Abstract
Recent research underscores the pivotal role of cellular organelles, such as mitochondria, the endoplasmic reticulum, and lysosomes, in maintaining cellular homeostasis. Their dynamic interactions are critical for metabolic regulation and stress response. Analysis of organelle proteomes offers valuable insights into their functions in both physiology and disease. Traditional proteomic approaches to studying isolated organelles are now complemented by innovative methodologies focusing on inter-organelle interactions. This review examines the integration of advanced proximity labeling technologies, including TurboID and split-TurboID, which address the inherent limitations of traditional techniques and enable precision proteomics of suborganelle compartments and inter-organellar contact sites. These innovations have led to discoveries regarding organelle interconnections, revealing mechanisms underlying metabolic processes such as cholesterol metabolism, glucose metabolism, and lysosomal repair. In addition to highlighting the advancements in TurboID applications, this review delineates the evolving trends in organelle research, underscoring the transformative potential of these techniques to significantly enhance organelle-specific proteomic investigations.
Keywords: Endoplasmic reticulum, Lysosome, Mitochondria, Proteomics
INTRODUCTION
Recent advancements in pathophysiological research have underscored the importance of exploring the function and structure of cellular organelles such as mitochondria, endoplasmic reticulum (ER), and lysosomes [1-3]. Notably, there has been a paradigm shift in research approaches, moving from the examination of isolated organelles towards an exploration of the dynamic interactions between them. This integrative perspective is pivotal for understanding the complexity of intracellular signaling and the regulation of metabolic processes essential for maintaining cellular homeostasis. These interactions between organelles are crucial for various cellular functions, including the transmission of calcium signals and stress responses within the cell. Key sites of organelle interconnection, such as mitochondria-ER, ER-lysosome, and lysosome-mitochondria junctions, play significant roles in pathophysiological processes.
For instance, in obesity, chronic enrichment of hepatic ER-mitochondria contact sites leads to calcium-dependent mitochondrial dysfunction, a condition that contributes to metabolic pathologies such as insulin resistance [4]. Similarly, increased mitochondria-ER contact has been shown to promote diabetic cardiomyopathy, as it leads to mitochondrial damage and impaired cardiac function due to calcium overload in cardiomyocytes [5]. Additionally, it has been reported that calcium transfer dysfunctions at the ER-lysosome interconnection are not only intricately linked to autophagic defects but may also underpin beta-cell damage [6]. The lysosome-mitochondria interconnection is also crucial for cholesterol transport and homeostasis [7].
However, the study of cellular organelles and their interconnections is hampered by the lack of methodologies capable of isolating specific regions free from nonspecific contaminants. To overcome these limitations, cutting-edge proximity labeling technologies using TurboID or split-TurboID have been developed [8,9]. TurboID is an engineered derivative of Escherichia coli biotin ligase (BirA) and split-TurboID are cleaved forms of TurboID, which are inactive when separated but regain activity upon reassembly. TurboID and split-TurboID generate reactive biotin species that covalently bond to proximal proteins. These biotinylated proteins can be enriched from the total cellular protein pool through streptavidin enrichment and subsequently subjected to quantitative and qualitative analysis via liquid chromatography–mass spectrometry (LC-MS). These advanced TurboID methods are expected to pave the way for spatial biology or molecular spatiomics [10,11].
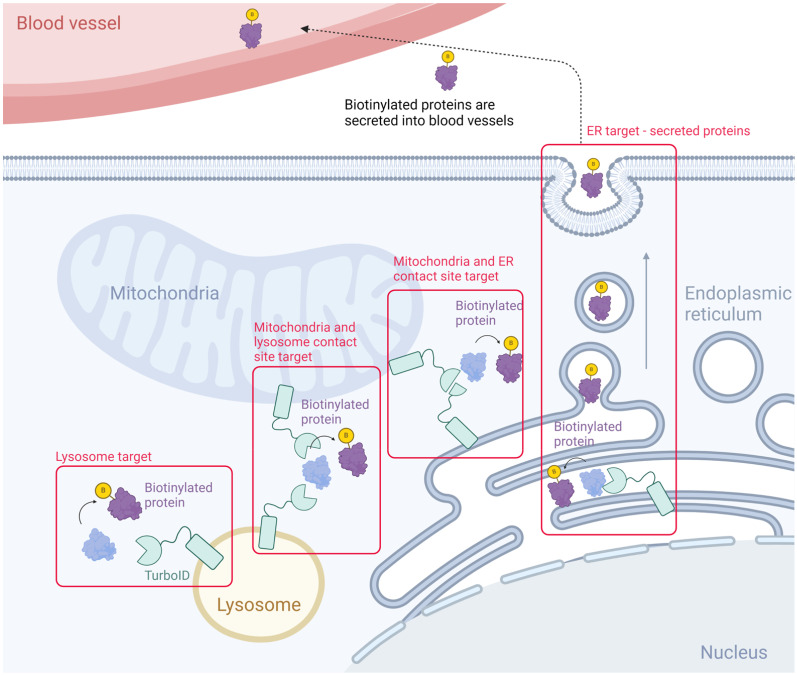
This review will present and comparatively analyze research articles that have employed TurboID technology to investigate individual cellular organelles and their interconnections, thus showing the current trends and advancements in TurboID research methodologies (Fig. 1, Table 1).
Fig. 1. Scheme for studying organelle proteomes with TurboID.
Specific organelle marker is utilized to achieve organelle specific localization of TurboID. Lysosome target: LAMP1, mitochondria and lysosome contact site target: TOM20 and LAMP1, mitochondria and ER contact site target: TOM20 and CB5. ER lumen target: SEC61B.
Table 1.
Recent TurboID-based organelle proteome studies
| Target organelle | Reference | TurboID construct | Main finding | |
|---|---|---|---|---|
| Single organelle | Lysosome | Shin et al. [12] | LYCHOS-TurboID (LYCHOS-TiD) | Binding between LYCHOS and GATOR1 |
| Lysosome | Tan et al. [13] | LAMP1-TurboID (Lyso-TurboID) | Accumulation of PI4K2A and ORP upon LMP | |
| ER | Kim et al. [14] | SEC61B-TurboID (iSLET) | Liver-specific secretory protein labeling | |
| ER | Wei et al. [15] | TurboID-KDEL (ER-TurboID) | 4-cell-type secretomes (hepatocyte, myocyte, pericyte and myeloid cell) | |
| ER | Wei et al. [19] | FLEx-ER-TurboID | 21-cell type secretomes of exercise training in mice | |
| Organelle contact | ER-mitochondria | Kwak et al. [21] | N-BirA-SEC61B (B1-SEC61B) Tom20-BirA-C (TOM20-B2) |
Identification of 115 MAM-specific proteins |
| ER-mitochondria | Cho et al. [22] | N-TOM20-TurboID (OMM-Tb(N)) TurboID-Cb5-C (Tb(C)-ERM) |
Identification of 101 ER-mitochondria contact proteins | |
| Mitochondria-lysosome | Kim et al. [7] | TOM20-TurboID LAMP1-TurboID TM4SF5-TurboID |
Identification of 63 MLCS proteins | |
| ER-Golgi | Yeerken et al. [23] | SEC31A-TurboID | Regulation of ER-to-Golgi transport via Nlp | |
| ER-Golgi | Kovács et al. [24] | OSBP-TurboID | Lipid exchange and cargo sorting via OSBP |
ER, endoplasmic reticulum; LMP, lysosomal membrane permeabilization; MAM, mitochondrial-associated membrane; MLCS, mitochondria-lysosome contact site; Nlp, ninein-like protein; OSBP, oxysterol binding protein.
SINGLE ORGANELLE
Lysosome and cholesterol
Lysosomes are recognized as intracellular nutrient sensors. It has been established that the Mechanistic Target of Rapamycin Complex 1 (mTORC1) senses nutrients like cholesterol and localizes to the lysosome, recruiting various proteins related to metabolic function, thus regulating lysosomal activity. However, the direct interactions between cholesterol and the mTORC1-scaffolding machinery have not been thoroughly elucidated, suggesting the existence of yet unidentified lysosomal nutrient sensing proteins.
Shin et al. [12] performed bioinformatic analysis and they suggested that the protein LYCHOS may regulate the activation of mTORC1 through cholesterol sensing. To elucidate the mechanism by which LYCHOS senses cholesterol and modulates mTORC1 function, the C-terminus of LYCHOS was fused with TurboID (LYCHOS-FLAG-TurboID) and introduced into cells to induce biotinylation of proteins binding to LYCHOS. Subsequent enrichment of biotinylated peptides derived from these cells, followed by mass spectrometry analysis, allowed for the construction of a proteome profile of proteins interacting with LYCHOS.
In their results, Shin et al. [12] revealed that GATOR1 subunits such as NPRL2, NPRL3, and DEPDC5 bind to LYCHOS, and they uncovered that interactions between LYCHOS and GATOR1 modulate mTORC1 signaling. The LYCHOS-TurboID provides a novel approach that overcomes the limitations of traditional methods for studying lysosomal proteomes, which may not adequately capture weak interactions.
Lysosome and repair
Lysosomal membrane permeabilization (LMP) has been identified as a pivotal factor in the etiology of various lysosome-associated disorders. In healthy cellular environments, a rapid rectification of LMP is imperative, yet the specific mechanisms underlying this reparative process remain insufficiently elucidated. Tan and Finkel [13] sought to explore the alterations in lysosomal protein expression in cells where LMP had been induced, utilizing these findings as a substrate to demystify the mechanisms governing lysosomal repair.
Tan and Finkel [13] engineered a cellular system to tag lysosomal proteins via biotinylation by expressing lysosome-targeted TurboID in 293T cells. Subsequent to the induction of lysosomal damage using LLOME (L-Leucyl-L-leucine methyl ester), biotinylated proteins were enriched, facilitating the selective extraction of lysosomal proteins. Comparative proteomic data between LLOME-treated and untreated cells were obtained using mass spectrometry analysis, thus enabling a differential profiling of lysosomal proteins upon LLOME-induced damage.
Utilizing proteome data, an upregulation of PI4K2A and the oxysterol-binding protein-related proteins ORP9 and ORP11 in the lysosome following LLOME treatment was observed. Subsequent assays corroborated the activation of PI4K2A-mediated PtdIns4P signaling in damaged lysosomes. The study delineated that PI4K2A fosters the accumulation of PtdIns4P in damaged lysosomes, which in turn recruits certain ORP family proteins. This recruitment is crucial in generating ER-lysosome membrane contact sites (MCSs), thereby facilitating the reparative mechanisms of the lysosome.
Tan and Finkel [13] deployed TurboID to overcome the limitations of existing methods for detecting lysosomal proteins. Through this novel approach, they successfully identified established markers of lysosomal damage, such as p62 and ESCRT subunits, thereby validating the methodology. Additionally, previously uncharacterized lysosomal proteins were discovered.
ER and liver-specific secretory proteins
Traditional methods for analyzing proteins secreted by specific cells have utilized in vitro cell culture systems or primary cell cultures derived from in vivo systems, followed by proteomic analysis of proteins secreted into the cell culture media. However, this approach carries technical limitations, including interference from predominant proteins existed in the cell culture media and from non-secreted proteins originating from damaged or dead cultured cells. To overcome these obstacles, there is a critical need for new analytical methods capable of analyzing proteins secreted by specific cells and tissues directly through in vivo system.
Kim et al. [14] sought to discover secreted proteins through biotin labeling by introducing biotin ligase TurboID expression vectors including TurboID-V5-KDEL and Sec61b-V5-TurboID, which are targeted to the ER lumen or ER membrane respectively. Initially introduced into cultured cells, the presence of biotin-labeled proteins in the cultured cell media was confirmed by enrichment using streptavidin mediated biotinylated protein enrichment, indicating successful biotinylation at secreted proteins by the introduced TurboID. Subsequently, this system was applied to liver tissue of a living mouse model, creating iSLET (in situ Secretory protein Labeling via ER-anchored TurboID) mice. Through various assays and mass spectrometry-based proteomic analysis, biotin-labeled secreted proteins expressed from the ER-targeted TurboID in the liver were detected in the bloodstream.
Kim et al. [14] demonstrated the identification of 27 biotinylated secreted proteins originating from the liver in the blood of iSLET mice. Furthermore, upon introducing iSLET into mice induced with insulin resistance by S961 administration, 20 liver-derived secreted proteins were distinctively identified in the insulin-resistant group. Furthermore, the proteins identified are known to be associated with insulin resistance, illustrating the successful application of iSLET in pathophysiological mouse models.
ER and tissue-specific secretory proteins
As we mentioned, secreted polypeptides play a crucial role in mediating intercellular or endocrine communication within biological systems. However, research methodologies to elucidate the cell type-specific regulatory mechanisms governing their secretion are markedly insufficient. Wei et al. [15] introduced a novel approach for discerning the secretome of specific cells within in vivo mouse models using adeno-associated virus (AAV), thereby providing a groundbreaking methodology for the exploration of intercellular signaling mechanisms.
Wei et al. [15] engineered AAV vectors to specifically express luminally oriented membrane-tethered TurboID, cytoplasmic oriented ER-localized TurboID, and luminally oriented ER-localized TurboID (Mem-TurboID, Cyto-TurboID, ER-TurboID) in hepatocytes, myocytes, pericytes, and myeloid cells. Biotin was administered to the mice through intraperitoneal injection or water administration to induce biotinylation of secreted proteins in the liver. Blood plasma derived from these mice was then subjected to streptavidin-mediated enrichment of biotinylated peptides, followed by mass spectrometry analysis to ascertain the tissue-specific secretome.
Through this secretome analysis, a total of 4,779 peptides corresponding to 303 proteins were identified as liver-derived secreted peptides. The proteins identified encompassed numerous classical secreted liver-derived plasma proteins, confirming that the in vivo system employed in the experiment functioned optimally. Additionally, this in vivo model system was utilized to illustrate the alterations in the secretome profile of secreted liver-derived plasma proteins induced by a high fructose, high sucrose (HFHS) diet. Notably, these secretome results specifically highlighted the unconventional secretion of BHMT as being upregulated by HFHS.
Additionally, beyond the hepatocyte-derived secretome, Wei et al. [15] established an in vivo system that enables the analysis of secretomes specifically expressed by myocytes, pericytes, and myeloid cells, demonstrating the capability to distinguish and compare the origins of proteins secreted in vivo. Overall, their research introduced a simplified process that enables the analysis of secretome originating from specific cells within a living biological system.
ER and exerkine
Numerous studies have shown that exercise has a positive impact on biological systems and can offer preventive and therapeutic effects against various diseases [16-18]. Despite growing interest in the role of proteins secreted into the bloodstream due to exercise, research has been limited due to the lack of efficient methodologies. Notably, there has been a lack of research extending beyond individual molecules to explore the organism-wide secretome response to physical activity.
Wei et al. [19] established in vivo system by introducing ER-TurboID, which induces biotinylation of secreted proteins, into specific tissues and cell types of mice expressing Cre recombinase via AAV transduction. The mouse model system, encompassing a total of 21 genotypes based on the specific tissue or cell type expressing ER-TurboID, is engineered to guarantee that biotinylated proteins are exclusively secreted into the bloodstream from the targeted tissues or cell types. Enrichment of biotinylated proteins from the derived mouse blood followed by mass spectrometry analysis enabled the selective analysis of the secretome from specific tissues and cell types. The tissues and cell types investigated by Wei et al. [19] include Pdgfra+ fibroblasts, vascular pericytes, endothelial cells, T cells, macrophages, adrenal tissue, smooth muscle, T and B cells, pancreas, brown adipose tissue, brain, gut, adipose tissue, kidney, bone muscle, heart, lung, liver, and muscle.
As a result, Wei et al. [19] revealed the secretion of 1,272 cell type-specific protein pairs from particular tissues or cell types into the bloodstream. Upon exercise training, it was discovered that 256 protein pairs exhibit significant changes, with 181 proteins showing substantial variance across 21 cell types. Furthermore, downstream analysis indicated that Pdgfra cells exhibit the most pronounced response to exercise. Additionally, Wei et al. [19] showed that the secreted enzyme CES2 plays a significant role in anti-obesity, anti-diabetic, and endurance-enhancing functions. Overall, Wei et al. [19] has established large-scale in vivo systems that enables the analysis of the secretome originating from various tissues and specific cell types in live mice.
ORGANELLE CONTACT
ER-mitochondria contact (split-BiolD)
Recent studies have elucidated that mitochondrial-associated membranes (MAMs) play a crucial role in regulating cellular physiology and are implicated in a multitude of metabolic disorders [20]. Previously, for the study of the MAM proteome, serial centrifugation techniques have been utilized traditionally; however, these methods exhibit several limitations such as low efficiency in purifying MAM proteins. In response to these challenges, there has been a consistent demand for the development of more accurate and efficient analytical methods capable of analyzing the MAM proteome.
Kwak et al. [21] developed a novel Contact-ID biotin labeling system by splitting the biotin ligase, BiolD (previous version of TurboID), into two fragments and fusing each to the ER membrane protein SEC61B and the outer mitochondrial membrane protein TOM20, respectively. Contact-ID system capitalizes on the colocalization of SEC61B and TOM20 at ER–mitochondria contact sites, triggering the assembly of BiolD fragments and thus activating biotinylation activity that labels proximate proteins. The system was introduced into HEK293 cells, and biotinylated peptides derived from these cells were enriched and analyzed via mass spectrometry to construct a proteome profile.
By employing mCherry-BioID-tagged cytosolic proteins as a negative control for comparative analysis, Kwak et al. [21] identified 115 MAM proteins. Among the discovered MAM proteins, KFBP8 was elucidated to play a role in the formation of MAMs and in calcium transport. Furthermore, they showed a novel approach for analyzing the topology of MAM proteins by investigating the biotinylation sites of the identified proteins.
In summary, Kwak et al. [21] provided a deeper insight into the diverse functions of MAM, such as in cholesterol metabolism and apoptosis processes, which are crucial for cellular homeostasis and health. In addition, the introduction of Contact-ID method represents a significant advance in the ability to profile proteins at organelle contact sites in living cells.
ER-mitochondria contact (split-TurboID)
Previous study has shown that biotin ligase (BiolD) can be split and reconstituted under specific conditions to measure spatial specificity through proximity labeling. However, split-BiolD exhibit several limitations, including comparatively longer labeling times. Therefore, the development of a more effective split enzyme system is needed.
Cho et al. [22] employed TurboID as the enzyme of choice for a split enzyme system. The study tested 14 TurboID split sites to identify the most optimal fragments. These split-TurboID fragments were then fused to FRB and FKBP, respectively, introduced into HEK293T cells, and rapamycin-dependent reconstitution was used to induce biotin proximity labeling, thus constructing a proteome for the ER–mitochondria contact sites. Cho et al. [22] identified 101 proteins at the ER–mitochondria contact site using a split-TurboID system fused with FRB and FKBP. Further downstream analysis has validated the reliability of the discovered ER–mitochondria contact site proteins. They successfully demonstrated that split-TurboID can analyze the proteome specific to the contact site between two organelles.
Mitochondria-lysosome contact
Transmembrane 4 L six family member 5 (TM4SF5) is a protein with four transmembrane domains and is expressed across various subcellular membranes. It is known to form complexes with a variety of other proteins, such as mTOR, and plays a role in diverse metabolic activities. However, the mechanisms by which TM4SF5 translocate to specific organellar MCSs and how its translocated presence impacts metabolism have not yet been definitively elucidated.
Kim et al. [7] utilized mCherry-TM4SF5-based fluorescence microscopy analysis to reveal the localization of TM4SF5 at mitochondria-lysosome contact sites (MLCSs). Additionally, cells were engineered to express TurboID conjugated to V5-tagged translocase of the outer membrane 20 (TOM20-V5-TurboID) and lysosomal associated membrane protein 1 (LAMP1-V5-TurboID) to biotinylate MLCSs proteins, followed by subsequent enrichment of biotinylated peptides and mass spectrometry analysis to identify the proteins of MLCSs. Furthermore, for identifying proteins recruited to MLCSs by TM4SF5, cells expressing TM4SF5-v5-TurboID were subjected to the same method described above to identify MLCS proteins, followed by comparative analysis.
Kim et al. [7] revealed that TM4SF5 localizes to MLCSs and that such localization is modulated by extracellular glucose levels. The research identified 63 MLCS proteins, which were categorized into TM4SF5-enriched MLCS proteins and those absent of TM4SF5. Among the TM4SF5-enriched MLCS proteins, FKBP8 was found to be the most abundant. Further investigations uncovered that TM4SF5, once translocated to MLCS, influences mitochondrial dynamics, such as mitochondrial fission and mitophagy, as well as cholesterol metabolism.
Kim et al. [7] not only identified proteins at the MLCSs but also revealed specific proteins whose location changed when the protein TM4SF5 translocated to MLCSs. By elucidating their functions, they illuminated the unique role of TM4SF5 at MLCSs and demonstrated that TM4SF5 facilitates the export of cholesterol from lysosomes to mitochondria, which subsequently leads to changes in mitochondrial metabolism.
ER to Golgi transport
The ER-to-Golgi transport process is essential for the proper distribution of proteins and membrane components within the cell. The mechanisms involved in the formation and movement of vesicles in this process are highly complex and many aspects remain unresolved. Ninein-like protein (Nlp) is an adaptor protein involved in the assembly and transport of some ER-to-Golgi vesicles. However, the exact role of Nlp in the transition and transport of these vesicles has not been fully elucidated.
To investigate the function of Nlp in ER-to-Golgi transport, Yeerken et al. [23] introduced the SEC31A-TurboID system into cells. This system allowed them to biotinylate proteins specifically binding to ER-to-Golgi vesicles. Through these approaches, the researchers confirmed that Nlp regulates the precise formation and movement of vesicles by directly interacting with SEC31A. They also discovered that Nlp interacts with SEC31A to facilitate the assembly and transport of vesicles containing specific proteins, such as β-Catenin and STING, enabling their efficient transit from the ER to the Golgi. Yeerken et al. [23] underscored the pivotal role of Nlp in ER-to-Golgi cargo trafficking, proposed an integrated transport model, and demonstrated TurboID's utility in dynamic organelle interaction studies, extending beyond static proteome analysis.
ER and trans-Golgi contact
The proper distribution of lipids within the cell, essential for maintaining cellular function, relies heavily on lipid exchange at the contact sites between the ER and trans-Golgi. This lipid exchange is mediated by various proteins and enzymes, particularly the oxysterol binding protein (OSBP), which is known to extract cholesterol from the ER and deliver it to the trans-Golgi. However, the mechanisms by which OSBP regulates protein sorting and the polarized distribution of plasma membrane cargo proteins between the ER and trans-Golgi have not been fully elucidated.
Kovács et al. [24] introduced the OSBP-TurboID system into cells and used OSBP inhibitors, ORPphilins, to inhibit the lipid exchange activity of OSBP. They then analyzed the biotinylated proteins proximal to OSBP. The researchers identified a total of 1,507 proteins in the vicinity of OSBP, including VAPA, VAPB, PI4KIIIβ, SAC1, TMED2, CERT1, and FAPP2, confirming that these proteins function cooperatively with OSBP at ER-trans-Golgi contact sites. They also observed that following ORPphilin treatment, the proportion of basolateral cargo proteins increased while the proportion of apical cargo proteins decreased. Kovács et al. [24] provided valuable insights into OSBP's role in lipid exchange and cargo sorting at ER–trans-Golgi contact sites, thereby expanding the application of TurboID in studying organelle transport pathways and cargo trafficking mechanisms.
CONCLUSION
The recent advancements in TurboID technology have played a crucial role in the study of cellular organelles, shedding light on the complexity and specificity of their functions and interconnections. TurboID has been successfully applied to individual organelles such as the lysosome, facilitating studies on cholesterol metabolism or lysosomal repair, as well as to the ER for dynamic tracking of tissue-specific secretory proteins. Techniques like Contact-ID and split-TurboID, designed for studying organelle contact sites, promise broader applications in investigating interactions, demonstrated by studies on ER-mitochondria and mitochondria-lysosome interactions.
TurboID technology has been applied across a spectrum of experimental models, ranging from individual cells to in vivo systems. Organelle studies using TurboID in this review have shown new dimensions of temporal control and specificity in mapping organelle interactions, demonstrating the adaptability of TurboID to diverse research needs. In this regard, beyond the application examples in the aforementioned organelles, TurboID technology can be a valuable tool for researchers studying various organelle proteomes, such as mitochondria-plasma membrane interactions, ER-plasma membrane interactions, and ER-lysosome interactions, which have recently attracted significant interest.
Recently, advanced analytical methods have been developed to detect true positive biotinylated sites labeled by TurboID. The newly introduced “Super-Resolution Proximity Labeling” [25,26] has shown clearer and more specific labeling, effectively minimizing background noise and false positives commonly seen in conventional approaches. In conclusion, the progress in TurboID technology for the study of individual organelles and their interactions will cause a paradigm shift in organelle biology. It provides a foundation for continuing to build our understanding of the intricate cellular communication network, along with the potential to discover new avenues for treating numerous diseases where organelle dysfunction plays a key role.
ACKNOWLEDGEMENTS
None.
Footnotes
FUNDING
This work was supported by the National Research Foundation of Korea (NRF-2022R1C1C2004982 to K.e.K.) and Yonsei Startup Research Project (2024-72-0021).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Bae Y, Kim GY, Jessa F, Ko KS, Han J. Gallic acid-mitochondria targeting sequence-H3R9 induces mitochondria-targeted cytoprotection. Korean J Physiol Pharmacol. 2022;26:15–24. doi: 10.4196/kjpp.2022.26.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghneim HK, Alfhili MA, Alharbi SO, Alhusayni SM, Abudawood M, Aljaser FS, Al-Sheikh YA. Comprehensive investigations of key mitochondrial metabolic changes in senescent human fibroblasts. Korean J Physiol Pharmacol. 2022;26:263–275. doi: 10.4196/kjpp.2022.26.4.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung Y, Kim W, Shin NK, Bae YM, Wie J. Unveiling the impact of lysosomal ion channels: balancing ion signaling and disease pathogenesis. Korean J Physiol Pharmacol. 2023;27:311–323. doi: 10.4196/kjpp.2023.27.4.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arruda AP, Pers BM, Parlakgül G, Güney E, Inouye K, Hotamisligil GS. Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nat Med. 2014;20:1427–1435. doi: 10.1038/nm.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Zhang X, Wen Y, Li S, Lu X, Xu R, Li C. Endoplasmic reticulum-mitochondria contacts: a potential therapy target for cardiovascular remodeling-associated diseases. Front Cell Dev Biol. 2021;9:774989. doi: 10.3389/fcell.2021.774989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen HT, Wiederkehr A, Wollheim CB, Park KS. Regulation of autophagy by perilysosomal calcium: a new player in β-cell lipotoxicity. Exp Mol Med. 2024;56:273–288. doi: 10.1038/s12276-024-01265-4. Erratum in: Exp Mol Med. 2024. doi: 10.1038/s12276-024-01265-4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JE, Park SY, Kwak C, Lee Y, Song DG, Jung JW, Lee H, Shin EA, Pinanga Y, Pyo KH, Lee EH, Kim W, Kim S, Jun CD, Yun J, Choi S, Rhee HW, Liu KH, Lee JW. Glucose-mediated mitochondrial reprogramming by cholesterol export at TM4SF5-enriched mitochondria-lysosome contact sites. Cancer Commun (Lond) 2024;44:47–75. doi: 10.1002/cac2.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. Erratum in: Nat Biotechnol. 2020;38:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho KF, Branon TC, Udeshi ND, Myers SA, Carr SA, Ting AY. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat Protoc. 2020;15:3971–3999. doi: 10.1038/s41596-020-0399-0. [DOI] [PubMed] [Google Scholar]
- 10.Choi CR, Rhee HW. Proximity labeling: an enzymatic tool for spatial biology. Trends Biotechnol. 2022;40:145–148. doi: 10.1016/j.tibtech.2021.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Kang MG, Rhee HW. Molecular spatiomics by proximity labeling. Acc Chem Res. 2022;55:1411–1422. doi: 10.1021/acs.accounts.2c00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin HR, Citron YR, Wang L, Tribouillard L, Goul CS, Stipp R, Sugasawa Y, Jain A, Samson N, Lim CY, Davis OB, Castaneda-Carpio D, Qian M, Nomura DK, Perera RM, Park E, Covey DF, Laplante M, Evers AS, Zoncu R. Lysosomal GPCR-like protein LYCHOS signals cholesterol sufficiency to mTORC1. Science. 2022;377:1290–1298. doi: 10.1126/science.abg6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan JX, Finkel T. A phosphoinositide signalling pathway mediates rapid lysosomal repair. Nature. 2022;609:815–821. doi: 10.1038/s41586-022-05164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim KE, Park I, Kim J, Kang MG, Choi WG, Shin H, Kim JS, Rhee HW, Suh JM. Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice. Nat Commun. 2021;12:5204. doi: 10.1038/s41467-021-25546-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei W, Riley NM, Yang AC, Kim JT, Terrell SM, Li VL, Garcia-Contreras M, Bertozzi CR, Long JZ. Cell type-selective secretome profiling in vivo. Nat Chem Biol. 2021;17:326–334. doi: 10.1038/s41589-020-00698-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho S, Lee H, Lee HY, Kim SJ, Song W. The effect of fibroblast growth factor receptor inhibition on resistance exercise training-induced adaptation of bone and muscle quality in mice. Korean J Physiol Pharmacol. 2022;26:207–218. doi: 10.4196/kjpp.2022.26.3.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sung DJ, Jeon YK, Choi J, Kim B, Golpasandi S, Park SW, Oh SB, Bae YM. Protective effect of low-intensity treadmill exercise against acetylcholine-calcium chloride-induced atrial fibrillation in mice. Korean J Physiol Pharmacol. 2022;26:313–323. doi: 10.4196/kjpp.2022.26.5.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SJ, Kim TW, Park TH, Lee IH, Jang EC, Kwon SC, Lee HJ, Choi JH, Lee JB. Thermotherapy as an alternative to exercise for metabolic health in obese postmenopausal women: focus on circulating irisin level. Korean J Physiol Pharmacol. 2022;26:501–509. doi: 10.4196/kjpp.2022.26.6.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei W, Riley NM, Lyu X, Shen X, Guo J, Raun SH, Zhao M, Moya-Garzon MD, Basu H, Sheng-Hwa Tung A, Li VL, Huang W, Wiggenhorn AL, Svensson KJ, Snyder MP, Bertozzi CR, Long JZ. Organism-wide, cell-type-specific secretome mapping of exercise training in mice. Cell Metab. 2023;35:1261–1279.e11. doi: 10.1016/j.cmet.2023.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giorgi C, Missiroli S, Patergnani S, Duszynski J, Wieckowski MR, Pinton P. Mitochondria-associated membranes: composition, molecular mechanisms, and physiopathological implications. Antioxid Redox Signal. 2015;22:995–1019. doi: 10.1089/ars.2014.6223. [DOI] [PubMed] [Google Scholar]
- 21.Kwak C, Shin S, Park JS, Jung M, Nhung TTM, Kang MG, Lee C, Kwon TH, Park SK, Mun JY, Kim JS, Rhee HW. Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proc Natl Acad Sci U S A. 2020;117:12109–12120. doi: 10.1073/pnas.1916584117. Erratum in: Proc Natl Acad Sci U S A. 2020;117:19605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho KF, Branon TC, Rajeev S, Svinkina T, Udeshi ND, Thoudam T, Kwak C, Rhee HW, Lee IK, Carr SA, Ting AY. Split-TurboID enables contact-dependent proximity labeling in cells. Proc Natl Acad Sci U S A. 2020;117:12143–12154. doi: 10.1073/pnas.1919528117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeerken D, Xiao W, Li J, Wang Y, Wu Q, Chen J, Gong W, Lv M, Wang T, Gong Y, Liu R, Fan J, Li J, Zhang W, Zhan Q. Nlp-dependent ER-to-Golgi transport. Int J Biol Sci. 2024;20:2881–2903. doi: 10.7150/ijbs.91792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovács D, Gay AS, Debayle D, Abélanet S, Patel A, Mesmin B, Luton F, Antonny B. Lipid exchange at ER-trans-Golgi contact sites governs polarized cargo sorting. J Cell Biol. 2024;223:e202307051. doi: 10.1083/jcb.202307051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YB, Jung M, Kim J, Charles A, Christ W, Kang J, Kang MG, Kwak C, Klingström J, Smed-Sörensen A, Kim JS, Mun JY, Rhee HW. Super-resolution proximity labeling reveals anti-viral protein network and its structural changes against SARS-CoV-2 viral proteins. Cell Rep. 2023;42:112835. doi: 10.1016/j.celrep.2023.112835. [DOI] [PubMed] [Google Scholar]
- 26.Shin S, Lee SY, Kang MG, Jang DG, Kim J, Rhee HW, Kim JS. Super-resolution proximity labeling with enhanced direct identification of biotinylation sites. Commun Biol. 2024;7:554. doi: 10.1038/s42003-024-06112-w. [DOI] [PMC free article] [PubMed] [Google Scholar]