Abstract
Galactofuranose (Galf) is a novel sugar absent in mammals but present in a variety of pathogenic microbes, often within glycoconjugates that play critical roles in cell surface formation and the infectious cycle. In prokaryotes, Galf is synthesized as the nucleotide sugar UDP-Galf by UDP-galactopyranose mutase (UGM) (gene GLF). Here we used a combinatorial bioinformatics screen to identify a family of candidate eukaryotic GLFs that had previously escaped detection. GLFs from three pathogens, two protozoa (Leishmania major and Trypanosoma cruzi) and one fungus (Cryptococcus neoformans), had UGM activity when expressed in Escherichia coli and assayed in vivo and/or in vitro. Eukaryotic GLFs are closely related to each other but distantly related to prokaryotic GLFs, showing limited conservation of core residues around the substrate-binding site and flavin adenine dinucleotide binding domain. Several eukaryotes not previously investigated for Galf synthesis also showed strong GLF homologs with conservation of key residues. These included other fungi, the alga Chlamydomonas and the algal phleovirus Feldmannia irregularis, parasitic nematodes (Brugia, Onchocerca, and Strongyloides) and Caenorhabditis elegans, and the urochordates Halocynthia and Cionia. The C. elegans open reading frame was shown to encode UGM activity. The GLF phylogenetic distribution suggests that Galf synthesis may occur more broadly in eukaryotes than previously supposed. Overall, GLF/Galf synthesis in eukaryotes appears to occur with a disjunct distribution and often in pathogenic species, similar to what is seen in prokaryotes. Thus, UGM inhibition may provide an attractive drug target in those eukaryotes where Galf plays critical roles in cellular viability and virulence.
In many microbes, the sugar galactofuranose (Galf) is an important constituent of glycoconjugates comprising major portions of the cell surface (17). In prokaryotes, Galf constitutes a key part of the mycobacterial cell wall and occurs in lipopolysaccharide (LPS) O-antigen domains, extracellular capsules, and polysaccharides (17, 19). In fungi such as Aspergillus, Galf is a major component within the cell wall and structural glycoproteins (13, 17). In pathogenic protozoa, Galf residues are key components of abundant surface glycosylphosphoinositol-anchored glycoconjugates, such as lipophosphoglycan (LPG) and glycoinositolphospholipids in Leishmania, and of mucins, glycosylphosphoinositol-anchored proteins, and lipids in Trypanosoma cruzi (6, 18). In contrast, Galf residues have not been found in humans or other metazoans (17), suggesting that inhibition of Galf synthesis could be an attractive target for chemotherapy in pathogens when its role(s) is critical for survival or virulence (17, 32).
The Galf synthetic pathway has been best studied for prokaryotes. Genetic and biochemical studies have shown that Galf arises through the action of UDP-galactopyranose mutase (UGM) (EC 5.4.99.9), which catalyzes the rearrangement of UDP-galactopyranose (Galp) to UDP-Galf, the substrate of cellular UDP-Galf transferases which participate in pathways such as LPS or cell wall biosynthesis (15, 16). The gene encoding UGM, GLF, was first located and hypothesized as such by Reeves and colleagues as part of genetic and structural studies of Escherichia coli K12 O antigen (27). It was then definitively identified and studied in E. coli, Klebsiella, and Mycobacteria (10, 15, 16). UGM is a flavin-dependent enzyme, and the E. coli enzyme structure has been solved (21). A detailed picture of the enzymatic mechanism involving a novel form of flavin-dependent catalysis has been developed (24). High-throughput assays for inhibitor screens have been developed, and a number of compounds showing activity against UGM activity and/or bacteria have been identified (22, 25, 28).
Less is known about the Galf synthetic pathway in eukaryotes. Previous efforts had not yielded the eukaryotic enzyme responsible for synthesis of their UDP-Galf substrate, although as in prokaryotes this was thought to arise by conversion of UDP-Galp to UDP-Galf (29). In the parasitic protozoan Leishmania, several genes encoding putative UDP-Galf transferases have been identified, including LPG1, which has been implicated in the synthesis of the core of the abundant surface glycoconjugate LPG (7, 20). Notably, the Leishmania genome encodes at least 6 candidate UDP-Galf transferases (34), and there are more than 20 related genes present in Trypanosoma cruzi (34) (unpublished data). No candidate UDP-Galf transferases have been reported in fungi, although they must exist given the number of Galf-containing glycoconjugates known in these organisms. In this report we have used a bioinformatics approach to identify the eukaryotic GLF gene family. We expressed four diverse members of this family in E. coli and demonstrated their activity in vivo and/or in vitro.
MATERIALS AND METHODS
PCR and construction of pET3 derivatives.
For Leishmania major, PCR oligonucleotides SMB2179 (5′-GCATGCCATATGAGCGCTGACAAGGTGGTCATAATC) and SMB2180 (5′-GCATGCGGATCCTACGAGGCCGTCGACGACCATGTGCA) (underlined bases represent sites added for subsequent cloning) were based upon the open reading frame (ORF) Lm18.0200 annotated in release 4.0 of the Leishmania major genome (www.genedb.org/genedb/leish). The PCR template was genomic DNA of L. major Friedlin (MHOM/IL/81/Friedlin) clone V1, and amplification was performed with 35 cycles of 30 s at 93°, 45 s at 50°, and 150 s at 68°C. For Trypanosoma cruzi, PCR oligonucleotides SMB2182 (5′-GCATGGCCATATGGCAGAATTATTGACACCGAAAATTG) and SMB2230 (5′-GCGATAGGATCCTCACATATCCTTCTGCAGTAGT) were based upon the second of two ORFs (Tc00.1047053511277.600 and Tc00.1047053507993.160) annotated in release 3.0 of the Trypanosoma cruzi genome project (www.genedb.org/genedb/tcruzi). The PCR template was DNA from the CL Brener strain of T. cruzi used for genome sequencing (kindly provided by D. Barthomoleu, The Institute for Genomic Research), and amplification was performed with 35 cycles of 30 s at 93°C, 45 s at 49°C, and 150 s at 68°C. For Cryptococcus neoformans, PCR primers Galf-S (5′-GGAATTCCATATGCCGTCCAGACTTGATTTT) and Galf-AS (5′-CGGGATCCCTACTTCAAGCGCCTCTCGA) were based upon ORF177.m02862 (The Institute for Genome Research; www.tigr.org/tdb/e2k1/cna1/index.shtml). Total RNA was isolated from C. neoformans serotype D strain JEC43α and converted to cDNA using the SuperScript First-Strand synthesis system as recommended by the manufacturer (Invitrogen, Carlsbad, CA). PCR amplification using HIFI Taq polymerase (Invitrogen, Carlsbad, CA) was performed with 30 cycles of 60 s at 94°C, 60 s at 55°C, and 120 s at 72°C. For C. elegans, gene predictions for the GLF ORF H04M03.4 were obtained from WormBase (http://www.wormbase.org). One internal region had not been determined experimentally, and we found by sequence analysis of several cDNAs (expressed sequence tags [ESTs] yk1442e06, yk1480e04, and yk1626a12 from Yuji Kohara, National Institute of Genetics, Mishima, Japan; GenBank accession no. DN856302-6; and ORF clone 10013@E10, obtained from Open Biosystems, Inc., Huntsville, AL) that the current gene model had missed a short intron, resulting in a predicted 9-amino-acid insertion (SVYCFLREV) not encoded by any of the three cDNAs sequenced. The C. elegans H04M03.4 ORF contained an internal NdeI site, requiring a series of PCR and cloning steps starting with the 10013@E10 cDNA template prior to insertion into the pET3A NdeI-BamHI sites.
All PCRs yielded products with the expected sizes, which as necessary were digested with NdeI and BamHI, ligated to pET-3A vector DNA previously digested with the same enzymes, and transformed into E. coli DH10B or DH5α, yielding pET3a-LmGLF, pET3a-CnGLF, pET3a-TcGLF, or pET3a-CeGLF (lab strains B5234, B5300, B5330, and B5425, respectively). The authenticity of candidate recombinants was confirmed by DNA sequencing (GenBank accession numbers AY900624, AY900625, AY900626, and BK005688, respectively). For T. cruzi GLF, our sequence corresponded well with the provisional Tc00.1047053511277.600 ORF, with three differences (GTG→ GCA/Val→Ala in the second codon and a silent A→G transition at nucleotide 1413). The first two differences may represent “cross priming” of the 5′ oligonucleotide used in PCR.
For rescue of Galf-dependent LPS biosynthesis, the pET-GLF constructs above were introduced into E. coli strain CWG288+pWQ70 (10), yielding strains B5364, B5363, B5365, and B5431, respectively. As described in more detail below, CWG288 contains a deletion of the E. coli rfb locus, while plasmid pWQ70 contains the rfb locus of Klebsiella pneumoniae bearing a deletion of the GLF gene (see reference 10 for the complete genotype and characterization of these). For enzymatic assays, two of the pET-GLF constructs were introduced into the E. coli host strain BL21(DE3) (B5304), yielding pET-LmGLF/BL21(DE3) and pET-CnGLF/BL21(DE3) (B5235 and B5303, respectively). Comparative phylogenetic analyses were conducted using MEGA version 3.0 (11).
LPS Western blotting.
For immunoblotting, cells were pelleted from 1 ml of an overnight E. coli culture, suspended in 50 μl of sample buffer (62.5 mM Tris, 2% sodium dodecyl sulfate, 10% glycerol, 2.5% β-mercaptoethanol, and 0.05% bromphenol blue), and boiled for 10 min. Samples were diluted fivefold further with sample buffer, and 5 μl was loaded on a discontinuous gel consisting of a 4% stacking gel above a 12.5% Tris-glycine separating gel (12). Fresh preparations gave the best results. Gels were run at 200 V for 40 min and electroblotted to nitrocellulose (Hybond-ECL; Amersham Biosciences) overnight at 54 mA. Western blotting was performed with rabbit anti-Klebsiella galactan I (generously provided by Whitfield and Clarke, U. Guelph) at a titer of 1:10,000 for 2 h at room temperature in 5% milk in TBST (10 mM Tris, 150 mM NaCl, 0.05% Tween 20 [vol/vol]). Binding was visualized with peroxidase-conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch, catalog no. 111-035-003) at a titer of 1:20,000 in TBST for 1 h at room temperature followed by detection by chemiluminescence (Western Lightning, Perkin-Elmer Life Sciences, catalog no. NEL100).
UGM enzyme assays.
One-liter cultures of E. coli BL21(DE3)/pET3a-CnGLF expressing Cryptococcus GLF (B5303), E. coli BL21(DE3)/pET3a-LmjGLF expressing Leishmania GLF (B5235), or E. coli BL21(DE3) without plasmid were grown overnight at 37°C in LB broth. Strains bearing pET3a-GLF constructs were grown in the presence of 100 μg/ml ampicillin. All cultures were induced by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside, followed by incubation at 22°C with shaking at 90 rpm for 3 h. Cultures were harvested by centrifugation, and cell pellets were resuspended in 20 ml 100 mM potassium phosphate buffer (pH 7.0) containing 0.15 M NaCl, 15% glycerol, 10 mM flavin adenine dinucleotide (FAD), 0.1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin A, and 1 μM leupeptin. The cells were broken by three passages through a French press and centrifuged at 20,000 × g for 40 min at 4°C. The supernatant fractions were frozen at −80°C until use.
UDP-Galf was synthesized enzymatically from UDP-Galp and purified as described previously (14). Activity assays were performed in 96-well PCR plates. Each well contained 50 μM UDP-Galf, 100 mM phosphate buffer at pH 7, 1 mM MgCl2, 15% glycerol, protein (80 to 10,000 ng E. coli extract expressing Cryptococcus GLF, 35 to 4,400 ng E. coli extract expressing Leishmania GLF, or 27,400 ng control E. coli BL21(DE3), and 20 mM sodium dithionite (to reduce the FAD to FADH2) in a 20-μl volume. FAD was added in various amounts depending on how much protein was added (final concentrations of 1.6 to 1,000 μM). Reactions were incubated at 30°C for 6 min and stopped by the addition of 40 μl 95% ethanol. Samples were analyzed by high-performance liquid chromatography (HPLC) as described previously (14), except that the solvent system was modified by elution of the sugar nucleotides for 10 min with 200 mM KH2PO4 followed by a 600 mM KH2PO4 wash. Yields of UDP-Galp were calculated from the relative fraction of UDP-Galf and UDP-Galp in the samples and the knowledge that 1 nmol of UDP-Galf was originally added to each well.
RESULTS
Identification of candidate GLFs.
Although the first prokaryotic GLF was identified in 1996, previous efforts to detect eukaryotic GLFs had not been successful. Recently the genomes of several eukaryotic pathogens known to synthesize Galf progressed towards completion, and we applied a combinatorial bioinformatics screen to these genomes in an effort to identify candidate GLFs. Briefly, these involved a combination of (i) BLAST searches with prokaryotic GLFs, (ii) searches for flavin binding domains, and (iii) appropriate phylogenetic distribution, specifically occurrence in fungi and protozoa known to synthesize Galf but absence from taxa lacking Galf. Since the presence of Galf has not been systematically addressed for many microbial and metazoan species, we applied the latter criterion conservatively. We focused these studies initially on the Leishmania major genome, using ORF predictions developed by the Leishmania genome project (release 4.0, August 2004; see www.genedb.org/genedb/leish).
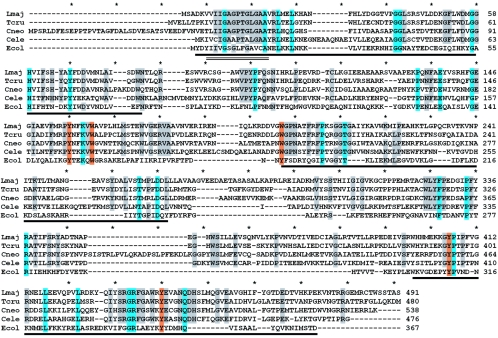
While many ORFs satisfied each of these criteria individually (albeit often quite weakly), a single candidate L. major ORF was identified in the combined screen. The Lm18.0200 GLF sequence predicted a 491-amino-acid protein, which showed a weak relationship to the Pfam family of flavin-containing amine oxidoreductases (PB010804; e = 6 × 10−4), including a potential FAD binding motif at amino acids 6 to 19 (Fig. 1, underlined by parallel lines). The three-dimensional structure of E. coli UGM has shown that the FAD binding domain actually encompasses a much larger portion of the protein (21), and limited sequence conservation homology was also evident throughout this region (Fig. 1, underlined by dark lines). BLAST searches of a variety of databases, including unfinished microbial genomes, revealed a number of strong BLAST hits (P < 10−40), most of which were annotated as hypothetical proteins although several were annotated as amine oxidases (Table 1; Fig. 2). Notably, in every case where sequence from organisms capable of Galf synthesis was analyzed, strong BLAST hits to Lm18.0200 were present. These included the protozoans Leishmania infantum and T. cruzi and the fungi Aspergillus, Cryptococcus, and Neurospora (Fig. 2; Table 1). Correspondingly, homologs of the candidate L. major GLF were not detected in vertebrates, nor in Trypanosoma brucei, Saccharomyces cerevisiae or Schizosaccharomyces pombe, species where Galf may be absent. Prokaryotic GLF/UGMs yielded weak BLAST scores (>0.2) across only the N-terminal portion of the Leishmania UGM, which undoubtedly contributed to the eukaryotic genes being overlooked previously.
FIG. 1.
Alignment of key eukaryotic and prokaryotic GLF proteins. The predicted proteins encoded by the L. major, T. cruzi, C. neoformans, and C. elegans GLF ORFs were aligned with that of E. coli using methods implemented in the Clustal program (4); the unedited alignment is shown. The amino acid positions are shown on the right, and asterisks mark 10-amino-acid intervals in the alignment. Residues identical in 5/5 sequences are shaded in blue, and residues identical in 4/5 sequences are shaded gray. Residues proposed as part of UDP-Galp binding site are shaded orange (all but one of these are 5/5 identities). The dark underlined regions correspond to the flavin-binding region of E. coli UGM (21); a smaller region showing homology to the Pfam family flavin-containing amine oxidoreductases (PB010804; residues 6 to 19 of the L. major UGM) is underlined by two parallel lines.
TABLE 1.
Genes showing strong similarity to eukaryotic GLFsa
| Species | Group | Species contains Gal?b | Gene | Annotationc | Blastp P value | % Identity |
|---|---|---|---|---|---|---|
| Leishmania major | Protists | YES | Lmj18.200 | UGM | (query) | 100 |
| Trypanosoma cruzi | Protists | YES | Tc00.1047053511277.600 | UGM | 10−160 | 58 |
| Tc00.1047053507993.160 | ||||||
| Aspergillus nidulans | Fungi | YES | EAA63683 | Hyp. | 10−117 | 45 |
| Neurospora crassa | Fungi | YES | EAA27372 | Hyp. | 10−117 | 44 |
| Magneporthe grisea | Fungi | UNK | EAA55038 | Hyp. | 10−115 | 44 |
| Gibberella zeae | Fungi | UNK | EAA75642 | Hyp. | 10−113 | |
| Cryptococcus neoformans | Fungi | YES | EAL19520 | UGM | 10−111 | 42 |
| Chlamydomonas rheinhardtii | Green algae | UNK | Table S2d | 10−110 | 42 | |
| Ustilago maydis | Fungi | UNK | UM03094 | Hyp. | 10−104 | 40 |
| Geobacter sulfurreducens | Eubacteria | UNK | AAR34886 | Flavin amine oxidase | 10−103 | 44 |
| Desulfovibrio vulgaris | Eubacteria | UNK | AAS94778 | Flavin amine oxidase | 10−91 | 40 |
| Desulfovibrio desulfuricans | Eubacteria | UNK | ZP00346806 | Protoporphyrinogen oxidase | 10−85 | 37 |
| Feldmannia irregularis virus | Algal virus | UNK | AAR26880 | Hyp. | 10−84 | 38 |
| Caenorhabditis briggsae | Nematodes | UNK | CAE72630 | Hyp. | 10−68 | 34 |
| Caenorhabditis elegans | Nematodes | UNK | AAD12787 | UGM | 10−68 | 34 |
| Ciona intestinalis | Nematodes | UNK | Table S2 | 10−62 | 31 | |
| Halocynthia roretzi | Urochordates | UNK | BAB20903 | HrTLCl | 10−46 | 26 |
| Pyrococcus horikoshii | Archea | UNK | B71153 | Hyp. | 10−27 | 27 |
| Pyrobaculum aerophilum | Archea | UNK | AAL62855 | Hyp. | 10−26 | 27 |
| Yersinia pseudotuberculosis | Eubacteria | PROB | CAB63294 | O-antigen WbyH | 10−18 | 22 |
| Methanococcus maripaludis | Archea | UNK | CAF30324 | Hyp. | 10−14 | 21 |
| Klebsiella pneumonia (prokaryotic GLF) | Eubacteria | YES | Q48485 | UGM | 0.2 | <10% (only N-term)e |
The genes are ordered based on their BlastP P value, based on searches with the L. major GLF query sequence.
YES, evidence showing occurrence of Galf in one or more glycoconjugates; PROB, Galf suspected to be present; UNK, not investigated or established.
Hyp., hypothetical protein. Annotations of L. major, T. cruzi, C. neoformans, and C. elegans GLFs reflect GenBank depositions arising from this work.
See the supplemental material.
N-term, N-terminal.
FIG. 2.
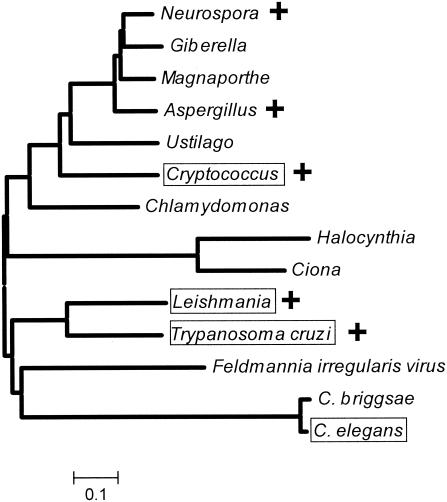
Phylogenetic tree of the eukaryotic GLF family ORFs. A minimum evolution tree for the predicted protein sequences of candidate eukaryotic GLFs was constructed using algorithms implicated in the MEGA3 software package (11). A “+” marks species where Galf has been detected; Galf has not been examined in the other taxa shown. Species where the GLF-encoded UGM activity was confirmed in this work are boxed. For information on the specific sequences analyzed, see Table 1 and Table S2 in the supplemental material. The scale at the bottom corresponds to the fraction of amino acid sequence difference.
Comparison of the predicted proteins encoded by the L. major, T. cruzi, and Cryptococcus neoformans GLFs to that of E. coli UGM/GLF (Fig. 1; Table 1) shows that the overall similarity to prokaryote GLFs is relatively low. Most regions of sequence conservation fall within the FAD binding domains determined previously from the structure of E. coli UGM (Fig. 1, underlined regions) (21). Significantly, genetic and structural analyses have identified a number of residues possibly involved in UDP-Galp binding, and with few exceptions these residues were identical in prokaryotic and eukaryotic GLF proteins (Fig. 1, residues shaded in orange). These residues were also conserved within the strong database hits noted elsewhere in this section (data not shown). The candidate eukaryotic GLF proteins showed a high degree of identity to each other (Fig. 1 and 2; Table 1).
While the correlation of GLF candidates with species known to carry out Galf synthesis was encouraging, strong BLAST hits were also obtained to sequences from a number of organisms not known to synthesize Galf, including the nematodes C. elegans and Caenorhabditis briggsae, the urochordate Halocynthia, and the algal Feldmannia irregularis virus (Table 1). A number of ESTs encoding partial GLFs were found in parasitic nematodes, including Brugia malayi, Onchocerca volvolus, Strongyloides stercoralis and Strongyloides ratti, Heterodura glycine, and Meloidogyne hapla and Meloidogyne arenaria (see Table S1 in the supplemental material), and we were able to assemble complete ORFs for the urochordate Ciona intestinalis and the alga Chlamydomonas reinhardtii (Table 1; see Tables S1 and S2 in the supplemental material). Several strong hits (P < 10−85) were obtained to eubacterial sequences annotated as “amine oxidases,” although enzymatic data supporting these assignments were lacking. Strong hits (P < 10−28 to 10−15) were also obtained with sequences from three archea (Pyrococcus horikoshii, Pyrobaculum aerophilum, and Methanococcus maripaludis) (Table 1). A phylogenetic tree depicting relationships among the eukaryotic candidates is shown in Fig. 2, and an alignment of the C. elegans ORF H04M03.4 is included in Fig. 1. As observed for the fungal and protozoal homologs, candidate flavin-binding regions and UDP-Galp binding residues were conserved in these predicted proteins (Fig. 2; also data not shown). Genes or proteins showing significant relationship were not observed in BLAST searches of the genomes of Giardia lamblia, Trichomonas vaginalis, Entamoeba, apicomplexans including Toxoplasma and Plasmodium, Tetrahymena, or several plant genomes including Arabidopsis.
The imperfect concordance between the occurrence of eukaryotic GLF sequences and Galf synthesis could arise from incomplete knowledge of Galf synthesis and/or an incorrect assignment of this gene family (or individual members therein) as encoding active UGMs. Thus, we sought confirmation that representative candidate GLF ORFs encoded UGM activity.
Rescue of Galf-dependent LPS synthesis in E. coli.
We developed an in vivo complementation assay for UDP-Galf synthesis in E. coli, adapted from work by Whitfield and colleagues (10). These authors showed that expression of the Klebsiella rfbKPO1 locus in an E. coli strain deleted for the endogenous LPS rfb locus led to the synthesis of LPS O1 antigen bearing the repeating unit [→3)-β-d-Galf-(1→3)-α-d-Galp-(1→]. Notably, inactivation of the Klebsiella GLF gene rfbDKPO1 abrogated LPS biosynthesis, which could be restored by episomal expression of KpGLF. We surmised that this would provide a rapid and convenient test for the activity of potential heterologous GLFs. Accordingly, we obtained the L. major, T. cruzi, C. neoformans, and C. elegans GLF ORFs by PCR and introduced them into the bacterial expression vector pET3a. These were then introduced into E. coli strain CWG288 (deleted for the endogenous rfb locus) containing plasmid pWQ70, which bears the Klebsiella rfbKPO1 locus with an inactivated GLF (rfbD) gene (10). LPS expression was monitored by Western blotting with rabbit anti-galactan I antibody, which recognizes a [Galf-Galp] repeating unit (5).
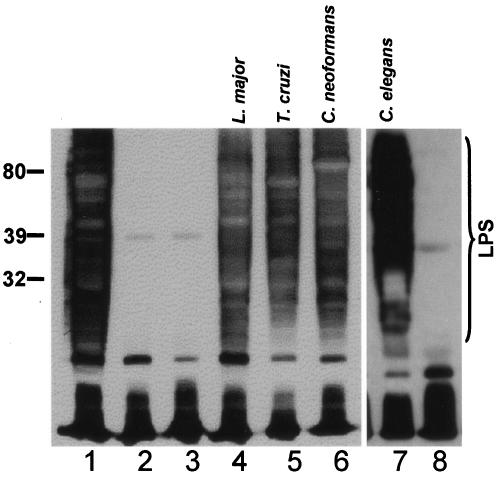
We confirmed that the intact Klebsiella rfb locus restored Galf-containing O1 LPS biosynthesis in E. coli strain CWG288 transformed with pWQ71, which bears an intact rfb locus, while plasmid pWQ70 bearing the inactivated GLF failed to do so (Fig. 3, lane 1 versus lane 2). Transformation of strain CWG288/pWQ70 with any of the four eukaryotic species' pET3a-GLF constructs (L. major, T. cruzi, C. neoformans, or C. elegans) resulted in synthesis of LPS reactive with anti-Klebsiella galactan I antisera, at levels comparable to those of the controls (Fig. 3, lanes 4 to 7). As expected, neither pET3a nor a frameshift mutant C. elegans GLF rescued LPS expression (Fig. 3, lanes 3 and 8, respectively). These data suggested that the eukaryotic candidate GLFs mediated Galf synthesis when expressed heterologously in E. coli.
FIG. 3.
In vivo rescue of Galf-dependent LPS synthesis in E. coli. Bacterial lysates were prepared and subjected to Western blot analysis with rabbit anti-Klebsiella galactan I antisera as described in Materials and Methods. The migration of molecular mass markers (in kDa) is shown on the left, and the region corresponding to migration of LPS is shown by the bracket on the right; the antiserum also reacts with low-molecular-weight material (perhaps lipid A and LPS core), but the identification of O antigen is unequivocal. All strains derive from the E. coli rfb deletion strain CWG288. Plasmids introduced were as follows: lane 1, pWQ71 (intact Klebsiella rfb locus; positive control); lane 2, pWQ70 (pWQ71 with deletion inactivating GLF); lane 3, pWQ70+pET3a (vector control); lane 4, pWQ70+pET3a-LmjGLF; lane 5, pWQ70+pET3a-TcGLF; lane 6, pWQ70+pET3a-CnGLF; lane 7, pWQ70+pET3a-CeGLF; and lane 8, pWQ70 + inactive mutant pET3a-CeGLF (clone 1.1, strain B3597, which contains a 1-nt deletion at position 129 introducing a premature stop amongst other PCR-generated mutations). The experiment shown in lanes 7 to 8 was performed separately from those shown in lanes 1 to 6.
While our data suggest that similar levels of LPS restoration were seen in the experimental versus control samples, some restraint is required in interpreting these data. As shown below, the eukaryotic UGMs are largely insoluble when expressed in E. coli. Thus, differences in the degree of eukaryotic GLF rescue of LPS expression may reflect either intrinsic differences in catalytic activity of the enzymes or “noise” arising from differences in expression and/or folding. Nonetheless, these data suggest that the eukaryotic UGMs are able to provide sufficient UDP-Galf under these conditions for LPS synthesis comparable to that of authentic bacterial UGMs.
UGM activity of L. major and C. neoformans GLFs.
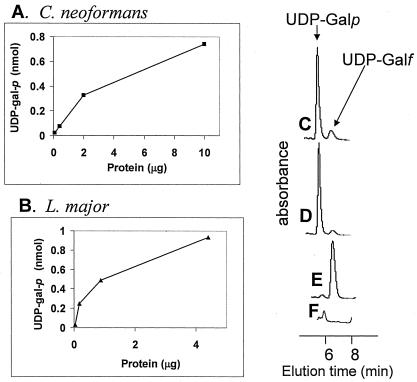
To confirm directly that candidate GLFs encoded UGM activity, we first expressed high levels of protein in E. coli using the pET3a system. While abundant expression was evident in sodium dodecyl sulfate-PAGE analysis, the majority of protein appeared in insoluble inclusion bodies (data not shown). Nonetheless, soluble protein extracts were prepared, and UGM activity was assayed in the reverse direction, following the conversion of UDP-Galf to UDP-Galp by HPLC (14). Since the E. coli strain BL21(DE3) lacks GLF, there was no endogenous background UGM activity in crude preparations (15) (Fig. 4E and F). In contrast, with extracts from cells expressing L. major or C. neoformans GLFs, the UDP-Galf substrate was converted to UDP-Galp (Fig. 4C and D) in a protein-dependent manner (Fig. 4A and B). These data establish that these eukaryotic GLFs encode proteins with UGM activity. In the future, studies of purified soluble and active eukaryotic UGMs will address the catalytic properties of these enzymes relative to those of prokaryotes in more detail.
FIG. 4.
UGM assays of eukaryotic GLFs expressed in E. coli. Protein-containing supernatants were prepared, and UGM activity was assayed by monitoring conversion of UDP-Galf to UDP-Galp by HPLC (15) as described in Methods. Panels A and B show the production of UDP-Galp as a function of added protein extract. (A) Extract from E. coli expressing C. neoformans GLF [BL21(DE3)/pET3a-CnGLF]; (B) Extract from E. coli expressing Leishmania GLF [BL2(DE3)/pET3a-LmjGLF]. Panels C-F show representative HPLC traces and controls used to generate the data in panels A and B. (C) The 10-μg protein point from (A); (D) the 4.4-μg protein point from (B); (E) 28 μg of protein incubated from E. coli BL21(DE3) incubated under standard assay conditions. In the standard assay, the total amount of UDP-Galf added was 1 nmol. Panel F shows 28 μg of protein incubated from E. coli BL21(DE3) incubated without UDP-Galf, showing that the small peak in (E) comes from endogenous UDP-Galp or UDP-Glc from E. coli.
DISCUSSION
By using a combinatorial bioinformatics approach incorporating criteria including sequence homology, protein motifs, and phylogenetic associations with Galf synthesis, we identified a candidate Leishmania GLF gene and from this a large family of potential eukaryotic GLFs. We used a rapid in vivo assay of GLFs expressed in E. coli as a first screen for UGM activity of several candidate eukaryotic proteins (Fig. 3), which were confirmed subsequently in enzymatic assays (Fig. 4). While the eukaryotic UGM/GLF proteins show some relationship to the previously described prokaryotic enzymes, the amino acid sequence divergence was extensive, and the correlation of potential GLF homologs with organisms known to synthesize Galf was imperfect. Perhaps for these reasons, eukaryotic GLFs had remained elusive despite efforts undertaken by many researchers.
Despite the extensive divergence, eukaryotic UGMs share key properties with those of prokaryotes. The eukaryotic proteins contain clear flavin binding motifs, and most importantly, a number of residues implicated by functional or structural criterion in substrate binding were conserved (Fig. 1). Preliminary analysis suggests that the eukaryotic UGMs can be readily modeled to the E. coli UGM structure determined previously, with a number of sequence insertions potentially mapping to external loops (data not shown). As in prokaryotes, eukaryotic UGMs lack obvious secretory signals and are likely to be cytoplasmic. Since the bulk of eukaryotic glycoconjugate synthesis typically occurs within the secretory pathway, this suggests that one or more nucleotide sugar transporters must recognize and transport UDP-Galf to lumenal compartments.
We found a number of genes with strong homology to GLFs encoding eukaryotic UGMs in species not known to synthesize Galf (Fig. 1 and 2; Table 1; see Tables S1 and S2 in the supplemental material). These predicted proteins also showed conservation of key UGM substrate-binding residues (Fig. 1, data not shown). Since the phylogenetic tree presented in Fig. 2 showed that members of both major branches of the eukaryotic GLF family had UGM activity, by parsimony we believe that most or all of these candidate GLFs encode proteins with UGM activity as well. Given the paucity of information on Galf in eukaryotes outside of fungi and protozoa, these new eukaryotic GLFs raise the possibility that Galf synthesis may occur more widely in species than previously supposed. For fungi such as Ustilago, Magneporthe, and Giberella, this was unsurprising, but the occurrence of eukaryotic GLFs in the nematodes C. elegans, the urochordates Ciona and Halocynthia, and the algae Chlamydomonas and algal virus Feldmannia (Fig. 1; Table 1; see Tables S1 and S2 in the supplemental material) was unanticipated. Notably, our data show that C. elegans GLF encodes a protein with UGM activity. Additionally, we found strong GLF homologs in a variety of unfinished genome and EST surveys, including Histoplasma capsulatum, which contains Galf within sphingolipids (2), and nematode parasites of humans (Brugia malayi and Onchocerca volvulus), animals (Strongyloides), and plants (Heterodura and Meloidogyne) (see Table S1 in the supplemental material) (3, 33).
Assuming that the presence of eukaryotic GLF within a species is predictive of Galf synthesis, our analyses suggest that the phylogenetic distribution of the GLF/Galf synthetic pathway shows some features reminiscent of those seen in prokaryotes. First, the GLF/Galf pathway occurs only sporadically, in widely divergent species, and second, it is often found in pathogenic/parasitic species from both the microbial and metazoan worlds. Why pathogens show an affinity for inclusion of Galf in their metabolomic repertoire is unknown; potential explanations might include the strong immunogenicity of Galf and/or its ability to adopt novel structural conformations (17, 31), both of which could contribute to microbial virulence and survival. Notably, the GLF relationships depicted in Fig. 2 or evident in Table 1 often do not closely follow those of the species involved. Potentially, the predilection of pathogens for Galf could be a contributing factor to the sporadic distribution of Galf synthesis among species, perhaps by lateral gene transfer mechanisms as seen in bacterial LPS O antigens (19). The presence of a GLF within the algal virus Feldmannia (Fig. 2) is interesting in this light, since viruses often can be transmitted laterally among species.
Galf-containing glycoconjugates are often dominant features of the surface of many protozoans and fungi, making Galf synthesis a potential target for chemotherapy. However, the role of Galf conjugates in the survival and virulence of eukaryotic microbes appear to vary greatly among species. For example, in fungi, Galf comprises only a small portion of the Cryptococcus capsule (30) but a major fraction of the abundant galactomannans in Aspergillus (13). Interestingly, in several global surveys of gene function using RNA interference approaches, inhibition of the C. elegans GLF encoded by gene H04M03.4 showed a variety of deleterious effects (1, 8, 23); future studies will be required to establish whether this involves Galf-containing glycoconjugates. Preliminary tests of a panel of prospective prokaryotic UGM inhibitors (22) suggest that they fail to inhibit the eukaryotic UGMs studied here (unpublished work), in keeping with the extensive amino acid sequence divergence (Fig. 1). Thus, it will be necessary to identify eukaryote-specific UGM inhibitors in the future. Interestingly, it has recently been suggested that several of the inhibitors of the Mycobacterium tuberculosis UGM may also act against bacteria independently of UGM (28).
For Leishmania, genetic studies suggest that parasites lacking Galf glycoconjugates, such as LPG and glycoinositolphospholipids, retain virulence in mammalian infections, although their ability to be transmitted by the insect vector sand fly is greatly reduced (26, 34, 35). Whether a similar conclusion pertains to Trypanosoma cruzi, where Galf-containing molecules are also highly abundant in infective stages, awaits confirmation (18). Given that the Leishmania and T. cruzi genomes encode numerous potential UDP-Galf transferases (34), genetic inactivation of their GLFs (which occur in one or two copies, respectively; Table 1) may offer an easier definitive test (9). Interestingly, T. brucei, which is thought to be evolutionarily more closely related to T. cruzi than Leishmania, lacks GLF and Galf-containing conjugates. While this could arise from evolutionary loss of GLF in this lineage, the complex disjunct distribution and relationships of GLF genes in other eukaryotic taxa suggest that other explanations cannot yet be ruled out.
In summary, we have identified a large eukaryotic GLF family and provided evidence that members of the two major groups within this family possess UGM activity. This in turn has permitted predictions about the occurrence of Galf synthesis among species and its frequent appearance within both microbial and metazoan pathogens. Future work will focus on exploitation of UGM-targeted inhibitors in chemotherapy and determination of the structure and role of Galf in those lineages for which information is currently lacking.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI31903 (S.M.B.), GM66303 (T.D.), AI33706, and AI057836 (M.R.M.).
We thank Natalia Akopyants for help with PCR amplification, Chad Rappleye, David Sanders, Sam Turco, Chris Whitfield, and Brad Clarke for helpful discussions and/or for providing E. coli strains and plasmids, Daniella Bartholomeu for T. cruzi cLBrener DNA, Yuji Kohara for providing C. elegans EST clones, and A. Capul, D. E. Dobson, and K. Zhang for comments on the manuscript. We also express our appreciation to the many genome and EST projects contributing data incorporated into Table 1 and into Tables S1 and S2 in the supplemental material.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Ashrafi, K., F. Y. Chang, J. L. Watts, A. G. Fraser, R. S. Kamath, J. Ahringer, and G. Ruvkun. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421:268-272. [DOI] [PubMed] [Google Scholar]
- 2.Barr, K., R. A. Laine, and R. L. Lester. 1984. Carbohydrate structures of three novel phosphoinositol-containing sphingolipids from the yeast Histoplasma capsulatum. Biochemistry 23:5589-5596. [DOI] [PubMed] [Google Scholar]
- 3.Blaxter, M., J. Daub, D. Guiliano, J. Parkinson, and C. Whitton. 2002. The Brugia malayi genome project: expressed sequence tags and gene discovery. Trans. R. Soc. Trop. Med. Hyg. 96:7-17. [DOI] [PubMed] [Google Scholar]
- 4.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, B. R., D. Bronner, W. J. Keenleyside, W. B. Severn, J. C. Richards, and C. Whitfield. 1995. Role of Rfe and RfbF in the initiation of biosynthesis of d-galactan I, the lipopolysaccharide O antigen from Klebsiella pneumoniae serotype O1. J. Bacteriol. 177:5411-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lederkremer, R. M., and W. Colli. 1995. Galactofuranose-containing glycoconjugates in trypanosomatids. Glycobiology 5:547-552. [DOI] [PubMed] [Google Scholar]
- 7.Huang, C., and S. J. Turco. 1993. Defective galactofuranose addition in lipophosphoglycan biosynthesis in a mutant of Leishmania donovani. J. Biol. Chem. 268:24060-24066. [PubMed] [Google Scholar]
- 8.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231-237. [DOI] [PubMed] [Google Scholar]
- 9.Kleczka, B., A.-C. Lamerz, H. Bakker, M. Wiese, R. Gerardy-Schahn, and F. H. Routier. 2004. Leishmania major UDP-galactopyranose mutase: characterisation and validation of a potential drug target. Glycobiology 14:abstr. 232.
- 10.Koplin, R., J. R. Brisson, and C. Whitfield. 1997. UDP-galactofuranose precursor required for formation of the lipopolysaccharide O antigen of Klebsiella pneumoniae serotype O1 is synthesized by the product of the rfbDKPO1 gene. J. Biol. Chem. 272:4121-4128. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Latge, J. P., H. Kobayashi, J. P. Debeaupuis, M. Diaquin, J. Sarfati, J. M. Wieruszeski, E. Parra, J. P. Bouchara, and B. Fournet. 1994. Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect. Immun. 62:5424-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, R., D. Monsey, A. Weston, K. Duncan, C. Rithner, and M. McNeil. 1996. Enzymatic synthesis of UDP-galactofuranose and an assay for UDP-galactopyranose mutase based on high-performance liquid chromatography. Anal. Biochem. 242:1-7. [DOI] [PubMed] [Google Scholar]
- 15.Nassau, P. M., S. L. Martin, R. E. Brown, A. Weston, D. Monsey, M. R. McNeil, and K. Duncan. 1996. Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J. Bacteriol. 178:1047-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan, F., M. Jackson, Y. Ma, and M. McNeil. 2001. Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J. Bacteriol. 183:3991-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedersen, L. L., and S. J. Turco. 2003. Galactofuranose metabolism: a potential target for antimicrobial chemotherapy. Cell Mol. Life Sci. 60:259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Previato, J. O., R. Wait, C. Jones, G. A. DosReis, A. R. Todeschini, N. Heise, and L. M. Previato. 2004. Glycoinositolphospholipid from Trypanosoma cruzi: structure, biosynthesis and immunobiology. Adv. Parasitol. 56:1-41. [DOI] [PubMed] [Google Scholar]
- 19.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan, K. A., L. A. Garraway, A. Descoteaux, S. J. Turco, and S. M. Beverley. 1993. Isolation of virulence genes directing surface glycosyl-phosphatidylinositol synthesis by functional complementation of Leishmania. Proc. Natl. Acad. Sci. USA 90:8609-8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders, D. A., A. G. Staines, S. A. McMahon, M. R. McNeil, C. Whitfield, and J. H. Naismith. 2001. UDP-galactopyranose mutase has a novel structure and mechanism. Nat. Struct. Biol. 8:858-863. [DOI] [PubMed] [Google Scholar]
- 22.Scherman, M. S., K. A. Winans, R. J. Stern, V. Jones, C. R. Bertozzi, and M. R. McNeil. 2003. Drug targeting Mycobacterium tuberculosis cell wall synthesis: development of a microtiter plate-based screen for UDP-galactopyranose mutase and identification of an inhibitor from a uridine-based library. Antimicrob. Agents Chemother. 47:378-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmer, F., C. Moorman, A. M. van der Linden, E. Kuijk, P. V. van den Berghe, R. S. Kamath, A. G. Fraser, J. Ahringer, and R. H. Plasterk. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLOS Biol. 1:E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soltero-Higgin, M., E. E. Carlson, T. D. Gruber, and L. L. Kiessling. 2004. A unique catalytic mechanism for UDP-galactopyranose mutase. Nat. Struct. Mol. Biol. 11:539-543. [DOI] [PubMed] [Google Scholar]
- 25.Soltero-Higgin, M., E. E. Carlson, J. H. Phillips, and L. L. Kiessling. 2004. Identification of inhibitors for UDP-galactopyranose mutase. J. Am. Chem. Soc. 126:10532-10533. [DOI] [PubMed] [Google Scholar]
- 26.Späth, G. F., L. Epstein, B. Leader, S. M. Singer, H. A. Avila, S. J. Turco, and S. M. Beverley. 2000. Lipophosphoglycan is a virulence factor distinct from related glycoconjugates in the protozoan parasite Leishmania major. Proc. Natl. Acad. Sci. USA 97:9258-9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tangallapally, R. P., R. Yendapally, R. E. Lee, K. Hevener, V. C. Jones, A. J. Lenaerts, M. R. McNeil, Y. Wang, S. Franzblau, and R. E. Lee. 2004. Synthesis and evaluation of nitrofuranylamides as novel antituberculosis agents. J. Med. Chem. 47:5276-5283. [DOI] [PubMed] [Google Scholar]
- 29.Trejo, A. G., G. J. Chittenden, J. G. Buchanan, and J. Baddiley. 1970. Uridine diphosphate alpha-d-galactofuranose, an intermediate in the biosynthesis of galactofuranosyl residues. Biochem. J. 117:637-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaishnav, V. V., B. E. Bacon, M. O'Neill, and R. Cherniak. 1998. Structural characterization of the galactoxylomannan of Cryptococcus neoformans Cap67. Carbohydr. Res. 306:315-330. [DOI] [PubMed] [Google Scholar]
- 31.Weller, C. T., M. McConville, and S. W. Homans. 1994. Solution structure and dynamics of a glycoinositol phospholipid (GIPL-6) from Leishmania major. Biopolymers 34:1155-1163. [DOI] [PubMed] [Google Scholar]
- 32.Weston, A., R. J. Stern, R. E. Lee, P. M. Nassau, D. Monsey, S. L. Martin, M. S. Scherman, G. S. Besra, K. Duncan, and M. R. McNeil. 1997. Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. Tuber. Lung Dis. 78:123-131. [DOI] [PubMed] [Google Scholar]
- 33.Wylie, T., J. C. Martin, M. Dante, M. D. Mitreva, S. W. Clifton, A. Chinwalla, R. H. Waterston, R. K. Wilson, and J. P. McCarter. 2004. Nematode.net: a tool for navigating sequences from parasitic and free-living nematodes. Nucleic Acids Res. 32(Database issue):D423-D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, K., T. Barron, S. J. Turco, and S. M. Beverley. 2004. The LPG1 gene family of Leishmania major. Mol. Biochem. Parasitol. 136:11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zufferey, R., S. Allen, T. Barron, D. R. Sullivan, P. W. Denny, I. C. Almeida, D. F. Smith, S. J. Turco, M. A. Ferguson, and S. M. Beverley. 2003. Ether phospholipids and glycosylinositolphospholipids are not required for amastigote virulence or for inhibition of macrophage activation by Leishmania major. J. Biol. Chem. 278:44708-44718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.