Abstract
Five diagnostic tests based on enzyme-linked immunosorbent assay (ELISA) technology for bovine paratuberculosis were evaluated by using individual serum or milk samples from 359 dairy cattle in seven paratuberculosis-free herds and 2,094 dairy cattle in seven Mycobacterium paratuberculosis-infected dairy herds. Three independent laboratories using three different culture procedures completed fecal cultures for M. paratuberculosis on these cattle and found 417 cows to be shedding M. paratuberculosis in their feces. An animal that was fecal culture positive for M. paratuberculosis by any of the three laboratories was considered a confirmed case of infection. The specificity of three ELISAs (two on serum and one on milk) was ≥99.8%. The specificity of the remaining two ELISAs, both done on serum, was 94.9 and 84.7%. Four of the five ELISAs evaluated produced similar sensitivity in detecting fecal culture-positive cattle (27.8 to 28.9%). Serum ELISA “D” had the lowest specificity (84.7%) and the highest sensitivity (44.5%), but if the cutoff value defining a positive test was changed from 125 to 250% (of the positive control) the sensitivity and specificity, 31.8 and 97.5%, respectively, were comparable to those of the other four assays. If the case definition for M. paratuberculosis infection was based on the culture results of a single laboratory instead of the combined results of three laboratories, ELISA sensitivity estimates were 45.7 to 50.0%. With the exception of ELISA D, assay agreement was high (kappa 0.66 to 0.85) for categorical assay interpretations (positive or negative), but linear regression of quantitative results showed low correlation coefficients (r2 = 0.40 to 0.68) due to the fact that ELISA results for some cows were high in one assay but low in another assay. Likelihood ratio analysis showed a direct relationship between the magnitude of ELISA result and the odds of a cow shedding M. paratuberculosis in its feces. If used judiciously and interpreted quantitatively, these ELISAs are useful tools in support of paratuberculosis control programs in dairy herds.
Johne's disease, caused by Mycobacterium paratuberculosis (also known as Mycobacterium avium subsp. paratuberculosis), has become a prevalent infectious disease problem for dairy cattle herds in most developed countries (23). Control programs require changes in herd management to limit opportunities for infection transmission to young stock coupled with diagnostic testing to identify the infected, or at least the most infectious, adult cattle (19). Because susceptibility of cattle to M. paratuberculosis is age dependent (8, 12), herd management changes focus on birthing and rearing methods of heifer calves destined to become members of the adult milking herd (19).
When within-herd infection rates are high, it is not economically feasible to cull (remove for slaughter) all test-positive cows. The net cost to cull and replace a Holstein dairy cow for a dairy producer is at least US$1,000 at today's cattle prices. Consequently, it is necessary to adopt testing strategies that provide both diagnostic and prognostic information. The owner needs to know which cows are most infectious and are not likely to survive another lactation; these cows need to be removed from the herd. It also would be helpful to know which infected cows are least infectious and are capable of sustaining another lactation and generating farm income.
Veterinary diagnostics for food animals are more strongly affected by end user economics than diagnostics for human diseases: there are no third-party payers, and profit margins in animal agriculture are small. Consequently, the most accurate and informative test results must be provided to the end users at the least cost. The diagnostic technology fulfilling this need is often based on antibody detection using enzyme-linked immunosorbent assay (ELISA) technology because of its low cost and high-throughput potential through automation. Several new ELISA kits for bovine paratuberculosis based on serum antibody detection have become available, and some companies have adopted this technology for milk samples. The purpose of the present study was to evaluate the sensitivity and specificity of five such assays.
MATERIALS AND METHODS
Dairy cattle herds.
Dairy cattle from 14 herds were included in the study. All herds were comprised of Holstein cattle, except one that had Jerseys. The uninfected population was comprised of 359 adult cattle from seven Minnesota dairy herds designated status level 4 according to the criteria of the U.S. Voluntary Bovine Johne's Disease Herd Status Program (35). Seven known M. paratuberculosis-infected Wisconsin herds, comprised of 2,094 adult cattle, were used to find cases of bovine paratuberculosis. These herds had no previous history of systematically testing for paratuberculosis or removal of test-positive cattle. The infected and noninfected herds were similar in many respects, including their standardized risk assessment scores for M. paratuberculosis infection transmission (Table 1).
TABLE 1.
Study herd characteristics
| Herd and ID codea | No. of lactating cows | Breed | Husbandry | RHAb | Risk assessment scorec | % Fecal culture positived |
|---|---|---|---|---|---|---|
| Noninfected herds | ||||||
| M | 68 | Holstein | Tie stall | 26,861 | 29 | 0.0 |
| N | 43 | Holstein | Stanchion | 23,000 | 69 | 0.0 |
| O | 48 | Holstein | Tie stall | 26,500 | 40 | 0.0 |
| P | 50 | Holstein | Tie stall | 23,500 | 46 | 0.0 |
| Q | 37 | Holstein | Stanchion | 22,000 | 74 | 0.0 |
| R | 56 | Holstein | Stanchion | 22,000 | 78 | 0.0 |
| S | 57 | Holstein | Stanchion | 25,300 | 53 | 0.0 |
| M. paratuberculosis-infected herds | ||||||
| B | 404 | Holstein | Freestall | 24,060 | 35 | 27.5C |
| C | 88 | Holstein | Stanchion | 24,835 | 43 | 14.8A |
| E | 308 | Holstein | Freestall | 23,900 | 48 | 22.7B |
| F | 261 | Jersey | Stanchion | 9,135 | 34 | 9.2A |
| G | 203 | Holstein | Freestall | 21,810 | 34 | 32.5C |
| J | 750 | Holstein | Freestall | 26,350 | 21 | 20.8B |
| K | 80 | Holstein | Stanchion | 22,820 | 26 | 9.5A |
ID, identification. Noninfected herds were all in Minnesota; infected herds were all in Wisconsin.
RHA, rolling herd average in pounds of milk per cow per year.
Risk assessment score based on Johne's disease assessment forms used in Minnesota. The Minnesota and Wisconsin herds were scored by different veterinarians.
Based on isolation of M. paratuberculosis by any one of three laboratories on aliquots of the same fecal sample collected at the same time as were the blood and milk samples. Superscript capital letters denote significant differences among herds; different letters indicate significant differences (P < 0.05).
Sample collection and processing.
Blood, milk, and fecal samples were collected simultaneously in most herds. For large herds, the work load required sample collection to be scheduled on different days; however, all samples within the herd were collected within 1 month. Blood was collected by tail vein venipuncture using anticoagulant-free Vacutainer tubes and needles Becton-Dickinson, Franklin Lakes, NJ). Milk was collected in 30-ml plastic vials during the regular milking process through the cooperation of the routine milk testing technician. Fecal samples were collected from the rectum by using disposable plastic examination gloves and transferred to Whirlpak plastic bags (Ft. Atkinson, WI). All samples were labeled and transported with refrigerant to a central laboratory for processing within 24 h of collection. On arrival, fecal samples were immediately divided into aliquots into four separate containers, packaged for shipment with refrigerant, and transported to cooperating laboratories so that the samples arrived within 48 h of collection. Milk samples were refrigerated and sent to the testing laboratory within 1 week. Blood samples were centrifuged, and the sera were harvested and then frozen until tested.
Fecal culture.
Fecal samples were tested fresh (not after freezing) for M. paratuberculosis by culture independently by three laboratories using different protocols. The University of Wisconsin laboratory decontaminated samples with 1% hexadecyl cetylpyridinium chloride (HPC) for 24 h, concentrated samples by filtration, and cultured the processed samples in modified BACTEC 12B media as previously described (5). The University of Minnesota laboratory decontaminated samples by using 0.75% HPC for 24 h, concentrated the samples by sedimentation for 72 h, and inoculated the processed samples onto four Herrold's egg yolk (HEY) agar slants containing mycobactin-J and antibiotics (BD Diagnostic Systems, Sparks, MD) as previously described (37). The University of Pennsylvania laboratory used 0.75% HPC for decontamination, double incubation, and centrifugation to concentrate samples and then inoculated one tube of the same commercial HEY medium as used by the Minnesota laboratory and three tubes of HEY without antibiotics but containing mycobactin-J (39). All M. paratuberculosis isolates were confirmed by PCR for IS900 and/or mycobactin dependency testing (2, 36). The fourth aliquot of feces was used for evaluation of a new direct PCR-based assay for M. paratuberculosis (to be reported in a separate publication).
All laboratories provided a score (0 to 4) to categorize the number of M. paratuberculosis organisms per gram of fecal sample tested. The laboratories using HEY converted the average number of M. paratuberculosis colonies seen per slant to scores as follows: 0, none; 1, 1 to 9 colonies; 2, 10 to 49 colonies; 3, 50 to 99 colonies; and 4, ≥100 colonies. BACTEC readings in weeks to positive were converted to scores as follows: 0, no M. paratuberculosis isolated after 12 weeks incubation; 1, >8 weeks; 2, 6.1 to 8 weeks; 3, 4.1 to 6 weeks; 4, ≤4 weeks. This system is based on previous studies showing a direct relationship between the time to detection and the number of M. paratuberculosis inoculated into the BACTEC vial (22). The mean score for the three laboratories was used to classify each animal's fecal shedding rate.
Antibody detection-based assays.
Four commercial ELISA kits, designated A, B, C and D, were evaluated for M. paratuberculosis serum antibody detection. All kits are self-contained having all necessary reagents, positive and negative control sera, and interpretation criteria. Kits A (HerdCheck M. paratuberculosis ELISA; IDEXX Laboratories, Inc., Westbrook, ME), B (ParaCheck; CSL/Biocor, Omaha, NE), and D (SERELISA ParaTB; Synbiotic Corp., San Diego, CA) are licensed by the U.S. Department of Agriculture (USDA). Kit C is produced in France (ELISA Paratuberculosis; Institut Pourquier, Montpellier, France). All antibody assays, except D, use a serum absorption step to remove antibodies that cross-react with Mycobacterium phlei a procedure that Yokomizo et al. demonstrated to increase M. paratuberculosis ELISA specificity (41, 42). All serum antibody assays were performed by University of Wisconsin laboratories according to manufacturers' instructions and interpreted as prescribed by the kit insert. The testing laboratories were blinded to fecal culture results.
A fifth ELISA (E) was based on detection of M. paratuberculosis antibody in milk samples. This in-house assay is not sold as a diagnostic kit but rather is offered as a service by a Michigan laboratory (Antel BioSystems, Inc., Lansing, MI). Milk ELISA results were provided by the testing laboratory without knowledge of the animal or herd M. paratuberculosis-infection status. Fewer cows were tested by ELISA on milk than on serum because not all cows in herds were lactating on the day of sampling.
Data analysis.
The case definition of a noninfected cow was any cow resident in any one of the seven Johne's disease program level 4 herds (35). The absence of M. paratuberculosis infection in these herds was confirmed by fecal culture of all adult cattle in each of the seven herds by all three participating laboratories. Assay specificity estimates were based on tests done on these cattle, i.e., all positive ELISA tests on cattle in these herds were considered to be false-positive tests.
The case definition of an infected cow was isolation of M. paratuberculosis from a fecal sample by any one of the three laboratories. The antibody detection-based ELISAs were compared for their ability to detect the cattle that were fecal culture positive at the time of sample collection. This is not a measure of true assay sensitivity based on comparison to an accepted “gold standard” because infected cattle shedding M. paratuberculosis in their feces when sampled (or shedding but not detected by any of the three cultures) obviously were not counted as cases. For simplicity, however, although the sensitivity estimates reported in the present study are “relative sensitivity” estimates, i.e., relative to fecal culture, we use the term “sensitivity” throughout this study. No prior studies have used M. paratuberculosis culture results for the same sample from multiple independent laboratories to increase the chances of identifying cases of infection. Consequently, to gauge the effect this more rigorous case definition had on assay sensitivity estimates, we also report estimates based on a single laboratory's fecal culture results, namely, those from the University of Minnesota. This laboratory was chosen because it used the culture method most commonly cited in prior publications in which fecal culture was used as the reference test for serological test evaluation and because this method had a M. paratuberculosis detection rate in between those of the other two laboratories that performed fecal culture. The rate of positive ELISAs for culture-positive cows was also calculated by the level of M. paratuberculosis shedding by using the mean fecal shedding score.
Sensitivity and specificity differences among herds were determined by using the Z-statistic for comparison of independent proportions (25). Agreement between pairs of assays was evaluated by McNemar's chi-square analysis and kappa calculation (25, 28). In similar pairwise fashion, linear regression was performed, and r2 values were reported as measures of the correlation in magnitude of quantitative test results (GraphPad InStat, version 3.00; GraphPad Software, San Diego, CA). Differences were considered significant at P < 0.05.
The likelihood ratio (LR) was determined for each of five ELISA result interpretation ranges for each of the five assays (30). The test manufacturer's cutoff for a positive test was used as the lower limit for one of the five test interpretation ranges. The LR was calculated as follows: LR = (percent infected cows with results in the range)/(percentage of noninfected cows with results in the range). Post-test M. paratuberculosis infection probabilities were calculated for each of five assay interpretation levels and six within-herd infection rates ranging from 1 to 30% by standard methods (30).
RESULTS
Fecal culture results confirmed that all program level 4 herds were not infected with M. paratuberculosis. For the seven infected dairy herds, the prevalence of M. paratuberculosis infections ranged from 9.2 to 32.5% (Table 1). There was no apparent relationship between herd infection rate, risk assessment score, or herd production level.
Diagnostic specificity among the five ELISAs evaluated ranged from 84.7% (95% CI: 82.9 to 86.5%) for assay D to 100% for assay C (Table 2). The three highest specificity assays—B, C, and E—had zero to two false-positive tests among the cattle tested, and assay specificities were not significantly different. The specificity of assays A and D varied significantly among herds ranging from 84.0 to 100% for assay A and 62.0 to 94.6% for assay D.
TABLE 2.
Specificity of antibody detection tests
| Herd | Neg/total (%)a
|
||||
|---|---|---|---|---|---|
| Test A | Test B | Test C | Test D | Test E | |
| M | 65/67 (97.0)A | 66/67 (98.5) | 67/67 (100.0) | 60/67 (89.6)A,1 | 66/67 (98.5) |
| N | 41/42 (97.6)A | 42/42 (100.0) | 42/42 (100.0) | 37/42 (88.1)A,1 | 42/42 (100.0) |
| O | 42/50 (84.0)B,C,1 | 50/50 (100.0) | 50/50 (100.0) | 31/50 (62.0)C,2 | 48/48 (100.0) |
| P | 49/50 (98.0)A | 50/50 (100.0) | 50/50 (100.0) | 45/50 (90.0)A,1 | 50/50 (100.0) |
| Q | 36/37 (97.3)A | 37/37 (100.0) | 37/37 (100.0) | 25/37 (67.6)B,1 | 37/37 (100.0) |
| R | 56/56 (100.0)A | 56/56 (100.0) | 56/56 (100.0) | 53/56 (94.6)A | 56/56 (100.0) |
| S | 53/57 (93.0)B,1 | 57/57 (100.0) | 57/57 (100.0) | 53/57 (93.0)A,1 | 52/52 (100.0) |
| Combined | 17/359 (95.26)2 | 358/359 (99.72) | 359/359 (100.00) | 349/359 (84.91)3 | 351/352 (99.72) |
Neg/total, number testing negative/total number tested (i.e., the specificity). Superscript letters A, B, and C indicate statistical comparisons among herds (within column). Different letters indicate significant differences in specificity (P < 0.05). Superscript numbers 1, 2, and 3 indicate statistical comparisons among test herds (within rows). Values without numerical superscripts are not significantly different, and different numbers indicate significant differences in specificity (P < 0.05). The percent ranges for tests A, B, C, D, and E are 84 to 100.0, 98.5 to 100.0, 100.0, 62.0 to 94.9, and 98.5 to 100.0, respectively.
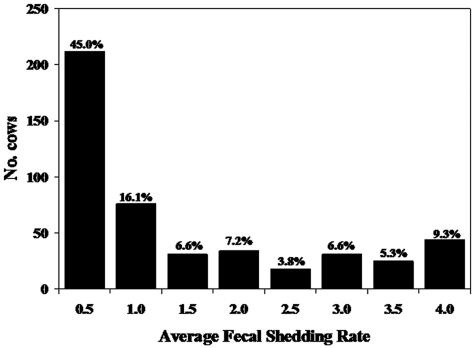
Among the 2,094 cattle tested, 417 (19.9%) were found to be infected with M. paratuberculosis by fecal culture. Mean fecal shedding rates among these infected cows were computed from the scores provided by the three fecal culture testing laboratories. The distribution of shedding rates among the culture-positive cows shows a preponderance of cows (45%) in the very low shedding category (Fig. 1).
FIG. 1.
Number and percentage of cows at each level of fecal shedding based on their average fecal shedding score for 443 M. paratuberculosis-infected cows among seven infected herds.
The ability of the ELISAs to detect culture-positive cows shedding at any level was not different for four of the five assays (27.95 to 28.92%; Table 3). Assay D's sensitivity was higher (44.5%; P < 0.05). When culture results from only the Minnesota laboratory were used to define cases of infection, assay sensitivities ranged from 45.7 to 62.7% (data not shown).
TABLE 3.
Percentage of cattle testing antibody positive compared to fecal culture results among the seven M. paratuberculosis infected dairy herds
| Fecal shedding scorea | Pos/total (%)b
|
Neg/total (%)b
|
|||
|---|---|---|---|---|---|
| Test A | Test B | Test C | Test D | Test E | |
| 0 | 92/1,459 (6.31)3 | 42/1,457 (2.88)2 | 21/1,454 (1.44)1 | 323/1,450 (22.28)4 | 50/1,355 (3.69)2 |
| 0.1-1.0 | 29/229 (9.61)1 | 22/229 (9.61)1 | 16/229 (6.99)1 | 50/175 (28.57)2 | 23/197 (11.68)1 |
| 1.1-2.0 | 12/68 (17.65)1 | 16/68 (23.53)1 | 13/68 (19.12)1 | 25/50 (50.00)2 | 17/65 (26.15)1 |
| 2.1-3.0 | 23/36 (63.89)1 | 20/36 (55.56)1 | 20/36 (55.56)1 | 14/25 (56.00)1 | 13/30 (43.33)1 |
| 3.1-4.0 | 63/82 (76.83)1 | 60/82 (73.17)1 | 67/82 (81.71)1 | 45/51 (88.24)2 | 52/72 (72.22)1 |
| All shedders combined | 120/415 (28.92)1 | 118/415 (28.43)1 | 116/415 (27.95)1 | 134/301 (44.52)2 | 105/364 (28.85)1 |
That is, the mean fecal shedding score range. Animals were only included in the “0” category if all three fecal culture tests were done and found to be negative.
Pos/total and Neg/total, number positive and number negative/total number tested, respectively. Superscript numbers 1, 2, 3, and 4 indicate statistical comparisons among tests for each fecal score (within rows). Different numbers indicate significant differences in the percentages of positive tests (P < 0.05).
There was a direct relationship between the level of fecal shedding of M. paratuberculosis and the percentage of positive assays (Table 3). Positive ELISAs were found for 6.9 to 28.6% (mean, 13.3%) of cows with low numbers of M. paratuberculosis in their feces (fecal scores > 0 to 1). At progressively higher fecal culture scores, the mean percentages of positive antibody assays for all five assays were 27.3, 54.9, and 78.4%, respectively.
With the exception of assay D, categorical result interpretation agreement among the assays was generally high: >86% of assays were in agreement when the sample was classified as positive or negative, no differences were noted by χ2 analysis, and kappa values were 0.66 to 0.85 (Table 4). Assay D was different from all other assays due to its higher sensitivity (44.5%) and lower specificity (84.7%), resulting in kappa values of 0.47 to 0.59 in pairwise analysis between ELISA D and each of the other four assays.
TABLE 4.
Pairwise comparison of antibody detection-based tests for paratuberculosis on fecal culture-positive cows
| Tests compared | No. of samples | % Agreement | χ2 valuea | Kappa statistic | Linear regression r2 value |
|---|---|---|---|---|---|
| A vs B | 415 | 90.6 | 0.026 | 0.77 | 0.583 |
| A vs C | 415 | 94.0 | 0.360 | 0.85 | 0.821 |
| A vs D | 300 | 80.4 | 40.700* | 0.59 | 0.677 |
| A vs E | 347 | 86.2 | 0.750 | 0.66 | 0.558 |
| B vs C | 415 | 92.3 | 0.125 | 0.81 | 0.589 |
| B vs D | 300 | 77.4 | 36.760* | 0.53 | 0.503 |
| B vs E | 347 | 87.3 | 1.455 | 0.68 | 0.384 |
| C vs D | 300 | 76.0 | 17.050* | 0.49 | 0.753 |
| C vs E | 347 | 87.6 | 0.581 | 0.69 | 0.509 |
| D vs E | 256 | 74.6 | 34.909* | 0.47 | 0.397 |
McNemar's χ2 value for correlated proportions. *, significant difference (P < 0.001).
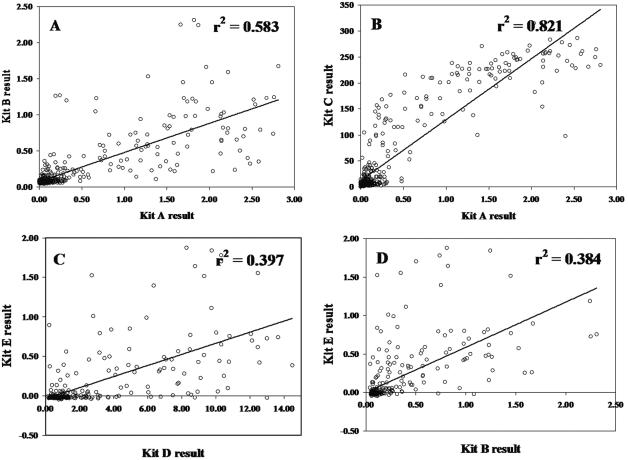
Although assay agreement was relatively high by categorical assay interpretation, a comparison of quantitative optical density results showed greater disparity. Scatter plots show that some samples had very high antibody levels according to one assay but very low antibody levels by a different assay (Fig. 2). Linear regression produced r2 values ranging from 0.384 to 0.821 (Table 4).
FIG. 2.
Scatter plots with linear regression lines comparing quantitative results for four pairs of ELISAs selected to show a range of correlations. (A) Kit A and kit B; (B) kit A and kit C; (C) kit D and kit E; (D) kit B and kit E.
For all five ELISAs the magnitude of the result was related to the likelihood that the tested animal was fecal culture positive (Table 5). For assays B, C, and E the LR (the odds of being fecal culture positive) for animals with results in the category just below the manufacturer's recommended ELISA cutoff was >3.2. For ELISA C, if the assay cutoff was lowered from 70 to 40 a modest increase in test sensitivity would result without affecting test specificity for the animals evaluated in the present study. If the cutoff for ELISA D was changed from the recommended value of 125 to 250% (percent the positive control), the sensitivity and specificity, 31.8 and 97.5%, respectively, were comparable to those of the other four assays. For ELISA A there was no significant difference between the rate of positive results among dairy cattle in the paratuberculosis-free herds and those that were fecal culture negative in the M. paratuberculosis-infected herds. Both ELISA A and D had the lowest specificity in herd O, being 84.0 and 62.0%, respectively.
TABLE 5.
LRs for five result ranges for each of the five assays evaluated
| Test (manufacturer's cutoff) | Result range | Infected cows
|
Noninfected cows
|
LRa | ||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| A (≥0.25) | <0.10 | 210 | 50.6 | 252 | 70.2 | 0.72 |
| 0.10-0.25 | 85 | 20.5 | 92 | 25.6 | 0.80 | |
| 0.26-0.40 | 21 | 5.1 | 12 | 3.3 | 1.51 | |
| 0.41-1.00 | 26 | 6.3 | 2 | 0.6 | 11.25 | |
| ≥1.00 | 73 | 17.6 | 1 | 0.3 | 63.15 | |
| B (≥0.100) | <0.00 | 159 | 38.1 | 286 | 79.7 | 0.48 |
| 0.000-0.050 | 116 | 27.8 | 65 | 18.1 | 1.54 | |
| 0.051-0.100 | 23 | 5.5 | 7 | 2.0 | 2.83 | |
| 0.101-0.500 | 63 | 15.1 | 1 | 0.3 | 54.24 | |
| ≥0.500 | 56 | 13.4 | 0 | 0.0 | ∞ | |
| C (≥70) | <10 | 213 | 51.4 | 317 | 88.3 | 0.58 |
| 10-20 | 51 | 12.3 | 36 | 1.0 | 1.23 | |
| 21-40 | 24 | 5.8 | 6 | 1.7 | 3.47 | |
| 41-70 | 10 | 2.4 | 0 | 0.0 | ∞ | |
| ≥70 | 116 | 28.0 | 0 | 0.0 | ∞ | |
| D (≥125) | 0-75 | 98 | 32.7 | 222 | 62.0 | 0.53 |
| 76-125 | 67 | 22.3 | 82 | 22.9 | 0.98 | |
| 126-250 | 40 | 13.3 | 45 | 12.6 | 1.06 | |
| 251-500 | 30 | 10.0 | 7 | 2.0 | 5.11 | |
| ≥500 | 65 | 21.7 | 2 | 0.6 | 38.78 | |
| E (≥0.10) | <0.00 | 213 | 58.7 | 291 | 82.7 | 0.71 |
| 0.00-0.05 | 42 | 11.6 | 58 | 14.8 | 0.78 | |
| 0.06-0.10 | 12 | 3.3 | 2 | 0.6 | 5.50 | |
| 0.11-0.15 | 13 | 3.6 | 1 | 0.3 | 12.61 | |
| ≥0.15 | 83 | 22.9 | 0 | 0.0 | ∞ | |
The values reported were derived from the original data and may differ slightly from the LRs calculated from the percentages in the table due to rounding.
For illustrative purposes, the post-test probabilities of M. paratuberculosis infection were calculated for one of the assays, ELISA B, at all five levels of interpretation and for within-herd infection prevalence ranging from 5 to 30% (Table 6). At the manufacturer's recommended cutoff of ≥0.100, the post-test probability of M. paratuberculosis infection was >90% when the within-herd prevalence was >10%. At the highest level of ELISA B result (i.e., ≥0.500) the post-test probability of infection was >90% when the within-herd prevalence was >5%.
TABLE 6.
Post-test probabilities of M. paratuberculosis infection for ELISA B at five levels of assay result and within-herd M. paratuberculosis infection prevalence ranging from 1 to 30%a
| ELISA result | LRb | Within-herd true prevalence (pretest infection probability) (%) at:
|
|||||
|---|---|---|---|---|---|---|---|
| 5% | 10% | 15% | 20% | 25% | 30% | ||
| <0.00 | 0.48 | 2.5 | 5.1 | 7.8 | 10.7 | 13.8 | 17.1 |
| 0.00-0.050 | 1.54 | 7.5 | 14.6 | 21.4 | 27.8 | 33.9 | 39.8 |
| 0.051-0.100 | 2.84 | 13.0 | 23.9 | 33.3 | 41.4 | 48.5 | 54.8 |
| 0.101-0.500 | 54.24 | 74.1 | 85.8 | 90.5 | 93.1 | 94.8 | 95.9 |
| ≥0.500 | ∞c | 91.3 | 95.7 | 97.2 | 98.0 | 98.5 | 98.8 |
The calculation steps were as follows: (i) pretest odds = (pretest probability)/[1 − (pretest probability)]; (ii) post-test odds = pretest odds × LR; and (iii) post-test probability = (post-test odds)/(post-test odds + 1).
LR for ELISA B from Table 5.
A value of 200 was used instead of dividing by ∞.
DISCUSSION
Bovine paratuberculosis is a chronic and infectious mycobacterial infection of the gastrointestinal tract (13). At least 22% of U.S. dairy herds are infected with M. paratuberculosis (38). Consolidation of the U.S. dairy industry and the ongoing practice of buying cattle without regard for the M. paratuberculosis infection status of either the purchased cattle or their herd of origin facilitates the spread of the infection among herds. The 20% fecal culture-positive rate seen among the seven infected dairy herds used in the present study is not unusual. Moreover, given the latency of this infection and the diagnostic sensitivity of fecal culture, the true infection rate in these herds is even higher.
For paratuberculosis diagnostics it can be challenging to evaluate an assay's specificity. In dairy cattle the time from infection, usually as a calf, to clinical disease varies from as short as 1 year to as much as 12 years, the upper limit of life span for dairy cattle on most farms (17). Prolonged disease latency makes it impossible to rely on a negative fecal culture as a “gold standard” to define absence of M. paratuberculosis infection for animals residing in known infected herds. Instead, the best standard for absence of M. paratuberculosis infection for an individual animal must be based on absence of infection from the entire herd in which the animal was raised. Moreover, absence of infection from a herd must be based on multiple annual negative tests of all adult cattle, as well as strict adherence to recommended management practices that prevent introduction of M. paratuberculosis-infected cattle into the herd. These standards are outlined in the Voluntary Bovine Johne's Disease Control Program (35) and served as the gold standard to qualify the adult cattle in seven dairy herds for ELISA specificity estimation in the present study.
The diagnostic specificity for three of the five ELISAs evaluated was ≥99.8% and not statistically different. This corroborates the findings of similar studies on the same ELISA kits (27). ELISAs A and D had significantly lower specificities, 94.9 and 84.7%, respectively (Table 2). The specificity rates of these two assays also varied among herds, whereas the other three ELISAs' specificity rates did not. Reasons for assay specificity differences among herds are not known but likely relate to qualitative or quantitative differences in the microbial flora the animals in those herds are exposed to, notably the presence of other mycobacteria, as well as the composition of antigens used as solid-phase antigens on ELISA plates and/or antigens contained in serum or milk absorption reagents. Herd-to-herd assay specificity variation is an important consideration in judging assay performance; it may even be necessary to include herds from different geographic areas (and therefore likely varied microbial flora) to obtain a full sense of the range of specificities displayed by the assay. Herd screening for possible M. paratuberculosis infection by ELISA demands use of high specificity assays to limit false-positive herd classifications and the time and effort spent trying to confirm the diagnosis (7, 16).
Among the fecal culture-negative cattle in the infected herds, <2.9% tested positive for serum antibody and <3.7% tested positive for antibody in milk for the three highest-specificity ELISAs. Arguably, these apparently ELISA false-positive cattle were truly infected with M. paratuberculosis, but fecal culture failed to detect the organism because the animal was not shedding it in feces on the day of sampling or was shedding at a level below the culture detection limits. M. paratuberculosis exposure, triggering antibody production without progressive infection, e.g., exposure as an adult when more infection resistant, is also a plausible explanation for ELISA positive findings in fecal culture-negative cattle. Regardless, this was a low-frequency occurrence for the high-specificity assays and of limited concern should these assays be used in a paratuberculosis control program.
The sensitivity of M. paratuberculosis fecal culture-positive dairy cattle detection by ELISA was the same for four of five assays and was related to the rigor of the case definition. If the sensitivity of these same assays were judged in comparison to the fecal culture results of a single laboratory (Minnesota Veterinary Diagnostic Laboratory), then the estimated sensitivities would be much higher, i.e., 45.7 to 50.0%, and in agreement with previous reports (6, 15, 21, 29, 31, 32, 39, 40). It is noteworthy that the diagnostic sensitivity of a new direct high-throughput PCR assay applied to fecal samples from cattle used in the present study, using the same single case definition, was also in this range (data not shown). Clearly, the fecal culture methodology and number of samples evaluated to establish a case definition affect the estimates of the sensitivity of other assays being evaluated and are a major study design issue to be considered when evaluating diagnostic tests for paratuberculosis. Given that antibody-based diagnostics detected fewer than one-third of all M. paratuberculosis-infected and excreting dairy cattle in a herd, the percentage of cattle testing positive could be multiplied by at least a factor of three to get a rough estimate of true within herd prevalence of infection. This is only applicable, of course, for herds that have not previously been testing for paratuberculosis and culling cattle based on test results.
The number of CFU of M. paratuberculosis in bovine fecal samples is considered a measure of the stage of infection in the animal. In the present study, the ability of an ELISA to detect an infected animal was directly related to the level of M. paratuberculosis shedding (Table 3), a finding consistent with previous reports (6, 32). Cattle with large numbers of M. paratuberculosis per gram of feces, so-called “heavy shedders,” were detected by the evaluated ELISAs >72% of the time. This is significant in that these animals logically represent the greatest risk for environmental contamination and thus infection transmission on a dairy farm. Evidence from field studies to support the contention that detection and management of the heavy shedders is critical to control of bovine paratuberculosis are lacking. However, both modeling and field data on human tuberculosis verify that focusing treatment and control efforts on the most infectious subset of Mycobacterium tuberculosis-infected persons can cause dramatic reductions in the prevalence of the infection in the general population (10). Whether the cattle shedding large numbers of M. paratuberculosis but missed by ELISAs quickly progress to clinical paratuberculosis and are therefore culled from the herd or remain in the herd and continue to spread infection is not known. Only well-designed and controlled longitudinal field studies will be able to answer these questions.
When ELISA results were judged dichotomously (positive or negative), the assays showed comparable accuracy and good agreement. ELISAs A, B, C, and E did not differ in classification of culture-positive cattle as test positive or negative (χ2, P > 0.05), and the kappa statistic indicated a high level of agreement (kappa ≥ 0.66; Table 4). Of these assays, B, C, and E were not significantly different in either sensitivity or specificity. ELISA D would have had comparable sensitivity and specificity estimates, i.e., 31.8 and 97.5%, respectively, if an assay cutoff of 250% (sample OD as a percentage of kit positive control) were used instead of 125% as recommended by the kit manufacturer (Table 5).
Scatter plots illustrate that serum or milk from individual cattle can respond very strongly when tested by one ELISA but not another. Representative plots are shown in Fig. 2, and linear regression correlation coefficients for all pairwise assay comparisons are listed in Table 4. Clearly, cattle can respond to M. paratuberculosis infection with serum antibodies detected by one ELISA kit but not in another. Since the formulation of the M. paratuberculosis solid-phase antigen and M. phlei serum absorption antigens used in these kits is proprietary, the nature of putative antigenic differences in kit components is not known. Identification of the diversity of M. paratuberculosis antigens or epitopes that induce antibody responses in cattle could lead to improved diagnostic kits. Purified single antigen assays will likely suffer from low diagnostic sensitivity.
LR analysis confirmed the earlier report that multilevel paratuberculosis ELISA interpretation is useful (4, 26). All five ELISAs evaluated showed a direct relationship between the magnitude of ELISA result and the likelihood the tested cattle were fecal culture-positive for M. paratuberculosis. LRs in combination with estimated within herd prevalence, i.e., the pretest probability of M. paratuberculosis infection, can be used to calculate the post-test probability of infection (shown for ELISA B in Table 6). For ELISAs A, B, C, and E, cattle with results in the highest of the five ELISA result ranges, so called “strong-positive” results, had a post-test probability of concurrent M. paratuberculosis fecal shedding of >90% if herds had a within herd prevalence >10%. For ELISAs B and C, a high (>90%) post-test probability of M. paratuberculosis infection was also possible for the next-lower level of assay result when herds had a >15% within-herd infection prevalence. For most M. paratuberculosis-infected commercial dairy herds, confirmatory testing of such animals by fecal culture would be neither necessary nor cost-effective. Post-test infection probabilities can be used in decision analysis models to optimize economic outcomes from decisions based on ELISA results. However, this requires estimation of the infection transmission risks for animals at each level of M. paratuberculosis shedding, a factor affected by on-farm management systems. Measurement of infection transmission risks for cows by stage of infection under different farm management conditions by well-controlled epidemiological field studies is an important next step in optimization of paratuberculosis control programs in dairy herds.
An important first step in a national paratuberculosis control program is segregation of cattle herds into infected and not infected categories (1, 18, 33). To halt the spread of M. paratuberculosis infection, the next step is to apply voluntary or mandatory biosecurity regulations to prevent movement of cattle from infected (or untested) herds into noninfected or at least test-negative herds. Rapid, low-cost, high-throughput assays are necessary if even a modest proportion of the 95.8 million cattle in the United States are to be tested (34). The use of high-specificity ELISAs avoids unnecessary follow-up testing of herds due to false-positive tests (7). Higher assay sensitivity than that available with current ELISAs for detection of infected individual animals would be desirable, but herd-level sensitivity is the more critical factor, and this can be improved by increasing the number of animals tested per herd and by focusing testing on the older animals since they have had sufficient time for a M. paratuberculosis infection to progress and induce antibody production (15).
ELISA E performed as well or better on milk samples than the ELISAs that used sera and are in agreement with a recent report on evaluation of this same commercial test (14). Assays that use milk samples are ideally suited for paratuberculosis control in dairy herds but impractical for use on beef cattle. Milk samples are normally collected by testing laboratories from all lactating cows in a herd on a monthly basis for measurement of somatic cell counts (a measure of udder health), plus milk fat and protein content. Milk testing laboratories are equipped with automated sample handing equipment that allows thousands of samples to be analyzed daily. In addition, electronic transmission of results downloaded to on-farm computers is common. These data are used on a daily basis for herd management. Milk testing laboratories could thus adapt paratuberculosis ELISA technology to expand the services they provide to dairy producers and more cost-effectively test dairy cattle. Unfortunately, at present there are no USDA-licensed kits validated for testing milk samples for antibodies to M. paratuberculosis.
Organism detection tests for paratuberculosis will remain a mainstay for the definitive diagnosis of paratuberculosis. Isolation of M. paratuberculosis in liquid or on solid medium not only provides the most sensitive and specific diagnostic method available but also offers the ability to complete molecular studies that increase our understanding of the epidemiology and microbial ecology of this pathogen (3, 39). Genetics-based diagnostic tests have not yet matched the accuracy of culture-based diagnostics for paratuberculosis and fail to provide viable organisms for further study (9, 11, 20, 24). If M. paratuberculosis becomes classified as a zoonotic pathogen and for adequate food safety it is decided that dairy and beef products only originate from paratuberculosis test-negative herds, the need for all types of paratuberculosis diagnostics on multiple types of animals and possibly humans will be enormous.
Acknowledgments
This project was funded in part by a cooperative agreement between the USDA-APHIS-Veterinary Services and the University of Wisconsin. Additional funding was provided by each of the cooperating laboratories and the Minnesota Board of Animal Health.
The dedicated efforts of the technical staff of the Johne's Testing Center, Vic Eggleston, Gail Thomas, Heather Cushing, Kelly Anklam, Brenna Kunkel, Melissa Hampton, Johnnie Ambrosy, the R. D. Schultz laboratory, and Terry Fyock at the University of Pennsylvania are sincerely appreciated. Also, Lindsey Aipperspach and the field veterinarians of the Minnesota Board of Animal health and the technicians of the various Dairy Herd Improvement associations who helped collect samples from dairy herds are gratefully acknowledged. We are especially indebted to the dairy herd owners who not only made their herds available for the present study but also assisted with sample collection and animal handling. We thank Becton Dickinson for the donation of a portion of the culture media used in the study and Philippe Pourquier of Institut Pourquier and Synbiotics for the donation of ELISA kits.
REFERENCES
- 1.Benedictus, G., J. Verhoeff, Y. H. Schukken, and J. W. Hesselink. 1999. Dutch paratuberculosis programme: history, principles and development, p. 2-15. In E. J. B. Manning and M. T. Collins (ed.), Proceedings of the Sixth International Colloquium on Paratuberculosis. International Association for Paratuberculosis, Madison, Wis.
- 2.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1989. Identification of a repetitive DNA sequence specific to Mycobacterium paratuberculosis. FEMS Microbiol. Lett. 60:175-178. [DOI] [PubMed] [Google Scholar]
- 3.Collins, M. T. 1996. Diagnosis of paratuberculosis, p. 357-371. In R. W. Sweeney (ed.), Paratuberculosis (Johne's disease). The W. B. Saunders Company, Philadelphia, Pa.
- 4.Collins, M. T. 2002. Interpretation of a commercial bovine paratuberculosis enzyme-linked immunosorbent assay by using likelihood ratios. Clin. Diagn. Lab. Immunol. 9:1367-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, M. T., K. B. Kenefick, D. C. Sockett, R. S. Lambrecht, J. McDonald, and J. B. Jørgensen. 1990. Enhanced radiometric detection of Mycobacterium paratuberculosis using filter concentrated fecal specimens. J. Clin. Microbiol. 28:2514-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dargatz, D. A., B. A. Byrum, L. K. Barber, R. W. Sweeney, R. H. Whitlock, W. P. Shulaw, R. H. Jacobson, and J. R. Stabel. 2001. Evaluation of a commercial ELISA for diagnosis of paratuberculosis in cattle. J. Am. Vet. Med. Assoc. 218:1163-1166. [DOI] [PubMed] [Google Scholar]
- 7.Donald, A. W., I. A. Gardner, and A. D. Wiggins. 1994. Cut-off points for aggregate herd testing in the presence of disease clustering and correlation of test errors. Prev. Vet. Med. 167-187.
- 8.Doyle, T. M. 1953. Susceptibility to Johne's disease in relation to age. Vet. Rec. 65:363-365. [Google Scholar]
- 9.Erume, J., J. Spergser, and R. Rosengarten. 2001. Rapid detection of Mycobacterium avium subsp. paratuberculosis from cattle and zoo animals by nested PCR. Afr. Health Sci. 1:83-89. [PMC free article] [PubMed] [Google Scholar]
- 10.Frieden, T. R. 2002. Can tuberculosis be controlled? Int. J. Epidemiol. 31:894-899. [DOI] [PubMed] [Google Scholar]
- 11.Garrido, J. M., N. Cortabarria, J. A. Oguiza, G. Aduriz, and R. A. Juste. 2000. Use of a PCR method on fecal samples for diagnosis of sheep paratuberculosis. Vet. Microbiol. 77:379-386. [DOI] [PubMed] [Google Scholar]
- 12.Hagan, W. A. 1938. Age as a factor in susceptibility to Johne's disease. Cornell Vet. 28:34-40. [Google Scholar]
- 13.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in Veterinary Medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrick, S. H., T. F. Duffield, D. F. Kelton, K. E. Leslie, K. D. Lissemore, and M. Archambault. 2005. Evaluation of enzyme-linked immunosorbent assays performed on milk and serum samples for detection of paratuberculosis in lactating dairy cows. J. Am. Vet. Med. Assoc. 226:424-428. [DOI] [PubMed] [Google Scholar]
- 15.Jakobsen, M. B., L. Alban, and S. S. Nielsen. 2000. A cross-sectional study of paratuberculosis in 1155 Danish dairy cows. Prev. Vet. Med. 46:15-27. [DOI] [PubMed] [Google Scholar]
- 16.Jordan, D. 1996. Aggregate testing for the evaluation of Johne's disease herd status. Aust. Vet. J. 73:16-19. [DOI] [PubMed] [Google Scholar]
- 17.Jubb, T., and J. Galvin. 2000. Herd testing to control bovine Johne's disease. Vet. Microbiol. 77:423-428. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy, D. J., and M. B. Allworth. 1999. Progress in national control and assurance programs for bovine Johne's disease in Australia, p. 19-26. In E. J. B. Manning and M. T. Collins (ed.), Proceedings of the Sixth International Colloquium on Paratuberculosis. International Association for Paratuberculosis, Madison, Wis.
- 19.Kennedy, D. J., and G. Benedictus. 2001. Control of Mycobacterium avium subsp. paratuberculosis infection in agricultural species. Rev. Sci. Tech. 20:151-179. [DOI] [PubMed] [Google Scholar]
- 20.Khare, S., T. A. Ficht, R. L. Santos, J. Romano, A. R. Ficht, S. Zhang, I. R. Grant, M. Libal, D. Hunter, and L. G. Adams. 2004. Rapid and sensitive detection of Mycobacterium avium subsp. paratuberculosis in bovine milk and feces by a combination of immunomagnetic bead separation-conventional PCR and real-time PCR. J. Clin. Microbiol. 42:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klausen, J., A. Huda, L. Ekeroth, and P. Ahrens. 2003. Evaluation of serum and milk ELISAs for paratuberculosis in Danish dairy cattle. Prev. Vet. Med. 58:171-178. [DOI] [PubMed] [Google Scholar]
- 22.Lambrecht, R. S., J. Carriere, and M. T. Collins. 1988. A model for analyzing growth kinetics of a slowly growing Mycobacterium sp. Appl. Environ. Microbiol. 54:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning, E. J. B., and M. T. Collins. 2001. Mycobacterium avium subsp. paratuberculosis: pathogen, pathogenesis, and diagnosis. Rev. Sci. Tech. 20:133-150. [DOI] [PubMed] [Google Scholar]
- 24.Marsh, I. B., and R. J. Whittington. 2001. Progress toward a rapid polymerase chain reaction diagnostic test for the identification of Mycobacterium avium subsp. paratuberculosis in faeces. Mol. Cell. Probes 15:105-118. [DOI] [PubMed] [Google Scholar]
- 25.Martin, S. W., A. H. Meek, and P. Willeberg. 1987. Measurement of disease frequency and production, p. 73-76. In S. W. Martin, A. H. Meek, and P. Willeberg (ed.), Veterinary epidemiology: principles and methods. Iowa State University Press, Ames, Iowa.
- 26.Naugle, A. L., W. P. Shulaw, W. J. A. Saville, T. E. Wittum, B. Byrum, and M. T. Collins. 2003. Calculation method for likelihood ratios dictates interpretation. Clin. Diagn. Lab. Immunol. 10:729-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitt, D. J., D. S. Pinch, A. Janmaat, and R. J. Condron. 2002. An estimate of specificity for a Johne's disease absorbed ELISA in northern Australian cattle. Aust. Vet. J. 80:57-60. [DOI] [PubMed] [Google Scholar]
- 28.Remington, R. D., and M. A. Schork. 1970. Chi-square tests for frequency data, p. 229-252. In Statistics with applications to the biological and health sciences. Prentice-Hall, Inc., Englewood Cliffs, N.J.
- 29.Ridge, S. E., I. R. Morgan, D. C. Sockett, M. T. Collins, R. J. Condron, N. W. Skilbeck, and J. J. Webber. 1991. Comparison of the Johne's absorbed EIA and the complement fixation test for the diagnosis of Johne's disease in cattle. Aust. Vet. J. 68:253-257. [DOI] [PubMed] [Google Scholar]
- 30.Sackett, D. L., R. B. Haynes, G. H. Guyatt, and P. Tugwell. 1991. The interpretation of diagnostic data, p. 69-152. In Clinical epidemiology: a basic science for clinical medicine. Little, Brown, and Company, Boston, Mass.
- 31.Stabel, J. R., S. J. Wells, and B. A. Wagner. 2002. Relationships between fecal culture, ELISA, and bulk tank milk test results for Johne's disease in US dairy herds. J. Dairy Sci. 85:525-531. [DOI] [PubMed] [Google Scholar]
- 32.Sweeney, R. W., R. H. Whitlock, C. L. Buckley, and P. A. Spencer. 1995. Evaluation of a commercial enzyme-linked immunosorbent assay for the diagnosis of paratuberculosis in dairy cattle. J. Vet. Diagn. Investig. 7:488-493. [DOI] [PubMed] [Google Scholar]
- 33.USAHA National Johne's Disease Working Group Certification Subcommittee. 1999. U.S. voluntary Johne's disease herd status program for cattle, p. 33-43. In E. J. B. Manning and M. T. Collins (ed.), Proceedings of the Sixth International Colloquium on Paratuberculosis. International Association for Paratuberculosis, Madison, Wis.
- 34.USDA. 2005. USDA economics, statistics, and market information system. USDA Agriculture Statistics Service, Washington, D.C. [Online.] http://usda.mannlib.cornell.edu/.
- 35.USDA-APHIS. 2002. Uniform program standards for the Voluntary Bovine Johne's Disease Control Program. USDA-APHIS, Washington, D.C. [Online.] http://www.aphis.usda.gov/vs/nahps/johnes/johnes-umr.pdf.
- 36.Vary, P. H., P. R. Andersen, E. Green, J. Hermon-Taylor, and J. J. McFadden. 1990. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne's disease. J. Clin. Microbiol. 28:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wells, S. J., S. M. Godden, C. J. Lindeman, and J. E. Collins. 2003. Evaluation of bacteriologic culture of individual and pooled fecal samples for detection of Mycobacterium paratuberculosis in dairy cattle herds. J. Am. Vet. Med. Assoc. 223:1022-1025. [DOI] [PubMed] [Google Scholar]
- 38.Wells, S. J., and B. A. Wagner. 2000. Herd-level risk factors for infection with Mycobacterium paratuberculosis in US dairies and association between familiarity of the herd manager with the disease of prior diagnosis of the disease and use of preventive measures. J. Am. Vet. Med. Assoc. 216:1450-1457. [DOI] [PubMed] [Google Scholar]
- 39.Whitlock, R. H., S. J. Wells, R. W. Sweeney, and J. Van Tiem. 2000. ELISA and fecal culture for paratuberculosis (Johne's disease): sensitivity and specificity of each method. Vet. Microbiol. 77:387-398. [DOI] [PubMed] [Google Scholar]
- 40.Yokomizo, Y. 1986. Evaluation on an enzyme-linked immunosorbent assay (ELISA) using Mycobacterium phlei-absorbed serum for the diagnosis of bovine paratuberculosis in a field study. Jpn. Ag. Res. Q. 20:60-67. [Google Scholar]
- 41.Yokomizo, Y., R. S. Merkal, and P. A. S. Lyle. 1983. Enzyme-linked immunosorbent assay for detection of bovine immunoglobulin G1 antibody to a protoplasmic antigen of Mycobacterium paratuberculosis. Am. J. Vet. Res. 44:2205-2207. [PubMed] [Google Scholar]
- 42.Yokomizo, Y., H. Yugi, and R. S. Merkal. 1985. A method of avoiding false-positive reactions in an enzyme-linked immunosorbent assay (ELISA) for the diagnosis of bovine paratuberculosis. Jpn. J. Vet. Sci. 47:111-119. [DOI] [PubMed] [Google Scholar]