Abstract
Asymmetric cell division in bacteria is achieved through cell polarization, where regulatory proteins are directed to specific cell poles. In Caulobacter crescentus, both poles contain a membraneless microdomain, established by the polar assembly hub PopZ, through most of the cell cycle, yet many PopZ clients are unipolar and transiently localized. We find that PopZ’s interaction with the response regulator CpdR is controlled by phosphorylation, via the histidine kinase CckA. Phosphorylated CpdR does not interact with PopZ and is not localized to cell poles. At poles where CckA acts as a phosphatase, dephosphorylated CpdR binds directly with PopZ and subsequently recruits ClpX, substrates, and other members of a protease complex to the cell pole. We also find that co-recruitment of protease components and substrates to polar microdomains enhances their coordinated activity. This study connects phospho-signaling with polar assembly and the activity of a protease that triggers cell cycle progression and cell differentiation.
Subject terms: Phosphorylation, Cellular microbiology, Cell signalling, Proteolysis
Asymmetric bacterial cell division involves the polar organizing protein PopZ recruiting cell fate regulators. Here, the authors show that, in Caulobacter crescentus, phosphorylation directly regulates client protein interactions with the PopZ hub.
Introduction
Many bacteria divide asymmetrically, generating two distinct cell types that can have vast differences in gene expression, morphology, and other behaviors1–3. This basic form of multicellularity was very likely to have arisen more than a billion of years before the emergence of eukaryotic life4. In rod-shaped bacteria, asymmetry is generally achieved through cell polarization, where distinct sets of regulatory proteins are directed to opposite cell poles, and subsequent cell division generates daughter cells that inherit different sets of regulatory factors and therefore different cell fates5,6. These systems depend on mechanisms that distinguish one pole from the other, and in many cases the associated molecular processes that drive this organization are not clear.
In Alphaproteobacteria, one of the key polarization factors is the polar organizing protein PopZ, which is required for the polar localization of many cell fate regulators7. In vitro, PopZ self-assemblies into oligomeric forms that undergo higher-order assembly into fibrils and larger structures8,9. Under some conditions, these assemblages behave as liquid-liquid phase-separated protein condensates and have the ability to interact with client proteins10,11. PopZ exhibits similar assembly and recruitment activities when expressed in Escherichia coli, where it accumulates as a single cytoplasmic focus12–14. In its natural context, PopZ and its associated proteins form three-dimensional structures that abut the membrane at cell poles15. Caulobacter crescentus PopZ is localized to both poles through most of the cell cycle, yet several of the regulatory proteins that depend on it for polar localization exhibit transient, unipolar localization16–18.
The N-terminal domain of PopZ is an interaction hub that binds to at least eleven different client proteins and recruits them from the cytoplasm to polar microdomains12,19. Including indirect interactions, PopZ serves as localization determinant for large interconnected protein networks12,16–18. For example, the protease ClpP and its partner ClpX are indirectly recruited to polar microdomains through interaction with the regulatory factor CpdR, which binds directly to PopZ12,17. CpdR is a cell cycle-regulated adaptor protein that delivers substrate proteins to ClpXP for timely degradation20. Some ClpXP substrates require one or more additional adaptor proteins, namely RcdA and PopA, and these are recruited to cell poles through interaction with PopZ at the same stage of the cell cycle as CpdR, which is known as the swarmer to stalked cell transition21. Thus, ClpP, ClpX, up to three adaptors, and the substrate proteins themselves22,23 are all co-recruited by PopZ into polar microdomains, where they become concentrated relative to bulk cytoplasm.
The question of how co-localization and concentration in PopZ microdomains affects the recruited proteins’ activities has not been conclusively resolved. Owing to the limited size of PopZ structures and the relative abundance of available clients, many PopZ-associated proteins are only partially localized in polar microdomains24,25. This raises the question of whether the levels of concentration and co-localization at cell poles are large enough to influence cell physiology. One study found that three different pole-localized ClpXP substrates were degraded more quickly in a ΔpopZ strain than in wildtype26, a phenotype that could occur if polar microdomains have an inhibitory effect on the activity of protease complexes. However, this is not easily reconciled with the observation that both ClpXP-mediated proteolysis and the polar accumulation of ClpXP complexes occur at the same stage of the cell cycle.
Two other unresolved questions concern the regulation of transient localization to cell poles and the mechanisms by which one PopZ microdomain is differentiated from another. These questions are exemplified by the members of the ClpXP proteolysis complex, which are localized to only one of the two poles, and only during the swarmer to stalked cell transition. Notably, both the CpdR and RcdA adaptor proteins are direct binding partners of PopZ12,19, which suggests that their interaction with the polar hub is subject to some form of regulation.
CpdR is a two-component response regulator whose phosphorylation state is regulated by the histidine kinase CckA, via an intermediary phospho-transfer protein ChpT. CckA exhibits kinase activity through the majority of the cell cycle, when CpdR and ClpXP protease complexes are neither pole-localized nor active in degrading substrates17,20,27. CckA works as a phosphatase during the swarmer to stalked cell transition, when CpdR is localized to one pole and ClpXP is active. CckA binds directly to PopZ and is localized to both cell poles through most of the cell cycle, where its activity is controlled through the influence of asymmetrically localized upstream regulators28.
Taking these observations together, we hypothesize that CckA kinase/phosphatase activity is a switch that regulates PopZ’s interaction with CpdR and the consequent recruitment of other members of ClpXP proteolysis complexes. In this work, we tested that hypothesis by modulating CpdR phosphorylation levels in C. crescentus, in a reconstituted E. coli system, and in vitro. We also asked if the concentration of ClpXP complex members at the cell pole could influence the overall rate of proteolysis for the entire cell, using a combination of cell imaging and computational modeling. The results explain how CpdR phosphorylation and de-phosphorylation influences polar asymmetry and cell differentiation during the C. crescentus cell cycle.
Results
CpdR-YFP localization is correlated with its phosphorylation state during the cell cycle
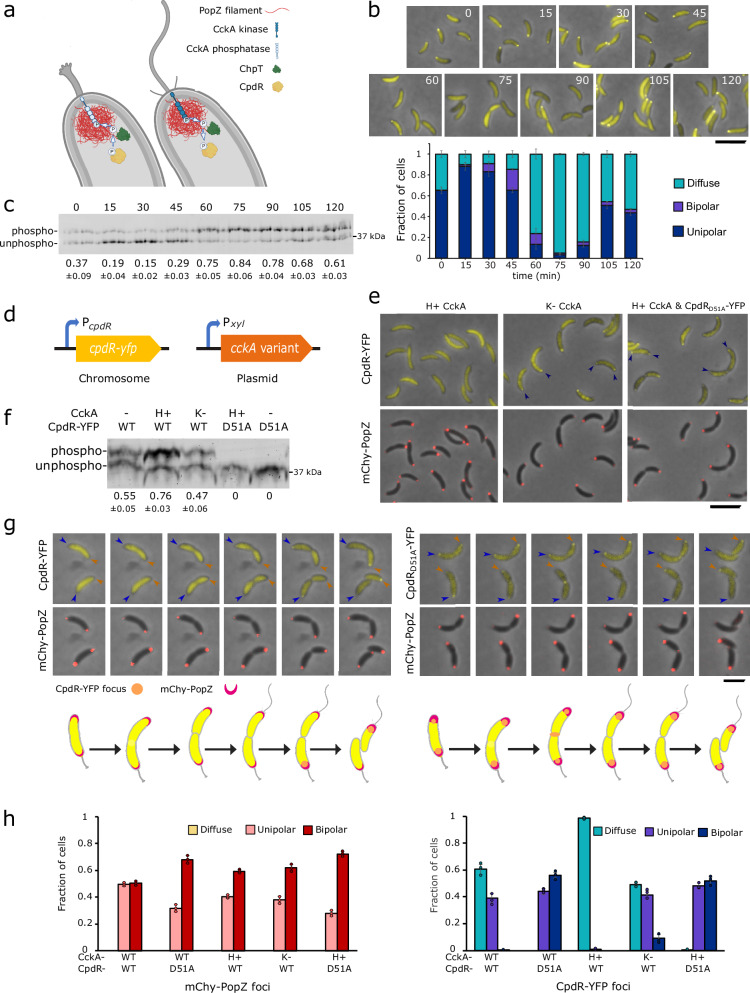
In wildtype C. crescentus, polar localization of CpdR-YFP temporally coincides with the time that it is dephosphorylated by CckA-ChpT phospho-transfer during the swarmer-to-stalked transition17,27 (Fig. 1a). We used cell cycle synchronization to quantify the relationship between CpdR-YFP localization and phosphorylation. After isolating cells at the swarmer cell (G0) stage, we counted the fraction of cells exhibiting polar CpdR-YFP localization (Fig. 1b) and quantified the relative levels of phosphorylated versus unphosphorylated CpdR-YFP over the course of the cell cycle (Fig. 1c). The highest and lowest degrees of polar localization temporally corresponded with the highest and lowest fractions of unphosphorylated CpdR-YFP, respectively. After cell division, CpdR-YFP remained diffuse in swarmer progeny but exhibited polar localization in stalked progeny, and correspondingly, we observed an intermediate level of CpdR-YFP phosphorylation in this heterogenous population (Fig. 1c). We conclude that there is a strong positive correlation between CpdR-YFP dephosphorylation and polar localization during the C. crescentus cell cycle.
Fig. 1. Correlations between CpdR’s phosphorylation state, polar localization, and co-localization with PopZ.
a CpdR phospho-signaling at stalked (left) and swarmer (right) cell poles, where CckA acts as a phosphatase or kinase, respectively. Graphics: BioRender. b C. crescentus cells expressing CpdR-YFP were synchronized and, at indicated time points over a cell cycle time course, aliquots were removed for observation. The plot shows the average frequencies of cells exhibiting zero, one, or two polar foci, and the error bar shows the range between the two biological replicates (n > 300/replicate/time point, scale bar = 5 μm). c CpdR-YFP phosphorylation levels in lysates from (b), observed by Phos-tag gel electrophoresis. The average intensities of the phosphorylated bands as a fraction of the sum of band intensities from both biological replicates are provided, along with the range between replicates. The replicate gel is provided in Supplementary Fig. 1a. d Genetic modifications for controlling CpdR-YFP phosphorylation. Single-copy cpdR-yfp is expressed from the native promoter, multicopy cckA variants are expressed from Pxyl without xylose induction. e Localization of CpdR-YFP or CpdRD51A-YFP and mChy-PopZ in different CckA signaling contexts. H+ and K− signify hyperactive kinase and kinase-deficient forms of CckA expressed from a multicopy plasmid, in addition to CckA expressed from the unmodified cckA locus. Arrowheads mark polar localization; scale bar = 5 μm. f CpdR-YFP phosphorylation levels in lysates from (e), observed using Phos-tag gel electrophoresis. The dash mark indicates no extrachromosomal copies of cckA. The average intensities of the phosphorylated bands as a fraction of the sum of band intensities from three biological replicates are provided, along with the standard deviation between replicates. Replicate gels are provided in Supplementary Fig. 1b. g Cells expressing CpdR-YFP or CpdRD51A-YFP and mChy-PopZ were observed during cell division, using time-lapse microscopy at 15 min intervals. The fluorescence levels of individual panels were adjusted differently to aid visualization. Scale bar = 2 μm; Graphics: BioRender. h Average frequencies of cells with diffuse, monopolar, and bipolar fluorescent foci, from strains imaged in (e, g) (n > 100/replicate, bar = standard deviation of three biological replicates). Source data and n values for (b, h) are provided as a Source Data file.
CckA kinase influences CpdR-YFP phosphorylation and its co-localization with PopZ
We asked if we could influence CpdR-YFP phosphorylation and localization by expressing mutant variants of CckA29 with differing levels of kinase/phosphatase activity from a plasmid (Fig. 1d–h). When we expressed a hyperactive-kinase (H+) form of CckA (G319E), the ratio of phosphorylated to unphosphorylated CpdR-YFP was relatively high, and the protein did not localize to cell poles. When we expressed a kinase deficient (K−) form of CckA (H322A), the ratio was substantially lower, and cells exhibited robust polar foci. We also observed transient localization of CpdR-YFP at the division plane, which likely arises from CpdR’s interaction with ClpXP protease complexes at this location30. To determine if polar localization is indirectly controlled by CckA signaling, we expressed a mutant form of CpdR-YFP (CpdRD51A-YFP) that cannot be phosphorylated17. Whether or not the hyperactive-kinase form of CckA was expressed in this context, CpdRD51A-YFP exhibited polar localization, and moreover, was usually localized to both of the cell poles in cells that also had a bi-polar distribution of mChy-PopZ (Fig. 1g, h). These results show that the kinase/phosphatase activity of CckA is closely correlated with the phosphorylation state of CpdR and its co-localization with PopZ at C. crescentus cell poles.
CpdR phosphorylation affects interaction with PopZ in E. coli
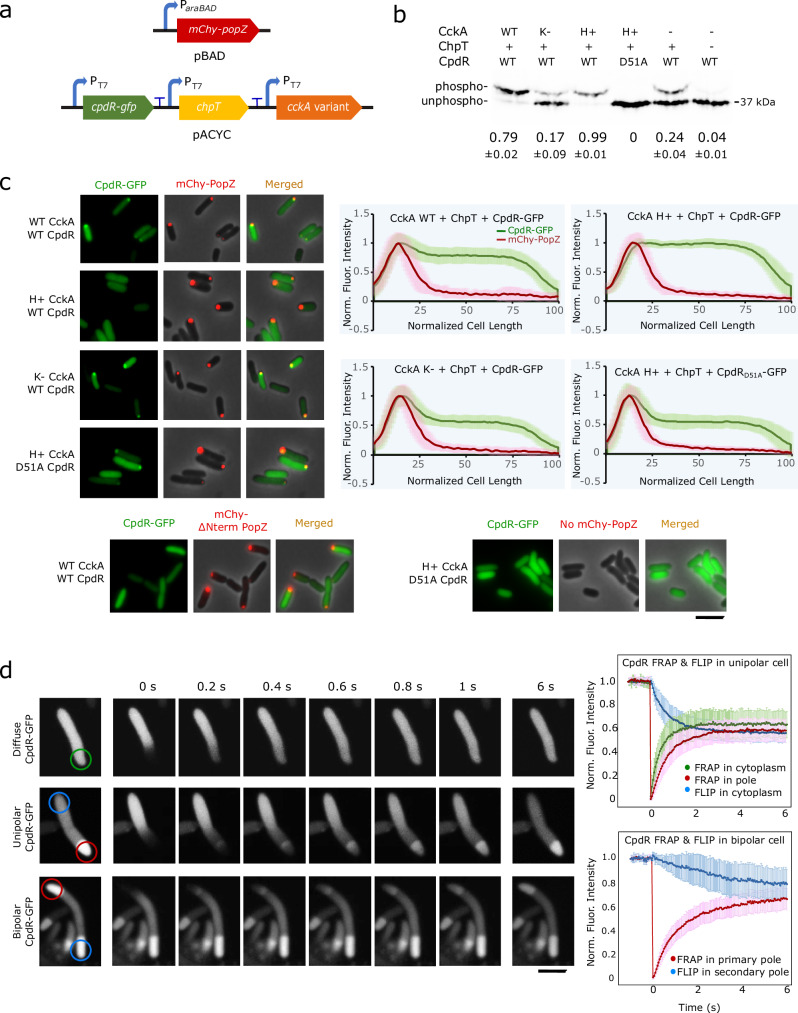
We reconstituted CpdR phosphorylation and studied polar localization in E. coli to determine whether CpdR’s phosphorylation state affects its interaction with the polar organizing protein PopZ. To do this, we co-expressed CpdR-GFP and mChy-PopZ together with the phospho-transfer protein ChpT and then added either a wildtype, kinase deficient, or hyperactive-kinase variant of CckA (Fig. 2a). Experiments were performed in a ∆clpXP mutant background31 to eliminate the possibility of interactions between C. crescentus CpdR and endogenous protease20. Combining ChpT and CpdR-GFP without any CckA resulted in a higher level of CpdR-GFP phosphorylation than CpdR-GFP alone (24% compared to 4%), indicating a background level of host-derived phosphorylation, which has been observed for some other heterologous response regulators in E. coli32. We could modulate CpdR-GFP phosphorylation by co-expressing hyperactive-kinase, wildtype, and kinase-deficient variants of CckA, which drove CpdR-GFP phosphorylation levels of 99%, 79%, and 17%, respectively (Fig. 2b). Under these conditions, we assessed interactions between CpdR-GFP and mChy-PopZ by observing these proteins’ sub-cellular localization patterns. Cells that expressed an N-terminal truncation mutant of PopZ or lacked mChy-PopZ did not accumulate CpdR-GFP in polar foci (Fig. 2c and Supplementary Fig. 2c), consistent with earlier studies which demonstrate that the N-terminal hub domain of PopZ is responsible for this interaction19. In cells expressing full-length mChy-PopZ, the presence or absence of ChpT itself did not affect CpdR-GFP’s co-localization with mChy-PopZ in polar foci (Supplementary Fig. 2b). With the further addition of wildtype CckA, we observed a similar degree of CpdR-GFP mChy-PopZ co-localization. In contrast, adding the hyperactive-kinase variant of CckA reduced the frequency of CpdR-GFP polar localization to zero (Fig. 2c). Conversely, strains expressing kinase-deficient CckA exhibited increased co-localization, and we found similarly elevated levels of co-localization when we expressed non-phosphorylatable CpdRD51A-GFP with hyperactive-kinase CckA. From these experiments, we conclude that phosphorylating CpdR inhibits its ability to co-localize with polar PopZ foci in E. coli.
Fig. 2. CpdR phosphorylation state influences CpdR-PopZ interactions in E. coli.
a Genes for reconstituting CpdR phosphorylation and PopZ interaction in E. coli. b CpdR-GFP phosphorylation levels in E. coli lysates, observed using Phos-tag gel electrophoresis. The average intensities of the phosphorylated bands as a fraction of the sum of band intensities from three biological replicates are provided, along with the standard deviation between replicates. Replicate gels are provided in Supplementary Fig. 2a. For CckA variants, WT = wildtype; H+ = hyperactive-kinase; K− = kinase-deficient. c mChy-PopZ and CpdR-GFP localization in E. coli cells. Normalized fluorescence intensities were plotted against cell length (n = 60, with 20 cells from 3 biological replicates. Lines trace mean value, shaded regions = SD). Scale bar = 5 μm. d FRAP and FLIP assay for CpdR-GFP in E. coli cells expressing PopZ. Recovery and loss of fluorescence were plotted against time in seconds (n = 20, Lines trace mean value, bar = standard deviation). Scale bar = 2 μm. Source data are provided as a Source Data file.
CpdR-PopZ interaction is highly dynamic
We consistently observed that a large fraction of CpdR is diffuse in the cell body, even when it is almost entirely dephosphorylated (Figs. 1b,g and 2c). We hypothesized that this is reflective of a weak and therefore highly transient interaction with PopZ, and tested this in FRAP experiments on E. coli cells co-expressing CpdR-GFP and mChy-PopZ (Fig. 2d). We found that PopZ-associated CpdR-GFP was rapidly replenished from the cytoplasmic pool after photobleaching, with a half-time of 0.61 s. This was slightly slower than the rate of recovery of CpdR-GFP diffusing through normal cytoplasm, at 0.36 s. During recovery, a wavefront of CpdR-GFP advanced through PopZ foci (Supplementary Video 1), which could occur if CpdR-GFP molecules are transiently held by interactions with PopZ. To obtain information on the off-rate, we created E. coli cells with mChy-PopZ foci at both cell poles. Bleaching CpdR-GFP at one pole resulted in rapid fluorescence recovery, with concomitant rapid fluorescence loss at the opposite pole. By contrast, the recovery rate of mChy-PopZ had a half-life of over 3 min (Supplementary Fig. 2d), indicating that associations among the scaffold molecules are more stable.
Direct interaction between CpdR and PopZ is controlled by phosphorylation
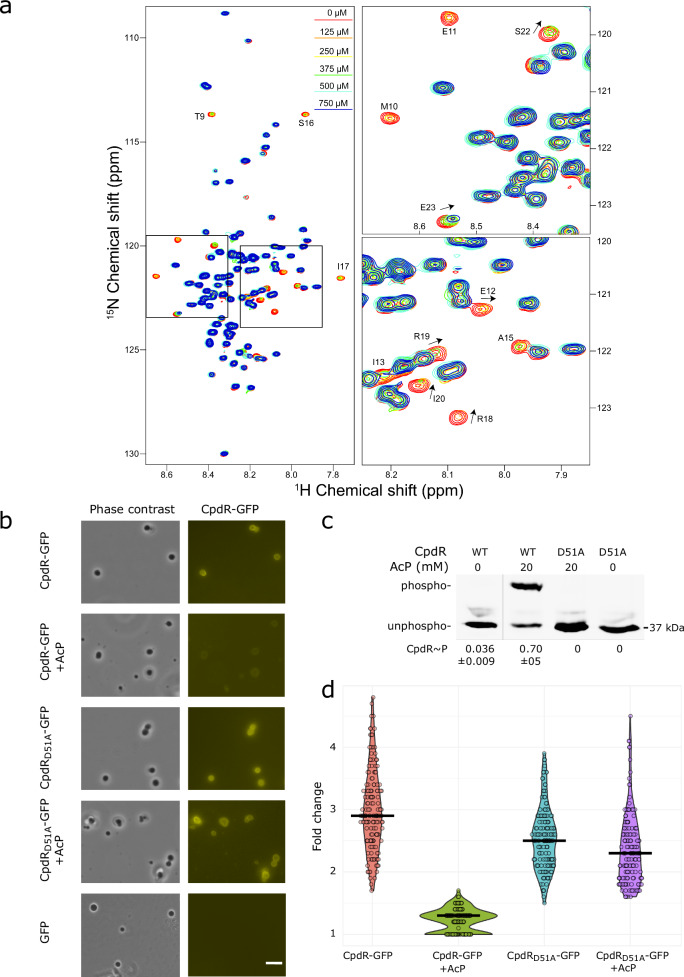
We used solution NMR spectroscopy to determine whether CpdR interacts directly with PopZ. CpdR-GFP was chosen as a ligand because the fusion protein was far more soluble than untagged CpdR at high concentration. The other ligand was C-terminally truncated PopZΔ134–177, which retains the N-terminal protein–protein interaction domain (Fig. 2c), but lacks the ability to self-assemble9. 15N-enriched PopZΔ134–177 (50 μM) was mixed with increasing concentrations of unlabeled CpdR-GFP (0–750 μM), and the spectra were analyzed for concentration-dependent changes (Fig. 3a). Most of the PopZΔ134–177 NMR peaks exhibited no significant changes, even at the highest concentration of CpdR-GFP, indicating residues whose local biochemical environments are not affected by the presence of this protein. A small number of PopZΔ134–177 residues displayed large chemical shift perturbations and significant peak broadening with increasing concentrations of CpdR-GFP. To ask if these effects are caused by interactions with GFP, we mixed 750 μM GFP with 50 μM of 15N-enriched PopZΔ134–177. No significant chemical shift perturbations were observed in this control experiment (Supplementary Fig. 3b), indicating that the large perturbations observed upon the addition of CpdR-GFP were induced by interactions with CpdR. The peaks exhibiting the most significant chemical shift perturbation and broadening are likely indicative of residues that interact directly with CpdR, whereas peaks with moderate effects chemical shifts may correspond to amino acids participating in secondary or indirect binding interactions (Supplementary Fig. 3a). PopZΔ134–177 residues E7-E23 exhibited the greatest changes, and these results are in good agreement with previous studies that identified the N-terminus of PopZΔ134–177 as the interaction site for RcdA and ChpT12,19.
Fig. 3. Direct interaction between CpdR and PopZ is inhibited by phosphorylation.
a Left: 1H-15N TROSY-HSQC NMR spectra overlay of 50 μM 15N-enriched PopZΔ134–177 with varying concentrations of CpdR-GFP: 0 μM (red), 125 μM (orange), 250 μM (yellow), 375 μM (green), 500 μM (cyan), and 750 μM (blue). Right: Enlarged regions highlight the changes observed with increasing CpdR-GFP concentration. Residues with the most significant perturbations are labeled, with arrows indicating the direction of the peak shift. b CpdR-GFP, CpdRD51A-GFP, or GFP alone was incubated with PopZ condensates. Phase contrast and YFP fluorescence channels are shown. +AcP = pre-incubation with acetyl phosphate. Scale bar = 10 μm. c CpdR-GFP phosphorylation levels in samples from (b), observed using Phos-tag gel electrophoresis. Figure panel shows a single gel from which irrelevant lanes are removed. The average intensities of the phosphorylated bands as a fraction of the sum of band intensities from 3 biological replicates are provided, along with the standard deviation between replicates. Replicate gels are provided in Supplementary Fig. 3a. d Ratios of the CpdR-GFP and CpdRD51A-GFP fluorescence intensities within condensates to outside condensates, imaged in YFP channel. Violin plot widths are proportional to the number of data points, bar shows population average. n = 150 condensates per sample (50 per biological replicate). Source data are provided as a Source Data file.
We asked if CpdR can also interact with macromolecular scaffolds comprised of full-length PopZ, and if this interaction can be influenced by CpdR phosphorylation. To do this, we generated phase-separated condensates of purified full-length PopZ10 and demonstrated that CpdR-GFP but not GFP alone partitions into the condensates (Fig. 3b). Next, we phosphorylated CpdR-GFP by pre-incubating it with acetyl phosphate (AcP), using CpdRD51A-GFP as a non-phosphorylatable receiver to control for the presence of AcP (Fig. 3c). We found that CpdR-GFP phosphorylation inhibited the partitioning of CpdR-GFP into PopZ condensates, while the same treatment had no such inhibitory effect on the CpdRD51A-GFP control, nor did it affect protein mobility in Phos-tag gel electrophoresis (Fig. 3c, d). These results suggest that CpdR phosphorylation reduced the partitioning of CpdR-GFP into PopZ condensates.
CpdR phosphorylation influences the localization of RcdA and ClpX
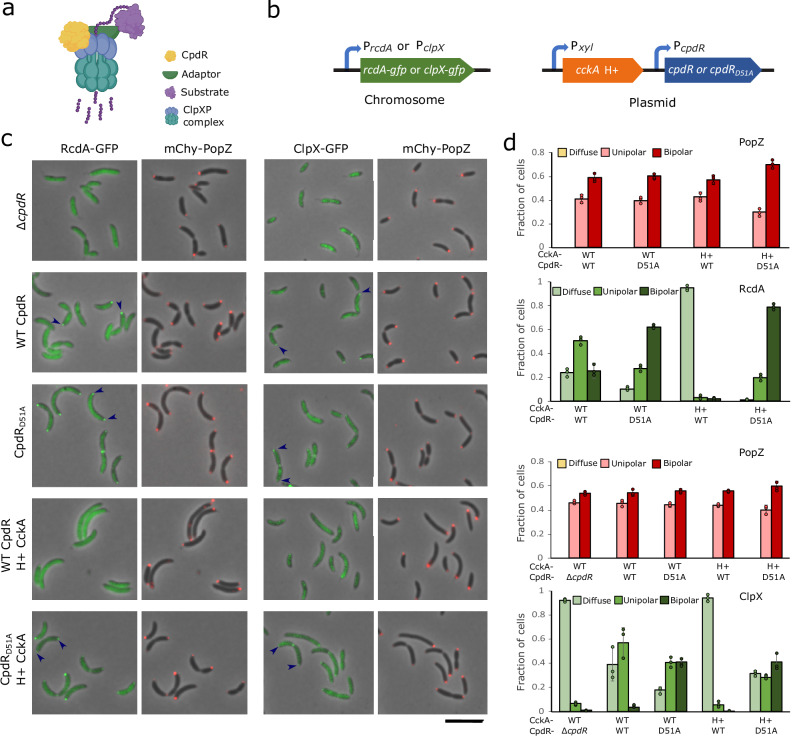
Since ClpX physically interacts with CpdR17, we hypothesized that we could control the localization of ClpXP complexes (Fig. 4a) by modifying the phosphorylation state of CpdR. To test this, we created strains in which either the ClpX-associated adaptor RcdA18 or ClpX itself were tagged with GFP and expressed from their endogenous promoters, and in which the native copy of popZ had been exchanged with a functional mChy-tagged version7 (Fig. 4b). In an otherwise wildtype genetic background, RcdA-GFP and ClpX-GFP both exhibited the expected pattern of transient co-localization with polar mChy-PopZ during the swarmer-to-stalked cell transition18,30 (Fig. 4c, d). In a ∆cpdR background, RcdA-GFP was almost completely diffuse, although many cells retained faint polar foci that were difficult to discern and quantify (Supplementary Fig. 4a). In agreement with an earlier report17, we also observed that CpdR is required for the polar localization of ClpX.
Fig. 4. Correlations between CpdR phosphorylation state and the localization of protease components.
a Diagram of CpdR-mediated substrate degradation by ClpXP. Depending on the substrate, an additional adaptor (green) could be RcdA and/or PopA. Graphics: BioRender. b Genetic modifications for observing RcdA -GFP or ClpXP -GFP in different CpdR phosphorylation contexts, built in a ∆cpdR; popZ::mChy-popZ C. crescentus strain background. c Localization of RcdA-GFP or ClpX-GFP and mChy-PopZ in different CpdR phosphorylation contexts. Arrowheads mark polar localization; scale bar = 5 μm. d The average frequencies of cells with diffuse, monopolar, and bipolar fluorescent foci in (c) (n > 100/replicate, bar = standard deviation of 3 biological replicates). Source data and n values for (d) is provided as a Source Data file.
To assess protein localization in cells where CpdR is always de-phosphorylated, we replaced endogenous cpdR with the cpdRD15A variant. RcdA-GFP was localized to both poles in cells that also had bi-polar mChy-PopZ foci, and, in agreement with an earlier report17, ClpX-GFP showed a similar, though less intensely localized pattern (Fig. 4c, d). To assess protein localization in cells where CpdR is always phosphorylated, we expressed the hyperactive variant of CckA. In these cells, RcdA-GFP and ClpX-GFP were diffuse. When we expressed hyperactive CckA in the cpdRD51A background, RcdA-GFP and ClpX-GFP exhibited bi-polar localization, indicating that CpdR de-phosphorylation is a controlling factor in these proteins’ polar localization. The differences in these strains’ localization patterns suggest that in normal cells, CpdR’s phosphorylation state is under strict temporal and spatial control, directing ClpXP complexes to one pole at the swarmer-to-stalked cell transition. Since ClpX is reported to play a key role in RcdA-GFP polar localization18, CpdR may promote RcdA localization indirectly, by recruiting ClpX to the pole. Additional RcdA may be localized to poles via direct contact with PopZ19.
Substrate proteolysis coincides with polar localization
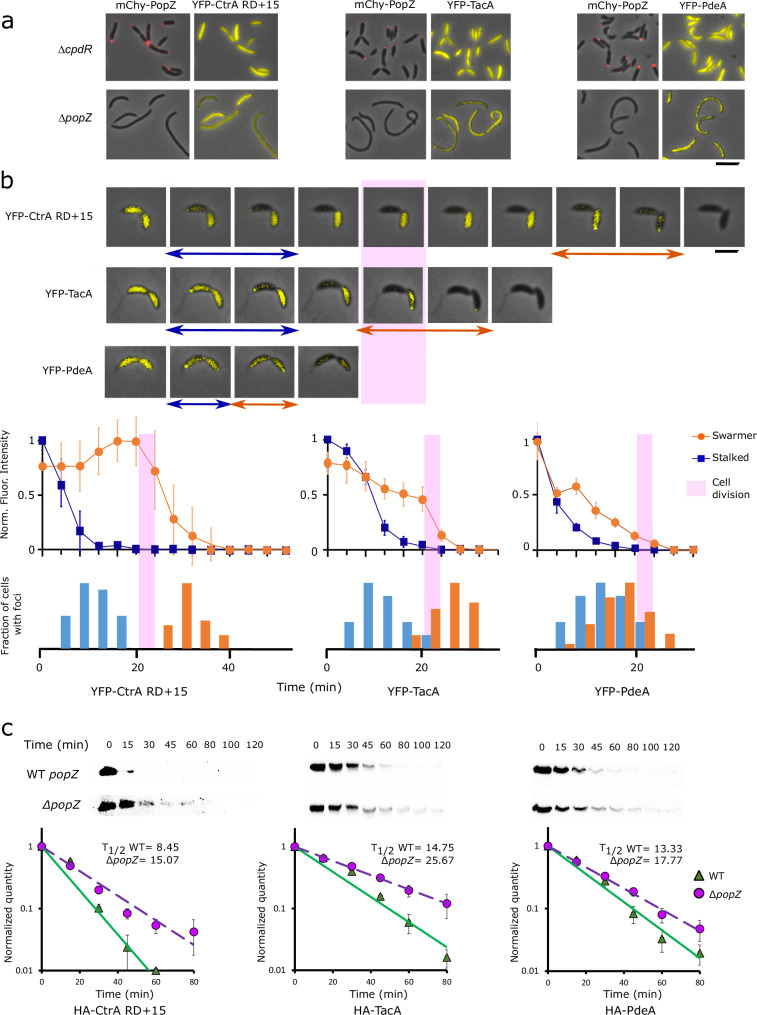
To assess the localization of proteolysis substrates, we induced the expression of YFP-tagged versions of PdeA, TacA, and CtrA RD + 1533 in wildtype and other genetic backgrounds and observed their localization patterns during the cell cycle. These three substrates were expressed from chromosomally integrated xylose-inducible promoters, and were chosen as representatives of CpdR-dependent, CpdR/RcdA-dependent, and CpdR/RcdA/PopA-dependent classes, respectively. Their localization patterns are consistent with earlier reports on PdeA and CtrA localization7,34, and add to the literature by showing that YFP-TacA localization depends on RcdA (Supplementary Fig. 4b), and that all three classes of substrates exhibit transient, unipolar localization in wildtype cells and diffuse localization in both ∆popZ and ∆cpdR backgrounds (Fig. 5a).
Fig. 5. Polar localization of CpdR substrates is correlated with increased rate of degradation.
a Localization of YFP-tagged substrates in ∆cpdR; popZ::mChy-popZ and ∆popZ C. crescentus strain backgrounds. Scale bar = 5 μm. b Time-lapse images of YFP-tagged substrate localization in a WT C. crescentus background, at 4 min intervals. Blue arrows mark frames with foci in stalked cell, orange arrows mark frames with foci in swarmer cell. Pink bar idicates the time of cell separation. After accounting for photobleaching and temporally alignging the cells with respect to the time of cell separation, average fluorescence intensities for stalked and swarmer cell bodies, normalized to maximum fluorescence intensity, were plotted against time (line graphs, with error bars showing standard deviation, n = 20 cells). The fractions of cells ehxhibiting a polar focus, normalized to the highest value observed, were plotted on the same time axis (bar charts). Scale bar = 2 μm. c Degradation of HA-tagged proteolysis substrates following inducer wash-out, observed by western blotting with α-HA antibody. Average band intensities from three separate experiments were plotted against time (graphs, bar = standard deviation). Source data are provided as a Source Data file.
We used time-lapse fluorescence microscopy to observe the spatial-temporal relationships between localization and proteolysis for the three substrate proteins (Fig. 5b). In stalked cell progeny, which are programmed to advance immediately to S-phase and subsequent stages of the cell cycle35–37, all three proteolysis substrates were cleared in the minutes before physical cell separation, and the highest frequency of polar localization was observed during this time period. Since cell separation lags the separation of progeny cells’ cytoplasm via inner membrane fusion by several minutes38, it is likely that proteolysis and polar localization occurred soon after compartmentalization. In swarmer cell progeny, which are developmentally delayed relative to stalked cell siblings, YFP-PdeA was often cleared concomitantly or within four minutes of its clearance from stalked cells, which coincided with the highest frequency of polar localization. The majority of YFP-TacA and YFP-CtrA RD + 15 were cleared ~2 min later, when their peak frequency in polar localization occurred. In a ∆popZ genetic background, none of the substrate proteins were localized to polar foci (Fig. 5b), and our measurements of YFP-CtrA RD + 15 fluorescence intensity (Supplementary Fig. 5a) suggest that it is cleared at a substantially slower rate than in a wildtype genetic background. Together, these observations provide additional evidence, now for multiple substrates and at high temporal and spatial resolution, that the polar localization of proteolysis substrates is closely correlated with their rapid proteolysis39.
PopZ facilitates rapid substrate degradation
To better understand the functional relationship between substrate protein degradation and polar localization, we performed two different types of experiments to assess the degradation rates in wildtype versus ∆popZ cells. In the first type of experiment, we blocked new protein synthesis by treating cells with chloramphenicol. Over subsequent time points, a subpopulation of cells in wildtype cultures retained large quantities of substrates for more than 60 min, but this was not observed in ∆popZ cultures (Supplementary Fig. 5b, c). We propose that chloramphenicol treatment interferes with the measurement of degradation rates by preventing cell cycle progression in swarmer cells, locking them into a stage of high substrate stability. ∆popZ strains, whose cell division is often uncoupled from the cell cycle7, do not appear to produce significant numbers of this cell type.
In the second type of experiment, we used inducer wash-out to block new substrate protein synthesis, allowing general protein synthesis and the cell cycle to continue (Fig. 5c and Supplementary Fig. 5a). Under these conditions, the three ClpXP substrates that we tested were degraded more rapidly in wildtype cells compared to ∆popZ. The difference was higher for substrates whose degradation is mediated by more adapter proteins: CtrA requires three adapters (CpdR, RcdA, and PopA), TacA requires two (CpdR and RcdA), and PdeA requires only CpdR23. Taking this together with the observation that the substrates, adaptors, and protease are concentrated in PopZ microdomains during proteolysis (Figs. 1g, 4c, and 5b), we propose that one of the functions of PopZ microdomains is to enhance the assembly of ClpXP-adaptor-substrate complexes.
A conceptual model for enhanced assembly of proteolysis complexes in PopZ microdomains
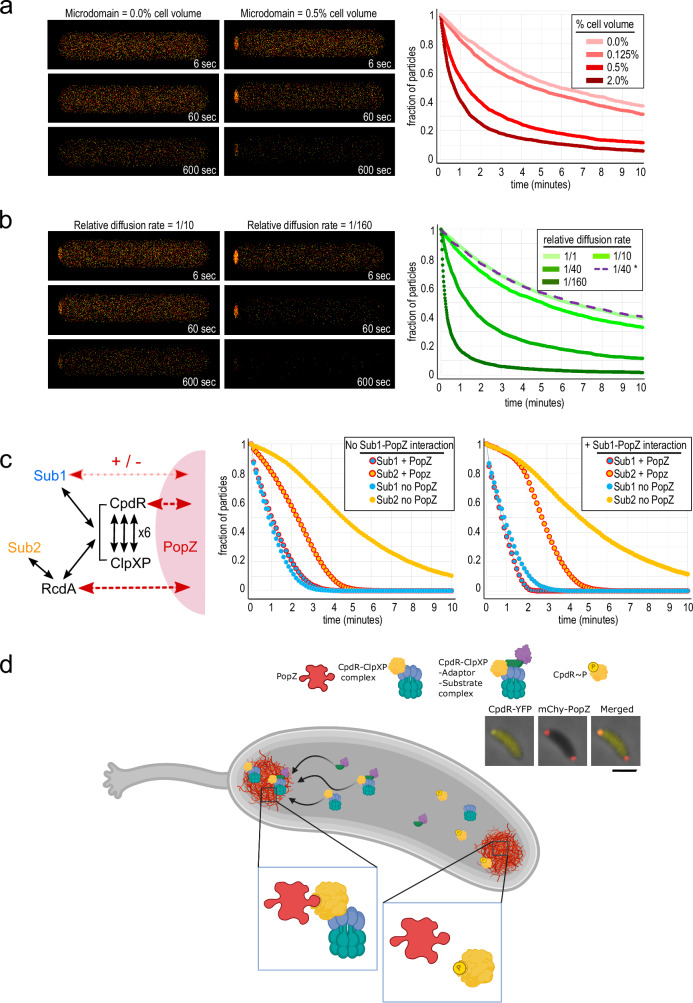
PopZ microdomains occupy ~0.5% of the cytoplasm15, and only a fraction of proteolysis substrates and protease components are found in this relatively small compartment. A question is whether PopZ microdomain could enhance proteolysis complex assembly and activity on a scale that is sufficient to affect the overall rate of degradation for the entire cell. Using the biochemical simulation program Smoldyn40, we developed a computational model to determine whether the physiological characteristics of the system, in terms of compartment sizes, protein localization, diffusion rates, rate of proteolysis, and number of substrate molecules, are compatible with the idea that PopZ microdomains enhance proteolysis.
We created a simplified model that considers interactions between two reactants, A and B, which diffuse within the cell and are eliminated after colliding. Using live-cell single-molecule tracking of pole-localized C. crescentus proteins to inform particle diffusion rates38, molecules A and B were allowed to exhibit Brownian motion in PopZ microdomains, but with a slower diffusion coefficient compared to normal cytoplasm. This had the effect of increasing local protein concentration, mimicking the effect of weak interactions with PopZ. In simulated cells with physiologically relevant values for microdomain size and particle diffusion rates, 50% of the reactants were degraded within 1.5 min, compared to more than 5 min in cells that lacked a polar microdomain (Fig. 6a and Supplementary Video 4). Increasing the number of substrate molecules in polar compartments, by either increasing compartment volume or lowering the rate of diffusion at the cell pole, had the effect of increasing reaction rate (Fig. 6a, b). Gains in reaction efficiency were subject to rational limits on microdomain volume and particle behavior (Supplementary Fig. 8a, b). For example, at extremely low diffusion coefficients, particles became “stuck” in polar microdomains and, although highly concentrated, collided less often. Further, concentrating only one of the two reactants in polar microdomains had no beneficial effect on the reaction rate (Fig. 6b).
Fig. 6. Conceptual models of substrate proteolysis in membraneless polar microdomains.
a, b Three-dimensional reaction-diffusion simulations with two types of particles, colored red and yellow, that disappear after colliding. Snapshots of cells from the indicated time during simulation are shown at left, and the number of particles remaining over time under different parameter conditions are shown at right. a Effect of varying the size of the polar microdomain while holding the particle diffusion rate at 1/40th the rate in bulk cytoplasm. b Effect of varying particle diffusion rates within polar microdomains while holding polar microdomain size at 0.5% of total cell volume, or of limiting polar concentration to only one reactant (dotted line). c A model of C. crescentus proteolysis that includes low-affinity interactions (black arrows) between substrates (Sub1 and Sub2), adaptors, and ClpXP protease, in which some proteins interact directly with PopZ (red arrows) and become concentrated in polar microdomains. Charts show the fractions of substrate particles remaining in simulations run with or without PopZ, either without (left) or with (right) a direct interaction between Sub1 and PopZ. d Localization of CpdR and associated ClpXP complexes as a consequence of asymmetric CckA signaling activity. Inset panels show fluorescence images of a C. crescentus stalked cell, where mChy-PopZ is localized to both poles and CpdR-YFP is localized to only the stalked pole. Scale bar = 10 μm. Source data are provided as a Source Data file. Graphics: BioRender.
We also created a more complex model, with physiological concentrations of protease and adaptors (Fig. 6c and Supplementary Video 4). Basing our model on known interactions with PopZ12, particles representing CpdR and RcdA interacted directly with polar microdomains, and, following in vivo observations25, they were concentrated in this compartment by reducing their polar diffusion coefficients to 1/40th the cytoplasmic rate. ClpXP, which was modeled as a hexamer41 that could bind up to six molecules of CpdR, was recruited to polar microdomains indirectly (Supplementary Fig. 8c). We added two model substrates, Sub1 analogous to PdeA in that it interacted with CpdR-primed ClpXP complexes for degradation20, and Sub2 analogous to TacA in that it required both CpdR and RcdA as adaptors for ClpXP degradation23. All protein–protein interactions were given the same association and dissociation rates, which corresponded to low-affinity binding (KD = 100 μM).
With these parameters, 14.4% of the total pool of CpdR was concentrated in polar microdomains (Supplementary Fig. 8c), which approximates our in vivo observations (17.1 ± 4.1% of CpdR-YFP was concentrated in polar foci at the 15 min time point in Fig. 1a). This model mimicked the pattern of substrate degradation in C. crescentus swarmer cells in that the single-adaptor substrate was degraded earlier than the dual-adaptor substrate (Fig. 5b). This is a sensible outcome, since three-component assemblies (Sub1, CpdR, and ClpXP) are more likely to form than four-component assemblies (Sub2, RcdA, CpdR, and ClpXP), and the limiting population of ClpXP particles is predominantly occupied by the more frequently occurring substrate complex. We observed that the proteolysis rate of the single-adaptor substrate was not strongly influenced by PopZ, while the time to 75% proteolysis for the dual-adaptor substrate absence was more than doubled in the absence of PopZ. We also found that the sequential timing of substrate degradation was sharpened and the influence of PopZ on Sub1 more closely resembled our in vivo data on PdeA degradation (Fig. 5c) if the model included physical interaction between Sub1 and PopZ, although the evidence for physical interaction between PdeA and PopZ is inconclusive12. In conclusion, the simulations provide support for the idea that the assembly and activity of protease complexes can be enhanced when multiple components are concentrated within PopZ microdomains, and that this can occur on a size and time scale that is relevant to proteolysis in vivo.
Discussion
In this work, we show that CpdR’s interaction with pole-localized PopZ is closely correlated with its phosphorylation state (Figs. 1–3), that changing CpdR phosphorylation level through CckA signaling or the expression of the CpdRD51A variant is sufficient to alter the localization of CpdR and associated ClpXP complexes between diffuse and bi-polar (Fig. 4), and that the polar accumulation of ClpXP substrates is positively correlated with proteolysis (Fig. 5). Together, these results suggest that CpdR-mediated recruitment of ClpXP to polar PopZ subdomains stimulates proteolytic activity (Fig. 6).
This study shows that phosphorylation directly regulates a client protein’s interaction with the PopZ hub. An earlier study showed that a chromosome segregation protein, ParA, loses affinity to PopZ after hydrolyzing ATP42. Other studies have found that a ClpX adaptor, PopA, interacts with PopZ when it is bound to cyclic di-GMP43,44. CpdR has a more basal position than PopA in the adaptor hierarchy that determines ClpXP substrate specificity, meaning that it targets a substantially larger number of substrates, including those that are targeted by PopA23. A third adaptor, RcdA, interacts directly with PopZ in E. coli co-expression experiments but requires dephosphorylated CpdR (Fig. 4c) or c-di-GMP bound PopA45 for polar localization in C. crescentus, where protein expression levels are lower. We hypothesize that RcdA itself has a relatively low affinity to PopZ that is increased by associating with other PopZ-interacting proteins, forming multiprotein complexes with high avidity. All three ClpX adaptors’ interactions with PopZ are post-translationally regulated, suggesting that this is a key leverage point in the regulation of proteolysis and cell cycle progression.
The mechanisms that control whether CckA works as a kinase or phosphatase are driving forces behind the localization of CpdR and associated members of ClpXP protease complexes. In stalked cells, where PopZ exhibits bipolar localization and CpdR/ClpXP are exclusively at the stalked pole (Figs. 1g, 4c), the differential activity of CckA at opposite poles29 may be sufficient to support this aspect of polar asymmetry. However, CckA control may not be sufficient to explain all aspects of protease localization and activity. Whereas three different substrates were localized and degraded simultaneously in stalked cell progeny, PdeA was localized and degraded several minutes earlier than TacA and CtrA in swarmer cell progeny (Fig. 5b). Some timing differences could be related to the expression of additional regulatory factors during pole remodeling at the swarmer to stalk transition. CtrA proteolysis requires a polar di-guanylate cyclase to activate PopA34, and TacA proteolysis may require the phosphatase-stimulating activity of SpmY, which is also recruited to the transitioning pole46.
The term “polar localization” is used here and in most other reports to communicate the idea that a protein is visibly concentrated at the cell poles relative to the bulk cytoplasm. But this term is highly imprecise and potentially misleading, as a protein may exhibit “polar localization” when a small fraction of the total is at a pole. For example, since both cell poles occupy ~0.1% of the total cytoplasm volume, only 1% of a protein that is 10-fold concentrated at cell poles would be pole-localized. Indeed, fluorescence images of many different proteins in C. crescentus, including CpdR, substrates, and other members of the ClpXP complex, suggest low levels of actual polar localization. How could a system benefit from having such a localization pattern?
An answer to this question may lie in the fact that many pole-localized proteins receive regulatory cues at a cell pole and carry out their activity in bulk cytoplasm. CpdR is dephosphorylated by CckA at the stalked cell pole, yet its targets are transcriptional regulators, chemotaxis regulators, and other proteins that work outside of PopZ microdomains. Thus, dynamic exchange between pole and cytoplasm is required for translating polar signaling into cytoplasmic activity. This is consistent with the CpdR-PopZ interaction dynamics we observed in E. coli (Fig. 2d) and the behavior of other polar regulators in C. crescentus25,47.
Low levels of polar localization coupled with highly dynamic exchange can have a substantial influence on entire populations of molecules on the whole-cell scale (Fig. 6a, b). Applying this to adaptors, substrates, and ClpXP complexes, we propose that their concentration in PopZ microdomains increases the frequency of intermolecular collisions and therefore enhances multiprotein complex assembly and overall proteolysis rate (Fig. 6c, d). Faster ClpXP-dependent proteolysis could make the cell cycle more robust by sharpening phase changes at the swarmer-to-stalked transition.
Methods
Bacterial plasmid and strain construction
All plasmids were constructed by Gibson cloning method, and descriptions are provided in Supplementary Table 1. Bacterial strains are listed in Supplementary Table 2. Fluorescent proteins mCherry (mChy), msfGFP or eYFP are described in FPbase.org.
E. coli vectors
Among the two multiple cloning sites of pACYC-duet (Invitrogen); from site 1 we used NcoI and NotI restriction sites for cloning cpdR-gfp and from site 2 we used NdeI and XhoI restriction sites for cloning chpT. For both CpdR-GFP and ChpT, a GTG start codon was used to moderate protein expression. In pCDF-duet (Invitrogen), cckA variants were cloned at site 1 bearing NcoI and NotI restriction site. These cckA variants were PCR amplified, including the T7 promoter, and cloned at NheI restriction site upstream of p15A ori in pACYC-duet. In a separate pACYC-duet plasmid, cpdR-gfp-6xhis was cloned at site 1.
C. crescentus vectors
Plasmid pBXMCS2 was used for cloning cckA variants in between NdeI and EcoRI restriction sites. NdeI and KpnI restriction sites were used in pMCS5, pXMCS5 and pVMCS6 vectors48. pVMCS6 transformants were selected on the basis of exhibiting constitutive rather than vanillate-inducible GFP signal, indicating that expression was driven by the native promoter. pXMCS5 transformants were selected on the basis of xylose-inducibility. In the host strain, mChy-PopZ complements the severe ∆popZ mutant phenotypes in the popZ::mChy-popZ strain background7.
Bacterial cell culture
E. coli cells were grown at 30 °C overnight in Luria–Bertani (LB) liquid media or at 37 °C in LB agar plate supplemented with 1.5% agar. Liquid cultures were grown on rotor if not mentioned otherwise and all the growth of bacterial culture was measured by absorption OD600. For E. coli strains, antibiotics were used at following concentrations: 50 μg ml−1 ampicillin, 20 μg ml−1 chloramphenicol, 50 μg ml−1 spectinomycin, 12 μg ml−1 oxytetracycline, 30 μg ml−1 streptomycin, 30 μg ml−1 kanamycin.
C. crescentus was grown at 28 °C in PYE liquid media or PYE agar plates supplemented with 1.5% agar. For antibiotic-resistant strains, antibiotics were used at the following concentrations: 2 μg ml−1 chloramphenicol, 25 μg ml−1 spectinomycin, 1 μg ml−1 oxytetracycline, 5 μg ml−1 streptomycin, 5 μg ml−1 kanamycin. C. crescentus plasmids were transformed via electroporation or genes with associated antibiotic markers were transduced using phage φCr30. Prior to analysis, stationary phase cells were diluted 50× in fresh PYE and grown until they reached an OD600 = 0.3. Xylose-inducible genes were induced by 0.2% final concentration of d-xylose for 2 h unless mentioned otherwise. C. crescentus synchronies were performed according to Toro et al.49 but in PYE media at 28 °C using GB#228.
Wide-field microscopy
Cells were immobilized on a 1% agarose gel pad and viewed with a Zeiss Axio Imager Z2 epifluorescence microscope equipped with a Hamamatsu Orca-Flash 4.0 sCMOS camera and a Plan-Apochromat 100×/1.46 Oil Ph3 objective. Zen 2 (Blue Edition) software was used for image capture and quantification. For fluorescence imaging, mChy was observed by excitation at 587 nm and emission at 610 nm, GFP by excitation at 488 nm and emission at 509 nm, and YFP by excitation at 508 nm and emission at 524 nm. Exposure times for imaging mChy, GFP and YFP-tagged proteins were 500 ms, 500 ms and 1000 ms, respectively, except that 50 ms exposures were used to image phase condensates. All microscopy was performed at ×1000 magnification.
For image quantitation, a cell was counted as having a polar focus if the appropriate fluorescence channel, when overlayed over the corresponding phase contrast image, displayed a clearly distinguishable increase in fluorescence intensity in the group of pixels in the area of the cell pole relative to the adjacent region of the cytoplasm. To quantify the fluorescence intensity of CpdR-YFP foci relative to cytoplasm, the spline tool in Zeiss Zen Blue software was used to manually outline the area of the polar fluorescence focus of CpdR-YFP and the remainder of the cell body in the YFP channel image, and the circle tool was used to outline a neighboring region to measure local background fluorescence. After subtracting the background signal, the YFP signal of the polar focus was divided by the YFP signal of the whole cell (n = 20 in each of 2 biological replicates, with standard deviation showing differences between cells).
Phos-tag gel assays
Buffer composition
Buffer A
0.5 M Tris HCl, 1.25 ml of 0.4% SDS (pH 6.8), 100 μl IgePal CA- 630, 1.5 ml glycerol, up to 9.5 ml H2O.
Buffer B
920 μl Buffer A, 50 μl 2-mercaptoethanol, 10 μl EDTA free protease-phosphatase inhibitor cocktail (HaltTM, Thermo Fisher), 20 μl lysozyme and 0.2 μl nuclease (CelLyticTM B plus kit, Sigma–Aldrich).
Sample buffer 10X
0.5 M Tris HCl, 1.25 ml of 0.4% SDS (pH 6.8), 5 ml glycerol, 10 mg bromophenol blue, up to 10 ml H2O.
Cell lysis
1 ml cells of OD600 0.6–0.8 were centrifuged at 9000 × g 2 min, washed 2× with LB or PYE and resuspended in 100 μl of freshly prepared Buffer B. After Incubating on a roller/rotor for 10 min in a 4 °C cold room, samples were centrifuged at 16,000 × g for 5 min at 4 °C and the cleared supernatant was transferred to a new tube and kept on ice. After adding 10 μl of 10× sample buffer, 30 μl sample was loaded onto a prepared Phos-tag gel in a cold room at 4 °C.
Phos-tag gels
Ten to twelve percent gels were prepared according to manufacturer’s instructions (Wako) with the exception of adding 40 μM MnCl2 and 15 μM Phos-tag reagent as final concentration. The samples were run at 100 V in a 4 °C cold room until the dye front eluted from the gel. In-gel GFP or YFP fluorescence was imaged in a gel doc (Biorad ChemidocTM MP) at 488 nm wavelength. Band intensities on gels were quantified using ImageJ 1.54 f.
E. coli co-expression assay and fluorescence quantitation
Stationary phase E. coli cells bearing pACYC-duet and pBAD plasmids were diluted 100× in fresh LB and grown on rotor at 30 °C until an OD600 of 0.3 (strain GB#1971–1977, Supplementary Table 1). Cells were induced with 0.2% arabinose for 3 h and 10 μM IPTG for the final 2 h to express mChy-PopZ and CpdR-GFP + ChpT + CckA, respectively. Image capture and quantification was performed as described by Nordyke et al.19. Cells with zero mChy-PopZ foci or foci at both poles were excluded from the analysis. Background-subtracted pixel intensities for each channel were measured along a straight line drawn lengthwise through mid-cell. Cubic spline interpolation was used to generate fluorescence intensity values for 100 equally spaced points along each line, then all points were normalized to 1 prior to averaging.
Photobleaching
An Olympus IX-81 inverted confocal microscope equipped with a Yokogawa spinning disk (CSU X1) and a sCMOS camera (Orca- FLASH 4.0; Hamamatsu) was used for confocal microscopy. Excitation wavelengths were controlled using an acousto-optical tunable filter (ILE4; Spectral Applied Research) and a 405 nm FRAP laser was used with 100 mW nominal power. MetaMorph 7.7 software (MetaMorph Inc.) was used for image acquisition. FRAP time-lapse images were acquired using a 60×, 1.35 NA oil objective at 50 ms time intervals. To create bipolar mChy-PopZ containing cells, E. coli cultures were supplemented with 30 μg ml−1 cephalexin along with 0.1 mM IPTG and 0.2% arabinose at OD600 = 0.3, and incubated for 4 h at 30 °C. To plot Fluorescence Recovery After Photobleaching (FRAP) and Fluorescence Loss in Photobleaching (FLIP), time-lapse images of 20 different cells from each type were acquired. Fluorescence intensity of the photobleached area, unbleached area and background area were quantified using FIJI software for all the time points. Normalization of fluorescence was performed by subtracting the background signal and multiplying by the photobleaching coefficient. Photobleaching coefficients were obtained using a neighboring unbleached cell as a reference. To calculate the photobleaching coefficient for Tn time point of the reference cell, the total cell fluorescence signal from the initial time point T1 was divided by the total cell fluorescence signal from Tn.
NMR protein–protein interactions
The expression and purification of 15N-enriched PopZΔ134–177 protein was described previously12,19. For CpdR-GFP fusion protein and GFP expression, E. coli precultures were grown overnight from a single colony at 37 °C and 255 rotation-per-min (RPM) in LB with 30 μg/mL chloramphenicol or 100 μg/mL ampicillin, respectively. LB was inoculated with 0.4% (v/v) E. coli preculture, grown to mid-log phase, and then induced for 10 h at 37 °C and 255 RPM with isopropyl ß-D-1-thiogalactopyranoside (IPTG) for CpdR-GFP and with 0.2% (w/v) l-arabinose for GFP. Cells were harvested via centrifugation at 7000 × g for 20 min at 4 °C and stored at −80 °C until further use. Cell pellets were resuspended in buffer containing 20 mM HEPES, 100 mM KCl, 2 mM MgCl2, and 20 mM imidazole at pH 7.5, supplemented with HaltTM Protease Inhibitor Cocktail and Benzonase® Nuclease. After three passes through a French Press at 1750 PSI, cell debris was separated by centrifugation at 27,000 × g for 30 min at 4 °C and the supernatant was passed through a 0.45 μm and then a 0.22 μm syringe filter. CpdR-GFP and GFP were purified from the filtered lysate by nickel affinity chromatography (HisTrapTM HP, 1 ml column) using an AKTA fast protein liquid chromatography system, using 20 mM HEPES, 100 mM KCl, 2 mM MgCl2, and 20 mM imidazole as wash buffer and supplementing with 500 mM imidazole for elution. The eluate was dialyzed overnight into 50 mM KH2PO4, 20 mM NaCl, 5 mM EDTA at pH 6.5 for NMR experiments. Samples were concentrated using a 30 kDa molecular weight cutoff centrifugal filter (Amicon Ultra), and protein concentration was estimated via UV-Vis spectrophotometry. Sample purity was confirmed by 12% SDS-PAGE and visualization with Coomassie brilliant blue.
Transverse Relaxation Optimized Spectroscopy 2D 1H-15N Heteronuclear Single Quantum Correlation (2D 1H-15N TROSY-HSQC) spectra were acquired using a 700 MHz Avance NEO NMR system equipped with a triple resonance helium-cooled cryoprobe. To characterize the binding of CpdR-GFP to PopZΔ134–177, a series of samples were prepared of 50 μM 15N-enriched PopZΔ134–177 and natural abundance “unlabeled” CpdR-GFP fusion protein of varying concentrations (0, 125, 250, 375, 500, and 750 μM). The samples were supplemented with D2O, NaN3, and trimethylsilylpropanesulfonate to a final concentration of 5% (v/v), 4 mM, and 0.2 mM, respectively. Each 2D 1H-15N TROSY-HSQC was acquired with 64 scans. The NMR datasets were processed using the NMRPipe software50 and analyzed with the POKY software suite51. The PopZΔ134–177 NMR spectra were assigned previously19, thus amino acids were readily identified in the data. Combined 1H and 15N chemical shift perturbations (ΔHN) were calculated using the equation 1: , where ΔH and ΔN are the chemical shift perturbations in ppm for 1H and 15N, respectively, and 0.15 is a scaling factor corresponding to the relative chemical shift dispersion in the 1H and 15N dimensions. Intensity perturbations were calculated as percentages of the 0 µM CpdR titration point peak intensities. Binding residues were determined by comparing the combined ΔHN chemical shifts and peak intensity perturbations for each residue. A chemical shift change larger than one standard deviation, corresponding to a continuous stretch of amino acids, was considered indicative of participation in binding.
PopZ condensates and in vitro phosphorylation
To prepare proteins, PopZ, CpdR-GFP, CpdRD51A-GFP were expressed using E. coli strains GB#169, GB#1969 and GB#1970. Cells were grown overnight to stationary phase, then diluted 100× in 1 L LB and grown back to OD600 = 0.6 in a 37 °C shaking incubator at 200 RPM. After inducing with 1 mM IPTG for 6 h, cells were pelleted by centrifuging at 5000 × g for 30 min, resuspended in 20 ml HMK buffer (20 mM HEPES, 2 mM MgCl2, 100 mM KCl pH 7.5) and lysed using a french pressure cell press (Sim-aminco) at 1000 psi pressure. Proteins were purified using HisPur cobalt resin (Thermo Scientific) according to the product literature batch purification method. Purity was assessed by SDS-PAGE and Coomassie blue staining, and yields were quantified by BCA protein assay kit (Thermo Scientific). Purified proteins were preserved in 100 μl aliquots at −80 °C after flash freezing in liquid nitrogen.
In vitro phosphorylation was performed with the indicated concentration of Acetyl phosphate (AcP) in HMK buffer with 20 mM MgCl2 and at a final concentration of 100 μM CpdR-GFP. Samples were incubated at 37 °C for 6 h. To form PopZ condensates, 20 mM MgCl2 was added to 50 μM purified PopZ for 10 min at room temperature. Next, PopZ condensates were mixed with 5 μM CpdR-GFP or GFP, and 7 μl of this mixture was transferred to a glass slide and covered with a coverslip. Imaging was performed immediately thereafter using phase contrast and the YFP fluorescence channels, with 50 ms exposure time, at ×1000 magnification.
Condensates of varying size were analyzed using Zeiss Zen Blue software. The linescan tool was used to draw a 10 μm line through the middle of each condensate, which was then separated into 160 bins of equal length. The fluorescence intensity of the bright ring of CpdR-GFP at the periphery of the condensate was obtained by taking the median value of the brightest 10 bins. The fluorescence intensity of the inner area of the condensates appeared to be affected by out-of-plane fluorescence, particularly in small condensates, and was therefore omitted from the analyses. The fluorescence intensity of the unbound pool of CpdR-GFP molecules was taken as the median value of the bins from outside the condensate. After subtracting background signal (observed in mounts without CpdR-GFP), we calculated the ratio of the fluorescence intensity of the bright peripheral ring of CpdR-GFP to the fluorescence intensity of the unbound CpdR-GFP molecules in the surrounding medium. To account for sample thickness, which altered fluorescence intensity from the out-of-focus pool above and below the condensates, we used the formula , Where R is peripheral ring intensity, P is the local unbound pool intensity, and A is the average intensity of the unbound pool across all condensates in the sample set. The value for A differed by less than 22% between samples.
ClpXP substrate degradation (fluorescence and western blotting)
WT and ∆popZ C. crescentus strains, expressing HA tagged CtrA RD + 15 (GB#1989, GB#1992), TacA (GB#1990, GB#1993), PdeA (GB#1991, GB#1994) were induced at OD600 = 0.3 with d-xylose in 3 ml volume each for 2 h. Cells were washed 3 times by centrifugation and resuspension in fresh PYE, with final adjustment to ~3 ml at the same OD600. Over the subsequent 2 h of growth on a rotary shaker at 28 °C, 300 μl sample aliquots were harvested and centrifuged at 9000 × g for 2 min. The cell pellets were resuspended in 30 μl SDS-PAGE sample buffer, flash frozen in liquid nitrogen, and preserved at −80 °C. Samples were loaded in a 10% SDS-PAGE gel and resolved at 100 V. After transferring to a PVDF membrane (0.45 μm pore size for HA-TacA and HA-PdeA samples, 0.22 μm pore size for HA-CtrA RD + 15 samples), Western blots were developed using an anti-HA mouse monoclonal primary antibody (Invitrogen 2-2.2.14) at 1:3000. Similar analyses were performed on strains expressing YFP-tagged CtrA RD + 15 (GB#1983, GB#1986), TacA (GB#1984, GB#1987), PdeA (GB#1985, GB#1988). Where chloramphenicol treatment was included, chloramphenicol was added at 30 μg ml−1 after washing away the inducer, and cells were incubated for a further 5 min at 28 °C before initiating the time course. Band intensities on gels were quantified using ImageJ 1.54 f.
Fluorescence imaging was performed at 4 min time intervals. Fluorescence quantification was performed using FIJI plugin MicrobeJ 5.13i. We corrected the fluorescence values in C. crescentus time-lapse experiments to account for photobleaching. To do this, we induced YFP expression in E. coli cells from the pBAD promoter to a YFP fluorescence intensity level that was no more than 2-fold different than the C. crescentus cells we wished to analyze. Next, we imaged the E. coli cells under the same time-lapse conditions that were used to observe C. crescentus. The total fluorescence of 20 E. coli cells were observed over 20 consecutive time points and photobleaching coefficients were obtained by the method described in the previous section. Following background subtraction, to obtain fluorescence values for C. crescentus observations, YFP photobleaching coefficients corresponding to the appropriate time intervals were multiplied by the whole cell C. crescentus fluorescence values.
Computational simulation
Smoldyn 2.71 simulations were run on a High Performance Computing cluster. Smoldith diffusion coefficients oyn files are provided in Supplementary Data 1, and a summary of the simulation parameters is provided in Supplementary Tables 3 and 4.
Size scaling
1 size unit in Smoldyn equaled 10 nm in real space. Rod-shaped cells were created from cylinders and hemispheres, with length and volume that correspond to ~2.84 µm and 0.61 fL, respectively, consistent with measurements of stalked cells52. The volume of polar PopZ compartments has been measured by 3D super-resolution fluorescence imaging53. A mid-range value of 3.4 × 106 nm3 was chosen for standard cell models, which corresponds to ~0.56% of the total cytoplasmic volume.
Time scale and diffusion rates
The parameters for diffusion coefficients are based on single-particle tracking studies in C. crescentus cells25. ChpT and CtrA were found to move sub-diffusively in bulk cytoplasm, with diffusion coefficients of 1.8 µm2/sec, which translates to 18 units2/ms in the simulations. ChpT and CtrA were observed to continue to exhibit Brownian diffusion while co-localized with PopZ at cell poles, but at substantially slower rates of 0.1 µm2/s and 0.01 µm2/s, respectively (corresponding to 1 units2/ms and 0.1 units2/ms). In simulated cells, the diffusion coefficient of all particles in bulk cytoplasm was set to 20 units2/ms, and the standard rate for particles that interact with PopZ in polar microdomains was set to 0.5 units2/ms. For all simulations, the time step was 0.1 ms, which corresponds to a resolution of ~20 nm40.
Number of particles
In simple models with only two types of particles, the number of each was set to 2500. For the more complex model, global measurements of gene transcription and translation rates in C. crescentus cells provided an accurate estimate of the total number of each protein24. To approximate the number of proteins following cell division, the total amount of protein produced per cell in PYE media was divided by two and rounded to the nearest hundred. Thus, the number of ClpXP hexamers was set at 1000, CpdR at 7000, and RcdA at 2200. Since many different proteins are degraded by ClpXP and altogether there are thousands of individual substrates, the number of CpdR-dependent and RcdA plus CpdR-dependent substrates was arbitrarily set at 8,000 each.
Reaction rates and binding kinetics
Smoldyn represents proteins as point-like particles, and updates each particle’s position over iterative time steps, assigning a random direction and a distance calculated from its diffusion coefficient. Interactions between particles are determined by proximity. If two reactive particles are positioned within a defined binding radius at the end of a time step, those particles will interact. If they remain associated for a time, they will diffuse as a combined particle and the timing of separation is defined by an off-rate. Reaction rates and binding equilibria are therefore determined by the combined influences of binding radii, particle concentrations, diffusion rates, and off-rates. For the simple two-particle model, an empirical process was used to determine a binding radius of 0.025, which yielded a reaction rate that roughly matched the observed rate of protein degradation in C. crescentus cells (Fig. 4b). For the complex model, Smoldyn calculated the binding radii from user-defined values for on-rate and off-rates. We used the same values for all particle interactions (1 × 104 M−1s−1/0.166 px3 ms−1 for on-rate and 10 s−1/0.01 ms−1 for off-rate), which are well within the range of measured rate constants for dimeric complexes54 and correspond to a weak association with a KD of 100 micromolar. All proteins were treated as monomers except for ClpXP, which was modeled as a hexamer that could bind up to six CpdR and/or CpdR-associated particles. With these parameters, the binding radius was 1.6 nm, which is less than the radius of the proteins being modeled, meaning that that they will not interact every time they collide. This could reflect conformational and geometric constraints on productive collisions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
The authors would like to thank Steve Andrews for comments and advice, and Jack C. Sylvester for technical assistance. K.V. was supported by the National Science Foundation Major Research Instrumentation Award (DBI-1828319) and an Institutional Development Award (IDeA) [CIBBR, P20GM113131] from the National Institute of General Medical Sciences of the National Institutes of Health. G.B. was supported by the National Institutes of Health under award numbers 2P20GM103432 and R01GM118792, and by the National Science Foundation under award number 2225849.
Author contributions
Author contributions: Y.M.A., L.M.B., K.V., and G.R.B. designed research and analyzed data; Y.M.A., L.M.B., and G.R.B. performed research; Y.M.A., K.V., and G.R.B. wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The datasets generated and analyzed during the current study are available in the source data file, or, in the case of microscope images of experimental replicates, available from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53395-y.
References
- 1.Poindexter, J. S. Biological properties and classification of the Caulobacter group. Bacteriol. Rev.28, 231–295 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Teeseling, M. C. F. & Thanbichler, M. Generating asymmetry in a changing environment: Cell cycle regulation in dimorphic alphaproteobacteria. Biol. Chem. 401, 1349–1363 (2020). [DOI] [PubMed]
- 3.Aldridge, B. B. et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science335, 100–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyons, N. A. & Kolter, R. On the evolution of bacterial multicellularity. Curr. Opin. Microbiol.24, 21–28 (2015). [DOI] [PMC free article] [PubMed]
- 5.Shapiro, L. & Losick, R. Protein localization and cell fate in bacteria. Science274, 712–718 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Lybarger, S. R. & Maddock, J. R. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol.183, 3261–3267 (2001). [DOI] [PMC free article] [PubMed]
- 7.Bowman, G. R. et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell134, 945–955 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toro-Nahuelpan, M., Plitzko, J. M., Schüler, D. & Pfeiffer, D. In vivo architecture of the polar organizing protein Z (PopZ) meshwork in the Alphaproteobacteria Magnetospirillum gryphiswaldense and Caulobacter crescentus: Analysis of PopZ microdomain organization via cryo-ET. J. Mol. Biol.434, 167423 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Bowman, G. R. et al. Oligomerization and higher-order assembly contribute to sub-cellular localization of a bacterial scaffold. Mol. Microbiol.90, 776–795 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saurabh, S. et al. ATP-responsive biomolecular condensates tune bacterial kinase signaling. Sci. Adv.8. https://www.science.org (2022). [DOI] [PMC free article] [PubMed]
- 11.Lasker, K. et al. The material properties of a bacterial-derived biomolecular condensate tune biological function in natural and synthetic systems. Nat. Commun.13, 5643 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes, J. A. et al. Caulobacter PopZ forms an intrinsically disordered hub in organizing bacterial cell poles. Proc. Natl. Acad. Sci. USA113, 12490–12495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergé, M. et al. Modularity and determinants of a (bi-) polarization control system from freeliving and obligate intracellular bacteria. Elife5, e20640 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebersbach, G., Briegel, A., Jensen, G. J. & Jacobs-Wagner, C. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell134, 956–968 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lew, M. D. et al. Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus. Proc. Natl. Acad. Sci. USA108, E1102–E1110 (2011). [DOI] [PMC free article] [PubMed]
- 16.Radhakrishnan, S. K., Thanbichler, M. & Viollier, P. H. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev.22, 212–225 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iniesta, A. A., McGrath, P. T., Reisenauer, A., McAdams, H. H. & Shapiro, L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. USA103, 10935–10940 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGrath, P. T., Iniesta, A. A., Ryan, K. R., Shapiro, L. & McAdams, H. H. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell124, 535–547 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Nordyke, C. T., Ahmed, Y. M., Puterbaugh, R. Z., Bowman, G. R. & Varga, K. Intrinsically disordered bacterial polar organizing protein Z, PopZ, interacts with protein binding partners through an N-terminal molecular recognition feature. J. Mol. Biol.432, 6092–6107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau, J., Hernandez-Alicea, L., Vass, R. H. & Chien, P. A phosphosignaling adaptor primes the AAA+ protease ClpXP to drive cell cycle-regulated proteolysis. Mol. Cell59, 104–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi, K. K. & Chien, P. Regulated proteolysis in bacteria: Caulobacter. Ann. Rev. Genet.50, 423–445 (2016). [DOI] [PMC free article] [PubMed]
- 22.Smith, S. C. et al. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc. Natl. Acad. Sci. USA111, 14229–14234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshi, K. K., Bergé, M., Radhakrishnan, S. K., Viollier, P. H. & Chien, P. An adaptor hierarchy regulates proteolysis during a bacterial cell cycle. Cell163, 419–431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aretakis, J. R., Gega, A. & Schrader, J. M. Absolute measurements of mRNA translation in caulobacter crescentus reveal important fitness costs of vitamin B 12 scavenging. mSystems4, e00170–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasker, K. et al. Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat. Microbiol.5, 418–429 (2020). [DOI] [PMC free article] [PubMed]
- 26.Joshi, K. K., Battle, C. M. & Chien, P. Polar localization hub protein PopZ restrains adaptor-dependent ClpXP proteolysis in caulobacter crescentus. J. Bacteriol.200, e00221–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biondi, E. G. et al. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature444, e00221–18 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Mann, T. H. & Shapiro, L. Integration of cell cycle signals by multi-PAS domain kinases. Proc. Natl. Acad. Sci. USA115, E7166–E7173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, Y. E., Tsokos, C. G., Biondi, E. G., Perchuk, B. S. & Laub, M. T. Dynamics of two phosphorelays controlling cell cycle progression in Caulobacter crescentus. J. Bacteriol.191, 7417–7429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams, B., Bhat, N., Chien, P. & Shapiro, L. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol. Microbiol.93, 853–866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaBreck, C. J. et al. Degradation of MinD oscillator complexes by Escherichia coli ClpXP. J. Biol. Chem.296, 100162 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garber, M. E., Fregoso, R., Lake, J., Kakouridis, A. & Mukhopadhyay, A. Pseudomonas response regulators produced in an E. coli heterologous expression host exhibit host-derived post-translational phosphorylation. Sci. Rep.12, 10336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan, K. R., Judd, E. M. & Shapiro, L. The CtrA response regulator essential for Caulobacter crescentus cell-cycle progression requires a bipartite degradation signal for temporally controlled proteolysis. J. Mol. Biol.324, 443–455 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Abel, S. et al. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell43, 550–560 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Childers, W. S. et al. Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol.12, e1001979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chong, T. N., Panjalingam, M., Saurabh, S. & Shapiro, L. Phosphatase to kinase switch of a critical enzyme contributes to timing of cell differentiation. mBio15, e0212523 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, C., Zhao, W., Duvall, S. W., Kowallis, K. A. & Childers, W. S. Regulation of the activity of the bacterial histidine kinase PleC by the scaffolding protein PodJ. J. Biol. Chem.298, 101683 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Judd, E. M., Ryan, K. R., Moerner, W. E., Shapiro, L. & Mcadams, H. H. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc. Natl. Acad. Sci. USA100, 8235–8240. [DOI] [PMC free article] [PubMed]
- 39.Ryan, K. R., Huntwork, S. & Shapiro, L. Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc. Natl Acad. Sci. USA101, 7415–7420 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews, S. S. Smoldyn: Particle-based simulation with rule-based modeling, improved molecular interaction and a library interface. Bioinformatics33, 710–717 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Fei, X. et al. Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate. Elife9, e52774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ptacin, J. L. et al. Bacterial scaffold directs pole-specific centromere segregation. Proc. Natl. Acad. Sci. USA111, E2046–E2055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozaki, S. et al. Activation and polar sequestration of PopA, a c-di-GMP effector protein involved in Caulobacter crescentus cell cycle control. Mol. Microbiol.94, 580–594 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Wang, J., Moerner, W. E. & Shapiro, L. A localized adaptor protein performs distinct functions at the Caulobacter cell poles. Proc. Natl. Acad. Sci. USA.10.1073/pnas.2024705118 (2020). [DOI] [PMC free article] [PubMed]
- 45.Duerig, A. et al. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev.23, 93–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janakiraman, B., Mignolet, J., Narayanan, S., Viollier, P. H. & Radhakrishnan, S. K. In-phase oscillation of global regulons is orchestrated by a pole-specific organizer. Proc. Natl. Acad. Sci. USA113, 12550–12555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matroule, J.-Y., Lam, H., Burnette, D. T. & Jacobs-Wagner, C. Cytokinesis monitoring during development: rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell118, 579–590 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Thanbichler, M., Iniesta, A. A. & Shapiro, L. A comprehensive set of plasmids for vanillate—and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res.35, e137 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toro, E., Hong, S.-H., Mcadams, H. H. & Shapiro, L. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc. Natl. Acad. Sci. USA105, 15435–15440 (2008). [DOI] [PMC free article] [PubMed]
- 50.Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Lee, W., Rahimi, M., Lee, Y. & Chiu, A. POKY: a software suite for multidimensional NMR and 3D structure calculation of biomolecules. Bioinformatics37, 3041–3042 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campos, M. et al. A constant size extension drives bacterial cell size homeostasis. Cell159, 1433–1446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gahlmann, A. et al. Quantitative multicolor subdiffraction imaging of bacterial protein ultrastructures in three dimensions. Nano Lett.13, 987–993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shammas, S. L., Rogers, J. M., Hill, S. A. & Clarke, J. Slow, reversible, coupled folding and binding of the spectrin tetramerization domain. Biophys. J.103, 2203–2214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
The datasets generated and analyzed during the current study are available in the source data file, or, in the case of microscope images of experimental replicates, available from the corresponding author upon request. Source data are provided with this paper.