. J. Lee, B. J. Kim, W. H. Kook, K. H. Park, and S. H. Lee. 2003. Identification of spotted fever group rickettsiae detected from Haemaphysalis longicornis in Korea. Microbiol. Immunol. 47:301-304. [DOI] [PubMed] [Google Scholar]
- 11.Leitner, M., S. Yitzhaki, S. Rzotkiewicz, and A. Keysary. 2002. Polymerase chain reaction-based diagnosis of Mediterranean spotted fever in serum and tissue samples. Am. J. Trop. Med. Hyg. 67:166-169. [DOI] [PubMed] [Google Scholar]
- 12.Mathis, A., R. Weber, H. Kuster, and R. Speich. 1997. Simplified sample processing combined with a sensitive one-tube nested PCR assay for detection of Pneumocystis carinii in respiratory specimens. J. Clin. Microbiol. 35:1691-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matto, J., M. Saarela, S. Alaluusua, V. Oja, H. Jousimies-Somer, and S. Asikainen. 1998. Detection of Porphyromonas gingivalis from saliva by PCR by using a simple processing method. J. Clin. Microbiol. 36:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okabayashi, T., K. Tsutiya, Y. Muramatsu, H. Ueno, and C. Morita. 1996. Serological survey of spotted fever group rickettsia in wild rats in Thailand in the 1970s. Microbiol. Immunol. 40:895-898. [DOI] [PubMed] [Google Scholar]
- 15.Raoult, D., and V. Roux. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10:694-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reizenstein, E., L. Lindberg, R. Mollby, and H. O. Hallander. 1996. Validation of nested Bordetella PCR in pertussis vaccine trial. J. Clin. Microbiol. 34:810-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sirisanthana, T., V. Pinyopornpanit, V. Sirisanthana, D. Strickman, D. J. Kelly, and G. A. Dasch. 1994. First cases of spotted fever group rickettsiosis in Thailand. Am. J. Trop. Med. Hyg. 50:682-686. [DOI] [PubMed] [Google Scholar]
- 18.Tamura, A., H. Urakami, and T. Tsuruhara. 1982. Purification of Rickettsia tsutsugamushi by Percoll density gradient centrifugation. Microbiol. Immunol. 26:321-328. [DOI] [PubMed] [Google Scholar]
- 19.Weller, S. J., G. D. Baldridge, U. G. Munderloh, H. Noda, J. Simser, and T. J. Kurtti. 1998. Phylogenetic placement of rickettsiae from the ticks Amblyomma americanum and Ixodes scapularis. J. Clin. Microbiol. 36:1305-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]