Abstract
We report the outcomes of the second session of the free online open-access workshop “Computational Applications in Secondary Metabolite Discovery (CAiSMD) 2022” that took place from 09 to 11 March 2022. The first session was held from 08 to 10 March 2021 and drew the attention of many early career scientists from academia and industry. The 23 invited speakers of this year’s workshop also came from academia and industry and 222 registered participants from five continents (Africa, Asia, Europe, South, and North America) took part in the workshop. The workshop highlighted the potential applications of computational methodologies in the search for secondary metabolites or natural products as drug candidates and drug leads. For three days, the participants of this online workshop discussed modern computer-based approaches for exploring NP discovery in the “omics” age. The invited experts gave keynote lectures, trained participants in hands-on sessions, and held round table discussions. These were followed by oral presentations during which much interaction between the speakers and the audience was observed. Selected applicants (early-career scientists) were offered the opportunity to give oral presentations (15 min) upon submission of an abstract. The final program available on the workshop website (https://indiayouth.info/index.php/caismd) comprised three keynote lectures, 14 oral presentations, two round table discussions, and four hands-on sessions. This meeting report also references internet resources for computational biology around secondary metabolites that are of use outside of the workshop areas and will constitute a long-term valuable source for the community.
Keywords: bioinformatics, chemoinformatics, secondary metabolites, open science, predictions, web tools
1. Introduction
Natural products (NPs) are not only the backbone of traditional medical systems but also of modern medicine, as many modern drugs are derived from natural sources. Apart from therapeutics, NPs are broadly utilized in nutrition and food technology, cosmetics, and biomaterial development, and are also used in multiple industrial processes [1]. NPs are lead compounds, which are frequently produced by plants and microbes as their secondary metabolites (SMs). However, producing large quantities of such compounds for industrial and clinical applications has been a persistent problem [2]. This has been compounded by the challenges linked to the isolation of NPs from complex mixtures and the difficulties often encountered in the total synthesis of SMs. During the past decade, SM discovery has been enhanced by the rapid progress in artificial intelligence and its applications [3]. Research in the field of NPs has therefore embraced the need for large-scale analysis of digitalized experimental data in the fields of metabolomics, transcriptomics, and genomics often referred to as the “omics” era [4]. This calls for the need for NP chemists to be properly trained in the new “omics” disciplines to be able to tackle the new challenges in the identification of SMs, elucidation of their structures, modes of action, and potential toxicities to enhance drug discovery from nature.
The “Computational Applications in Secondary Metabolite Discovery” (CAiSMD) was conceived to be an annual event that brings together scientists from the fields of bioinformatics and chemoinformatics who find interest in the computational methodologies and applications that are important for the discovery of NPs or their transformation into drug leads [5]. Currently, the event is purely online, although there are plans underway to get a hybrid meeting that combines face to face participation as well as a virtual participation.
The current series of virtual workshops is intended to introduce participants to modern computer-based approaches and tools for the exploration of the NPs and “omics” world. Most of the tools (software, web servers, databases, etc.), methods, and results presented to the participants were recent (dating from 2019 and later). The focus was on bioinformatics, chemoinformatics, NP chemistry, computational drug design, and genomic analysis, with applications in drug discovery. The cost-free workshop was conducted in English and open to the entire scientific community. M.Sc. and Ph.D. students, postdoctoral researchers, and early-career scientists were the target group of the workshop. Selected participants could submit an abstract indicating if they wish to give 15-min oral presentations.
All sessions with oral presentations and the parallel hands-on sessions (HS) were accessible through Zoom. All digital references are summarized in Table 1 and the final program and hand-out sessions are available on the website (https://caismd.indiayouth.info/). Since the workshop was intended in large part to attract early-career researchers, two formats were included, that is parallel hands-on sessions and round table discussions, partially led by postdoctoral scientists.
Table 1:
Important web links shared during the CAiSMD 2022 workshop.
| Speaker (lecture) | Group website (web link to tools presented) | Reference |
|---|---|---|
| Ozlem Tastan Bishop (KL 01) Email: o.tastanbishop@ru.ac.za |
Research Group: https://rubi.ru.ac.za | [6, 7] |
| Tools: South African Natural Compounds Database (SANCDB) https://sancdb.rubi.ru.ac.za/ | ||
| Dušanka Janežič (KL02) Email: dusanka.janezic@upr.si |
Research group: https://www.famnit.upr.si/sl/zaposleni-in-sodelavci/dusanka.janezic/ | [10, 11] |
| Tools: | ||
| ProBiS tools: http://insilab.org and https://probis.nih.gov | ||
| Pieter Dorrestein (KL03) Email: pdorrestein@ucsd.edu |
Research group: http://dorresteinlab.ucsd.edu | [12] |
| Tools: GNPS: Global natural products social molecular Networking (https://dorresteinlab.ucsd.edu/gnps) | ||
| Kiran K. Telukunta (OP01) Email: kiran.telukunta@indiayouth.info |
Research group: Tarunavadaanenasaha Muktbharatonnayana Samstha (TMS) Foundation (https://indiayouth.info/index.php) | [13, 14] |
| Tools: Galaxy tutorials (https://galaxyproject.org/learn/) | ||
| Victor Chukwudi Osamor (OP02) Email: vcosamor@gmail.com; victor.osamor@covenantuniversity.edu.ng |
Research group: https://covenantuniversity.edu.ng/Profiles/Osamor-Victor-Chukwudi#.YGYag1UzbIU | [15] |
| Tools: OsamorSoft (not available) | ||
| Mary A. Chama (OP03) Email: machama@ug.edu.gh |
Research group: | |
| Tools: not available | ||
| Samuel A. Egieyeh (OP04) Email: segieyeh@uwc.ac.za |
Research group: https://www.uwc.ac.za/study/all-areas-of-study/schools/school-of-pharmacy/people | [17] |
| Tools: not available | ||
| Ya Chen (OP05) Email: ya.chen@univie.ac.at |
Research group: The computational drug discovery and design group (COMP3D) at University of Vienna (https://comp3d.univie.ac.at/) | [18] |
| Tools: New E-resource for drug discovery (https://nerdd.univie.ac.at/) Source code: https://github.com/anyachen/RingSystems | ||
| Miquel Duran-Frigola (OP06) Email: miquel@ersilia.io |
Research group: https://ersilia.io/ | |
| Tools: Chemical Checker (https://pypi.org/project/chemicalchecker/) | ||
| Akachukwu Ibezim (OP07) Email: akachukwu.ibezim@unn.edu.ng |
Research group: not available | |
| Tools: not available | ||
| Daniel M Shadrack (OP08) Email: mshadrack@sjut.ac.tz; dmshadrack@gmail.com |
Research group: not available | |
| Tools: not available | ||
| Jean Moto Ongagna (OP09) Email: jean.monfils@yahoo.fr |
Research group: | |
| Tools: not available | ||
| Mai M. Farid (OP10) Email: mainscience2000@gmail.com |
Research group: not available | |
| Tools: Not available | ||
| Jude Betow (OP11) Email: betow.jude@ubuea.cm |
Research group: University of Buea Center for drug discovery (www.ub-cedd.org) | |
| Tools: not available | ||
| Lucie Karelle Djogang (OP12) Email: luciekarelledjogang@yahoo.fr |
Research group: Theoretical chemistry at the University of Yaoundé I | |
| Tools: not available | ||
| Lucas Paul (OP13) Email: lucaspaul33@gmail.com; lucasp@nm-aist.ac.tz |
Research group: not available | |
| Tools: not available | ||
| Pierre Valery Kemdoum Sinda (OP14) Email: psindakemdoum@gmail.com |
Research group: not available | |
| Tools: not available | ||
| Kai Blin (HS01) Email: kblin@biosustain.dtu.dk |
Research group: https://orbit.dtu.dk/en/persons/kai-kristof-blin | [] |
| Tools: https://antismash.secondarymetabolites.org/
https://antismash-db.secondarymetabolites.org/ https://mibig.secondarymetabolites.org/ | ||
| Pankaj Mishra (HS02) Email: pankaj@uresearcher.com |
Research group: https://uresearcher.com | |
| Tools: https://uresearcher.com/courses | ||
| Darshana Joshi (HS03) Email: darshanajoshi762@gmail.com; Alanis Tanya Edwin (HS03) |
Research group: https://indiayouth.info/index.php/our-programs/life-sciences-wing |
[26, 27] |
| Kiran K. Telukunta (HS03) Email: kiran.telukunta@indiayouth.info |
Tools: https://training.galaxyproject.org/training-material/topics/transcriptomics/tutorials/rna-seq-vizwith-heatmap2/tutorial.html | |
| Daniel M. Shadrack (HS04) Email: mshadrack@sjut.ac.tz; dmshadrack@gmail.com |
Research group: not available | |
| Tools: not available | ||
| Thommas Musyoka (HS04) Email: musyoka.thommas@ku.ac.ke |
Research group: not available | |
| Tools: not available | ||
| Fidele Ntie-Kang (HS04) Email: fidele.ntie-kang@ubuea.cm |
Research group: University of Buea Center for drug discovery (www.ub-cedd.org) | [28, 29] |
| Tools: ANPDB compound database (http://african-compounds.org/anpdb/) |
2. Workshop contents
2.1. Keynote lectures (KLs)
The organizers took the initiative to start identifying, inviting, and corresponding with key experts in the field who could provide inputs in the form of keynote lectures, oral presentations, and online hands-on training tutorials. All information regarding deadlines and registration was published on the workshop website from which interested applicants could download an abstract template and make all relevant uploads. Three keynote lectures (KLs) were given during the workshop, each lasting 45 min. The first keynote lecture was given by Özlem Tastan Bishop on day one who introduced some of the novel approaches to computational drug discovery using natural compounds: Identification of allosteric drug targeting sites; searching allosteric modulators via natural compounds, i.e. from the South African Natural Compounds Database (SANCDB) [6, 7]; and understanding the allosteric mechanisms of these modulators in the presence of evolutionary mutations. The case study was SARS-CoV-2 main protease (Mpro). The research group identified six potential allosteric modulators from SANCDB against Mpro [8, 9]. They observed that the stability of the potential hit compounds drastically changed in the presence of some of the mutations. Additionally, in the presence of some of the mutations, the allosteric communication path between the allosteric ligand binding site and the active site was lost. Collectively, the computational approaches that they established in this study offered routes for novel rational drug discovery methods and provided computationally feasible platforms to identify key functional residues implicated in allosteric signalling in the presence of allosteric modulators. Since the establishment of SANCDB, the database has attracted significant interest in diverse domains including natural product research, drug discovery, cheminformatics, and machine learning. In the last two years, SANCDB has also been screened by their research group and others against SARS-CoV-2 drug targets.
The second KL was presented by Dušanka Janežič. Her team developed new methodological solutions for the prediction and study of protein binding sites on the protein databank (PDB) scale, based on graph theoretical approaches, combined with molecular dynamics simulations. The special focus was on the development of new algorithms for the prediction of protein binding sites (ProBiS) [10, 11] and new web tools for modeling pharmaceutically interesting molecules - ProBiS Tools (algorithm, database, web server). The ProBiS Tools are the first to allow the identification of interactions between protein structures, the prediction of ligand selectivity and binding, and the monitoring of the effects of conserved waters and sequence variants on ligand binding, to surpass human involvement in drug design. All ProBiS Tools are freely available to the academic community at http://insilab.org and https://probis.nih.gov.
On the third day, Pieter Dorrestein highlighted the latest mass spectrometry-based tools in his KL, including the research group’s crowdsource molecular annotation platform and repository scale analysis, to study the chemistry of the diet and microbiome associated with the host (plants, animals, humans) in relation to ecosystem health information as well as understanding the chemistry of human, environmental, or ecological habitats. This was to establish the roles microbes play and their relationship to the chemistries in our bodies proportional to human health. The speaker also presented the Global Natural Products Social Molecular Networking (GNPS) [12], a web-based mass spectrometry ecosystem that aims to be an open-access knowledge base for community-wide organization and sharing of raw, processed or identified tandem mass (MS/MS) spectrometry data, developed by his group.
2.2. Oral presentations (OPs)
There were 14 scheduled OPs. The lecture topics included presentations of both novel computational methodologies as well as recent results. The first two oral presentations OP1 and OP2 were done on the second day of the workshop. OP1 was presented by Kiran K Telukunta who introduced the Galaxy framework [13] and gives an exordium to open-source research and opens the door for accelerated research. The organizers planned to insert a hands-on session in the next edition of the CAiSMD workshop which will guide participants on how to use the Galaxy framework for visualization analysis [14]. OP2 was given by Victor Chukwudi Osamor and collaborators. They highlighted a Computational Investigation of Natural Products as Lead Compounds on Drug Repurposing using Clustering and Fingerprinting Algorithms [15]. They studied natural molecules from the African natural compounds database and traditional drugs from the PubChem database. A total of 4936 small molecules were used in the experiment, containing 12 existing drugs for cancer and 4924 natural compounds. The ChemmineR cheminformatics package in R was used to generate the fingerprints for each molecule and perform other preprocessing tasks, the molecular similarity between molecules was then calculated using the Tanimoto coefficient with a cutoff of 0.5. For the clustering, the agglomerative hierarchical and K-means clustering techniques were adopted, and the result was visualized using dendrograms. Their result, based on the similarity property principle, showed that drugs with similar molecular structures are likely to have the same properties. This may indicate that they have similar properties and can be further tested as potential drug candidates. In the study, the dataset was reduced to 482 molecules after conducting the structural molecular similarity measurement.
On day three, there were 12 oral presentations (OP3-OP14). Mary A. Chama and co-workers discussed on in silico and in vitro studies of the cytotoxicity and the mode of action of the compounds dichapetalins A and M. She reported that both dichapetalins were isolated by column chromatography and identified using NMR and mass spectrometry (OP3). The PIDGINv2 [16] was used for target prediction, while molecular docking was carried out using the GOLD v5.4 software suite. Cytotoxicity measurements were done against the MCF-7 cells via the MTT assay and target validation was carried out with expression studies with qPCR. From the result, the IC50 of dichapetalin M was higher (4.71 μM and 3.95 μM) for 48 and 72 h of treatment, respectively, compared to curcumin (with an IC50 of 17.49 μM and 12.53 μM for 48 h and 72 h of treatment, respectively). The in vitro expression studies with qPCR confirmed an antagonistic effect of dichapetalin M on PXR (NR112) signaling; supporting the PXR signaling pathway as a possible mode of action of dichapetalin M. The findings suggest that dichapetalin M could be a lead compound for a potential anticancer drug.
Samuel A. Egieyeh gave a talk that focused on molecular insights into the interaction between the spike protein of the Wildtype, Delta, and Omicron variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [17] and the human angiotensin converting enzyme 2 (hACE2). He examined the potential for the design of fusion inhibitors (OP4). His team used a combination of three-dimensional protein model characterization, protein-protein docking, molecular docking, and molecular dynamics to identify potential differences in the electronic potential distribution, solvation energy, interaction energy, interacting residues, and dynamics of the mutants (Delta and Omicron) and wild type spike proteins. Pharmacophore modeling was used to predict potential fusion inhibitors. In the studies, the differences in the protein properties, electronic potential distribution, solvation energy, interaction energy, interacting residues, and dynamics between the mutants (Delta and Omicron) and wild-type spike proteins were observed, and identify potential fusion inhibitors by pharmacophore modelling of the receptor binding motif of the spike proteins.
Ya Chen gave a presentation on cheminformatics analysis of ring systems in NPs (OP5) [18]. They compiled comprehensive, curated data sets of NPs, synthetic compounds, and approved drugs, from which ring systems were extracted. Cheminformatics approaches were employed to analyze the structural and physicochemical properties of the NP ring systems, and the coverage of NP ring systems by readily purchasable, synthetic compounds. In addition to common 2D physicochemical properties, such as molecular weight and the number of nitrogen/oxygen atoms, they also considered the 3D shape and electronic properties. Importantly, they deployed a new algorithm to automatedly extract ring systems from chemical structures and a new method to maximize the use of the available stereochemical information on ring systems. The source code used in this analysis was also made available. From the study, the diversity of NP ring systems and their importance to drug discovery were quantified. The study showed the structural diversity of NP ring systems (which are more complex and more of them are macrocycles) compared to the ring systems observed in synthetic compounds.
OP6 was given by Miquel Duran-Frigola et al., who explained how the Ersilia Model Hub can be deployed in the form of a fully functional, comprehensive virtual screening cascade that is coupled with medicinal chemistry, parasitology, and ADME experimental pipelines. The main asset of Ersilia Open-Source Initiative (EOSI; https://ersilia.io) is the Ersilia Model Hub (a free, online, open-source platform) where scientists can browse through a catalog of AI/ML models and select the ones that are relevant to their research and run predictions online. They gathered, in a single resource, two classes of models. They collect models developed by third parties and available in scientific publications. The second class of models was developed in-house and in collaboration with research groups that operate in the so-called Global South. Special emphasis was put on AI/ML models that can handle natural product(-like) compounds. Akachukwu Ibezim and co-workers evaluated the trypanocidal activity and mode of action of a steroid from Vitex simplicifolia leaves (OP7). V. simplicifolia leaves were collected, identified, extracted with methanol, and fractionated using dichloromethane (DCM) following the procedures as described in their earlier work [19]. They used combined vacuum liquid chromatography (VLC) with silica gel (230–400 mesh, 30 × 30 cm, 500 g) as the stationary phase and a semi-preparative HPLC (Dionex P580) system coupled to a photodiode array detector (UVD340S) to isolate, purify, and characterize the obtained phytochemical. The plant fraction and compound were assayed against Trypanosoma brucei rhodesiense (STIB 900), Trypanosoma cruzi bloodstream forms, and L6 cells. They employed Swiss Target Prediction and GeneCard databases to retrieve drug targets for the isolate and the two pathogens and used the Draw Venn Diagram platform analysis to identify the common targets of both the disease and the isolate. Lastly, bioinformatics tools and techniques were applied to predict the biological target(s) of the isolate. The methanol leaf extract of V. simplicifolia showed moderate trypanocidal activity against T. b. rhodesiense (IC50 = 14.2 μg/ml). The DCMF brought a 56.25 % reduction in parasitaemia at 100 mg/kg in T. b. brucei-infected mice. Further chromatographic separation of DCMF with the gradient of DCM: methanol yielded a steroid identified as ajugasterone C. The isolate was effective against T. b. rhodesiense (IC50 = 10.12 ± 0.3) and T. cruzi (IC50 = 46.05 ± 1.5), however, it impact on the mammalian skeletal myoblast cell line (L6 cells) raises toxic concern (IC50 > 100 μg/ml) (SIT.b.r > 9.80 and SI T. cruzi > 2.01). Through bioinformatics manipulations, they found that the isolate interacts with Trypanosoma cysteine cathepsins (CatL and CatB) to exhibit activity. Also, interaction with adenosine kinase (ADK) was identified regarding T. b. rhodesiense. These mechanisms of action are valuable in optimization exercises. They observed that the isolate had some toxicity concerns though phytochemicals are known to have a high safety profile.
Daniel M Shadrack exposed the participants to the concept of polypharmacology, where one natural product can have multiple targets, and then focused on the efficacy of natural products in an aqueous environment (OP8). He emphasized solubility and aggregation issue of drug molecules and how this relates to the efficacy of natural products. Finally, some success stories of chemo-bioinformatics approaches in drug design were highlighted.
The remaining OPs were dedicated to the “Young Investigators Session” and comprised seven presentations mostly from Ph.D. students and postdocs. Jean Moto Ongagna gave a presentation (OP9) on the insights into intramolecular interactions of transition metal-bis-(N-heterocyclic carbene) complexes and topological unraveling the C–H Bond activation from QTAIM/ELF/EDA/NBO and BET Theory. The bond properties and electronic structure of novel tetrahedral and square planar CH2-bridged bis-(N-heterocyclic carbene) transition metals halides were described. They indicated that some of these complexes facilitate the catalytic activity of the oxidative activation of the C–H bonds of hydrocarbons. In a context where natural gas, mainly lower alkanes (methane 60 %, propane 5 %, and LPG) constitute a huge resource of fossil fuels and feedstocks worldwide, the design and implementation of such compounds on the direct oxidation of alkanes are worthy [20]. These types of complexes are widely used in the catalytic petrochemical conversion of lower alkane into alcohol because of their more stable metal–ligand bond. It is important to note that metal–halide bonds play a decisive role in these reactions. These factors can be fine-tuned to achieve an improved catalytic activity. Finally, they aimed to propose oxidative activation reaction mechanisms for these complexes to facilitate their synthesis.
OP10 dealt with investigating the effect of trigonelline on memory function in Alzheimer’s disease transgenic model mouse, 5XFAD which overexpressed mutated APP and PS1 genes [21, 22] presented by Mai M. Farid. They observed that the oral administration of trigonelline for 14 days significantly enhanced object recognition and object location memories. In addition, trigonelline significantly ameliorated axonal and dendrite atrophy in amyloid β-treated cortical neurons. After the oral administration of trigonelline, they isolated plasma and cerebral cortex at 30 min, 1 h, 3 h, and 6 h. According to the result obtained using LC-MS/MS, trigonelline was detected in both plasma and cortex from 30 min after, suggesting good penetration of trigonelline into the brain. Analysis of target proteins of TGN in neurons by a drug affinity responsive target stability (DARTS) method identified that creatine kinase B-type (CKB) is a direct binding protein of TGN. Treatment with a CKB inhibitor cancelled the TGN-induced axonal and dendritic growth. These results suggest that trigonelline could be a promising therapeutic candidate for AD. During OP11, Jude Betow presented a search for schweinfurthins and other SMs from the Cameroonian medicinal plant Macaranga occidentalis (Euphorbiaceae) and evaluated the possible anticancer activities of the SMs. Six compounds were isolated, labeled MOC1–MOC6 which were then analyzed using GC-MS and 1H NMR analysis. They observed that compounds MOC1 and MOC2 were mixtures i.e., MOC1: 9,12-octadecadienoic acid-(Z,Z)-methyl ester (methyl linoleate) and 9-octadecenoic acid-E-methyl ester (methyl-E-oleate); MOC2: phytosterols; β-sitosterol acetate, γ-sitosterol, β-stigmasterol and campesterol; and MOC3: a pentacyclic triterpenoid 28-norolean-17-en-3-ol. From the results obtained, none of the compounds falls under the class of schweinfurthins. They planned to use GC-MS and spectroscopic analysis to determine the structures of the other isolated compounds (MOC4–MOC6), together with the unidentified compounds on the GC-MS chromatogram (RT:17.847, RT:20.142, and RT:27.967) and their subsequent submission for anticancer screening.
Next, Lucie K. Djogang gave a talk based on molecular docking and in silico ADMET predictions of amodiaquine derivatives as antimalarial agents (OP12). She explored in silico absorption, distribution, metabolism, excretion, and toxicity (ADMET) to study some substituted amodiaquine compounds and predicted the potential interaction modes and binding affinities of the designed ligands with the enzymes of different pathways (folate and glycolytic). Through this study, she observed that all the ligands modeled, interacted with all three enzymes, and the human and Plasmodium falciparum TPI bind with amodiaquine derivatives using two distinct binding sites and residues. Moreover, the top-scoring ligand AmoJ shows a high binding affinity with all the receptors compared to amodiaquine and other derivatives. During OP13, Lucas Paul investigated the influence of intermolecular and intramolecular hydrogen bonding and the influence of polar and nonpolar solvents on the physical properties of linamarin. Furthermore, they performed solvation free‐energy and electronic structure analysis. A detailed analysis showed intermolecular hydrogen bonding between polar solvents (water, MeOH and DMSO) and the hydroxyl oxygens of linamarin. According to the findings, water exhibits the strongest interaction with linamarin’s functional groups among the investigated solvents, and from the solvation-free energy calculations, DMSO is the best solvent since it prefers to interact with linamarin over itself, while water prefers to interact with itself. The solute-solvent interactions are strongest between linamarin and DMSO, the solvent-solvent interactions are strongest in water. As a result, the solvation free‐energy calculations reveal that linamarin solvation is most favorable in DMSO. The last presentation (OP14) was given by Pierre V. K. Sinda, who had isolated secondary metabolites responsible for the hepatoprotective effect of the ethanol extract of Pentaclethra macrophylla stem bark and studied their structure-activity relationship. P. macrophylla Benth (Mimosaceae) is a medicinal plant commonly used in Cameroon to treat several diseases such as itching and liver diseases. In the study, hydrogen peroxide (H2O2)-induced lipid peroxidation to hepatocyte membranes’ model was used to successively assess the hepatoprotective-bioguided fractionation of the ethanolic extract of P. macrophylla. For the in vivo hepatoprotective test, mice were treated orally with the ethyl acetate (EtOAc) fraction of the ethanol extract and subjected to d -galactosamine/lipopolysaccharide-induced (GaIN/LPS) hepatotoxicity. Blood samples were collected for alanine aminotransferase (ALAT), aspartate aminotransferase (ASAT), TNF-α, and IL-1β assays. The ethanol extract was suspended in distilled water and successively extracted with EtOAc and n-BuOH to yield the EtOAc and n-BuOH fractions as well as the residual aqueous fraction. The hepatoprotective test showed that the EtOAc fraction was the most effective (IC50: 3.214 μg/mL) compared to silymarin used as reference (IC50: 117.4 μg/mL). Its fractionation by column chromatography yielded three active sub-fractions which were purified to give one monoglyceride, one carboxylic acid, two steroids, two tannins, and one terpenoid. The structures were established based on their mass spectrometry, 1D and 2D NMR data, and by comparison of the data with those of related compounds present in the literature. Compound 11-O-galloyl bergenin (IC50:1.8 μg/mL) was the most effective. In vivo, the EtOAc fraction significantly reduced the serum level of ALAT, ASAT, and TNF-α, and increased the liver protein content. Thus, this fraction could further be submitted for more investigations as lead for liver diseases.
2.3. Hands-on sessions (HS)
Contrary to the last 2021 edition with five parallel hands-on sessions (HS) of 90 min each, this year had only four hands-on sessions, split into two categories; bioinformatics and chemoinformatics. Each category had one session on day 1 and one session on day 2, while day 3 was dedicated to a “Test Yourself” (TY) session, during which participants were expected to practice by themselves on exercises that had been left by the instructors during the HS sessions proper. The TY lasted 45 min, while each HS lasted 90 min.
During HS01, Kai Blin taught participants how “antibiotics and Secondary Metabolites Analysis Shell” (antiSMASH) [23] predicts the biosynthetic gene clusters (BGCs) on the topic “Genome Mining using antiSMASH, and the antiSMASH and MIBiG databases”. Participants gained some experience in running antiSMASH and interpreting the results of such runs and learned how to use the antiSMASH database [24] and the Minimum Information about a Biosynthetic Gene cluster (MIBiG) database [25] to gain additional insights into the predicted BGCs.
HS02 was conducted by Pankaj Mishra. This session was focused on how to build individual machine learning-based tools to classify and predict the biological activities of small molecules. Participants also learned how to build machine learning-based virtual screening tools to rapidly search millions of molecules. The skills learned can be applied to other areas with different data as well. Generally, participants were trained on how to install software packages; how to collect bioactivity data; how to process bioactivity data; how to process chemical structures; how to carry out molecular feature engineering; how to build, train and evaluate bioactivity classification models; how to prepare virtual a screening dataset; how to conduct machine learning-based virtual screening; and how to select chemical hits.
During the third HS jointly coordinated by Darshana Joshi, Alanis T. Edwin, and Kiran K. Telukunta, the participants were trained on the topic “Visualization of RNA-Seq with Galaxy Framework”. They used heatmaps to visualize the differentially expressed genes in the samples [26]. The current galaxy training tutorial examines the expression profiles of basal and luminal cells in the mammary gland of a virgin, pregnant and lactating mice [27] using the heatmap2 tool available in the Galaxy. Participants were trained on how to generate a heatmap in which the top genes differentially expressed in the luminal cells from the pregnant mice versus the luminal cells from the lactating mice will be plotted. Heatmaps generated from the heatmap2 tool in the Galaxy were used to visualize the differential expression of genes in RNA-Seq samples.
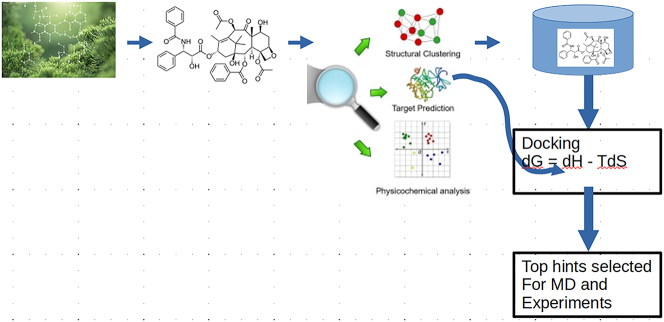
The last hands-on session (HS04) was focused on virtual screening for the fast and cheap identification of bioactive natural products. Daniel M. Shadrack, Thommas M. Musyoka, and Fidele Ntie-Kang trained participants on how to explore web tools that permit the search of databases including similarity and sub-structure searching for privileged scaffolds. The first part of the session which lasted about 50 min, participants were taught how to perform virtual screening from large libraries (focusing on natural products libraries from African sources), e.g. the South African Natural Compounds Database (SANCDB – https://sancdb.rubi.ru.ac.za/) [6,7], which is a collection of 1012 compounds derived from South African natural sources. Since its inception in 2015, the database has been used for various machine learning and in silico virtual drug screening studies with a recent study identifying several potential hits against severe acute respiratory syndrome coronavirus 2 (SARS-COV-2). As part of a recent update, a unique feature incorporating the compound dataset analogs from two leading commercial databases (Molport and Mcule) was included. The feature will not only allow users to explore a larger chemical space during screening but also allow them to seamlessly purchase compounds for their biological studies. Participants were introduced to the database with an emphasis on how they can obtain compounds for both their virtual screening and biological studies. The second part of the session (approximately 20 min) was focused on natural product databases originating from the regions of Northern [28] and East Africa [29] (http://africancompounds.org/anpdb/), as shown in Figure 1.
Figure 1:
Virtual screening protocol used during the HS04 for the identification of potent natural products from African medicinal plants.
2.4. Round table discussions
There were two round table discussions (RTDs); RTD01 during the middle of day 2 and RTD02 at the end of the workshop. RTD01 was focused on the novel tools and methods presented during day 1 and part of day 2. Among the interesting questions that emerged were which tools would be the best to address the increasing volume of data on secondary metabolites, genomic sequences, transcriptomes, as well as which tools progressively integrate updated data and databases, whether the results obtained from clustering data using the Galaxy platform would solve the current issues of constantly increasing volume of data, etc. During the RTD02 that followed the early career researcher’s session and led to the close of the workshop, the most important questions were raised to the organizers about the frequency of the CAiSMD workshop series and the need to couple the online sessions with face-to-face meetings. All participants agreed that the workshop contents were quite enriching but differences in time zones between participants would suggest a more harmonized data sharing platform. A compendium of computational tools used during the workshop was compiled and summarized in Table 1.
3. Conclusions
Computational tools and methods continue to play a vital role in the discovery and applications of SMs in several scientific disciplines, including drug discovery and the pharmaceutical industry, in general. With these tools and methods dispersed in the scientific literature and platforms and with the constantly increasing volume of scientific data stored in databases, it becomes imperative to drill students and early career scientists on these approaches and update their knowledge on currently available tools. Initially conceived as an annual event, the CAiSMD2022 brought together 23 invited speakers of this year’s workshop who also came from academia and industry and 222 registered participants from five continents (Africa, Asia, Europe, South, and North America). Subsequent events are planned every two years, meaning that the next sessions are planned in March 2024. It is intended that the currently online workshop will eventually become a hybrid event including both physical presence and online participation.
Acknowledgments
The authors would like to thank the editors XYZ for their guidance and review of this article before its publication. The TMS Foundation is acknowledged for supporting the workshop for its online presence and facilitating the participants to find the content about CAiSMD by hosting the information about speakers and events. The workshop organizers also acknowledge some technical support from the IT team of the Technische Universität Dresden, Germany.
Supplementary information
The workshop slides and materials for the hands-on sessions are available for free download from the website (https://indiayouth.info/index.php/caismd/downloads).
Abbreviations
- ANPDB
African Natural Products Database
- GNPS
The Global Natural Product Social Molecular Networking
- HS
hands-on session
- KL
keynote lecture
- NP
natural product
- OP
oral presentation
- RTD
round table discussion
- SANCDB
South African Natural Compounds Database
Footnotes
Research ethics: Not applicable
Author contributions: Conception, FNK and JLM; Writing of text: FNK, DBE, JLM, Editing, all authors. All authors agreed on the final version before submission.
Competing interests: The authors declare no conflicting financial interests
Research funding: We acknowledge financial support from the Bill & Melinda Gates Foundation through the Calestous Juma Science Leadership Fellowship awarded to Fidele Ntie-Kang (grant award number: INV-036848 to University of Buea). FNK also acknowledges joint funding from the Bill Bill & Melinda Gates Foundation and LifeArc (award number: INV-055897 and Grant ID: 10646) under the African Drug Discovery Accelerator program. FNK acknowledges further funding from the Alexander von Humboldt Foundation for a Research Group Linkage project. KB and TW were supported by grant NNF20CC0035580 of the Novo Nordisk Foundation.
Data availability: Not applicable
Contributor Information
Fidele Ntie-Kang, Email: fidele.ntie-kang@ubuea.cm.
Ya Chen, Email: ya.chen@univie.ac.at.
Jonathan A. Metuge, Email: Jonathan.metuge@aamu.edu.
Özlem Tastan Bishop, Email: o.tastanbishop@ru.ac.
Jutta Ludwig-Müller, Email: jutta.ludwig-mueller@tu-dresden.de.
References
- 1.Atanasov AG, Waltenberger B, Pferschy-Wenzig EM, Linder T, Wawrosch C, Uhrin P, et al. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol Adv. 2015;33:1582–614. doi: 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA, Voigt CA. Synthetic biology to access and expand nature’s chemical diversity. Nat Rev Microbiol. 2016;14:135–49. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–29. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 4.van Santen JA, Kautsar SA, Medema MH, Linington RG. Microbial natural product databases: moving forward in the multi-omics era. Nat Prod Rep. 2021;38:264–78. doi: 10.1039/d0np00053a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ntie-Kang F, Telukunta KK, Fobofou SAT, Chukwudi Osamor V, Egieyeh SA, Valli M, et al. Computational applications in secondary metabolite discovery (CAiSMD): an online workshop. J Cheminform. 2021;13:64. doi: 10.1186/s13321-021-00546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatherley R, Brown DK, Musyoka TM, Penkler DL, Faya N, Lobb KA, et al. Tastan Bishop O. SANCDB: a South African natural compound database. J Cheminform. 2015;7:29. doi: 10.1186/s13321-015-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diallo BN, Glenister M, Musyoka TM, Lobb K, Tastan Bishop O. SANCDB: an update on South African natural compounds and their readily available analogs. J Cheminform. 2021;13:37. doi: 10.1186/s13321-021-00514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheik Amamuddy O, Afriyie Boateng R, Barozi V, Wavinya Nyamai D, Tastan Bishop O. Novel dynamic residue network analysis approaches to study allosteric modulation: SARS-CoV-2 Mpro and its evolutionary mutations as a case study. Comput Struct Biotechnol J. 2021;19:6431–55. doi: 10.1016/j.csbj.2021.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheik Amamuddy O, Verkhivker GM, Tastan Bishop O. Impact of early pandemic stage mutations on molecular dynamics of SARS-CoV-2 Mpro . J Chem Inf Model. 2020;60:5080–102. doi: 10.1021/acs.jcim.0c00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konc J, Janezic D. ProBiS algorithm for detection of structurally similar protein binding sites by local structural alignment. Bioinformatics. 2010;26:1160–8. doi: 10.1093/bioinformatics/btq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konc J, Lešnik S, Škrlj B, Janežič D. ProBiS-dock database: a web server and interactive web repository of small ligand-protein binding sites for drug design. J Chem Inf Model. 2021;61:4097–107. doi: 10.1021/acs.jcim.1c00454. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol. 2016;34:828–37. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–44. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syme A, Soranzo N. A short introduction to galaxy (galaxy training materials) [Online] 2021. https://training.galaxyproject.org/training-material/topics/introduction/tutorials/galaxy-introshort/tutorial.html Accessed 07 Mar 2022. [Google Scholar]
- 15.Osamor IP, Osamor VC. OsamorSoft: clustering index for comparison and quality validation in high throughput dataset. J Big Data. 2020;7:48. doi: 10.1186/s40537-020-00325-6. [DOI] [Google Scholar]
- 16.Mervin LH, Bulusu KC, Kalash L, Afzal AM, Svensson F, Firth MA, et al. Orthologue chemical space and its influence on target prediction. Bioinformatics. 2018;34:72–9. doi: 10.1093/bioinformatics/btx525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rees-Spear C, Muir L, Griffith SA, Heaney J, Aldon Y, Snitselaar JL, et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34:108890. doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Rosenkranz C, Hirte S, Kirchmair J. Ring systems in natural products: structural diversity, physicochemical properties, and coverage by synthetic compounds. Nat Prod Rep. 2022;39:1544–56. doi: 10.1039/d2np00001f. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim MA, Mohammed A, Isah MB, Aliyu AB. Anti-trypanosomal activity of African medicinal plants: a review update. J Ethnopharmacol. 2014;154:26–54. doi: 10.1016/j.jep.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Munz D, Strassner T. Propane activation by palladium complexes with chelating bis (NHC) ligands and aerobic cooxidation. Angew Chem Int Ed Engl. 2014;53:2485–8. doi: 10.1002/anie.201309568. [DOI] [PubMed] [Google Scholar]
- 21.Farid MM, Yang X, Kuboyama T, Tohda C. Trigonelline recovers memory function in Alzheimer’s disease model mice: evidence of brain penetration and target molecule. Sci Rep. 2020;10:16424. doi: 10.1038/s41598-020-73514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farid MM, Nagase T, Yang X, Nomoto K, Kuboyama T, Inada Y, et al. Effects of Trigonella foenum-graecum seeds extract on Alzheimer’s disease transgenic model mouse and its potential active compound transferred to the brain. Jpn J Food Chem Saf. 2021;28:63–70. [Google Scholar]
- 23.Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49:W29–35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blin K, Shaw S, Kautsar SA, Medema MH, Weber T. The antiSMASH database version 3: increased taxonomic coverage and new query features for modular enzymes. Nucleic Acids Res. 2021;49:D639–43. doi: 10.1093/nar/gkaa978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kautsar SA, Blin K, Shaw S, Navarro-Muñoz JC, Terlouw BR, van der Hooft JJJ, et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020;48:D454–8. doi: 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle M. Visualization of RNA-seq results with heatmap2 (galaxy training materials) [Online] . 2021. https://training.galaxyproject.org/training-material/topics/transcriptomics/tutorials/rna-seq-vizwith-heatmap2/tutorial.html Accessed 09 Mar 2022.
- 27.Fu NY, Rios AC, Pal B, Soetanto R, Lun AT, Liu K, et al. EGF-mediated induction of Mcl-1 at the switch to lactation is essential for alveolar cell survival. Nat Cell Biol. 2015;17:365–75. doi: 10.1038/ncb3117. [DOI] [PubMed] [Google Scholar]
- 28.Ntie-Kang F, Telukunta KK, Döring K, Simoben CV, Moumbock AAF, Malange YI, et al. NANPDB: a resource for natural products from northern African sources. J Nat Prod. 2017;80:2067–76. doi: 10.1021/acs.jnatprod.7b00283. [DOI] [PubMed] [Google Scholar]
- 29.Simoben CV, Qaseem A, Moumbock AFA, Telukunta KK, Günther S, Sippl W, et al. Pharmacoinformatic investigation of medicinal plants from East Africa. Mol Inf. 2020;39:e2000163. doi: 10.1002/minf.202000163. [DOI] [PMC free article] [PubMed] [Google Scholar]