Abstract
Phlebotomus argentipes is the established vector of leishmaniasis in the Indian sub-continent. Antibodies to sand fly salivary antigens are biomarkers for vector-host exposure in leishmaniasis-endemic regions. Ph. argentipes transmits Leishmania donovani in Sri Lanka, primarily causing cutaneous leishmaniasis (CL). Our study compared the performance of salivary gland homogenate (SGH) from a lab-reared local strain of Ph. argentipes females to a composite recombinant salivary biomarker (rPagSP02 + rPagSP06) in a CL-endemic population. Sera from 546 healthy individuals, 30 CL patients, and 15 non-endemic individuals were collected. Western blot analysis of Ph. argentipes SGH identified immunogenic bands between 15 kDa and 67 kDa, with bands of predicted molecular weight ∼of 15 kDa (SP02) and ∼28–30 kDa (SP06) as the major antibody targets. Indirect ELISAs using SGH or rPagSP02 + rPagSP06 antigens showed high sensitivity (96.7%) and specificity (100%), detecting comparable seropositivity in endemic populations. rPagSP02 + rPagSP06 exhibited enhanced discriminatory ability, supported by a strong positive correlation (r = 0.869) with SGH. Our findings indicate that the composite rPagSP02 + rPagSP06 salivary biomarker effectively identifies Ph. argentipes exposure in individuals living in Sri Lanka, showing promising potential for use in surveillance. These findings should be further validated to confirm the epidemiological applications in leishmaniasis-endemic regions.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-77666-2.
Keywords: Cutaneous leishmaniasis, Phlebotomus argentipes, Salivary antigen, Vector exposure, Surveillance, Sri Lanka
Subject terms: Immunology, Biomarkers
Introduction
Leishmaniasis, a neglected vector-borne disease, poses significant public health challenges worldwide1,2. A total of 102 countries and 5 continents have reported endemic leishmaniasis transmission2. The disease is caused by Leishmania protozoan species and is transmitted by phlebotomine sand flies3. Phlebotomus argentipes is vector of cutaneous leishmaniasis (CL) and visceral leishmaniasis (VL) in the Indian subcontinent (ISC), with Leishmania donovani as the causative agent4–6.
The World Health Organization (WHO) has set long-term targets for the global elimination of neglected tropical diseases (NTDs), with VL being a primary focus in their 2030 roadmap for NTD eradication. As part of integrated approaches, 56 countries aimed to eliminate VL by 2025, and 64 countries by 20307–9. However, despite the significant reduction in VL incidence in India, Nepal, and Bangladesh through elimination programs in endemic areas, new cases of CL caused by L. donovani have emerged primarily in non-endemic regions10–16. Persistence of reservoirs due to atypical Leishmania species-phenotype associations pose a challenge for achieving the elimination targets. Sri Lanka was the first country to report CL due to L. donovani in the ISC17,18, with similar subsequent reports from several other countries, the most recent being from India19–21. The elimination initiative has not yet outlined the tools for assessing and maintaining its success, apart from the use of clinical outcomes to measure progress16.
The salivary proteins of sand flies represent promising biomarkers that induce a specific antibody response in both humans and animals22. These proteins hold significant potential as a tool for quantifying exposure to sand fly bites and the risk of contracting leishmaniasis23–27. Studies conducted in India, Nepal and Bangladesh demonstrated the use of anti-salivary antibodies to estimate human exposure to Ph. argentipes bites28–32. A combination of two recombinant Ph. argentipes salivary proteins, rPagSP02 + rPagSP06, was recently reported as a reproducibly sensitive and specific biomarker to measure human-vector exposure in a VL endemic population from India32.
Here, we sought to test the immunogenicity and applicability of the composite salivary biomarker (rPagSP02 + rPagSP06), based on Indian Ph. argentipes, as a reliable marker of vector exposure in an endemic population in Sri Lanka.
Results
Study population
The characteristics of the study population, including living conditions are shown in Table 1. The study participants belonged to varying age groups, with a mean age of 49 ± 15 years. The majority of participants were females (n = 361/546, 66.0%). Occupationally, the participants represented a range of sectors, with the largest group being farmers (n = 92/546, 16.8%). Among the participants, 29.1% (n = 161/546) reported traveling outside their local area within the past 6 months. Housing characteristics revealed that most houses had walls constructed of bricks only (n = 378/546, 69.2%) and tiled roofs (n = 372/546, 68.2%). The participants primarily slept on beds (n = 523/546, 95.8%), and a significant proportion used mosquito nets (n = 407/546, 74.5%). It was notable that 46.0% (n = 251/546) of participants reported using insect repellents. A high proportion (n = 383/546, 70.1%) of participants engaged in outdoor activities during daytime, whereas only a very small minority were active during dusk (14.1%) or dawn (10.8%) when the likelihood of being bitten by sand flies is higher. The presence of cats and dogs or both in the vicinity was reported by 54.6% (n = 298/546) of participants. Regarding the surroundings, the presence of outdoor latrines, storerooms, and cattle sheds was common (n = 338/546, 61.8%) (Table 1).
Table 1.
Demographic characteristics and living conditions of study population.
| Characteristic feature/s | Mean ± SD¶/Percentage/Ratio Endemic participants n = 546 |
||
|---|---|---|---|
| Age (mean ± SD¶) | 49 ± 15 years | ||
|
18 to 37 years 38 to 57 years 58 to 77 years 78 to 97 years |
139 (25.3%) 228 (41.7%) 172 (31.5%) 8 (1.5%) |
||
| Gender (M: F) | 185: 361 (1: 2) | ||
| Marital status | |||
| Married | 368 (67.4%) | ||
| Unmarried | 178 (32.6%) | ||
| Other | 0 | ||
| Education | |||
| No proper education | 20 (3.7%) | ||
| Grade 1 to 5 | 38 (7.0%) | ||
| Grade 6 to 11 | 322 (59.0%) | ||
| Ordinary level | 153 (28.0%) | ||
| Advanced level | 10 (1.8%) | ||
| Degree or above | 3 (0.5%) | ||
| Occupation | |||
| Farmer | 92 (16.8%) | ||
| Self-employed | 26 (4.8%) | ||
| Government-sector worker | 9 (1.6%) | ||
| Private sector | 22 (4.0%) | ||
| Volunteer worker | 4 (0.7%) | ||
| Retired | 18 (3.3%) | ||
| Non-working or not employed | 375 (68.7%) | ||
| Past travel history (within 6 months) | |||
| Yes | 161 (29.1%) | ||
| No | 385 (70.5%) | ||
| Walls of the house constructed of | |||
| Brick only | 378 (69.2%) | ||
| Brick and plaster | 161 (29.5%) | ||
| Other | 7 (1.3%) | ||
| Roof of the house constructed of | |||
| Tiles | 372 (68.2%) | ||
| Asbestos | 131 (24.0%) | ||
| Other | 43 (7.8%) | ||
| Location of participant’s sleeping area | |||
| On the floor | 23 (4.2%) | ||
| On the bed | 523 (95.8%) | ||
| Use of insect repellants | |||
| Yes | 251 (46.0%) | ||
| No | 295 (54.0%) | ||
| Use of mosquito nets | |||
| Yes | 407 (74.5%) | ||
| No | 139 (25.5%) | ||
| Outdoor activity time | Working hours | ||
| Less than 1 h | 1 to 3 h | More than 3 h | |
| 4 am to 8 am | 28 (5.1%) | 31(5.8%) | 0 |
| 8 am to 4 pm | 130 (23.8%) | 67 (12.3%) | 186 (34.0%) |
| 4 pm to 8 pm | 29 (5.3%) | 37 (6.8%) | 11(2.0%) |
| 8 pm to 4 am | 0 | 0 | 27 (4.9%) |
| Presence of domestic and peri domestic animals | |||
| Cats/dogs or both | 298 (54.6%) | ||
| Poultry/farm animals | 14 (2.6%) | ||
| None | 234 (42.8%) | ||
| Presence of other buildings and features within 50 m from the house | |||
| Outdoor latrines, store rooms and cattle sheds | 338 (61.8%) | ||
| Jungles, shrubs, paddy lands, water bodies | 93 (17.1%) | ||
| Dump sites, compost pits | 43 (7.9%) | ||
| None | 72 (13.2%) | ||
Standard deviation.
Identification of immunogenic antigens in saliva of the localPh. argentipesstrain
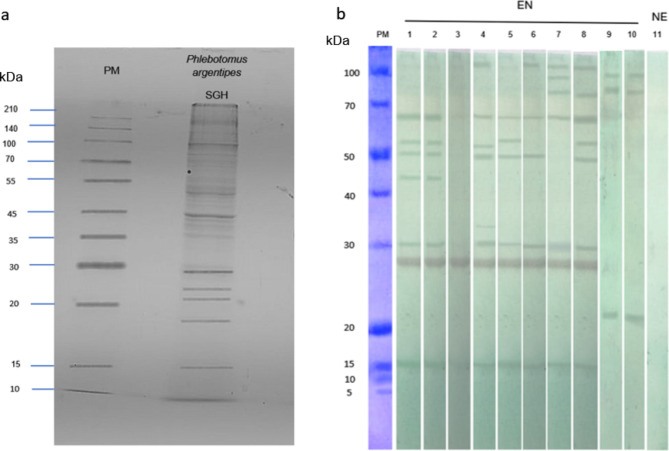
SDS-PAGE revealed a diverse range of protein bands for the Ph. argentipes SGH of field-collected females from Sri Lanka, with molecular weights ranging from ∼10 kDa to ∼140 kDa (Fig. 1a). Western blot analysis of serum samples from individuals from a CL endemic region in Sri Lanka had a strong reactivity against Ph. argentipes SGH, identifying multiple immunogenic bands at approximately ∼15 kDa, ∼28 kDa, ∼30 kDa, ∼45 kDa, ∼50 kDa, ∼55 kDa, and ∼67 kDa (Fig. 1b). Notably, the ∼28–30 kDa and ∼15 kDa bands emerged as major targets of the immune response, along with the ∼67 kDa band prominently recognized in seven out of eight endemic individuals (Fig. 1b). Importantly, non-endemic control sera showed no reactivity with SGH of Ph. argentipes (Fig. 1b).
Fig. 1.
Identification of antigenic Phlebotomus argentipes salivary proteins. (a) The SGH protein profile of Ph. argentipes. 12% SDS–PAGE gel stained by Coomassie Blue. (b) Western blots showing the reactivity of IgG in endemic sera against Ph. argentipes salivary gland antigens (lane 1–8), or Culex spp. salivary gland antigens (lane 9–10: against sera of the endemic individuals screened in lanes 1 and 8). Lane 11: a non-endemic serum sample screened against Ph. argentipes salivary gland antigens. PM: protein marker; SGH: salivary gland homogenate; kDa: Kilodaltons; EN: endemic sera; NE: Non-endemic sera. The uncropped gel image and blot image can be found as supplementary Fig. S1 and Fig. S2 online.
Crucially, when we tested the sera from subjects 1 and 8, that were strongly reactive against Ph. argentipes SGH (Fig. 1b), against SGH of Culex spp., mosquitoes that are prevalent in the same area, we did not observe cross-reactivity to Ph. argentipes immunodominant proteins, indicating the specificity of the immune response.
Comparison of anti-saliva antibody responses to SGH and rPagSP02 + rPagSP06
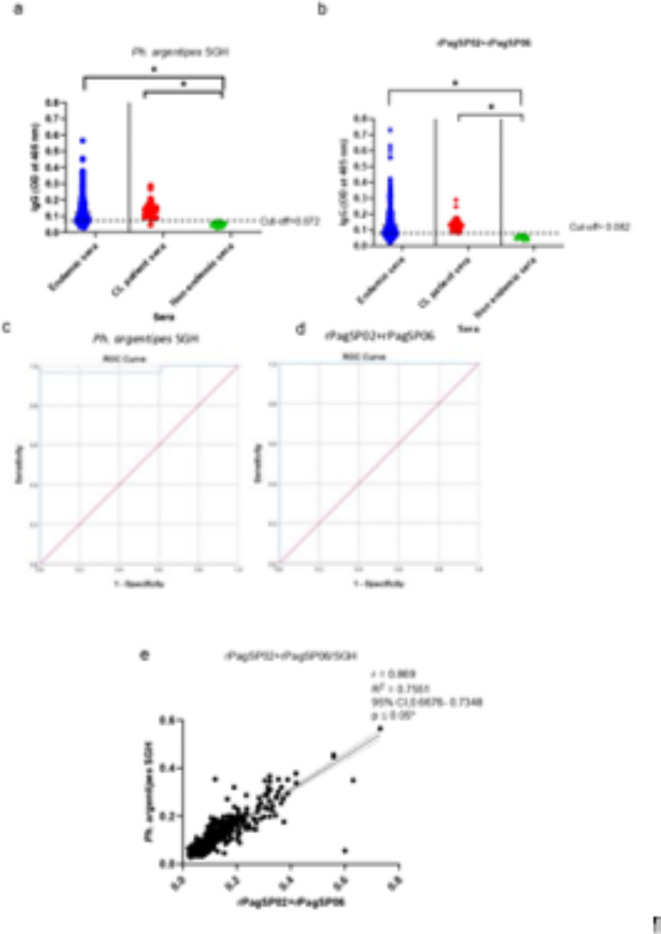
Next, we compared the antibody response to SGH and rPagSP02 + rPagSP06 using indirect ELISA. The antibody levels of healthy and CL patients from the endemic area were significantly higher compared to non-endemic controls for both SGH and rPagSP02 + rPagSP06 (Fig. 2a, b). The cut-off values determined based on the receiver operating characteristic (ROC) curve for these antigens were 0.072 and 0.082, respectively (Fig. 2a, b). The ROC curve analysis showed a sensitivity of 96.7% and specificity of 100% for both antigens (Fig. 2c, d). The rPagSP02 + rPagSP06 antigen had a discriminatory ability, with an area under the curve (AUC) value of 1.000 in the ROC analysis. The SGH antigen also exhibited a high discriminatory power, with an AUC value of 0.979. Furthermore, a strong positive correlation (r = 0.869, 95% CI) was observed between SGH and rPagSP02 + rPagSP06, as determined by Pearson’s rank correlation test (Fig. 2e). In the endemic population consisting of healthy individuals and CL patients (n = 546), both antigens exhibited a considerable proportion of seropositive individuals, with a marginal increase in seropositivity rates observed for the rPagSP02 + rPagSP06 antigen (65.2% versus 63.2% for SGH) (Table 2.). Notably, the exposure levels were similar between CL cases and endemic controls, highlighting the widespread exposure to sand fly bites in both groups. The mean absorbance values corresponding to the mean level of anti-saliva IgG antibodies also followed a similar pattern, with moderately higher values for the rPagSP02 + rPagSP06 antigen (0.130 ± 0.090 versus 0.119 ± 0.072 for SGH). Among the CL patients’ sera (n = 30), both antigens were recognized in 29 individuals and exhibited a high sensitivity and specificity with seropositivity rates of 96.6% (Table 2.). The mean absorbance values in the CL patients’ sera were comparable for both rPagSP02 + rPagSP06 and SGH, with moderately higher values for the former (0.136 ± 0.044) (Table 2.). The mean levels of anti-saliva IgG antibodies were very low in the control non-endemic group with the antigen combination and SGH giving comparable values (0.048 ± 0.010 for rPagSP02 + rPagSP06 antigen and 0.049 ± 0.010 for SGH) (Table 2.). The agreement between the two ELISA tests for two antigens was substantial, with a Kappa value of 0.684 (0.617–0.752 CI at 95%). This indicates a strong consistency between the two assays in detecting the same samples as positive or negative. A total of 345 sera from endemic individuals were classified as positive by both ELISAs, while 128 sera were classified as negative by both tests. In 18 cases, ELISA 1 identified the sera as positive, whereas ELISA 2 classified them as negative. Conversely, 55 sera were classified as negative by ELISA 1 and positive by ELISA 2. The Kappa coefficient was calculated to assess the level of agreement between the two tests beyond chance.
Fig. 2.
The rPagSP02 + rPagSP06 composite biomarker of exposure to Ph. argentipes exhibits high sensitivity and specificity in Sri Lanka. (a) Anti-salivary IgG antibodies against 2 µg/mL of salivary gland homogenate (SGH) from Sri Lankan Ph. argentipes, cut-off at 0.072. (b) Anti-salivary IgG antibodies against 1 µg/mL of rPagSP02 + rPagSP06 composite recombinant salivary antigen of Ph. argentipes, cut-off at 0.082. The cut-off value of the indirect ELISA was calculated using a receiver operating characteristic (ROC) curve with OD values of non-endemic (NE) individuals (negative controls, n = 15) and sera from clinically confirmed CL-positive patients (Positive controls, n = 30). Antigens were tested against serum samples (1:50 dilution) from 546 individuals living in endemic areas of CL, 30 newly diagnosed CL patients and 15 non-endemic individuals. (c,d) Receiver operating characteristic (ROC) curve for indirect ELISA of SGH, area under the curve (AUC) value equals to 0.979 (c) and rPagSP02 + rPagSP06, AUC value equals to 1.000 (d). (e) Pearson’s rank correlation test between Ph. argentipes SGH and rPagSP02 + rPagSP06. Correlation coefficient r = 0.869 at 95% CI. A two-tailed p < 0.05 as considered statistically significant.
Table 2.
Comparison of the performance of rPagSP02+rPagSP06 and SGH as biomarkers of exposure to Ph. argentipes
| Sera | rPagSP02 + rPagSP06 antigenα ELISA 1 |
SGHβ ELISA 2 |
||
|---|---|---|---|---|
| Seropositive | Seronegative | Seropositive | Seronegative | |
| Endemic population (n = 546) | 356 (65.2%) | 190 (34.8%) | 345 (63.2%) | 201 (36.8%) |
|
Mean absorbance ± SD¶ |
0.130 ± 0.090 | 0.119 ± 0.072 | ||
|
CL patients’ sera (n = 30) |
29 (96.6%) | 1 (3.4%) | 29 (96.6%) | 1 (3.4%) |
|
Mean absorbance ± SD¶ |
0.136 ± 0.044 | 0.138 ± 0.052 | ||
|
Non endemic (n = 15) |
0 (0%) | 15 (100%) | 0 (0%) | 15 (100%) |
|
Mean absorbance ± SD¶ |
0.048 ± 0.010 | 0.049 ± 0.010 | ||
¶Standard deviation, αComposite recombinant salivary antigen, βPrepared from a local strain.
Discussion
Salivary proteins secreted by sand flies play crucial roles in facilitating blood feeding by exerting anti-coagulant properties25,33–35, while specific proteins have the capability to elicit an immune response36–39,30,31. Moreover, these salivary proteins hold the potential to be used in predicting disease prevalence and for estimation of the level of sand fly bites and vector exposure22,26,40,41. Developing a highly specific and sensitive biomarker to detect human exposure to Ph. argentipes bites is of utmost importance in regions endemic to leishmaniasis in the ISC, particularly as tools for monitoring vector control.
The protein repertoire of Ph. argentipes saliva from our study aligns with findings by another report, which identified salivary proteins of Ph. argentipes, including a D7-related protein (GenBank ID: ABA12141), an antigen 5-related protein (GenBank ID: ABA12137), and an apyrase (GenBank ID: ABA12135)38. Moreover, the authors also discovered three PpSP15-like proteins (GenBank ID: ABA12133, ABA12139, and ABA12134), and a 33 kDa-sized protein of unknown function (GenBank ID: ABA12140), which are comparable in size to proteins detected in our study.
We identified seven immunogenic salivary proteins with molecular weights of approximately 15 kDa, 28 kDa, 30 kDa, 45 kDa, 50 kDa, 55 kDa, and 67 kDa, which are similar to those found in a previous study on the salivary antigens of Indian Ph. argentipes32. Many of the salivary proteins we identified had previously been characterized via Edman degradation42. The protein with a size of approximately 15 kDa exhibits similarity to both PpSP15 from Ph. papatasi and PagSP02 from Ph. argentipes. The size of this antigen is comparable to the component of the composite recombinant antigen (rPagSP02)32. Meanwhile, the proteins at approximately 28 kDa-30 kDa are likely D7-related proteins, possibly including PagSP06, and the size of this antigen is comparable to the component of the recombinant antigen (rPagSP06)32. The recent comparison study of the immunogenic profiles of SGH from Indian and Sri Lankan Ph. argentipes revealed that the ∼28–30 kDa immunogenic band contained PagSP05, PagSP06, and PagSP17, according to Electrospray Ionization/Liquid Chromatography/Tandem Mass Spectrometry43. Notably, the presence of PagSP06 could be a key component in the immunogenic role of this band. Additionally, the proteins at approximately 45 kDa could be related to yellow proteins42. However, immunogenic proteins of approximately 50 kDa and 55 kDa have not been reported before. PagSP06 from Ph. argentipes has a predicted molecular weight of 32 kDa, but forms doublets at around 67 kDa32. Of note, both PagSP02 and PagSP06 from Ph. argentipes were not recognized by sera from individuals bitten by Ph. papatasi32. Unlike in India, there are no records of Ph. papatasi from Sri Lanka according to past literature44–49.
A PpSP32-like protein (PagSP06) was considered as a biomarker of Ph. argentipes exposure in humans in Bangladesh30. A more recent study demonstrated that combining PagSP06 and PagSP02 improves performance of the biomarker, resulting in a noticeable decrease in cross-reactive antibodies32. Notably, in our experiments, no cross-reaction was observed with the salivary proteins of Culex spp. Given that Culex mosquitoes are the most abundant insect species in the study area, the absence of cross-reactivity underscores the species specificity of Ph. argentipes salivary antigens.
Salivary proteins from sand flies offer valuable information for assessing vector control strategies and gaining a better understanding of vector dynamics27,29. The findings of this present study demonstrate the utility of anti-salivary IgG antibodies as serological markers for assessing exposure to bites of Ph. argentipes, the sand fly vector associated with transmission of CL in Sri Lanka. The clear difference between endemic and non-endemic samples strongly supports the marker’s potential use in identifying new endemic areas. Similar to India32, the evaluation of the composite recombinant antigen (rPagSP02 + rPagSP06) using indirect ELISA proved to be as effective as SGH in detecting anti-salivary IgG antibodies in Sri Lanka, providing an alternative that is less labor intensive and more reproducible. Interestingly, the level of exposure and the percentage of individuals testing positive for vector exposure were comparable to, or even higher than, those reported in previous studies conducted in various endemic regions such as Bangladesh in August 2014 and India in November 201930,32. Of note, our study was conducted during a period when the sand fly density was high. The limited number of non-endemic controls is a limitation of this work and future studies should include a larger control group to enhance validity.
In conclusion, the composite rPagSP02 + rPagSP06 antigen proved to be an effective approach for assessing direct human-vector contact specific to Ph. argentipes sand flies in Sri Lanka. The composite biomarker demonstrated high discriminatory power, a strong correlation with SGH, and promising diagnostic potential in identifying vector exposure in both endemic residents and CL patients. Further research and validation are essential to comprehensively explore the clinical and epidemiological applications of these antigens in leishmaniasis. Continued follow-up studies are crucial to better understand the nature of persistent pattern of antibodies. By demonstrating the high sensitivity and specificity of the rPagSP02 + rPagSP06 composite biomarker in Sri Lanka, this study reinforces its validity for use in surveillance studies in the ISC, providing a much-needed tool to assess efficacy of interventions and changes in exposure to Ph. argentipes bites post-VL elimination.
Methods
Study area
The study area, Ambalantota in the Southern province of Sri Lanka, was chosen due to its endemicity for CL infection. The selection was informed by recent CL prevalence data from the Medical Officer of Health (MOH) unit in Ambalantota for the years 2018, 2019, and 202149. Areas were classified as endemic or non-endemic based on case incidence data spanning the past 18 years (2001 to 2019). An annual incidence rate of less than 1 case per 1,000 people signified non-endemicity, whereas 10 cases or more per 1,000 people indicated an endemic classification50.
Collection of human sera
Sera were collected from healthy individuals aged 18 and above who were naturally exposed to sand fly bites (n = 546) that resided in the Ambalantota MOH area, an endemic CL area. Individuals with a history of CL were excluded from the study. Demographic data and information on living conditions were recorded using a standardized and previously-validated questionnaire during a house-to-house survey, following informed consent. Blood collection, with 3 mL per individual, was carried out on a voluntary basis and the sera obtained were stored at -70 °C until further use. Negative control serum samples (n = 15) were obtained from healthy donors from non-endemic areas of CL and CL sera (n = 30) collected in a previous Island wide survey and used as positive controls.
Salivary gland dissection
Ph. argentipes (Sri Lankan strain) female sand flies (aged between 5 and 7 days), maintained at the insectary facility at the Department of Parasitology, Faculty of Medicine, University of Colombo, Sri Lanka, under controlled conditions of 26 °C temperature, 75% humidity, and fed on 30% sucrose solution and Culex spp. mosquitoes, collected in cattle-baited net traps, were dissected on a glass slide using fine needles in cold phosphate-buffered saline (PBS) with a pH of 7.4. Their salivary glands (20 gland pairs for sand flies, 10 gland pairs for mosquitoes) were stored in 1.5 mL micro-tubes containing PBS at -70 °C.
Preparation of salivary gland homogenate (SGH)
The dissected salivary glands of Ph. argentipes (40 pairs in 40 µL PBS) and salivary glands of Culex spp. (20 pairs in 20 µl PBS) were frozen in liquid nitrogen for 2 min and then thawed in a hot water bath maintained at 37 °C for a minute. After thawing, the sample was centrifuged at 14,000 rpm for 1 min. The resulting pellet was sonicated for 2 min and then centrifuged again at 8,000 rpm for 2 min. The supernatant was carefully separated from the pellet.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis and western blotting
The sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting were conducted according to published protocols32 to identify immunogenic salivary proteins of Ph. argentipes. SGH proteins of Ph. argentipes (40 µg per lane) and Culex spp. (40 µg per lane) were separated on a 12% SDS-PAGE gel under non-reducing conditions. The separated protein bands were then transferred to a nitrocellulose membrane and cut into strips. These strips were blocked overnight at 4˚C in 5% non-fat milk diluted in Tris-buffered saline with 0.05% Tween 20 (TBS-Tw) and subsequently incubated for 3 h with human sera from 8 endemic individual of the study and 2 non-endemic individuals (diluted 1:80 in TBS-Tw). After washing with TBS-Tw, the strips were incubated with peroxidase-conjugated Goat anti-human IgG antibody (diluted 1:4,000 in TBS-Tween) for 1 h. The chromogenic reaction was developed using a TMB substrate solution. Molecular weight protein markers were used for estimating the sizes of the protein bands.
Indirect ELISA to detect anti-saliva IgG antibodies
Flat-bottom 96-well polyvinyl chloride untreated microplates (HiMedia, Cat No. RM1239) were coated with 50 µL of 2 µg/mL of Ph. argentipes SGH or 1 µg/mL of composite antigen rPagSP02 + rPagSP06. Briefly, the antigens were diluted in carbonate-bicarbonate buffer, pH 9.6, and the plates were incubated overnight at 4 °C. The plates were blocked for 2 h at room temperature with 200 µL of 20% horse serum prepared in Tris-buffered saline with 0.05% Tween 20 (TBST). Hundred microliters of sera were diluted at 1:50 in TBST with 5% horse serum and incubated for 1 h at 37 °C. A secondary alkaline phosphatase-conjugated goat anti-human IgG (H + L) (Novus Biologicals, CO, USA) was incubated at 1:10,000 for 1 h at 37 °C. Hundred microliters of p-nitrophenyl phosphate liquid substrate (Mabtech, Stockholm, Sweden) were added to all wells and incubated for 1 h at room temperature, and the optical density (OD) values were recorded at a 405 nm wavelength using an ELISA plate reader (BioTek Epoch, Vermont, USA). The results were expressed as optical density (OD). Duplicate wells were maintained for each sample throughout the assay. The correlation between SGH and composite recombinant salivary antigen was estimated.
Statistical analysis
Statistical analysis and graphs were conducted using GraphPad Prism 8.0 software. The characteristic features of the study population were analyzed using standard descriptive statistics. Pearson’s rank correlation test was used to determine the p-value and the correlation coefficient (r) at a 95% confidence interval (CI). Cohen’s Kappa statistic was used to assess the agreement between the two ELISA tests. A p-value less than 0.05 was considered statistically significant.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge Mr. D. Sunil Shantha, Mr. M.P. Ariyapala, and Mr. Anura Mahakumara for invaluable field assistance, and the Department of Parasitology, Faculty of Medicine, University of Colombo, for logistical support.
Author contributions
SBP performed the experiments, analyzed the data, and wrote the manuscript. NS, SS, SD, EI, JGV, SK and NDK conceptualized the study, interpreted the data, and edited the manuscript. SS and NS designed and supervised the field study. EI, JGV and SK provided the recombinant composite antigen for the study, and SD was involved in the synthesis of the recombinant product. NDK supervised the overall project, managed collaborations, interpreted the data, and edited the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Funding
This research was supported by the National Institute of Allergy and Infectious Diseases, NIH, under Award Number U01AI136033. The content is solely the authors’ responsibility and does not necessarily represent the official views of the NIH.
Data availability
Data supporting the conclusions of this article are included within the article. The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
Ethics permission was obtained from the Ethics Review Committee of the Faculty of Medicine, University of Colombo, Sri Lanka (EC-21-074), and informed, written consent was obtained from all study participants. The study was conducted in accordance with the approved guidelines of the declaration of Helsinki.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yamey, G. <ArticleTitle Language=“En”>The world’s most neglected diseases. BMJ. 325, 176–177 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Leishmaniasis fact sheets. (2023). https://www.who.int/news-room/fact-sheets/detail/leishmaniasis
- 3.Bailey, M. S. & Diana, N. J. Cutaneous leishmaniasis. Clin. Dermatol.25, 203–211 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Singh, O. P., Singh, B., Chakravarty, J. & Sundar, S. Current challenges in treatment options for visceral leishmaniasis in India: a public health perspective. Infect. Dis. Poverty. 5, 1–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maroli, M., Feliciangeli, M. D., Bichaud, L., Charrel, R. N. & Gradoni, L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med. Vet. Entomol.27, 123–147 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Lukeš, J. et al. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc. Natl. Acad. Sci.104, 9375–9380 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.United Nations. Resolution adopted by the General Assembly on 25 September 2015. transforming our world: the 2030 agenda for sustainable development. chrome (2015). http://-extension://vefaidnbmnnnibpcajpcglclefindmkaj/https://documents-dds-nyv un.org/doc/UNDOC/GEN/N15/291/89/PDF/N1529189.pdf?vOpenElement
- 8.World Health Organization. Ending the neglect to attain the Sustainable Development Goals: a road map for neglected tropical diseases 2021–2030. (2020). https://www.who.int/neglected_diseases/RevisedDraftNTD-Roadmap-23Apr2020.pdf
- 9.Ruiz-Postigo, J. A. et al. Global leishmaniasis surveillance: 2019–2020, a baseline for the 2030 roadmap/Surveillance mondiale de la leishmaniose: 2019–2020, une periode de reference pour la feuille de route a l’horizon 2030. Wkly Epidemiol. Rec.96(35), 401–420 (2021). [Google Scholar]
- 10.Pandey, K. et al. Forty years (1980–2019) of visceral leishmaniasis in Nepal: trends and elimination challenges. Trans. R Soc. Trop. Med. Hyg.117, 460–469 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Dial, N. J. Science and surveillance in the visceral leishmaniasis elimination programme in India (Doctoral dissertation, London School of Hygiene & Tropical Medicine). (2022). 10.17037/PUBS.04664932
- 12.Thakur, L. et al. Atypical leishmaniasis: A global perspective with emphasis on the Indian subcontinent. PLoS Negl. Trop. Dis.12, e0006659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostyn, B. et al. Transmission of Leishmania donovani in the hills of Eastern Nepal, an outbreak investigation in Okhaldhunga and Bhojpur districts. PLoS Negl. Trop. Dis.9, e0003966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mondal, D. et al. Enhanced case detection and improved diagnosis of PKDL in a Kala-azar-endemic area of Bangladesh. PLoS Negl. Trop. Dis.4, e832 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandey, K. et al. Emergence of cutaneous leishmaniasis in Nepal. Trop. Med. Health. 49, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundar, S., Singh, O. P. & Chakravarty, J. Visceral leishmaniasis elimination targets in India, strategies for preventing resurgence. Expert Rev. Anti Infect. Ther.16, 805–812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karunaweera, N. D., Pratlong, F., Siriwardane, H. V., Ihalamulla, R. L. & Dedet, J. P. Sri Lankan cutaneous leishmaniasis is caused by Leishmania donovani zymodeme MON-37. Trans. R Soc. Trop. Med. Hyg.97, 380–381 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Siriwardana, H. Y. et al. Leishmania donovani and cutaneous leishmaniasis, Sri Lanka. Emerg. Infect. Dis.13, 476–478 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar, N. P., Srinivasan, R., Anish, T. S., Nandakumar, G. & Jambulingam, P. Cutaneous leishmaniasis caused by Leishmania donovani in the tribal population of the Agasthyamala Biosphere Reserve forest, Western Ghats, Kerala, India. J. Med. Microbiol.64, 157–163 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Thakur, L. et al. Leishmania donovani infection with atypical cutaneous manifestations, Himachal Pradesh, India, 2014–2018. Emerg. Infect. Dis.26, 1864–1869 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lata, S., Kumari, S., Das, R., Pasi, S. & Dhiman, R. C. Typical and atypical cutaneous leishmaniasis in Himachal Pradesh (India). Heliyon. 7, e07282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coutinho-AbreuIV. & ValenzuelaJ.G. Comparative evolution of sand fly salivary protein families and implications for biomarkers of vector exposure and salivary vaccine candidates. Front. Cell. Infect. Microbiol.8, 290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Moura, T. R. et al. Enhanced Leishmania braziliensis infection following pre-exposure to sandfly saliva. PLoS Negl. Trop. Dis.1, e84 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marzouki, S. et al. Salivary antigen SP32 is the immunodominant target of the antibody response to Phlebotomus papatasi bites in humans. PLoS Negl Trop Dis. 6, e (2012). (1911). [DOI] [PMC free article] [PubMed]
- 25.Rohoušová, I. & Volf, P. Sand fly saliva: effects on host immune response and Leishmania transmission. Folia Parasitol.53, 161–171 (2006). [PubMed] [Google Scholar]
- 26.Carvalho, A. M. et al. Seroconversion to Lutzomyia intermedia LinB-13 as a biomarker for developing cutaneous leishmaniasis. Sci. Rep.7, 3149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veysi, A. et al. Human immune response to Phlebotomus sergenti salivary gland antigens in a leishmaniasis-endemic focus in Iran. Pathog Glob Health. 114, 323–332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clements, M. F. et al. Measurement of recent exposure to Phlebotomus argentipes, the vector of Indian visceral leishmaniasis, by using human antibody responses to sand fly saliva. Am. J. Trop. Med. Hyg.82, 801–807 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gidwani, K. et al. Serological markers of sand fly exposure to evaluate insecticidal nets against visceral leishmaniasis in India and Nepal: a cluster-randomized trial. PLoS Negl. Trop. Dis.5, e1296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sumova, P. et al. PpSP32-like protein as a marker of human exposure to Phlebotomus argentipes in Leishmania donovani foci in Bangladesh. Int. J. Parasitol.51, 1059–1068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed, G. et al. Exploring new immunological insight on SP15 (∼ 14 kDa) family protein in saliva of Indian sand-fly (Phlebotomus argentipes) in experimental visceral leishmaniasis. Cell. Immunol.332, 51–57 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Iniguez, E. et al. Composite Recombinant Salivary Proteins Biomarker for Phlebotomus argentipes Provides a Surveillance Tool Postelimination of Visceral Leishmaniasis in India. J. Infect. Dis.226, 1842–1851 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribeiro, J. M. & Francischetti, I. M. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu. Rev. Entomol.48, 73–88 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Francischetti, I. M. Platelet aggregation inhibitors from hematophagous animals. Toxicon. 56, 1130–1144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdeladhim, M., Kamhawi, S. & Valenzuela, J. G. What’s behind a sand fly bite? The profound effect of sand fly saliva on host hemostasis, inflammation and immunity. Infect. Genet. Evol.28, 691–703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh, K. N. & Mukhopadhyay, J. The effect of anti-sandfly saliva antibodies on Phlebotomus argentipes and Leishmania donovani. Int. J. Parasitol.28, 275–281 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Pandey, R. K. & &Prajapati, V. K. Exploring sand fly salivary proteins to design multiepitope subunit vaccine to fight against visceral leishmaniasis. J. Cell. Biochem.120, 1141–1155 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Martín-Martín, I., Molina, R. & Jiménez, M. Identifying salivary antigens of Phlebotomus argentipes by a 2DE approach. Acta Trop.126, 229–239 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Aronson, N. E. et al. Antibody Responses to Phlebotomus papatasi Saliva in American Soldiers with Cutaneous Leishmaniasis Versus Controls. Front. Trop. Dis.2, 766273 (2022). [Google Scholar]
- 40.Gomes, R. & Oliveira, F. The immune response to sand fly salivary proteins and its influence on Leishmania immunity. Front Immunol.3, 110 (2012). [DOI] [PMC free article] [PubMed]
- 41.Willen, L. et al. Field study of the improved rapid sand fly exposure test in areas endemic for canine leishmaniasis. PLoS Negl. Trop. Dis.13, e0007832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson, J. M. et al. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC Genom.7, 1–23 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piyasiri, S. B. et al. Comparative Analysis of Phlebotomus argentipes Vector of Leishmaniasis in India and Sri Lanka. Microorganisms. 12, 1459. 10.3390/microorganisms12071459 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naotunne, T. D. et al. Cutaneous leishmaniasis in Sri Lanka. An imported disease linked to the Middle East and African employment boom. Trop. Geogr. Med.42, 72–74 (1990). [PubMed] [Google Scholar]
- 45.Athukorale, D. N., Seneviratne, J. K., Ihalamulla, R. L. & Premaratne U.N. Locally acquired cutaneous leishmaniasis in Sri Lanka. J. Trop. Med. Hyg.95, 432–433 (1992). [PubMed] [Google Scholar]
- 46.Ozbel, Y. et al. Distribution and ecological aspects of sand fly (Diptera: Psychodidae) species in Sri Lanka. J. Vector Ecol.36, S77–S86 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Gajapathy, K. et al. Molecular identification of potential leishmaniasis vector species within the Phlebotomus (Euphlebotomus) argentipes species complex in Sri Lanka. Parasite Vectors. 6, 1–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senanayake, S. A., Abeyewicreme, W., Dotson, E. M. & Karunaweera, N. D. Characteristics of phlebotomine sandflies in selected areas of Sri Lanka. Southeast. Asian J. Trop. Med. Public. Health. 46, 994–1004 (2015). [PMC free article] [PubMed] [Google Scholar]
- 49.Wijerathna, T. & Gunathilaka, N. Morphological identification keys for adults of sand flies (Diptera: Psychodidae) in Sri Lanka. Parasites Vectors. 13, 1–3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karunaweera, N. D. et al. Spatiotemporal distribution of cutaneous leishmaniasis in Sri Lanka and future case burden estimates. PLoS Negl. Trop. Dis.15, e0009346 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the conclusions of this article are included within the article. The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.