Abstract
Sequencing of a library of small RNAs from Giardia intestinalis identified a novel class of small sense and antisense RNAs homologous to the retroposon family GilT/Genie1 that is located at certain telomeres. These small RNAs may contribute to silencing GilT expression via the RNA interference pathway.
In recent years, our understanding of how eukaryotic gene expression is regulated has been fundamentally changed by the discovery of the RNA interference (RNAi) pathway (11), whereby small noncoding 20- to 30-nucleotide (nt) RNAs control gene expression at both the transcriptional and posttranscriptional levels (reviewed in references 1, 6, 14, 16, and 17). The RNAi pathway is an ancient trait of eukaryotes, and it has been demonstrated throughout the eukaryotic lineage from protozoa to humans. However, there are a few notable exceptions: the genomes of the yeast Saccharomyces cerevisiae, of the apicomplexan parasite Plasmodium falciparum, and of the trypanosomatid protozoa Trypanosoma cruzi and Leishmania major are devoid of the genes that are the hallmark of the RNAi pathway (26). The current evidence suggests that the RNAi mechanism was lost independently several times during eukaryotic evolution.
Genome sequencing of Giardia intestinalis, an intestinal protozoan parasite that is regarded by many as an early divergent eukaryote, reveals the presence of two genes that are central to the RNAi pathway (16), namely, a member of the Argonaute protein family (accession numbers AY142143 and EAA43025) and a Dicer-like enzyme (accession numbers AY142144 and EAA41574). In addition, a gene with the features of an RNA-dependent RNA polymerase (RdRp) has been annotated (accession numbers AF293414 and AAM73688). To gather evidence that G. intestinalis possesses a functional RNAi pathway, we investigated whether small noncoding RNAs are found in Giardia. Towards this end, we generated a library of 20- to 30-nt small RNAs using as starting material size-fractionated RNA derived from Giardia trophozoites. For the construction of the library we essentially followed the protocol previously established in our laboratory for cloning small 20- to 30-nt RNAs from Trypanosoma brucei (8, 25) but with the important modification that the cloning of the small RNAs was directional. This was achieved by ligating the 3′ ends of the small RNAs to the adaptor RNA oligonucleotide 5′P-CUGUAGGAUCCAUCAAU-idT3′ (DpnII recognition sequence is underlined). Upon digestion of double-stranded (ds) cDNA with the restriction enzyme DpnII, which cleaves on either side of the sequence GATC, a six-nucleotide identification bar code remained attached to the 3′ end of the small RNA. After cloning, we determined the sequences of the small RNA fragments, which we refer to as tags. The sizes of the tags varied from 20 to 29 nt, with the following distribution: 6% 20 nt, 12% 21 nt, 14% 22 nt, 8% 23 nt, 12% 24 nt, 11% 25 nt, 9% 26 nt, 9% 27 nt, and 14% 28 to 29 nt.
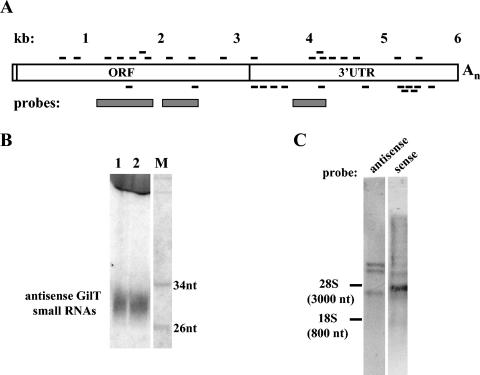
By using the BLAST algorithm, we could identify 403 sequences in the current Giardia genome database (Table 1). As with any library made from size-selected RNAs, most tags (74%) were derived from structural RNAs, namely, ribosomal RNAs, tRNAs, and small nuclear RNAs (Table 1). Seventeen percent of the tags matched with identified open reading frames (ORFs), with putative ORFs, or with putative intergenic regions in proximity to ORFs. The few tags corresponding to annotated ORFs had the sense polarity, and thus were probably derived from degradation fragments of putative mRNAs. In the case of the putative ORFs, strand assignment was uncertain because, in most cases, ORFs were present on both strands where the tags localized. Of note is the fact that 33 tags or 8% of the tags (19 sense and 14 antisense tags) were homologous to the GilT/Genie 1 element (Table 1 and Fig. 1A), one of three retroposon families that inhabit the Giardia genome (5, 7). Following the nomenclature proposed by Arkhipova and Morrison (5), GilT and GilM elements are non-long terminal repeat retroposons with long open reading frames that have the potential to code for a reverse transcriptase and associated nucleic-acid-binding and restriction-like endonuclease domains. The coding region common to both elements is followed by an unusually long 3′ untranslated region of about 2 to 3 kb and a poly(A) stretch. In contrast, GilD elements have multiple deletions in the coding region, suggesting that they are no longer active. Interestingly, separate arrays of GilT and GilM elements are located at distinct telomeres, and the 5′ end of the most telomeric element, which is often truncated, is directly fused to the telomeric repeats with the sequence (TAGGG)n. These retroposons, by inserting at chromosome ends, may function to protect telomeres from degradation (5). The protein-coding regions of GilT and GilM elements are about 50% identical (5); thus, the two elements can be easily distinguished at the DNA level. Among the tags we sequenced, we found matches only to the GilT sequences, and it appeared that these tags were distributed relatively uniformly throughout the length of the element (Fig. 1A; see also Fig. S1 in the supplemental material), indicating that the entire element is transcribed to generate sense and antisense transcripts. Furthermore, we identified three tags corresponding to transcripts from telomeric repeats with the structure 5′(CCCUA)n3′ (see Fig. S1 in the supplemental material). This provides evidence that the bottom strand of the telomeric repeats is transcribed. In contrast, even using relaxed parameters for the BLAST sequence comparisons, we did not find tags corresponding to the GilM element. Whether GilM elements are transcribed is at present not known.
TABLE 1.
Distribution of Giardia intestinalis small RNA tags
| Genetic element | No. of identified tags (% of total) |
|---|---|
| Ribosomal RNAs (28S, 18S, 5.8S, and 5S) | 50 (12) |
| tRNAs | 244 (61) |
| Small nuclear RNAs | 4 (1) |
| Retroposon GilT | 33 (8) |
| Telomeric repeats | 3 (1) |
| ORFs and intergenic regions | 69 (17) |
| Total number of tags identifieda | 403 |
BLAST searches were performed at the Giardia Genome Database (www.mbl.edu/Giardia) and included contig consensus sequences as well as all sequence reads not included in the assembly. Over 600 tags could not be assigned.
FIG. 1.
(A) Schematic representation of the GilT element and distribution of small sense and antisense GilT tags. Sense and antisense tags are indicated as dashes above and below the GilT drawing, respectively. The sizes and positions of the tags are indicative only and not to scale (for precise locations and sequences of the tags, see Fig. S1 in the supplemental material). The positions of the probes used in the Northern blots are indicated. UTR, untranslated region. (B) Identification of GilT small RNAs in G. intestinalis. Total RNA from G. intestinalis was enriched for small RNAs as described previously (25). Two different RNA samples (lanes 1 and 2) were fractionated on a 15% sequencing gel and analyzed by Northern blotting with a radiolabeled sense riboprobe mixture covering the regions indicated in panel A. M, end-labeled MspI-digested pBR322 molecular weight marker. (C) Large transcripts from GilT elements. Total Giardia RNA (10 μg) from trophozoites was fractionated by electrophoresis on a 1.2% agarose-formaldehyde gel and analyzed by Northern blotting with a sense or antisense radiolabeled GilT probe prepared by asymmetric PCR from the regions indicated in panel A.
To verify the existence of GilT small RNAs, we hybridized a Northern blot of Giardia RNA enriched for small RNAs with a sense GilT-specific probe (Fig. 1B) and detected a species of small RNA of about 30 nt representing antisense transcripts. The same-sized RNA species reacted with an antisense RNA probe (data not shown), confirming that both sense and antisense GilT small RNAs are present in Giardia. The apparent molecular weights of these transcripts corresponded to lengths that were a few nucleotides longer than the lengths of the cloned tags (20 to 29 nt). It is possible that RNAs longer than 30 nt are underrepresented in our library, since we used size-fractionated RNA as a starting material. Alternatively, the GilT small RNAs could have an anomalous electrophoretic mobility due to a high G+C content. Next, we analyzed the presence of large GilT transcripts in total RNA by conventional Northern blotting using GilT sense and antisense probes (Fig. 1C). The antisense probe revealed three prominent transcripts, one smaller than and two larger than 3,000 nt. These transcripts showed that certain GilT elements are transcribed, possibly to produce mRNAs. Antisense transcripts were also detected, but they appeared heterogeneous in size. Although there was a hybridizing band with a molecular weight similar to that of 28S rRNA, we cannot discount the possibility that there was cross-hybridization with this abundant rRNA. Nevertheless, the results indicate that both sense and antisense GilT transcripts are expressed in Giardia. This was not surprising, considering that sense and antisense transcription has been documented for several Giardia loci (10).
What might be the origin and function of GilT small RNAs? One attractive possibility is that they represent small RNAs analogous to the small interfering RNAs (siRNAs), which are the hallmark of the RNAi pathway and function as guides for triggering degradation of homologous transcripts (9). Naturally occurring siRNAs are thought to originate from long dsRNAs and are enriched in sequences derived from both strands of repeated-sequence families, including retroposons, transposons, and other repeats (2-4, 8, 12, 15, 20, 22, 29). Since these repeated-sequence families are often associated with heterochromatic regions, the corresponding siRNAs are likely to function in guiding heterochromatin formation, as first revealed by studies with Schizosaccharomyces pombe (18, 27, 28). In addition, in certain instances, the ablation of the RNAi pathway can lead to transposon mobilization (13, 22, 24) and often results in the upregulation of transcripts from mobile elements and other repeats (4, 21, 23). The cleavage of these transcripts triggered by the homologous siRNAs would inhibit movement of the corresponding elements, thus aiding in the maintenance of genome integrity (19). In the case of Giardia, we hypothesize that long sense and antisense GilT transcripts anneal to form dsRNA or are converted by the Giardia RdRp-like enzyme into dsRNA, which would then be cleaved by the Giardia Dicer-like enzyme to produce the small sense and antisense RNAs we have documented by cloning and Northern analysis. By analogy with the function of repeat-derived siRNAs in other systems, GilT small RNAs may assist in silencing expression of the corresponding retroposon transcripts to inhibit retroposition and/or function in heterochromatin formation at those telomeres where GilT elements reside. In the latter case, telomeric silencing of GilT elements might spread to genes that are upstream from GilT elements, thus affecting their expression. Once the sequences of the Giardia subtelomeric genes are known, it will become possible to analyze whether these genes have any special expression features.
Supplementary Material
Acknowledgments
This work was supported in part by grant AI56333 from the National Institutes of Health to E.U. Work performed by E.U. at the Marine Biological Laboratory (MBL) in Woods Hole, MA, was supported by the Baxter Postdoctoral Fellowship Fund, the Erik B. Fries Endowed Fellowship, the John O. Crane Fellowship Fund, the Charles R. Crane Fellowship Fund, the H. Burr Steinbach Memorial Fellowship Fund, and the James A. and Faith Miller Fellowship Fund. C.T.'s stay at the MBL was made possible by the generous support of the Baxter Postdoctoral Fellowship Fund, the Erik B. Fries Endowed Fellowship, the John O. Crane Fellowship Fund, and the H. Burr Steinbach Memorial Fellowship Fund.
We are indebted to Stephen Hajduk for hosting E.U. and C.T. in his laboratory, to Andrew McArthur for his invaluable support in bioinformatics analysis, to all the members of the Program in Global Infectious Diseases in Woods Hole, MA, for making our experience fruitful and enjoyable, and to Huafang Shi for superb technical assistance.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Ambros, V. 2004. The functions of animal microRNAs. Nature 431:350-355. [DOI] [PubMed] [Google Scholar]
- 2.Aravin, A. A., M. S. Klenov, V. V. Vagin, F. Bantignies, G. Cavalli, and V. A. Gvozdev. 2004. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 24:6742-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravin, A. A., M. Lagos-Quintana, A. Yalcin, M. Zavolan, D. Marks, B. Snyder, T. Gaasterland, J. Meyer, and T. Tuschl. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337-350. [DOI] [PubMed] [Google Scholar]
- 4.Aravin, A. A., N. M. Naumova, A. V. Tulin, V. V. Vagin, Y. M. Rozovsky, and V. A. Gvozdev. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11:1017-1027. [DOI] [PubMed] [Google Scholar]
- 5.Arkhipova, I. R., and H. G. Morrison. 2001. Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead. Proc. Natl. Acad. Sci. USA 98:14497-14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baulcombe, D. 2004. RNA silencing in plants. Nature 431:356-363. [DOI] [PubMed] [Google Scholar]
- 7.Burke, W. D., H. S. Malik, S. M. Rich, and T. H. Eickbush. 2002. Ancient lineages of non-LTR retrotransposons in the primitive eukaryote, Giardia lamblia. Mol. Biol. Evol. 19:619-630. [DOI] [PubMed] [Google Scholar]
- 8.Djikeng, A., H. Shi, C. Tschudi, and E. Ullu. 2001. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs. RNA 7:1522-1530. [PMC free article] [PubMed] [Google Scholar]
- 9.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmendorf, H. G., S. M. Singer, and T. E. Nash. 2001. The abundance of sterile transcripts in Giardia lamblia. Nucleic Acids Res. 29:4674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ketting, R. F., T. H. Haverkamp, H. G. van Luenen, and R. H. Plasterk. 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 14.Lippman, Z., and R. Martienssen. 2004. The role of RNA interference in heterochromatic silencing. Nature 431:364-370. [DOI] [PubMed] [Google Scholar]
- 15.Llave, C., K. D. Kasschau, M. A. Rector, and J. C. Carrington. 2002. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14:1605-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meister, G., and T. Tuschl. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343-349. [DOI] [PubMed] [Google Scholar]
- 17.Mello, C. C., and D. Conte, Jr. 2004. Revealing the world of RNA interference. Nature 431:338-342. [DOI] [PubMed] [Google Scholar]
- 18.Noma, K., T. Sugiyama, H. Cam, A. Verdel, M. Zofall, S. Jia, D. Moazed, and S. I. Grewal. 2004. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 36:1174-1180. [DOI] [PubMed] [Google Scholar]
- 19.Plasterk, R. H. 2002. RNA silencing: the genome's immune system. Science 296:1263-1265. [DOI] [PubMed] [Google Scholar]
- 20.Reinhart, B. J., and D. P. Bartel. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297:1831. [DOI] [PubMed] [Google Scholar]
- 21.Shi, H., A. Djikeng, C. Tschudi, and E. Ullu. 2004. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol. Cell. Biol. 24:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sijen, T., and R. H. Plasterk. 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426:310-314. [DOI] [PubMed] [Google Scholar]
- 23.Svoboda, P., P. Stein, M. Anger, E. Bernstein, G. J. Hannon, and R. M. Schultz. 2004. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 269:276-285. [DOI] [PubMed] [Google Scholar]
- 24.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 25.Tschudi, C., A. Djikeng, H. Shi, and E. Ullu. 2003. In vivo analysis of the RNA interference mechanism in Trypanosoma brucei. Methods 30:304-312. [DOI] [PubMed] [Google Scholar]
- 26.Ullu, E., C. Tschudi, and T. Chakraborty. 2004. RNA interference in protozoan parasites. Cell. Microbiol. 6:509-519. [DOI] [PubMed] [Google Scholar]
- 27.Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi, S. I. Grewal, and D. Moazed. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 29.Zilberman, D., X. Cao, and S. E. Jacobsen. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716-719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.