Abstract
Amino acids in the environment of Saccharomyces cerevisiae can transcriptionally activate a third of the amino acid permease genes through a signal that originates from the interaction between the extracellular amino acids and an integral plasma membrane protein, Ssy1p. Two plasma membrane-associated proteins, Ptr3p and Ssy5p, participate in the sensing, which results in cleavage of the transcription factors Stp1p and Stp2p, removing 10 kDa of the N terminus of each of them. This confers the transcription factors with the ability to gain access to the nucleus and activate transcription of amino acid permease genes. To extend our understanding of the role of Ptr3p and Ssy5p in this amino acid sensing process, we have isolated constitutive gain-of-function mutants in these two components by using a genetic screening in which potassium uptake is made dependent on amino acid signaling. Mutants which exhibit inducer-independent processing of Stp1p and activation of the amino acid permease gene AGP1 were obtained. For each component of the SPS complex, constitutive signaling by a mutant allele depended on the presence of wild-type alleles of the other two components. Despite the signaling in the absence of inducer, the processing of Stp1p was more complete in the presence of inducer. Dose response assays showed that the median effective concentration for Stp1p processing in the mutant cells was decreased; i.e., a lower inducer concentration is needed for signaling in the mutant cells. These results suggest that the three sensor components interact intimately in a complex rather than in separate reactions and support the notion that the three components function as a complex.
Differences in nutrient availability from one environment to another and changes in nutrient abundance that occur as a consequence of growth present fundamental challenges to free-living microbes. Prokaryotic and eukaryotic microorganisms have evolved nutrient-sensing systems that detect changes in extracellular nutrient concentrations and initiate signal transduction responses that alter the expression of relevant nutrient transporter genes (19). In the yeast Saccharomyces cerevisiae, for example, distinct sensing systems have been described for phosphate (15) and for glucose and amino acids (reviewed in reference 8).
In the sensing pathway described here, S. cerevisiae cells respond to extracellular amino acids by transcriptional induction of some of the amino acid transporters, such as Agp1p. Genetic studies have played a key role in identifying and dissecting key components of the pathway. Due to overlapping specificities of amino acid transporters, a mutant screening designed to identify mutations conferring resistance to inhibitors of branched-chain amino acids was successful in identifying the upstream factors that mediate amino acid signaling in this organism. Thus, SSY1, SSY5, and PTR3 were found to be required for the induction of downstream amino acid permease genes by amino acids (6, 10, 21, 23, 24) and encode the components of the so-called (12) SPS (Ssy1-Ptr3-Ssy5) sensor. Ssy1p seems to be the actual amino acid-sensing molecule. Although it is a membrane protein resembling an amino acid permease, Ssy1p seems not to transport amino acids, yet it is required for amino acid-stimulated transcription of the inducible downstream permease genes. The discovery of mutations in SSY1 that confer constitutive signaling in the absence of extracellular amino acids (14) revealed that, even if Ssy1p might normally be able to transport small amounts of amino acids, transported amino acids seem not to constitute the intracellular signal that mediates downstream induction of the responsive permease genes.
Ssy1p, Ptr3p, and Ssy5p are believed to be part of a membrane-associated signaling complex (12, 24). Consistent with this idea, both Ssy5p and Ptr3p remain essential for amino acid signaling even in cells expressing the SSY1 signaling constitutive alleles (14). A key function of the SPS sensor is the activation of the transcription factors Stp1p (22) and Stp2p, which bind to the promoters (27) and induce increased transcription of the responsive amino acid permease genes. Andréasson and Ljungdahl (3, 4) found that activation of these transcription factors involves the proteolytic removal of an inhibitory amino terminal region. This processing normally requires all three components of SPS, but Abdel-Sater et al. (1) found that overexpression of an N-terminally tagged version of Ssy5p causes constitutive processing of Stp1p and suggested that Ssy5p is a serine protease that carries out the cleavage. Full-length Stp1p and Stp2p are retained in the cytoplasm in a manner dependent on the amino terminus of these transcription factors and on the function of the ASI1 through ASI3 genes (13). Removal of the amino terminus of Stp1p or Stp2p by SPS allows its transport to the nucleus and the consequent transcriptional response (3).
Although Ssy5p has been proposed to be the protease that cleaves Stp1p and Stp2p, other details of the molecular function of SPS remain largely unknown. For example, how does Ssy1p recognize amino acids? Is transport of amino acid by Ssy1p part of the conformational change that activates the SPS? Do Ssy1p, Ssy5p, and Ptr3p actually form a complex? What are the functional dependency relationships between members of the SPS? What specific roles does each member play?
We previously devised a genetic screening that allowed the identification of mutations in SSY1 that result in constitutive signaling (14). The observation that Ssy5p and Ptr3p remain essential for signaling in these cells is consistent with a model in which all three proteins function interdependently or with one in which Ssy5p and Ptr3p function downstream of Ssy1p through separate interactions. The isolation of mutants containing constitutively signaling forms of Ssy5p and/or Ptr3p could help to distinguish between these alternatives by testing whether they are functionally dependent on Ssy1p. Establishing these relationships would also allow us to place additional constraints on a functional model of the SPS sensor and perhaps provide clues to its structural constraints. In this report, we describe the results of a genetic screening designed to identify constitutively signaling alleles of SSY5 and PTR3. The isolated alleles result in amino acid-independent processing of Stp1p and thus account for the constitutive expression of SPS-responsive permease genes. We have tested the dependency of these gain-of-function mutations on each of the components of the SPS sensor, and we discuss the significance of the results with regard to the mechanism of SPS signaling.
MATERIALS AND METHODS
Media.
Standard glucose-based media synthetic dextrose (SD; synthetic minimal), synthetic complete (SC), and yeast-peptone-dextrose complex media were prepared as described previously (31). However, amino acid concentrations in SC medium were as specified previously elsewhere (17). Selections for kanR-marked strains were made on solid yeast-peptone-dextrose complex medium supplemented with 300 μg/liter G418 (Sigma).
Yeast strains.
Strains M5077 (ssy5Δ) and M5078 (ptr3Δ) were created in the genetic background of strain M4955, which was designed (14) to grow on SD medium in an amino acid signaling-dependent way (Table 1). They were constructed using the loxP-kanMX-loxP gene disruption technology with plasmid pUG6 (18) as follows. For deletion of PTR3, the primer pair PTR3pUG6.for (5′ ATC AAT GAT TAC CTT ATC AGC ACT GAA AAG ATA CCC GTA AAA TTT AGC ACT TCG TAC GCT GCA GGT CGA C 3′) and PTR3pUG6.rev (5′ GAT AGT GTT TTC TTT TAA TAC CTG TAT ACC AGA ACC TTA AAC ATA CGT ATG CAT AGG CCA CTA GTG GAT CTG 3′) was used for PCR amplification of the deletion cassette from plasmid pUG6. For deletion of SSY5, the primer pair SSY5pUG6.for (5′ GTA CAG AAA ACG TAA ATA TAC AAT AAA GGT TGA ATA AAC ATA CTA GAT ATT TCG TAC GCT GCA GGT CGA C 3′) and SSY5pUG6.rev (5′ AAT GGG TTA AAT AAC TTC AAA AAG GCA AAT CAT CCA TCT AGT TGT GGA TCG CAT AGG CCA CTA GTG GAT CTG 3′) was used. Strain M4955 was transformed with the PCR-generated deletion cassettes, and selection on G418 yielded strains M5077 (ssy5Δ) and M5078 (ptr3Δ).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| M4955 | MATatrk1::HIS3 trk2::HIS3 ura3 trp1 pAGP1-KAT1 | 14 |
| M5077 | M4955 ssy5::kanMX | This work |
| M5078 | M4955 ptr3::kanMX | This work |
| M4054 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ | 17 |
| M4723 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ptr3Δ | P. S. Nielsen |
| M4724 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ssy5Δ | P. S. Nielsen |
| M4871 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ssy1Δ | P. S. Nielsen |
| M5359 | M4054 AGP1::PAGP1-lacZ | This work |
| M5360 | M4723 AGP1::PAGP1-lacZ | This work |
| M5361 | M4724 AGP1::PAGP1-lacZ | This work |
| M5380 | M4871 AGP1::PAGP1-lacZ | This work |
| M5443 | M4723 STP1::ZZ-kanMX | This work |
| M5444 | M4724 STP1::ZZ-kanMX | This work |
| M5445 | M4871 STP1::ZZ-kanMX | This work |
| M5447 | M4054 STP1::ZZ-kanMX | This work |
Strains M5359, M5360, M5361, and M5380 were constructed by transformation of M4054, M4723, M4724 and M4871, respectively, with plasmid pPEP17 linearized with BmgBI, which cuts the AGP1 promoter 387 bp upstream from the start codon. Integrants were selected on G418.
Stp1p was tagged (29) at the C terminus in the genome with an immunoglobulin G-binding domain (Z) of protein A (36). A PCR fragment encoding a doublet of domain Z, a kanMX cassette and 50-bp overlaps to STP1, was amplified from plasmid pFZ (36) using primers 5′-AGA CAC AAC AAT ATT TGA ATT TTT ACA ATG ACA ACT TTG GGT CAC AAT TTG GAG CAG GGG CGG GTG C-3′ and 5′-CAT CGG CTT TCC AAT ATG ATA CCC TTA TTT TTA TCC CGT GTT ATA TTT AAG GTC GAC GGT ATC GAT AAG-3′. Strains M4723, M4724, M4871, and M4054 were transformed with the resulting PCR product, selecting on G418, and yielding strains M5443, M5444, M5445, and M5447, respectively. Correct integration was verified by PCR using primers 5′-TGG TTT TAG CAG ACG CGA TAC-3′ and 5′-GAT ACT AGC TCA TCT TCT TCG TCG A-3′.
Plasmids.
pPEP10 was constructed as follows. SSY5 promoter (495-bp) and SSY5 terminator (540-bp) fragments were produced by PCR using primer pair SSY5-3 (5′CCA AGC TTA CTT GTG GCT GAT ACG C 3′) and SSY5-4 (5′ GGG TTC GTA TCA TGA CTG TTT GGG 3′) and primer pair SSY5-5 (5′ CGG GAT CCA TTC AGG CGC ATG GAT CTT GAC C 3′) and SSY5-6 (5′ GCT CTA GAC TGT GAA CCA AGG TAC CTT CG 3′), respectively. The SSY5 promoter PCR fragment contains 296 bp of the promoter region and 155 bp of the coding region, whereas the SSY5 terminator PCR fragment contains 148 bp of the coding region and 376 bp of the terminator region. The fragments were digested with HindIII-BamHI and XbaI-BamHI, respectively, and inserted into pRS316 digested with HindIII-XbaI. Plasmid pPEP10 can be linearized with BamHI, which cuts between the promoter and terminator, allowing recombination with a PCR-mutagenized fragment of the entire SSY5 open reading frame (ORF).
pPEP11 construction.
PTR3 promoter (406-bp) and terminator (417-bp) fragments were produced by PCR using primer pair PTR3-3 (5′ GGA ATT CCT TAA ACC AAC TTG GCT ACC G 3′) and PTR3-4 (5′ CGG GAT CCG TAT TGC ACA TGT GAT TCG 3′) and primer pair PTR3-5 (5′ CGG GAT CCG GCA ACA AGC TTT ATA TTC TCG AC 3′) and PTR3-6 (5′ GCT CTA GAG TGC ACC CCA TCT AAA CGA AAC 3′), respectively. The PTR3 promoter PCR fragment contains 254 bp of the promoter region and 140 bp of the coding region, whereas the PTR3 terminator PCR fragment contains 114 bp of the coding region and 287 bp of the terminator region. The fragments were digested with EcoRI-BamHI and XbaI-BamHI, respectively, and inserted into pRS316 digested with EcoRI-XbaI. Plasmid pPEP11 can be linearized with BamHI, which cuts between the promoter and terminator, allowing recombination with a PCR-mutagenized fragment of the entire PTR3 ORF.
The AGP1-lacZ reporting plasmid pPEP15 was created by inserting a 995-bp AGP1 promoter fragment, made by PCR using the primers AGP1-1 (5′ GTC GAC GAG CGC CTT TAC CTC AAC CTA CCA TGG 3′) and AGP1-2 (5′ TGT GCG AAG CTA TCT TTG TCT ATA TTA GCG TGC 3′), into the SalI-EcoRI sites of plasmid pYC-Z130 (28). The 518-bp FseI CEN-ARS fragment was removed from pPEP15 to form the integrating AGP1-lacZ-reporting plasmid pPEP17.
Plasmids pSSY5 and pPTR3 (23; also referred to as pMB5 and pMB2, respectively [9a]) contain a 3-kb HindIII-SacII fragment with SSY5 and a 5.4-kb XbaI and KpnI fragment with PTR3, respectively, inserted into pRS316 (CEN/URA3) (32).
Mutagenesis of the SSY5 and PTR3 coding regions.
Libraries of PCR fragments of SSY5 and PTR3 with randomly introduced mutations were constructed with Mutazyme polymerase (Stratagene) and primers SSY5-1 (5′ GTA CTG GTG TAA ACT CGA TAT ACC G 3′) and SSY5-2 (5′ CCA TCT AGT TGT GGA TCA ATG TCC 3′) and primers PTR3-1 (5′ GGT ACG AAA TAC ACA ACT GAT AGG CG 3′) and PTR3-7 (5′ CTT TTA ATA CCT GTA TAC CAG AAC C 3′), using plasmids pSSY5 and pPTR3, respectively, as templates and the conditions recommended by the manufacturer. Custom DNA sequencing of the isolated mutant plasmids was carried out by MWG Biotech (Germany). We observed a difference between the sequence of the SSY5 gene cloned (23) from the YCp50 library (30) and the sequence in the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/). Thus, the A nucleotide in codon 685 (GAC) of the SGD sequence was absent in our data. Deletion of the A results in a Ssy5p that is 12 amino acids longer than the version in the SGD. The extended Ssy5p has a C terminus that is identical to those of other Saccharomyces species (SGD).
Isolation of constitutively signaling SSY5 and PTR3 mutants.
Strain M5077 (trk1Δ trk2Δ ssy5D) harboring plasmid pAGP1-KAT1 (14) was transformed with 0.5 μg pPEP10 linearized with BamHI and 0.5 μg Mutazyme-amplified SSY5 ORF by using a Gietz lab yeast transformation kit and procedure II (Genomics ONE, Buffalo, NY). The transformation reaction was plated on SC medium without uracil and tryptophan but supplemented with 100 mM KCl to allow expression of the inserted SSY5 alleles. After 20 h at 30°C, transformants were replica plated to SD medium. After 2 to 4 days at 30°C, fast-growing, potentially constitutive SSY5 mutants appeared on the SD plates. Mutants were streaked to single colonies on SD plates, and plasmid DNA was rescued using a yeast DNA extraction reagent kit (Pierce, Rockford, Illinois). Escherichia coli strain DH5α was transformed with the yeast DNA preparations, and SSY5-containing plasmids were identified by colony PCR screening using primers SSY5-3 (5′-CCA AGC TTA CTT GTG GCT GAT ACG C-3′) and SSY5-4 (5′-GGG TTC GTA TCA TGA CTG TTT GGG-3′). The plasmids were reintroduced into yeast strain M5077 and analyzed for their growth phenotype on SD plates and on SD plates supplemented with 0.2 mM leucine or 100 mM KCl.
PTR3 mutants were isolated in the same manner, using strain M5078 (trk1Δ trk2Δ ptr3Δ) as the screening strain and pPEP11 linearized with BamHI as the receptor plasmid. The PTR3 ORF mutant library was generated with Mutazyme as described above. PTR3-containing E. coli DH5α transformants were identified using primers PTR3-3 (5′-GGA ATT CCT TAA ACC AAC TTG GCT ACC G-3′) and PTR3-4 (5′-CGG GAT CCG TAT TGC ACA TGT GAT TCG-3′).
β-Galactosidase activity was determined by using exponential-phase yeast cells harvested from cultures inoculated at an optical density at 600 nm (OD600) of 0.2 from overnight SD cultures and grown to an OD600 of 0.8 in SD medium. Soluble protein extracts were prepared from cell pellets using the Y-PER reagent procedure from Pierce. The β-galactosidase activity of the lysates was then determined (5, 10).
Qualitative β-galactosidase assays were carried out by resuspending cells grown on SD plates supplemented with leucine (2 mM) in buffer Z (5, 10) containing 0.2% (wt/vol) Na-N-lauroyl-sarcosine and 0.2 mg/ml X-Gal (5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside). Cell suspensions were incubated at 30°C overnight or until a blue color was detectable.
Dose-response analysis.
Yeast cultures were grown exponentially overnight at 30°C in SD medium. When an OD600 of 0.3 to 0.5 was reached, 5-ml aliquots were exposed to leucine at a concentration ranging from 5 nM to 1 mM. Samples of 1.5 ml were withdrawn after 10 min, and proteins were extracted as described below.
Western analysis.
Proteins were extracted under denaturing conditions with NaOH and β-mercaptoethanol from exponentially growing yeast cells and precipitated with trichloroacetic acid (9). Proteins were separated on NuPAGE 4 to 12% Bis-Tris gradient gels (Invitrogen) and blotted onto Invitrolon polyvinylidene difluoride membranes (Invitrogen). Protein blots were incubated with a complex of horseradish peroxidase antibody and horseradish peroxidase (Rabbit PAP Z 0113; DakoCytomation) in a 1:10,000 dilution in primary antibody diluent (Invitrogen), and chemiluminescence was detected using ECL Plus (Amersham Biosciences) and a Storm 860 apparatus. Quantification of chemiluminescence was carried out with ImageQuant software, version 5.0.
RESULTS
Genetic screening for constitutively signaling SSY5 and PTR3 mutants.
Constitutively signaling SSY5 and PTR3 mutants were isolated using the KAT1 potassium channel reporter system for amino acid sensing developed by Gaber et al. (14). This reporter system makes use of a yeast host strain that is impaired in potassium import by deletion of the TRK1 and TRK2 genes and a reporter construct in which transcription of the gene for the Arabidopsis thaliana KAT1 potassium channel is controlled by a target promoter of SPS signaling. The selected promoter normally controls the expression of the AGP1 amino acid permease gene.
trk1Δ trk2Δ cells grow poorly on standard minimal medium (SD medium), in which the potassium concentration is 7 mM. However, when these cells contain the reporter plasmid (pAGP1-KAT1), normal growth is restored by addition of leucine, since this amino acid induces SPS sensor-mediated activation of the AGP1 promoter, which leads to production of the KAT1 potassium channel and, consequently, increased potassium import.
Similar to the previous isolation of a mutant SSY1 allele that constitutively activates the AGP1 promoter (14), here, we present the isolation of constitutive SSY5 and PTR3 mutants using the KAT1 reporter system. The SSY5 ORF was subjected to mutagenesis by error-prone PCR amplification as described in Materials and Methods. Yeast strain M5077 (trk1Δ trk2Δ ssy5D/pAGP1-KAT1) was transformed with a mixture of the mutagenized DNA and linearized pPEP10, a centromere-based, URA3-based plasmid that contains sequence overlaps with the mutagenized DNA. Subsequent homologous recombination in vivo ensured insertion of the mutagenized copies of the SSY5 ORF into pPEP10. Among the approximately 9,000 uracil-independent transformants screened, 24 potentially constitutive SSY5 mutants were identified by their early appearance on SD plates. Plasmid DNA was isolated from seven of these mutants and reintroduced into strain M5077 (trk1Δ trk2Δ ssy5Δ/pAGP1-KAT1). The transformants grew rapidly on SD medium, indicating that this trait was associated with the plasmid-borne mutant SSY5 allele. DNA sequencing revealed that the mutant plasmid pSSY5-6 harbored a single base pair substitution in the SSY5 ORF, whereas each of the other mutants contained alterations at several positions. The mutation in plasmid pSSY5-6 was a transition from G to A in codon 512 (GAA to AAA), resulting in a Glu-to-Lys substitution (E512K) in Ssy5p.
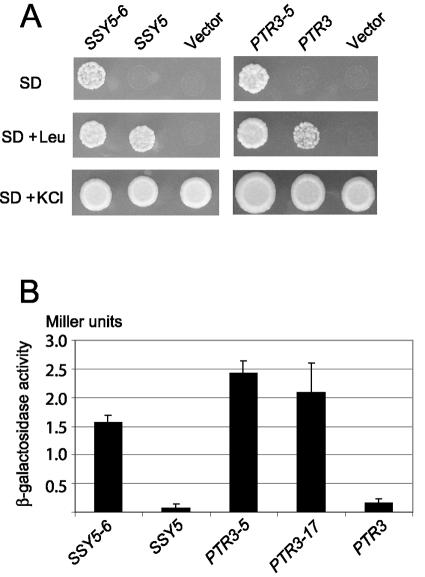
The M5077/pSSY5-6 transformant was analyzed for its growth phenotype by using SD medium and SD medium supplemented with 0.2 mM leucine or 100 mM KCl. M5077/pSSY5-6 cells formed colonies within 3 days of incubation at 30°C on SD medium, whereas M5077 transformed with plasmid pSSY5 (carrying the wild-type SSY5 gene) or the vector without insert (pRS316) exhibited almost no growth (Fig. 1A, left panel). On SD medium supplemented with leucine, both M5077/pSSY5-6 and M5077/pSSY5 cells grew normally, whereas M5077/pRS316 failed to grow, due to the absence of a functional SSY5 gene in this strain. As expected, all three strains grew on SD medium supplemented with KCl. Our interpretation of these results is that the SSY5-6 mutation causes activation of the AGP1 promoter through the SPS signaling pathway, independent of the presence of extracellular amino acid.
FIG. 1.
SSY5 and PTR3 can be mutated to activate the AGP1 promoter in the absence of amino acids. (A) Suspensions of strains M5077 (trk1Δ trk2Δ ssy5Δ/pAGP1-KAT1) or M5078 (trk1Δ trk2Δ ptr3Δ/pAGP1-KAT1) transformed with the centromere-based plasmid pRS316 carrying the indicated SSY5 or PTR3 allele or without insert (vector) were spotted onto minimal medium (SD), minimal medium with 0.2 mM leucine (SD + Leu), or minimal medium with 100 mM KCl (SD + KCl) and incubated for 3 days at 30°C. (B) β-Galactosidase activity in cell extracts of strains M5360 (ptr3Δ agp1::Pagp1-lacZ-KanMX) and M5361 (ssy5Δ agp1::Pagp1-lacZ-KanMX) transformed with the centromere-based plasmid pRS316 carrying the indicated PTR3 and SSY5 alleles and grown in minimal medium (SD medium). Standard deviation error bars are shown.
By using an analogous approach, PTR3 ORF DNA amplified by mutagenic PCR was introduced together with linearized pPEP11 into yeast strain M5078 (trk1Δ trk2Δ ptr3Δ/pAGP1-KAT1) to identify constitutive alleles of PTR3. Eight potentially constitutive PTR3 mutants were obtained. Plasmids harboring these mutations were isolated and reintroduced into strain M5078, and growth on SD confirmed the presence of mutant PTR3 alleles on the plasmid. DNA sequencing revealed that the plasmids pPTR3-5 and pPTR3-17 each contained a single base pair exchange in the PTR3 ORF, whereas the rest of the mutants were affected at multiple sites. The PTR3-5 mutant had acquired a G-to-A transition in codon 439 (CAA to CGA), resulting in a Glu-to-Arg substitution (Q439R), whereas the PTR3-17 mutant has a C-to-A transversion at codon 435 (ACA to AAA), resulting in a Thr-to-Lys substitution (T435K).
The growth phenotypes of M5078 cells harboring pPTR3-5, pPTR3, or the pRS316 vector are shown in Fig. 1A (right panel). The PTR3-5 allele conferred upon M5078 cells the ability to grow on SD medium in the absence of amino acids, whereas M5078/pPTR3 and M5078/pRS316 cells failed to grow. Similar results were obtained with PTR3-17 (data not shown), indicating that both mutant PTR3 alleles suppress the low-potassium phenotype of M5078 cells, presumably by amino acid-independent activation of the AGP1::KAT1 reporter.
The SSY5 and PTR3 mutants activate the AGP1 promoter in the absence of extracellular amino acids.
To determine if the SSY5 and PTR3 mutants confer amino acid-independent activation of the AGP1 promoter, plasmids pSSY5-6, pPTR3-5, and pPTR3-17 were introduced into M5361 (ssy5Δ AGP1::lacZ) and M5360 (ptr3Δ AGP1::lacZ) cells, and β-galactosidase levels in these cells grown in SD were measured and compared to those expressing wild-type SSY5 and PTR3 alleles (Fig. 1B). The levels of β-galactosidase in the mutant cells were significantly higher than those in the wild-type cells, showing that the mutant SSY5 and PTR3 alleles constitutively activate the AGP1 promoter. When the cells were grown in SD supplemented with leucine, the wild-type and the mutant SSY5 and PTR3 alleles activated the AGP1 promoter to the same extent as that observed with cells expressing mutant alleles in the absence of leucine (data not shown).
Constitutive SSY5 and PTR3 alleles confer increased Stp1p processing in the absence of amino acids.
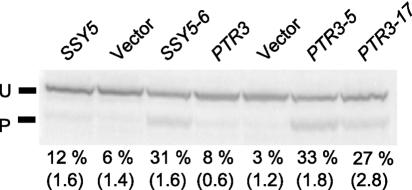
Proteolytic removal of ca. 10 kDa of the N terminus of the Stp1p (or Stp2p) transcription factor is a necessary step for the induction of amino acid-responsive permease genes (3). To analyze the SSY5 and PTR3 mutants in a more direct way than using the AGP1::lacZ reporter, we examined the effect on Stp1p processing by Western blot analysis of cells in which Stp1p is produced as a fusion protein with a C-terminal addition of a 15.5-kDa tandem doublet of an immunoglobulin G-binding domain (Z) of Staphylococcus aureus protein A (see Materials and Methods). Stp1p processing in ssy5Δ and ptr3Δ cells transformed with plasmids that express either wild-type or constitutive mutant alleles of SSY5 or PTR3, respectively, was measured (Fig. 2). In M5444 (ssy5Δ, STP1::ZZ) cells expressing a plasmid-borne wild-type SSY5 gene, the steady-state level of processed Stp1-ZZ is ∼12% of total Stp1p-ZZ in the absence of amino acids. Similarly, in wild-type cells (strain M5447; STP1::ZZ), Stp1 processing is 14% (data not shown). However, expression of SSY5-6 in M5444 (ssy5Δ) resulted in an 2.5-fold increase in Stp1p processing. In M5443 (ptr3Δ) cells harboring the plasmid-borne wild-type PTR3 gene, the steady-state level of processed Stp1p was 8% of total Stp1p-ZZ. However, cells expressing PTR3-5 or PTR3-17 exhibited processed Stp1p-ZZ levels of 33% and 27%, respectively. Stp1p-ZZ processing in M5445 (ssy1Δ) cells expressing the constitutive SSY1-102 allele (14) was found to be 48%, whereas cells harboring the wild-type SSY1 allele have a processing level of only 2% (data not shown). Collectively, our results show that the constitutive SSY5, PTR3, and SSY1 mutants have increased steady-state levels of processed Stp1p in the absence of extracellular amino acids.
FIG. 2.
The constitutive SSY5 and PTR3 mutants exhibit increased levels of Stp1p processing in the absence of amino acids. Strains M5444 (ssy5Δ STP1-ZZ, first three lanes) and M5443 (ptr3Δ STP1-ZZ, last four lanes) transformed with centromere-based plasmid pRS316 carrying the indicated PTR3 and SSY5 alleles were grown in minimal medium (SD medium), and total protein was extracted and subjected to Western analysis of Stp1p-ZZ processing. The percentages of processed Stp1-ZZ in proportion to total Stp1-ZZ are indicated. The standard errors of the means of at least two experiments are shown in parentheses. U, unprocessed Stp1p-ZZ; P, processed Stp1p-ZZ.
Constitutive SSY5 and PTR3 mutants confer a gain of function.
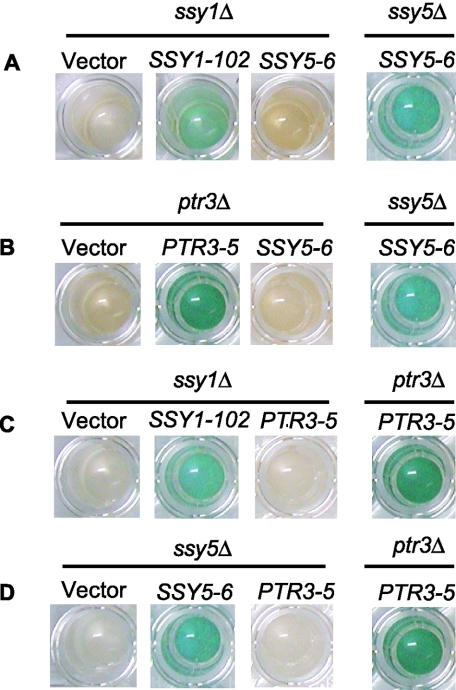
To test for dominance, the constitutive SSY5 and PTR3 alleles were introduced on centromere-based plasmids into strain M5359, which is wild type with regard to the SPS sensor components but has an AGP1 promoter-lacZ construct integrated at the AGP1 locus. The levels of β-galactosidase activity in these transformants grown in the absence of leucine are presented in Fig. 3A. The β-galactosidase activity in M5359 cells expressing the SSY5 and PTR3 mutant alleles was significantly increased compared to control cells with wild-type SSY5 and PTR3 alleles on the plasmids. Thus, the SSY5 and PTR3 mutants confer gain of function in SPS-mediated signaling.
FIG. 3.
The constitutive SSY5 and PTR3 mutants exhibit gain of function (dominance). (A) β-Galactosidase activity in extracts of wild-type strain M5359 (agp1::Pagp1-lacZ-KanMX) transformed with the centromere-based plasmid pRS316 carrying the indicated PTR3 and SSY5 alleles and grown in minimal medium (SD medium). (B) Percentage of processed Stp1-ZZ in proportion to total Stp1-ZZ in wild-type strain M5447 (STP1::ZZ) transformed with same plasmids. Standard deviation error bars are shown.
We also supplemented these data with the more direct analysis of Stp1p processing in wild-type (M5447, STP1::ZZ) cells expressing the various constitutive alleles (Fig. 3B). The level of Stp1p processing in the mutants was significantly increased compared to that in wild-type cells. Moreover, it can be seen that similar levels of Stp1p processing are obtained when the mutant alleles are expressed either in the M5447 wild-type strain or in the ssy5Δ and ptr3Δ deletion strains (compare Fig. 3B with Fig. 2).
Constitutive signaling by SSY5 or PTR3 mutants is dependent on all components of the SPS sensor.
Amino acid signaling normally requires all three SPS sensor components, Ssy1p, Ptr3p, and Ssy5p (6, 10, 21, 23, 24). To test whether the constitutive SSY5 mutant is able to circumvent SSY1 and PTR3 function, SSY5-6 was introduced into strains M5380 (ssy1Δ) and M5360 (ptr3Δ) harboring an integrated AGP1::lacZ reporter. Amino acid signaling in these cells was revealed by the blue coloration that resulted from β-galactosidase activity in X-Gal-containing buffer (Fig. 4). The absence of detectable β-galactosidase activity in SSY1- or PTR3-deleted cells expressing SSY5-6 shows that the mutant Ssy5 protein is dependent on Ssy1p and Ptr3p.
FIG. 4.
Constitutive signaling by the SSY5-6 and PTR3-5 mutants is dependent on the two other components of the SPS sensor. Strains carrying the indicated deletions and transformed with the centromere-based plasmid pRS316 carrying the indicated mutant genes were grown in SD medium and permeabilized. Exposure to the substrate X-Gal yielded a blue color where activation of the AGP1 promoter had resulted in the presence of β-galactosidase. (A) Strains M5380 (ssy1Δ agp1::lacZ) and M5361 (ssy5Δ agp1::lacZ). (B) Strains M5360 (ptr3Δ agp1::lacZ) and M5361. (C) Strains M5380 and 5360. (D) Strains 5361 and 5360.
We similarly failed to detect β-galactosidase activity in M5380 (ssy1Δ) and M5361 (ssy5Δ) expressing the PTR3-5 and PTR3-17 mutations (Fig. 4). These results show that constitutive signaling by the isolated PTR3 mutants is unable to bypass the need for functional Ssy1p and Ssy5p; in other words, the constitutive signaling in these mutants is dependent on SSY1 and SSY5.
Constitutive SSY5, PTR3, and SSY1 mutants require less inducer to give strong signaling.
To further characterize the constitutive SSY5 and PTR3 mutants presented in this paper and the SSY1-102 mutant previously described (14), we analyzed the pattern of Stp1p processing in response to different concentrations of leucine. The data collected from these experiments were used to estimate a median effective concentration (EC50), which is the concentration of leucine in the medium that results in 50% processed Stp1 protein; the EC50 value is also a reflection of the apparent Kd for leucine binding to Ssy1p. The resulting dose-response curves for a wild-type strain and strains with the Ssy5-E512K, Ptr3-Q439R, Ptr3-T435K, and Ssy1-T382K.mutant sensor components are shown in Fig. 5.
FIG. 5.
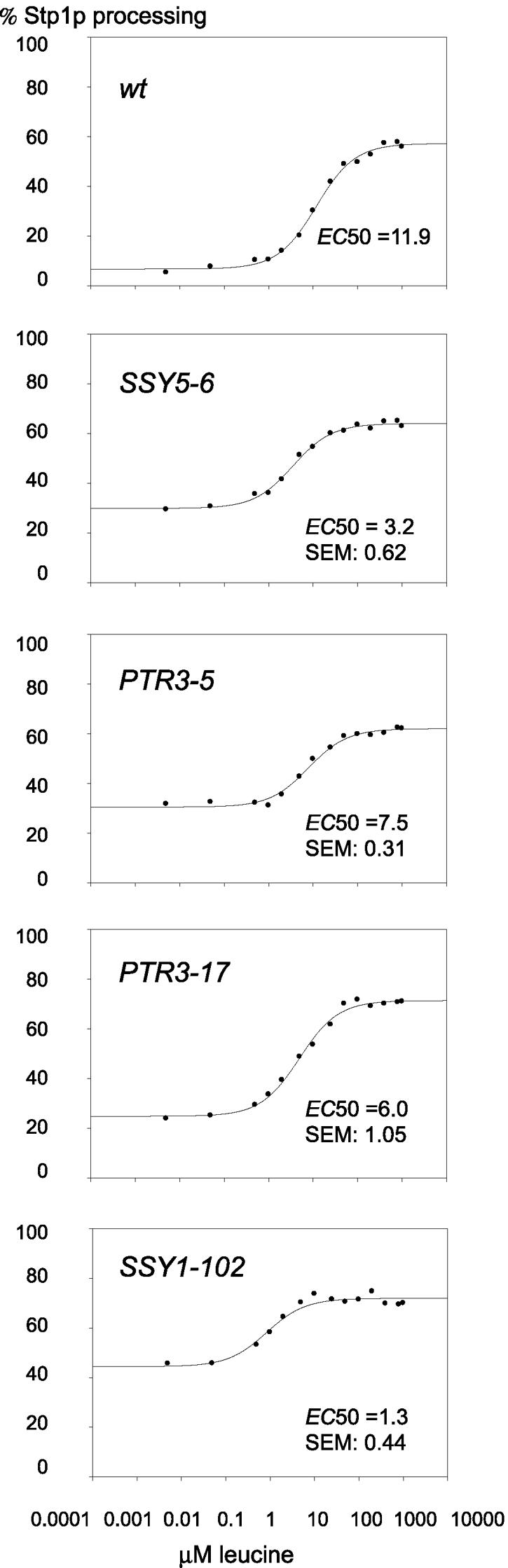
The constitutive SPS sensor mutants require less inducer to give strong signaling. Cells growing exponentially in SD medium were exposed to leucine at various concentrations, and Stp1-ZZ processing was monitored. EC50 (apparent Kd) was calculated using SigmaPlot 2000 by fitting the data to y = y0 + ax/(b + x), where b is EC50. In each panel for the mutants, the indicated values for the mean and the standard error of the mean for EC50 are derived from three experiments carried out in identical ways on different days; data points and the fitted curve represent one of the three experiments. The indicated mutations were present as inserts in the centromere-based and URA3-based vector pRS316 in ura3 strains deleted for the corresponding wild-type (wt) SSY5, PTR3, or SSY1 gene, respectively. In the upper panel, the chromosomal SSY5 deletion was complemented by a wild-type SSY5 gene harbored in pRS316 (pSSY5).
To obtain a control EC50 value for leucine in a SPS sensor wild-type background, we used strain M5444 (ssy5Δ) complemented by the wild-type SSY5 allele on a pRS316 centromere-based plasmid (pSSY5). As shown in Fig. 5, the EC50 value was determined to be 11.9 μM; the same EC50 value was also found using wild-type strain M5447 in which the three SPS-encoding genes are present in one copy at their normal chromosomal location (for three experiments, one of which is shown in Fig. 5 [upper panel], the EC50 was 12.25 and the standard error of the mean was 0.92). It may be noted that the Stp1p processing reaches a level of only about 60 to 70%. Plating for single colonies on uracil-containing medium and replica plating of these onto uracil-free medium showed that this is due to the loss of pSSY5 in about one-third of the cells in the culture, a value typical for the pRS316 vector (32). In contrast, Stp1p processing typically reaches 100% in the EC50 determinations carried out with wild-type strain M5447 (29). Since we obtain the same EC50 value for the M5444/pSSY5 and M5447 cells, we consider the system in which the gene for a SPS sensor component is deleted on the chromosome and complemented by the corresponding wild-type gene or constitutive mutant gene on a plasmid to be a reliable experimental system for determining EC50 values.
Compared to the wild type, a 3.7-, 1.6-, 2.0-, and 9.0-fold reduction in EC50 is observed for Ssy5-E512K, Ptr3-Q439R, Ptr3-T435K and Ssy1-T382K, respectively. The most dramatic reduction in EC50 is found for Ssy1-T382K, which is believed to interact directly with the extracellular inducer. A more moderate reduction in EC50 is found for the SSY5 and PTR3 mutant proteins. Nevertheless, we conclude that the leucine concentration needed to induce a strong signaling response is reduced in each of the mutants.
Thus, the mutational changes of Ptr3p and Ssy5p make these proteins more prone to trigger the onset of Stp1p processing, and they also increase the apparent affinity between Ssy1p and extracellular ligand, presumably by affecting an equilibrium between a nonsignaling and a signaling conformation. This effect supports the notion that the Ssy1, Ssy5, and Ptr3 proteins interact intimately in a complex.
DISCUSSION
In S. cerevisiae, the ability to sense the presence of extracellular amino acids and to initiate signal transduction, leading to increased transcription of amino acid permease genes, depends on the function of several genes, including SSY1, PTR3, and SSY5 (6, 10, 12, 21, 23, 24). While Ssy1p resembles members of the amino acid permease family, it is apparently unable to transport significant quantities of amino acids. Thus, Ssy1p is believed to act as a receptor, initiating signal transduction in a manner dependent on Ptr3p and Ssy5p. That these three proteins are not only functionally but also physically associated is supported by various experimental data. Firstly, several tagged versions of Ptr3p and Ssy5p were localized to the plasma membrane despite their lack of typical membrane-spanning domains (12, 20, 24). Secondly, overexpression of the cytoplasmic, N-terminal domain of Ssy1p inhibits sensor function, a finding consistent with formation of defective, nonsignaling complexes consisting of the Ssy1p N-terminal domain and protein(s) normally binding to Ssy1p (7, 12). Thirdly, two-hybrid experiments are consistent with the existence of a complex between the three proteins. Thus, Ptr3p was found to interact with itself and with Ssy5p (7), and Ssy1p was found to interact with Ptr3p (35). It is important to note, however, that all of these data address primarily the interactions necessary for signaling but are not critically concerned with whether Ssy1p, Ptr3p, and Ssy5p actually function in a complex and what the functional properties of such a complex could be. The present work sheds some light on this issue.
The putative complex comprising Ssy1p, Ptr3p, and Ssy5p has been termed the SPS sensor (12). The three proteins mediate amino acid-dependent activation of the transcription factors Stp1p and Stp2p (3, 4), and their individual roles are beginning to emerge through analysis of genetically altered members of the putative complex. The notion that extracellular amino acid directly interacts with Ssy1p gained support from the isolation of constitutively signaling and hyperresponsive SSY1 mutants (14). Thus, whether or not Ssy1p can normally transport small amounts of amino acid, imported amino acids per se are not required for signaling. These mutations also significantly increased the apparent affinity for amino acids, providing further support for the hypothesis that Ssy1p functions by direct interaction with extracellular amino acids. Concerning Ssy5p, recent evidence suggests that this protein is a trypsin-like serine protease whose activity is required for the cleavage and consequent activation of Stp1p and Stp2p (1, 2). So far, the function of Ptr3p is not known; however, a scan of the Ptr3 protein sequence against the protein signatures in the InterPro databases using the InterProScan tool (26), suggests that Ptr3p may contain WD40 repeats (25, 33) in the C-terminal region. A comparison of the Ptr3p sequence to 10 WD40 repeat models in the Superfamily server (16) aligned the Ptr3p sequences from residues K364 to C605 with the models, suggesting five WD40 repeats in Ptr3p. Since repeated WD40 motifs are known to act as a site for protein-protein interactions (16, 25), the proposed WD40 motifs may be involved in interactions with Ssy1p and Ssy5p that regulate the activation of the Ssy5p proteolytic properties upon binding of amino acids to Ssy1p. Interestingly, the constitutive PTR3 mutants result in the amino substitutions T435K and Q439R, which map within the proposed WD40 repeat region. Whether Ptr3p indeed belongs to the WD40 repeat superfamily must await structural data for the protein.
In this report, using essentially the same genetic screening that yielded SSY1-constitutive alleles, we show that gain-of-function mutations in PTR3 and SSY5 that confer robust activation of a downstream amino acid permease gene, even in the absence of extracellular amino acids, can be isolated.
We obtained several different constitutive alleles of both PTR3 and SSY5 (described above and data not shown) and, in each case, activation of the downstream reporter genes depended on the presence of the complete SPS sensor. Thus, PTR3-5 and PTR3-17 exhibited no activation in ssy1Δ or ssy5Δ cells, and SSY5-6 exhibited no activation in ssy1Δ or ptr3Δ cells. These results support the hypothesis that the three proteins actually function in a complex, since they are not easily reconciled with a model in which sequential signaling steps between the three proteins are separable.
The dose-response relationships for signaling in the mutants are highly relevant for the understanding of the interactions in the complex. Notably, we have shown that in the PTR3 and SSY5 constitutive mutants, the apparent affinity for leucine, as measured by the EC50 for induction of Stp1p processing, was altered. The EC50s for PTR3-5 and PTR3-17 cells (7.5 and 6.0 μM, respectively) and for SSY5-6 cells (3.2 μM) were reduced compared to that for wild-type cells (11.9 μM), although not to the extent observed with cells expressing the constitutive SSY1-102 (1.3 μM). If Ptr3p and Ssy5p functioned individually downstream of Ssy1p, one would not expect constitutive mutations to affect the EC50 in this manner. Rather, these results are easily explained by assuming that Ssy1p, Ptr3p, and Ssy5p function in a highly interdependent, conformationally coordinated manner.
The evidence suggesting that Ssy5p acts as the protease that activates Stp1p and Stp2p (1, 2) is consistent with Ssy5p functioning as the most downstream component of the SPS sensor. If so, one might expect that some constitutive alleles of SSY5 could confer activity independent of Ssy1p and Ptr3p. Fortuitously, Per Ljungdahl et al. generated a constitutively active allele of SSY5 that functions even in the absence of the other members of the SPS sensor, not through point mutation but by the addition of six tandem copies of the hemagglutinin epitope tag (HA6) to the amino terminus of Ssy5p (2).
The different behavior of HA6-Ssy5p and Ssy5-E512K (encoded by SSY5-6) is thought provoking and may be accounted for as follows. The proteolytic activity of Ssy5p appears to be internally inhibited. The inhibition can be counteracted by interaction with Ssy1p and Ptr3p in a way dependent on extracellular amino acid binding to Ssy1p. In this picture, HA6-Ssy5p is not properly inhibited and can act without Ssy1p and Ptr3p. Ssy5-E512K, on the other hand, is affected in the interaction with the partners in the complex. Interestingly, the Ssy5-E512K substitution is positioned in the protease region of Ssy5p close to amino acid D545, which has been proposed to be part of the trypsin-like serine protease catalytic triad (H465, D545, and S640) in Ssy5p (1, 2). An understanding of how this mutation confers constitutivity and maintains Ssy1p and Ptr3p dependence must await interaction studies of the SPS sensor components.
The recent finding that casein kinase I is necessary for the endoproteolytic processing of Stp1p (1, 34) adds to the complexity of the signal transduction mechanism of the SPS sensor. In addition, the involvement of Grr1p, a component of the SCFGrr1 ubiquitin ligase complex, remains a puzzling issue in the cascade of steps leading from the recognition of amino acids at the plasma membrane to Stp1p cleavage (1, 4, 11, 20, 34). We expect that further use of gain-of-function mutants will be useful for shedding more light on this intriguing signal transduction system.
To summarize, the present work provides two major findings that support the notion that Ssy1p, Ptr3p, and Ssy5p function as a dynamic complex. First, the constitutive mutants need the other two components to function; second, the EC50 for signaling is affected not only by SSY1-102 but also by PTR3-5, PTR3-17, and SSY5-6. We suggest that all three components participate in a conformational shift of the full complex between a signaling conformation and a nonsignaling conformation. In this way, PTR3-5, PTR3-17, and SSY5-6 can be mutations that affect an equilibrium between the two conformations. Such mutations are indeed expected to affect the EC50 for signaling. We wish to emphasize that our results, by their very nature, cannot be regarded as proof of the existence of the complex of Ssy1p, Ptr3p, and Ssy5p. However, the previous two-hybrid and other data suggesting the existence of such a complex have, by the present and very different approach, been supported and complemented with data that say more about dynamics and functionality than can two-hybrid interactions.
Acknowledgments
We thank Lisbeth F. Petersen for excellent technical assistance and Helge A. Andersen, Per O. Ljungdahl, and Claes Andréasson for helpful discussions. Furthermore, we thank Claes Andréasson for drawing our attention to the discrepancies between yeast Ssy5p C-terminal sequences.
REFERENCES
- 1.Abdel-Sater, F., M. El Bakkoury, A. Urrestarazu, S. Vissers, and B. André. 2004. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell. Biol. 24:9771-9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andréasson, C. 2004. Ph.D. thesis. Karolinska University Press, Stockholm, Sweden.
- 3.Andréasson, C., and P. O. Ljungdahl. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16:3158-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andréasson, C., and P. O. Ljungdahl. 2004. The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full SPS-sensor control. Mol. Cell. Biol. 24:7503-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. Green Publishing Associates and Wiley Interscience, New York, N.Y.
- 6.Barnes, D., W. Lai, M. Breslav, F. Naider, and J. M. Becker. 1998. PTR3, a novel gene mediating amino acid-inducible regulation of peptide transport in Saccharomyces cerevisiae. Mol. Microbiol. 29:297-310. [DOI] [PubMed] [Google Scholar]
- 7.Bernard, F., and B. André. 2001. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol. Microbiol. 41:489-502. [DOI] [PubMed] [Google Scholar]
- 8.Boles, E., and B. André. 2004. Role of transporter-like sensors in glucose and amino acid signalling in yeast. Top. Curr. Genet. 9:121-153. [Google Scholar]
- 9.Brandt, A. 1991. Pulse labeling of yeast cells as a tool to study mitochondrial protein import, p. 369-376. In A. M. Tartakoff (ed.), Methods in cell biology, vol. 34. Vectorial transport of proteins into and across membranes. Academic Press, Inc., San Diego, Calif. [DOI] [PubMed]
- 9a.Bruun, M. B. 1997. M.S. thesis. University of Copenhagen, Copenhagen, Denmark.
- 10.Didion, T., B. Regenberg, M. U. Jørgensen, M. C. Kielland-Brandt, and H. A. Andersen. 1998. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27:643-650. [DOI] [PubMed] [Google Scholar]
- 11.Eckert-Boulet, N., B. Regenberg, and J. Nielsen. 2005. Grr1p is required for transcriptional induction of amino acid permease genes and proper transcriptional regulation of genes in carbon metabolism of Saccharomyces cerevisiae. Curr. Genet. 47:139-149. [DOI] [PubMed] [Google Scholar]
- 12.Forsberg, H., and P. O. Ljungdahl. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21:814-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsberg, H., M. Hammar, C. Andréasson, A. Molinér, and P. O. Ljungdahl. 2001. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 158:973-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaber, R. F., K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2003. Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot. Cell 2:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giots, F., M. C. V. Donaton, and J. M. Thevelein. 2003. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47:1163-1181. [DOI] [PubMed] [Google Scholar]
- 16.Gough, J., K. Karplus, R. Hughey, and C. Chothia. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313:903-919. [DOI] [PubMed] [Google Scholar]
- 17.Grauslund, M., T. Didion, M. C. Kielland-Brandt, and H. A. Andersen. 1995. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1269:275-280. [DOI] [PubMed] [Google Scholar]
- 18.Güldener, U., S. Heck, T. Fiedler, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holsbeeks, I., O. Lagatie, A. Van Nuland, S. Van de Velde, and J. M. Thevelein. 2004. The eukaryotic plasma membrane as a nutrient-sensing device. Trends Biochem. Sci. 29:556-563. [DOI] [PubMed] [Google Scholar]
- 20.Huh, W-K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 21.Iraqui, I., S. Vissers, F. Bernard, J.-O. de Craene, E. Boles, A. Urrestarazu, and B. André. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jørgensen, M. U., C. Gjermansen, H. A. Andersen, and M. C. Kielland-Brandt. 1997. STP1, a gene involved in pre-tRNA processing in yeast, is important for amino acid uptake and transcription of the permease gene BAP2. Curr. Genet. 31:241-247. [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen, M. U., M. B. Bruun, T. Didion, and M. C. Kielland-Brandt. 1998. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast 14:103-114. [DOI] [PubMed] [Google Scholar]
- 24.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19:5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madrona, A. Y., and D. K. Wilson. 2004. The structure of Ski8p, a protein regulating mRNA degradation: implications for WD protein structure. Protein Sci. 13:1557-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, D. Barrell, A. Bateman, D. Binns, M. Biswas, P. Bradley, P. Bork, P. Bucher, R. R. Copley, E. Courcelle, U. Das, R. Durbin, L. Falquet, W. Fleischmann, S. Griffiths-Jones, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, R. Lopez, I. Letunic, D. Lonsdale, V. Silventoinen, S. E. Orchard, M. Pagni, D. Peyruc, C. P. Ponting, J. D. Selengut, F. Servant, C. J. A. Sigrist, R. Vaughan, and E. M. Zdobnov. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31:315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jørgensen, R. J. Planta, M. C. Kielland-Brandt, and H. A. Andersen. 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264:613-622. [DOI] [PubMed] [Google Scholar]
- 28.Olesen, K., P. Franke Johannesen, L. Hoffmann, S. Bech Sørensen, C. Gjermansen, and J. Hansen. 2000. The pYC plasmids, a series of cassette-based yeast plasmid vectors providing means of counter-selection. Yeast 16:1035-1043. [DOI] [PubMed] [Google Scholar]
- 29.Poulsen, P., B. Wu, R. F. Gaber, K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2005. Amino acid sensing by Ssy1. Biochem. Soc. Trans. 33:261-264. [DOI] [PubMed] [Google Scholar]
- 30.Rose, M. D., P. Novick, J. H. Thomas, D. Botstein, and G. R. Fink. 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60:237-243. [DOI] [PubMed] [Google Scholar]
- 31.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 32.Sikorsky, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 34.Spielewoy, N., K. Flick, T. I. Kalashnikova, J. R. Walker, and C. Wittenberg. 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24:8994-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, Srinivasan, M., P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 36.Whyte, J. R. C., and S. Munro. 2001. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev. Cell 4:527-537. [DOI] [PubMed] [Google Scholar]