Abstract
Protein phosphatase 2A (PP2A) catalytic subunit can be covalently modified at its carboxy terminus by phosphorylation or carboxymethylation. Determining the effects of these covalent modifications on the relative amounts and functions of different PP2A heterotrimers is essential to understanding how these modifications regulate PP2A-controlled cellular processes. In this study we have validated and used a novel in vivo assay for assessing PP2A heterotrimer formation in Saccharomyces cerevisiae: the measurement of heterotrimer-dependent localization of green fluorescent protein-PP2A subunits. This assay relies on the fact that the correct cellular localization of PP2A requires that it be fully assembled. Thus, reduced localization would occur as the result of the inability to assemble a stable heterotrimer. Using this assay, we determined the effects of PP2A C-subunit phosphorylation mimic mutations and reduction or loss of PP2A methylation on the formation and localization of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers. Collectively, our findings demonstrate that phosphorylation and methylation of the PP2A catalytic subunit can influence its function both by regulating the total amount of specific PP2A heterotrimers within a cell and by altering the relative proportions of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers up to 10-fold. Thus, these posttranslational modifications allow flexible, yet highly coordinated, regulation of PP2A-dependent signaling pathways that in turn modulate cell growth and function.
Protein phosphatase 2A (PP2A) is a major eukaryotic serine/threonine phosphatase that is highly conserved from yeast to human. PP2A activity has been functionally linked to a variety of cellular processes including cell cycle progression, DNA replication, transcription, RNA splicing, and translation (9, 17, 35). PP2A is able to participate in such a wide array of processes because of its inherent variability. It usually exists as a heterotrimer composed of a structural (A) subunit, a regulatory/targeting (B-type) subunit, and a catalytic (C) subunit (6, 14, 15, 28). In the budding yeast Saccharomyces cerevisiae, the structural A subunit is encoded by one gene, TPD3 (33, 34). The catalytic C subunits are encoded by two highly related genes, PPH21 and PPH22, which appear to be functionally redundant (22, 29), and there are single representatives of only two classes of B-type subunits: the B class, encoded by CDC55 (8), and the B′ class, encoded by RTS1 (26, 27). In contrast, five different classes of B-type subunits have been reported in mammals, with each class possessing multiple isoforms (9). Four are encoded by the cellular genome, designated PR55 (B), PR61 (B′), PR72 (B"), and PR93/110 (striatin family; putative B‴), while one can be provided by viral genomes (9, 21). Thus, PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers represent multimeric forms of PP2A conserved from yeast to mammals.
Posttranslational modification of PP2A subunits is an important means by which cells regulate PP2A activity and function. The catalytic subunits can be regulated by at least three modifications: methylation of the conserved C-terminal leucine (L369 in Pph21p), phosphorylation of a conserved tyrosine two residues from the C terminus (Y367 in Pph21p), and phosphorylation of an unidentified threonine (2, 7, 9, 19, 20, 32, 36-38 and references therein). Recently we and others have begun to dissect the effects of these modifications on the assembly of PP2AB/Cdc55p and PP2AB'/Rts1p heterotrimers. C-subunit methylation enhances the stable formation of PP2AB′/Rts1p heterotrimers in yeast (36) and has been reported to be required for PP2AB′ heterotrimer formation in mammalian cells based on experiments in vitro (32). Methylation is also necessary for efficient formation of PP2AB/Cdc55p heterotrimers in both yeast and mammalian cells (32, 36-38). In addition, mutations that mimic phosphorylation of the catalytic subunit inhibit stable formation of PP2AB/Cdc55p heterotrimers in both yeast and mammalian cells (19, 20, 36), suggesting that phosphorylation inhibits the formation of PP2AB/Cdc55p heterotrimers.
Despite progress in understanding the regulation of these two PP2A heterotrimers by covalent modifications, key questions remain unanswered due to limitations of current assays. For example, we could not quantitate the importance of methylation for stable formation of PP2AB′/Rts1p heterotrimers due to the instability of Rts1p in lysates (36). Thus, it is not known whether there is a similar reduction in both PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers or a differential reduction that alters their relative amounts. Furthermore, due to similar technical reasons, we were unable to determine if mutations mimicking phosphorylation of the catalytic subunit affect stable PP2AB′/Rts1p heterotrimer formation (36). Thus, no data exist for either yeast or mammalian cells as to whether there is differential regulation of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers by phosphorylation or methylation.
Another important aspect of the control of PP2AB′/Rts1p and PP2ACdc55 heterotrimer formation is the fact that in S. cerevisiae, the cellular concentration of Rts1p is approximately 12 times that of Cdc55p (6). Furthermore, the A subunit, Tpd3p, is limiting for trimer formation, since the total amount of B-type subunits (Rts1p plus Cdc55p) is fourfold greater than the amount of Tpd3p (6). Thus, competition for B-type subunit binding to the AC dimer must occur. Modifications of the C subunit that stabilize or destabilize B-subunit binding to the AC dimer would clearly have an effect on this competition, especially if these effects were different for the two B subunits. However, because of the limitations of coimmunoprecipitation assays for analyzing Rts1p complexes, some other way of assessing in vivo associations of the altered C subunits with the other PP2A subunits was required to investigate the relative effects of covalent modifications on the formation of stable PP2A heterotrimers.
In this study, we present the details of a novel in vivo assay based on heterotrimer-dependent localization of green fluorescent protein (GFP)-PP2A subunits, its validation, and its use to assess the in vivo efficiency of PP2A heterotrimer formation in cells expressing Pph21p C-terminal mutants or lacking PP2A methylation. Our results both validate this assay and confirm our previous findings that methylation, and likely phosphorylation, of catalytic-subunit C-terminal residues regulates PP2AB/Cdc55p heterotrimer formation. Moreover, they also establish for the first time the extent to which these Pph21p mutants or loss of methylation affect the stable formation of PP2AB′/Rts1p heterotrimers in vivo. Finally, they reveal a new role for the PP2A methyltransferase, Ppm1p, in regulating intracellular targeting of Cdc55p to the bud tip.
MATERIALS AND METHODS
Strains, plasmids, and media.
All strains used are derivatives of W303 and are listed in Table 1. Plasmids used are listed in Table 2. The ppm1Δ strains were generated by crossing MSG66, MSG107, and MSG167 (6) with BY4741 ppm1::KAN (Research Genetics). Each strain was backcrossed with the parental W303 strain five times. Standard protocols were used for yeast growth and media (24) and for recombinant DNA methodologies (25). For microscopic visualization of fluorescently tagged proteins, cells were grown in YPD (1% yeast extract, 2% bactopeptone, 2% glucose) or synthetic medium supplemented with the appropriate amino acids and 2% glucose and then washed and viewed in synthetic medium with the appropriate amino acids and 2% glucose. Yeast transformations were performed using lithium acetate as described previously (11).
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| CY1145 | MATaade2-1 trp1-1 leu2-3,112 pph21::URA3 pph22::HIS | 16 |
| MSG129 | MATα ade2-1 trp1-1 leu2-3,112 pph21::URA3 pph22::HIS3 RTS1-GFP | 6 |
| MSG139 | MATα ade2-1 trp1-1 leu2-3,112 pph21::URA3 pph22::HIS3 GFP-TPD3 | 6 |
| MSG152 | MATα ade2-1 trp1-1 leu2-3,112 pph21::URA3 pph22::HIS3 GFP-CDC55 | 6 |
| MSG261 | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 ppm1::KAN GFP-TPD3 | This study |
| MSG248 | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 ppm1::KAN RTS1-GFP | This study |
| MSG257 | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 ppm1::KAN GFP-CDC55 | This study |
TABLE 2.
Plasmids
| Name | Description | Source or reference |
|---|---|---|
| CY1678 | PPH21, CEN vector/TRP1 | 16 |
| CY1624 | HA-PPH21, CEN vector/TRP1 | 4 |
| pMG528 | PPM1, CEN vector/TRP1 | This study |
| pRS 314-T364A | PPH21 (T364A), CEN vector/TRP1 | 36 |
| pRS 314-T364D | PPH21 (T364D), CEN vector/TRP1 | 36 |
| pRS 314-Y367F | PPH21 (Y367F), CEN vector/TRP1 | 36 |
| pRS 314-Y367E | PPH21 (Y367E), CEN vector/TRP1 | 36 |
| pRS 314-L369Δ | PPH21 (L369Δ), CEN vector/TRP1 | 36 |
| pMG424 (pRS314-L369A) | PPH21 (L369A), CEN vector/TRP1 | This study |
pMG528 was constructed by PCR amplifying a 3.0-kb fragment of PPM1, including 1.4 kb upstream and 0.6 kb downstream from the open reading frame, from W303 genomic DNA. This PCR product was digested with SacI and XbaI and ligated into YCp22.
pMG424 was constructed via QuickChange mutagenesis (Stratagene, La Jolla, CA) using CY1678 as the template and the following primers: GACGCCAGATTACTTTGCCTGAGTATGTATAC and GTATACATACTCAGGCAAAGTAATCTGGCGTC (base changes are shown in bold). This method introduced the leucine-to-alanine change and introduced a novel DdeI site. pMG433 was generated exactly as was pMG424, except that CY1624 was used as the template. The other PPH21 mutant plasmids were previously described (36).
Microscopic analysis and quantification of localization.
In Results, we present differences obtained by quantitating the percentage of cells with proper localization of GFP-PP2A subunits. Fluorescence signal in all cells was visualized on a BX60 microscope (Olympus, Tokyo, Japan) using an enhanced green fluorescent protein filter set (Chroma Technology, Brattleboro, VT). Fields of representative cells were captured with a charge-coupled device (CCD) camera (Olympus) with Magnafire software (Olympus) and analyzed using Photoshop (Adobe Systems, Mountain View, CA). For each field, cells in the appropriate cell cycle stage (see Results) were scored as having or lacking the type of localization in question. The percentage of localization of GFP-Cdc55p, Rts1p-GFP, and GFP-Tpd3p at the bud tip and bud neck in cells expressing PPH21 was standardized to 100%. The percentages of localization in cells expressing PPH21 mutants were corrected accordingly. The percentages of cells exhibiting localization and standard deviation were determined using Excel (Microsoft, Redmond, CA). We counted a minimum of 200 cells/experiment and performed each experiment multiple times. Initial experiments were performed blind. Some subsequent experiments were not performed blind, because differences were obvious. Moreover, some counts were repeated by multiple people, and results differed by no more than five percentage points. To support the validity of this method, we used some of the same captured fields of cells and measured the intensity of the signal at the bud tip and bud neck of all cells at the appropriate cell cycle stage using NIH Image J. We did not use this method to measure the fluorescence intensity at the kinetochore, because unlike the bud tip and bud neck, the kinetochore is not a defined landmark visible in all cells. The results obtained were very similar (data not shown), demonstrating the validity of our method.
Protein isolation, electrophoresis, and Western analysis.
The extraction of total proteins by solubilizing cells in 1.8 M NaOH-5% β-mercaptoethanol or by lysis with glass beads, the separation of proteins by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and procedures used for Western analysis have been previously described (26, 36). Protein extracts containing equal amounts of protein as assayed by a Bio-Rad protein assay kit were separated using 10% SDS-PAGE. An affinity-purified rabbit polyclonal antibody specific for Pph21p and Pph22p was used to visualize wild-type and mutant Pph21p proteins in cell lysates. This antibody was raised (Proteintech, Inc.) against two Pph21p, Pph22p-specific peptides (EVDENHNRQFLQYDPSVRPGE and KPGSSGIADHKSSKPLE) cross-linked to keyhole limpet hemocyanin via an added amino-terminal cysteine residue using a Pierce Imject conjugation kit. This antibody recognized yeast C subunits in lysates and did not visualize a similar band in a PPH21/PPH22 double-knockout strain (see Fig. 2). Protein concentrations were visualized using horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch) and enhanced chemiluminescence. The amounts of the various C subunits were quantitated using a Bio-Rad Fluor S-Max chemilumimager that directly measures band intensities without the use of film via a supercooled CCD camera that provides linear data over 4.8 orders of magnitude. This method yielded highly reproducible results that did not vary with image capture times. For each experiment, the amounts of the mutant Pph21p proteins were normalized to the amount of wild-type Pph21p and expressed as a percentage. The mean and standard deviation of results for three separate colonies are presented.
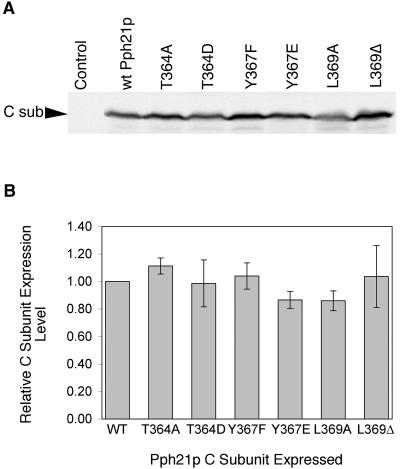
FIG. 2.
Relative expression levels of wild-type and mutant Pph21p. (A) MSG139 cells expressing no exogenous Pph21p/Pph22p (Control) and MSG139 cells expressing exogenously introduced wild-type Pph21p or one of the various Pph21p mutants were grown in SD (Trp−) medium at 30°C to early log phase (optical density at 600 nm, ≈1.0). Cells were lysed, and equal amounts of total protein were analyzed by SDS-PAGE, transferred to nitrocellulose, and then immunoblotted with affinity-purified rabbit anti-Pph21p/Pph22p polyclonal antibody (DP79/80) to allow comparison of C subunit (C sub) expression levels. (B) The C-subunit levels in three different colonies of wild-type and mutant Pph21p-expressing cells were quantitated using a Bio-Rad Fluor S-Max chemilumimager that directly measures band intensities without the use of film via a supercooled CCD camera that provides linear data over 4.8 orders of magnitude. This method yielded highly reproducible results that did not vary with image capture times. The expression levels of the mutant Pph21p C subunits are shown relative to that of wild-type C subunit. Error bars reflect ± standard deviation.
RESULTS
PP2A subunit localization as an in vivo assay for heterotrimer formation.
An idea for a novel quantitative assay for heterotrimer formation arose from our recent studies defining the dynamics and rules governing PP2A subunit localization in S. cerevisiae using PP2A subunits fused to the GFP (6). We found that PP2AB′/Rts1p heterotrimers and PP2AB/Cdc55p heterotrimers have specific localizations within cells that can be easily visualized by fluorescence microscopy (Fig. 1). Importantly, each of these heterotrimeric PP2A species has a unique localization that can be monitored to evaluate proper heterotrimer formation and localization. PP2AB′/Rts1p heterotrimers, but not PP2AB/Cdc55p heterotrimers, localize to the kinetochore of small budded cells (6). Conversely, PP2AB/Cdc55p heterotrimers, but not PP2AB′/Rts1p heterotrimers, localize to the bud tip of small and medium budded cells (6). Proper localization of GFP-Tpd3p and Rts1p-GFP is completely dependent on heterotrimer formation, while GFP-Cdc55p can maintain substantial localization independently of its assembly into a heterotrimer (6). Thus, decreased formation of stable PP2AB′/Rts1p heterotrimers will cause a proportionate decrease in localization of GFP-Tpd3p and Rts1p-GFP to the kinetochore. Decreased formation of PP2AB/Cdc55p heterotrimers, on the other hand, will result in a proportionate decrease in GFP-Tpd3p localization to the bud tip and a lesser decrease in GFP-Cdc55p to the same location. Therefore, quantified GFP-Tpd3p localization at the kinetochore and bud tip can be used as an assay to measure the formation of stable PP2AB′/Rts1p or PP2AB/Cdc55p heterotrimers. Localization of Rts1p-GFP and GFP-Cdc55p can be further used to confirm the GFP-Tpd3p localization results.
FIG. 1.
Schematic showing localization of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers to bud tip, bud neck, and kinetochore. Arrows indicate localization of the two different heterotrimers to specific and common subcellular sites. The subunit composition of each heterotrimer is shown in parentheses.
As described below, we also assayed localization of GFP-PP2A subunits to the bud neck of postmitotic cells, where both PP2AB′/Rts1p and PP2AB/Cdc55p heterotrimers localize (6). This provided a second subcellular site for assaying localization of both PP2AB′/Rts1p and PP2AB/Cdc55p heterotrimers. Similar results obtained for the same heterotrimer at two distinct subcellular locations strengthened the likelihood that the observed effects were on heterotrimer formation rather than on a more specific mechanism, such as targeting.
To assess the levels of cell cycle-dependent localization of PP2A subunits, cells from asynchronous cultures were divided into four classes based on cell cycle morphology: nonbudded, small/medium budded, large budded with one nucleus (pretelophase), and large budded with two nuclei (posttelophase). Each class was scored for either having or lacking a characteristic localization pattern of a particular GFP-PP2A subunit. Table 3 summarizes the various GFP-PP2A subunits followed in this study. The percentage of cells in each class showing the trademark localization pattern of that subunit was then calculated. This methodology gave us a quantified level of GFP-PP2A localization that reflects the level of stable PP2A heterotrimers. For ease of comparison, the relative localization levels were expressed as percentages of wild-type control levels. We previously found that analyzing asynchronous cultures in this manner gives results similar to those from determining subunit localization patterns from synchronous cultures (6). Moreover, an advantage of assaying asynchronous populations is that it allowed us to rule out the possibility that reduced localization was simply due to alterations in cell cycle timing of PP2A heterotrimer localizations rather than reduced heterotrimer formation. Finally, we also quantified the pixel intensity of localized GFP subunits for a subset of cells using NIH Image J (data not shown; see Material and Methods). We found that these results were in excellent agreement with our quantitation method based on counting cells with characteristic localization; thus, no bias or subjectivity was present in our counts. Furthermore, given that the pixel intensity quantitation method was very tedious and did not offer any additional insight over our more straightforward and simple method, we conducted all other experiments using our approach. As seen below and in Table 4, our results strongly support the validity of this assay for measuring effects on the formation and proper localization of stable PP2A heterotrimers in vivo.
TABLE 3.
Summary of the various GFP-PP2A subunits in this study, their localizations, and which PP2A heterotrimer they monitor
| GFP-PP2A subunit | Subcellular location | PP2A heterotrimer responsible for this localization | % Localization if no stable heterotrimer is formed |
|---|---|---|---|
| GFP-Tpd3p | Bud tip | PP2AB/Cdc55p | 0 |
| GFP-Tpd3p | Bud neck | PP2AB/Cdc55p and PP2AB′/Rts1p | 0 |
| GFP-Tpd3p | Kinetochore | PP2AB′/Rts1p | 0 |
| Rts1p-GFP | Bud neck | PP2AB′/Rts1p | 0 |
| Rts1p-GFP | Kinetochore | PP2AB′/Rts1p | 0 |
| GFP-Cdc55p | Bud tip | PP2AB/Cdc55p | ≈30-40 (see Fig. 3C) |
| GFP-Cdc55p | Bud neck | PP2AB/Cdc55p | ≈30-40 (see Fig. 5B) |
TABLE 4.
Summary of previous and current data on PP2A C-subunit mutants
| Mammalian
|
S. cerevisiae
|
|||||
|---|---|---|---|---|---|---|
| Mutation in C subunita | Methylation level (%)a | B-subunit bindinga | C subunit (Pph21p)b | Binding of Cdc55p (co-IP)b | Binding of Cdc55p (in vivo assay)c | Binding of Rts1p (in vivo assay)d |
| None (309 aa)e | 94 ± 3 | +++ | WT (369 aa)e | +++ | +++ | +++ |
| T304A | 98 ± 0.4 | ++++ | T364A | ++ | ++ | +++ |
| T304D | 75 ± 11 | − | T364D | − | − | +++ |
| Y307F | 43 ± 18 | ++++ | Y367F | +++ | ++ | ++ |
| Y307E | ≤5 | − | Y367E | − | − | − |
| L309Af | Reduced | Reduced | L369A | NT | + | ++ |
| L309Δ | 0 | − | L369Δ | NT | − | − |
Data for mammalian C-subunit mutants except the L309A mutant are taken from reference 38. C-subunit mutations are generally referred to by the position mutated preceded by the wt amino acid and followed by the introduced residue. Abbreviations for the amino acid residues are as follows: A, Ala; D, Asp; E, Glu; F, Phe; L, Leu; T, Thr, and Y, Tyr; Δ, deletion of residue. ++++, more than wild type level; +++, 100 to 71% of wild type level; −, <15% of wild-type level.
Data for yeast C-subunit coimmunoprecipitations (co-IP) taken from reference 36. C-subunit mutants are referred to as described in footnote a above. +++, >50% of wild-type level; ++, 49 to 25% of wild-type level; +, 24 to 10% of wild-type level; −, <10% of wild-type level in at least two of three localization assays; NT, not tested.
Wild-type mammalian C subunit has 309 amino acids (aa), whereas wild-type yeast has 369.
The Pph21p mutants used in this study are expressed at similar levels.
To validate our assay and gain additional insights into the role of phosphorylation and methylation in regulating PP2A heterotrimer formation and function, we first chose to examine the effect on the localization of PP2AB/Cdc55p heterotrimers of a set of carboxy-terminal mutants designed to mimic phosphorylation and/or block phosphorylation or methylation. The last six carboxy-terminal PP2A C-subunit residues, TPDYFL, are conserved from yeast to human. This sequence contains known and putative phosphorylation sites and the site of methylation, the carboxy-terminal leucine. Table 4 shows the S. cerevisiae Pph21p mutants used in both the previous and current studies, the analogous mammalian PP2A C-subunit mutants, and a brief summary of the previously reported effects of these mutations on methylation of the C subunit (mammalian cells only) and the formation of stable B subunit (Cdc55p)-containing heterotrimers. Specifically, the mutants were generated by mutation of a threonine residue (T364 in Pph21p) and a tyrosine residue (Y367 in Pph21p) near the carboxy terminus or by mutation or deletion of the carboxy-terminal leucine (L369 in Pph21p). The threonine was mutated to a conserved alanine (T364A) to prevent phosphorylation or to an aspartate (T364D) to mimic phosphorylation, while the tyrosine was changed to a conserved phenylalanine (Y367F) or a nonconserved glutamate (Y367E) for the same reasons. The C-terminal leucine was mutated to alanine (L369A) to reduce methylation (1) or was deleted (L369Δ) to abolish methylation (38).
Because proper PP2A localizations depend upon the cellular stoichiometry of the wild-type and mutant C subunits (M. S. Gentry and R. L. Hallberg, unpublished results), we first measured the levels of the various wild-type and mutant Pph21p proteins. Each of the gene constructs used contained the normal PPH21 promoter and was carried on a CEN vector. Western analysis (Fig. 2) of these different proteins showed that each Pph21p mutant was expressed at a level similar to that of wild-type Pph21p expressed from the same vector. This was true even for Pph21p(L369Δ), which was previously reported not to be detectable via Western analysis when it was hemagglutinin tagged (36), indicating that the presence of the N-terminal hemagglutinin epitope tag uniquely destabilizes this mutant. Thus, our data indicate normal stability for all the mutant proteins studied.
Pph21p mutants mimicking phosphorylation or blocking methylation greatly inhibit PP2AB/Cdc55p heterotrimer formation and localization to the bud tip.
To help validate our assay, we analyzed the T364A, T364D, Y367F, and Y367E PP2A C-subunit mutants for stable PP2AB/Cdc55p heterotrimer formation, because these same mutants had been assayed previously for stable PP2AB/Cdc55p heterotrimer formation by a coimmunoprecipitation assay (Table 4). Largely because of technical limitations of coimmunoprecipitation assays, the L369A and L369Δ C-subunit mutants (Table 4) had not been previously assayed in yeast for stable PP2AB/Cdc55p heterotrimer formation. Therefore, we also analyzed these two mutants to determine their effect on the formation of stable PP2AB/Cdc55p heterotrimers.
We assayed PP2AB/Cdc55p heterotrimer bud tip localization in wild-type or mutant Pph21p-expressing MSG139 and MSG152 strains in which the endogenous TPD3 or CDC55 gene had been respectively replaced with a gene encoding either a GFP-Tpd3p or a GFP-Cdc55p fusion protein. These strains are deleted for both PPH21 and PPH22, and thus PP2AB/Cdc55p heterotrimer formation and localization depend solely on the competence of plasmid-expressed Pph21p proteins.
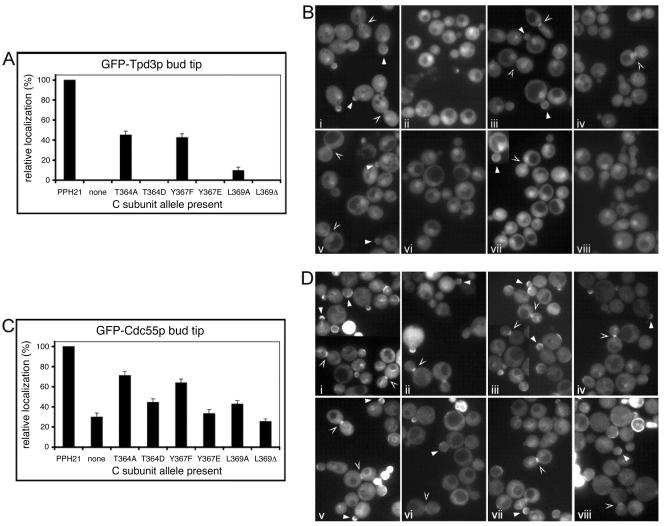
The results shown in Fig. 3A and B show that the percentage of cells with Cdc55p-mediated GFP-Tpd3p bud tip localization was dramatically decreased for cells expressing pph21 (L369A) (to ∼10% of wild-type) and abolished in cells expressing pph21 (T364D), pph21 (Y367E), or pph21 (L369Δ). Only the conserved pph21 (T364A) and pph21 (Y367F) mutants supported substantial levels (40 to 45% of wild type) of localization of GFP-Tpd3p to the bud tip (Fig. 3A). These results parallel those obtained previously using coimmunoprecipitation of PP2A complexes from S. cerevisiae and mammalian cell extracts (Table 4), indicating that our in vivo complex formation assay was functioning as we had predicted. In addition, they reveal effects of the C-subunit mutations not previously measurable, such as the absolute requirement for a residue at position 369 in Pph21p for PP2AB/Cdc55p heterotrimer formation in yeast and the fact that the L369A C subunit still retains the ability to form some stable PP2AB/Cdc55p heterotrimers.
FIG. 3.
PP2AB/Cdc55p bud tip localization. GFP-Tpd3p and GFP-Cdc55p bud tip localizations were visualized in MSG139 and MSG152, respectively. Cells were grown at 30°C to early log phase. Multiple fields of cells were examined and photographs taken of each field. Cells were divided into four classes: nonbudded, small/medium budded, large budded with one nucleus, and large budded with two nuclei (determined by 4′,6′-diamidino-2-phenylindole). Cells in each class were then scored as having or lacking the trademark localization in question. In this and subsequent figures, arrows denote kinetochore localization; closed arrowheads denote bud tip localization; and open arrowheads denote bud neck localization. Some panels contain cells from more than one field from the same experiment. (A) Quantified GFP-Tpd3p bud tip localization. (B) MSG139 (GFP-Tpd3p) cells expressing PPH21, no C subunit (none), or PPH21 mutants. i, PPH21 from CY1678; ii, empty pRS314 vector; iii, PPH21 (T364A) from pRS314-T364A; iv, PPH21 (T364D) from pRS314-T364D; v, PPH21 (Y367F) from pRS314-Y367F; vi, PPH21 (Y367E) from pRS314-Y367E; vii, PPH21 (L369A) from pMG424; viii, PPH21 (L369Δ) from pRS314-L369Δ. (C) Quantified GFP-Cdc55p bud tip localization. (D) MSG152 (Cdc55p-GFP) cells expressing PPH21, no C subunit (none), or PPH21 mutants. i, PPH21 from CY1678; ii, empty pRS314 vector; iii, PPH21 (T364A) from pRS314-T364A; iv, PPH21 (T364D) from pRS314-T364D; v, PPH21 (Y367F) from pRS314-Y367F; vi, PPH21 (Y367E) from pRS314-Y367E; vii, PPH21 (L369A) from pMG424; viii, PPH21 (L369Δ) from pRS314-L369Δ.
Since formation of PP2AB/Cdc55p heterotrimers increases the localization of GFP-Cdc55p to the bud tip (6), the extent of GFP-Cdc55p localization provides an additional tool to evaluate the level of stable PP2AB/Cdc55p heterotrimers in cells. Therefore, to confirm our results obtained with GFP-Tpd3p, we examined GFP-Cdc55p localization at the bud tip in cells expressing the various Pph21p mutants. Since GFP-Cdc55p can still localize to some extent when heterotrimer formation is prevented (6), we expected at most to see a partial reduction in the percentage of cells exhibiting GFP-Cdc55p bud tip localization due to expression of Pph21p mutants. Indeed, 29% of control cells lacking any Pph21p protein still showed some Cdc55p bud tip localization (Fig. 3C, second column), indicating that the maximum reduction in localization we should expect to see with any mutant would be approximately 70%. Consistent with the Tpd3p localization results, the cells expressing pph21 (T364D), pph21 (Y367E), pph21 (L369A), and pph21 (L369Δ) did decrease GFP-Cdc55p localization to the bud tip by ∼60 to 75% (Fig. 3C). Also consistent with the Tpd3p results, cells expressing pph21 (T364A) and pph21 (Y367F) displayed a nearly wild-type localization of GFP-Cdc55p to the bud tip (Fig. 3C).
Taken together, these results validate the use of our localization assay for measuring the level of stable PP2A heterotrimer formation in yeast cells and indicate that Cdc55p-directed PP2A localization and thus the formation of PP2AB/Cdc55p heterotrimers is dramatically decreased in cells expressing Pph21p C-terminal mutants that mimic phosphorylation, inhibit methylation, or lack an amino acid at position 369. Conversely, conserved mutations that do not mimic but should instead block phosphorylation do not abolish localization, indicating that phosphorylation at these sites is not necessary for proper PP2AB/Cdc55p heterotrimer formation or localization.
Mutations that mimic phosphorylation at position 364 or alter leucine 369 affect localization of PP2AB′/Rts1p heterotrimers much less than they affect localization of PP2AB/Cdc55p heterotrimers.
It is not known whether phosphorylation and methylation of the PP2A catalytic subunit affect the in vivo levels of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers to the same extent or whether there are differences between the effects on these two heterotrimers. This knowledge is critical to understanding the relative effect of phosphorylation and methylation on different PP2A pathways. The relative effect on these two heterotrimeric PP2A forms could not be determined previously because the instability of Rts1p in yeast lysates precluded the use of a coimmunoprecipitation assay to determine the abilities of the various mutants to support stable PP2AB′/Rts1p heterotrimer formation or to quantitate the importance of PP2A methylation for assembly of PP2AB′/Rts1p heterotrimers (36). We therefore used our localization assay to analyze the abilities of our set of carboxy-terminal Pph21p mutants to form stable PP2AB′/Rts1p heterotrimers that localize properly in vivo. Since Tpd3p and Rts1p are completely interdependent in establishing and/or maintaining their kinetochore localization (Table 3) (6), we used both GFP-Tpd3p and Rts1p-GFP localization at the kinetochore as an in vivo assay to monitor the formation of stable PP2AB′/Rts1p heterotrimers. For these assays, MSG139 and MSG129 cells in which the endogenous TPD3 or RTS1 genes have been respectively replaced with genes encoding a GFP-Tpd3p or Rts1p-GFP fusion protein were used. Of note, the MSG139 (GFP-Tpd3p) cells are the same cells in which Cdc55p-directed Tpd3p localization was assayed; therefore, the relative effects of each individual mutant on Cdc55p and Rts1p localization should be most readily compared in this strain, since the levels of mutant Pph21p expression and conditions are identical.
The results of the analysis are shown in Fig. 4A and B. Uniformly, the percent localization obtained by quantitating Tpd3p was always strikingly similar to that obtained by quantitating Rts1p localization. This similarity strengthens confidence in the results, especially given that these values were obtained from two separate strains expressing two different GFP-fusion proteins. As expected, control cells lacking any Pph21p expression (Fig. 4A, “none'”) showed no localization of either Rts1p or Tpd3p to the kinetochore.
FIG. 4.
PP2AB′/Rts1p kinetochore localization. All cells were treated and analyzed as described in the legend to Fig. 3. GFP-Tpd3p and Rts1p-GFP kinetochore localizations were visualized in MSG139 and MSG129, respectively. (A) Quantified GFP-Tpd3p and Rts1p-GFP kinetochore localizations. The black bars represent GFP-Tpd3p kinetochore localization. The white bars represent Rts1p-GFP kinetochore localization. (B) MSG129 cells expressing PPH21, no C subunit (none), or PPH21 mutants. i, PPH21 from CY1678; ii, empty pRS314 vector; iii, PPH21 (T364A) from pRS314-T364A; iv, PPH21 (T364D) from pRS314-T364D; v, PPH21 (Y367F) from pRS314-Y367F; vi, PPH21 (Y367E) from pRS314-Y367E; vii, PPH21 (L369A) from pMG424; viii, PPH21 (L369Δ) from pRS314-L369Δ. Some panels contain cells from more than one field from the same experiment. Symbols are as defined in the legend to Fig. 3.
The Pph21p mutants, on the other hand, varied in their ability to support the proper localization of Rts1p-containing heterotrimers. As observed for Cdc55p heterotrimer localization to the bud tip, pph21 (T364A) and pph21 (Y367F) both supported substantial PP2AB′/Rts1p heterotrimer localization to the kinetochore, indicating that phosphorylation of these residues is not necessary for PP2AB′/Rts1p heterotrimer formation and localization. However, the amount of PP2AB′/Rts1p heterotrimer localization for pph21 (T364A) was higher than that for PP2AB/Cdc55p, indicating that PP2AB/Cdc55p heterotrimer formation is more sensitive than PP2AB′/Rts1p heterotrimer formation to conservative changes at position 364. As observed for PP2AB/Cdc55p heterotrimers, mutation of tyrosine 367 to mimic phosphorylation (pph21 [Y367E]) or deletion of the carboxy-terminal leucine (pph21 [L369Δ]) abolished PP2AB′/Rts1p heterotrimer formation and localization. Thus, PP2AB′/Rts1p heterotrimer formation and localization are also likely regulated by phosphorylation of tyrosine 367 and require an intact amino acid at position 369. Interestingly, while pph21 (L369A) supported only a small amount (<10%) of Cdc55p localization to the bud tip, it supports substantially more (∼40%) localization of PP2AB′/Rts1p holoenzyme to the kinetochore, providing the first evidence that stable formation of PP2AB′/Rts1p heterotrimers may be less dependent on C-subunit methylation than is stable formation of PP2AB/Cdc55p heterotrimers. Finally, the most striking difference found was with the phosphorylation mimic mutant at position 364, pph21 (T364D). Although this mutant supported no PP2AB/Cdc55p heterotrimer formation and localization (Fig. 3A, T364D), it allowed substantial PP2AB′/Rts1p stable heterotrimer formation (almost 60% of the wild-type level) (Fig. 4A). This result suggests that phosphorylation at this position would not greatly affect PP2AB′/Rts1p heterotrimer formation but will have a strong differential effect on the relative levels of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers.
PP2A bud neck localization in cells expressing Pph21p mutants confirms bud tip and kinetochore results.
Our bud tip and kinetochore results provide the first evidence of a differential regulation of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimer formation by phosphorylation and methylation. To further investigate differential regulation of these heterotrimers, we examined the effects of our mutants on Tpd3p, Rts1p, and Cdc55p localization at the bud neck, a subcellular localization where both PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers are found (6).
For this assay, the most straightforward result to interpret is that of Rts1p-GFP, because its localization is completely dependent on PP2AB′/Rts1p heterotrimer formation (Fig. 5A, compare PPH21 to “none”). The levels of Rts1p-GFP localization at the bud neck in cells expressing the different mutants parallel well the levels of Rts1p-GFP localization to the kinetochore in the same cells (compare Fig. 5A with Fig. 4A). The only exception is that pph21 (T364D) shows a greater (∼75%) reduction in PP2AB′/Rts1p heterotrimer localization to the bud neck than to the kinetochore (∼40%), suggesting that phosphorylation of this residue would have a differential effect on PP2AB′/Rts1p heterotrimer localization to different subcellular locations.
FIG. 5.
PP2AB′/Rts1p and PP2AB/Cdc55p bud neck localizations. All cells were treated and analyzed as described in the legend to Fig. 3. GFP-Tpd3p, Rts1p-GFP, and GFP-Cdc55p bud neck localizations were visualized in MSG139, MSG129, and MSG152 cells, respectively. (A) Quantified Rts1p-GFP bud neck localization. (B) Quantified GFP-Cdc55p bud neck localization. (C) Quantified GFP-Tpd3p bud neck localization.
For Cdc55p localization to the bud neck, results with the mutants (Fig. 5B) must be compared to the control level of GFP-Cdc55p at the bud neck in the absence of PP2AB/Cdc55p heterotrimer formation (Fig. 5B, “none”). In agreement with our bud tip localization results, pph21 (T364A) and pph21 (Y367F) supported substantial additional localization of PP2AB/Cdc55p heterotrimers to the bud neck, while pph21 (Y367E), pph21 (L369Δ), and pph21 (T364D) abolished formation of these heterotrimers (Fig. 5B). Moreover, comparison of GFP-Cdc55p (Fig. 5B) and Rts1p-GFP (Fig. 5A) bud neck localization provides further evidence for differential regulation of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimer formation: in cells expressing pph21 (T364D), PP2AB/Cdc55p heterotrimer bud neck localization was abolished (i.e., it was the same as in control cells lacking any heterotrimers; compare the T364D and “none” columns in Fig. 5B), while PP2AB′/Rts1p heterotrimers retained >20% of their wild-type level of bud tip localization (compare the T364D and “none” columns in Fig. 5A).
GFP-Tpd3p localization to the bud neck is completely dependent on heterotrimer formation (Fig. 5C, “none”), but its presence at the bud neck represents the contribution of both PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers. GFP-Tpd3p bud neck localization was consistent with the Rts1p-GFP and GFP-Cdc55p results: no localization of GFP-Tpd3p to the bud neck was seen with Pph21p mutants pph21 (Y367E) and pph21 (L369Δ), which cannot form either PP2AB/Cdc55p or PP2AB′/Rts1p heterotrimers (Fig. 5C), while substantial localization was seen with mutants that supported both PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimer formation. Finally, as expected, GFP-Tpd3p localization of mutants that supported no PP2AB/Cdc55p heterotrimer localization to the bud neck (pph21 [T364D] and pph21 [L369A]) mirrored Rts1p-GFP localization to the bud neck (Fig. 5C).
Taken as a whole, these data corroborate our bud tip and kinetochore localization results. The major localization results with the various Pph21p mutants are summarized in the last two columns of Table 4. Comparison of the effects of these mutants on stable formation of PP2AB/Cdc55p heterotrimers (Table 4, next-to-last column) and on PP2AB′/Rts1p heterotrimers (Table 4, last column) supports the idea that phosphorylation of T364 would differentially regulate these two types of heterotrimers while phosphorylation of Y367 would simultaneously reduce them both. Moreover, the data in Table 4 indicate that, like Y367, L369 is essential for the stable formation of both types of heterotrimers. Finally, the differential effect of L369A on stable formation of the PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers (Table 4) suggests, but does not prove, that there may also be a differential effect of methylation on PP2A heterotrimer formation in yeast.
PP2AB′/Rts1p and PP2AB/Cdc55p heterotrimer levels are reduced to different degrees in ppm1Δ cells lacking C-subunit methylation.
The results we obtained above with L369A suggested that methylation may differentially regulate the formation of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers. To test this more directly, we conducted experiments in which we modulated PP2A methylation without mutation of the PP2A catalytic subunit. In yeast, Ppm1p is at least the primary, and probably the only, methyltransferase modifying the catalytic subunits of PP2A (12, 36, 37). PP2A AC dimers, PP2ACdc55 heterotrimers, and PP2AB′/Rts1p heterotrimers are all destabilized to some degree in cells lacking catalytic subunit methylation due to disruption of PPM1 (36, 37). However, as mentioned previously, we were unable to quantitate the extent to which the formation of stable PP2AB′/Rts1p heterotrimers was affected in ppm1Δ cells (36) and thus could not compare the relative importance of methylation for the formation of stable PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers. We therefore used our localization assay to address this important question by deleting PPM1 in strains expressing the various GFP-PP2A subunit fusion proteins and scoring cells as having or lacking characteristic GFP-PP2A subunit localization patterns.
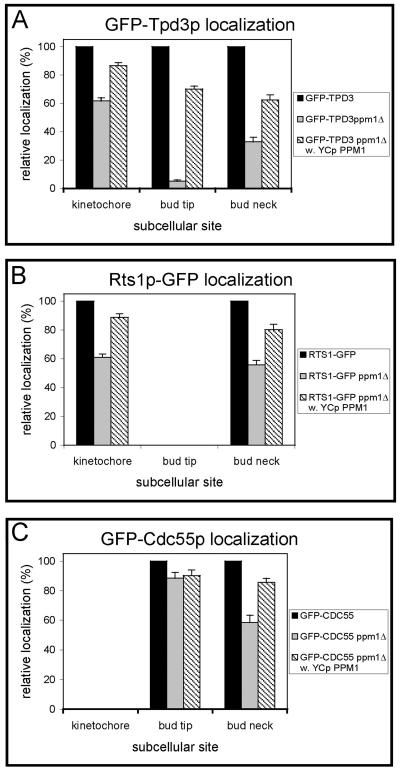
We first examined GFP-Tpd3p localization in ppm1Δ cells (MSG261). Most strikingly, GFP-Tpd3p localization to the bud tip decreased by 20-fold (95%) in ppm1Δ cells (Fig. 6A), indicating that PP2A methylation is required for stable PP2AB/Cdc55p heterotrimer formation. On the other hand, the percentage of cells with GFP-Tpd3p kinetochore localization decreased by less than twofold (40%) in ppm1Δ cells compared to the level in wild-type cells (Fig. 6A), indicating that PP2A methylation enhances, but is not absolutely required for, stable PP2AB′/Rts1p heterotrimer formation. Similarly, GFP-Tpd3p bud neck localization decreased by three- to fourfold (70%) in ppm1Δ cells (Fig. 6A), reflecting a decrease in both types of heterotrimers. When PPM1 expressed from its endogenous promoter on a CEN plasmid was reintroduced into ppm1Δ cells, GFP-Tpd3p localizations returned to near-wild-type levels (Fig. 6A, striped bars). This demonstrates that the defects seen are due to loss of Ppm1p function. These data establish for the first time that Rts1p-directed GFP-Tpd3p localization is only moderately reduced in ppm1Δ cells lacking PP2A methylation, while Cdc55p-directed GFP-Tpd3p localization is nearly abolished.
FIG. 6.
PP2AB′/Rts1p and PP2AB/Cdc55p localizations in ppm1Δ cells. All cells were treated and analyzed as described in the legend to Fig. 3. GFP-Tpd3p, Rts1p-GFP, and GFP-Cdc55p bud neck localizations were visualized in MSG261, MSG248, and MSG257 cells, respectively. (A) Quantified GFP-Tpd3p kinetochore, bud tip, and bud neck localizations. (B) Quantified Rts1p-GFP kinetochore and bud neck localizations. (C) Quantified GFP-Cdc55p bud tip and bud neck localizations.
We next examined Rts1p-GFP localization in ppm1Δ cells (MSG248). Rts1p-GFP localization at the kinetochore and bud neck decreased less than two-fold (40 to 45%) in ppm1Δ cells versus the level in wild-type cells (Fig. 6B). As expected, Rts1p-GFP localization levels appeared very similar to that of GFP-Tpd3p at the kinetochore (Fig. 6A, gray bar). As with GFP-Tpd3p localization, when PPM1 carried on a CEN plasmid was introduced into ppm1Δ cells, Rts1p-GFP localizations returned to near-wild-type levels (Fig. 6A, striped bars).
Finally, we examined GFP-Cdc55p localization in ppm1Δ cells (MSG257), which lack PP2A methylation (36). Since GFP-Cdc55p can localize independently of heterotrimer formation, we expected only a partial inhibition of GFP-Cdc55p localization in ppm1Δ cells. Based on the fact that loss of PPM1 has nearly the same effect as the T364D mutant on the formation of PP2AB/Cdc55p heterotrimers, we expected about a twofold decrease in GFP-Cdc55p localization to both the bud neck and the bud tip. Consistent with this prediction, GFP-Cdc55p localization to the bud neck was reduced by 42% in ppm1Δ cells compared to the level in wild-type cells (Fig. 6C). PPM1 carried on a CEN plasmid largely restored the loss of GFP-Cdc55p localization to the bud neck seen in ppm1Δ cells (Fig. 6C, striped bar). Surprisingly, however, GFP-Cdc55p localization to the bud tip was reduced by less than 10% in ppm1Δ cells instead of the expected twofold decrease. Thus, while PP2A methylation is essential for efficient formation and proper localization of stable PP2AB/Cdc55p heterotrimers and enhances Cdc55p localization to the bud neck, it is not required for efficient localization of Cdc55p to the bud tip. Moreover, loss of PPM1 actually enhances Cdc55p localization to the bud tip in the absence of substantial heterotrimer formation, indicating a novel role for Ppm1p in regulating Cdc55p bud tip localization.
DISCUSSION
In order to understand how cells regulate the diverse functions of PP2A through covalent modification, it is necessary to understand the relative effects that these modifications have on different holoenzyme forms of PP2A. Different PP2A heterotrimers share the catalytic (Pph21p/Pph22p) and structural (Tpd3p) subunits, and so covalent modifications of the catalytic subunit may affect B-type regulatory subunit associations directly or may affect them indirectly because of competition for these shared subunits (6). In this study we have described and used a novel assay to monitor PP2A heterotrimer formation and function: measurement of heterotrimer-dependent localization of GFP-PP2A subunits. Some advantages of this novel assay over in vitro assays are, first, that it avoids artifactual association of subunits that might have been compartmentalized in vivo; second, that it circumvents instability issues with Rts1p that occur upon cell lysis; third, that it yields well-controlled comparative data for effects on PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimer formation in the same cells; and fourth, that it measures not only the formation of specific PP2A complexes but also their ability to localize properly.
Effects of mutation and loss of methylation on PP2AB/Cdc55p heterotrimer formation and localization.
The results of the current study both confirm and extend our earlier findings indicating that methylation, and probably phosphorylation, regulates the stable formation of PP2AB/Cdc55p heterotrimers. In the present study, we have shown for the first time that mutation or loss of the Pph21p carboxy-terminal leucine, the site of methylation resulted in a near or complete loss, respectively, of PP2AB/Cdc55p heterotrimer formation in vivo. These results, which are consistent with results obtained using analogous mutants in mammalian cells (Table 4) or in Pph22p (5), show that the carboxy-terminal Pph21p leucine is also critical in yeast Pph21p for obtaining stable PP2AB/Cdc55p heterotrimers. Furthermore, our in vivo results with ppm1Δ cells confirm previous in vitro results from our lab and another group showing that PP2A methylation is essential for efficient formation of PP2AB/Cdc55p heterotrimers (36, 37). Moreover, we extend these findings by showing effects on in vivo localization of these heterotrimers.
Results from previous coimmunoprecipitation analysis of phosphorylation mimic mutants were vulnerable to the possibility that low-affinity in vivo associations might not survive the in vitro processing necessary for that approach. However, our current results clearly show that there is no detectable functional PP2AB/Cdc55p heterotrimer formation in vivo in cells expressing phosphorylation mimic Pph21p mutants. Thus, phosphorylation is not required for PP2AB/Cdc55p heterotrimer assembly but rather is predicted to block it. Phosphorylation at tyrosine 367 may reduce PP2AB/Cdc55p heterotrimer formation in large part indirectly by affecting methylation, given that acidic substitution of this residue in mammalian PP2A abolishes methylation (38). However, Y367E is completely defective for PP2AB/Cdc55p heterotrimer localization in yeast, while loss of methylation caused by deletion of PPM1 leaves a small but significant amount of residual heterotrimer formation. Deletion of leucine 369 also completely ablates PP2AB/Cdc55p heterotrimer formation instead of leaving some residual heterotrimer formation. The more-severe effects of Y367E and L369Δ compared to loss of methylation suggest that these mutations have effects on PP2AB/Cdc55p heterotrimer formation that are independent of methylation. These results are consistent with the idea that both Y367 and L369 may make essential contacts with Cdc55p during PP2AB/Cdc55p heterotrimer formation. A direct test of this hypothesis will likely await crystallization of PP2AB/Cdc55p heterotrimers.
A surprising observation from this study was that Ppm1p plays a role in regulating Cdc55p localization to the bud tip that is independent of heterotrimer formation. Although a previous study found an effect of Rts1p on Cdc55p localization (6), differences in Rts1p heterotrimer levels cannot explain our current result, because mutants like the T364D mutant have a reduction in stable PP2AB′/Rts1p heterotrimers similar to that of ppm1Δ cells yet do not have near-wild-type levels of GFP-Cdc55p localization. More likely, an additional, unknown mechanism explains this observation. Although the data are not sufficient to determine presently what this mechanism is, they suggest that Ppm1p contributes either directly or indirectly to reducing Cdc55p localization to the bud tip and that deletion of PPM1 relieves this inhibition. This function is not via Pph21p and Pph22p, because their deletion greatly reduces Cdc55p bud tip localization. Although other possible explanations exist, we favor the possibility that the effect may be mediated via an alternate substrate of Ppm1p, perhaps another PP2A-like phosphatase.
Effects of mutation and loss of methylation on PP2AB′/Rts1p heterotrimer formation and localization.
Our results demonstrate that PP2AB′/Rts1p heterotrimer formation and localization are affected by methylation and likely also by phosphorylation. Using our in vivo assay, we show for the first time that loss of PP2A methylation results in less than a twofold reduction in stable PP2AB′/Rts1p heterotrimers in vivo. Thus, methylation provides the cell with a mechanism to modulate the formation and function of PP2AB′/Rts1p heterotrimers, but it is not necessary for substantial PP2AB′/Rts1p heterotrimer formation. This finding seems to be in contrast to previous results from in vitro approaches (32). Tolstykh and colleagues found that purified B′ heterotrimers contained only methylated C subunits, suggesting that methylation was very important for the formation of these B′ heterotrimers. We suggest that the difference between our in vivo result and their in vitro result is probably due to loss of lower-affinity PP2A complexes during lysis and purification of PP2A heterotrimers. Our results clearly show that in vivo, substantial PP2AB′/Rts1p heterotrimer formation can occur in the absence of methylation.
While loss of methylation reduced PP2AB′/Rts1p heterotrimer formation only less than twofold, loss of leucine 369 abolished it, indicating that this residue directly or indirectly functions in PP2AB′/Rts1p heterotrimer formation independently of methylation. This result is important because it indicates that carboxy-terminal leucine C-subunit deletion mutants cannot be used experimentally to determine the importance of PP2A methylation for PP2A function (as has previously been done). Interestingly, while the phosphorylation mimic mutant T364D only mildly affected PP2AB′/Rts1p heterotrimer formation, a phosphorylation mimic mutant only three residues away (Y367E) abolished Rts1p localization to the kinetochore and the bud neck. It is intriguing that both tyrosine 367 and leucine 369 are required for binding of Cdc55p and Rts1p and for stable methylation, especially in light of the fact that these same residues are conserved with related phosphatases, such as PP4, which is also methylated (13), and Sit4p, which has been reported to bind the A and B subunits of PP2A (18).
Differential effects of PP2A methylation and phosphorylation on formation of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers.
An exciting finding of this study is that methylation regulates the formation of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers differently in S. cerevisiae. These differential effects may well be one of the ways in which regulatory subunit binding to the A subunit is controlled in the presence of competing excess B-type subunits. The stable formation of both trimeric forms of PP2A is enhanced by methylation; however, methylation to normal levels (∼60%) (36) enhances stable formation of PP2AB/Cdc55p heterotrimers 20-fold, while it enhances stable formation of PP2AB′/Rts1p heterotrimers only less than twofold. Thus, cells have the capacity via changes in methylation not only to reduce or enhance the levels of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers but also to cause at least a 10-fold shift in the relative amounts of PP2AB/Cdc55p and PP2AB′/Rts1p trimeric complexes. Previous in vitro results from several different laboratories, including our own, gave no indication that methylation or phosphorylation might affect PP2AB/Cdc55p heterotrimers more severely than PP2AB′/Rts1p heterotrimers. Our new data show for the first time the differential effects of methylation, and likely phosphorylation, on these two PP2A B-type subunit families.
Because of the high conservation of PP2A subunits and their regulation, yeast has been a useful model system for gaining insights into PP2A regulation in mammalian cells. Alterations in PP2A activity and PP2A substrates have been found in both Alzheimer's disease and cancer (10, 31). PP2AB heterotrimers are a major source of Tau phosphatase activity in the brain (30) and are the only form of PP2A targeted by E4orf4, an adenovirus protein capable of selectively killing a variety of cancer cells in a PP2A-dependent manner (23). The results in this report provide important evidence that disruption of PP2A methylation strongly targets PP2A ABC heterotrimer formation while only modestly affecting AB′C heterotrimer formation. Together with previous data, this indicates that drugs modulating PP2A methylation would represent the most selective means available to date for regulating PP2A trimers containing the B subunit. Thus, our current results have important implications for the possibility of modulating specific PP2A functions by targeting the PP2A methyltransferase and methylesterase enzymes with drugs, including possible applications in Alzheimer's and cancer therapies.
Differential effects of PP2A methylation on formation and function of other PP2A complexes.
In addition to the positive effects of PP2A methylation on the stable formation of PP2AB/Cdc55p and PP2AB′/Rts1p heterotrimers, there is evidence for positive effects of demethylation on the formation of other PP2A complexes. Loss of PP2A methylation due to deletion of PPM1 increases the amount of the TOR signaling protein, Tap42p, bound to Pph21p and Pph22p (37), and the mammalian homolog of Tap42p, α4, demonstrates increased binding in vivo to a mutant that lacks methylation (3, 38). Moreover, striatin family (putative B‴) PP2A complexes show increased binding in vivo to C-subunit mutants that are deficient in methylation (38). Interestingly, the Pph21p mutants (Y367E and L369Δ) shown in this study to be the most defective in binding both Cdc55p (B subunit) and Rts1p (B′ subunit) correspond to the two mammalian PP2A C-subunit mutants (Y307E and L309Δ) that show the most enhanced (10- to 15-fold) complex formation with striatin family members (38). Thus, our new data support the idea that demethylation- or phosphorylation-induced disassembly of B and B′ complexes results in an increased effective concentration of C subunit that enhances complex formation with striatin family members and perhaps Tap42p/α4. Overall, methylation likely increases and decreases the formation of a wide variety of PP2A complexes in a differential manner in multiple species. Together, these alterations in amounts and proportions of different PP2A complexes would result in a coordinated change in multiple phenotypes that in turn affect cell growth and function.
In summary, although coimmunoprecipitation assays are useful for studying aspects of PP2A complex formation in vivo, they do not reproduce the actual conditions in a cell. The amounts of the various PP2A complexes in vivo at any given time are a function of a combination of relative B-type subunit affinities for A/C heterodimers and local available PP2A subunit concentrations. These factors in turn can be regulated dynamically by events such as modification of the C subunit and subcellular compartmentalization. The in vivo assay we validated and used in this study for analysis of PP2A complexes provides a means to evaluate the results of the combined effects of these complex variables. This assay should be very useful as well in the future for elucidating other mechanisms by which cells regulate PP2A complex formation and function.
Acknowledgments
This work was supported by a National Institutes of Health Grant to D.C.P. (CA57327) and an NSF Grant to R.L.H. (MCB-0113355).
We thank Liz Hallberg for excellent technical assistance, the SU/UMU yeast club for helpful comments, and Anita Corbett, Courtney Plattner, Monica McQuoid, Carlos Moreno, Steven Moody, and Sankhavaram Panini for critical reading of the manuscript.
Under agreements between Upstate Biotechnology, Inc., Santa Cruz Biotechnologies, Stratagene, Inc., and Emory University, David Pallas is entitled to a share of sales royalty received by the university from these companies. In addition, this author serves as a consultant to Upstate Biotechnology, Inc. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict-of-interest policies.
REFERENCES
- 1.Bryant, J. C., R. S. Westphal, and B. E. Wadzinski. 1999. Methylated C-terminal leucine residue of PP2A catalytic subunit is important for binding of regulatory Balpha subunit. Biochem. J. 339:241-246. [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, J., B. L. Martin, and D. L. Brautigan. 1992. Regulation of protein serine-threonine phosphatase type-2A by tyrosine phosphorylation. Science 257:1261-1264. [DOI] [PubMed] [Google Scholar]
- 3.Chung, H., A. C. Nairn, K. Murata, and D. L. Brautigan. 1999. Mutation of Tyr307 and Leu309 in the protein phosphatase 2A catalytic subunit favors association with the alpha 4 subunit which promotes dephosphorylation of elongation factor-2. Biochemistry 38:10371-10376. [DOI] [PubMed] [Google Scholar]
- 4.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10:1904-1916. [DOI] [PubMed] [Google Scholar]
- 5.Evans, D. R., and B. A. Hemmings. 2000. Mutation of the C-terminal leucine residue of PP2Ac inhibits PR55/B subunit binding and confers supersensitivity to microtubule destabilization in Saccharomyces cerevisiae. Mol. Gen. Genet. 264:425-432. [DOI] [PubMed] [Google Scholar]
- 6.Gentry, M. S., and R. L. Hallberg. 2002. Localization of Saccharomyces cerevisiae protein phosphatase 2A subunits throughout mitotic cell cycle. Mol. Biol. Cell 13:3477-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, H., and Z. Damuni. 1993. Autophosphorylation-activated protein kinase phosphorylates and inactivates protein phosphatase 2A. Proc. Natl. Acad. Sci. USA 90:2500-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Healy, A. M., S. Zolnierowicz, A. E. Stapleton, M. Goebl, A. A. DePaoli-Roach, and J. R. Pringle. 1991. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 11:5767-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janssens, V., and J. Goris. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353:417-439.11171037 [Google Scholar]
- 10.Janssens, V., J. Goris, and C. Van Hoof. 2005. PP2A: the expected tumor suppressor. Curr. Opin. Genet Dev. 15:34-41. [DOI] [PubMed] [Google Scholar]
- 11.Kaiser, S., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics, p. 234. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Kalhor, H. R., K. Luk, A. Ramos, P. Zobel-Thropp, and S. Clarke. 2001. Protein phosphatase methyltransferase 1 (Ppm1p) is the sole activity responsible for modification of the major forms of protein phosphatase 2A in yeast. Arch. Biochem. Biophys. 395:239-245. [DOI] [PubMed] [Google Scholar]
- 13.Kloeker, S., J. C. Bryant, S. Strack, R. J. Colbran, and B. E. Wadzinski. 1997. Carboxymethylation of nuclear protein serine/threonine phosphatase X. Biochem. J. 327:481-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kremmer, E., K. Ohst, J. Kiefer, N. Brewis, and G. Walter. 1997. Separation of PP2A core enzyme and holoenzyme with monoclonal antibodies against the regulatory A subunit: abundant expression of both forms in cells. Mol. Cell. Biol. 17:1692-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, X., A. Scuderi, A. Letsou, and D. M. Virshup. 2002. B56-associated protein phosphatase 2A is required for survival and protects from apoptosis in Drosophila melanogaster. Mol. Cell. Biol. 22:3674-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, F. C., and K. T. Arndt. 1995. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 14:2745-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millward, T. A., S. Zolnierowicz, and B. A. Hemmings. 1999. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem. Sci. 24:186-191. [DOI] [PubMed] [Google Scholar]
- 18.Nickels, J. T., and J. R. Broach. 1996. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 10:382-394. [DOI] [PubMed] [Google Scholar]
- 19.Ogris, E., D. M. Gibson, and D. C. Pallas. 1997. Protein phosphatase 2A subunit assembly: the catalytic subunit carboxy terminus is important for binding cellular B subunit but not polyomavirus middle tumor antigen. Oncogene 15:911-917. [DOI] [PubMed] [Google Scholar]
- 20.Ogris, E., I. Mudrak, E. Mak, D. Gibson, and D. C. Pallas. 1999. Catalytically inactive protein phosphatase 2A can bind to polyomavirus middle tumor antigen and support complex formation with pp60(c-src). J. Virol. 73:7390-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pallas, D. C., L. K. Shahrik, B. L. Martin, S. Jaspers, T. B. Miller, D. L. Brautigan, and T. M. Roberts. 1990. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell 60:167-176. [DOI] [PubMed] [Google Scholar]
- 22.Ronne, H., M. Carlberg, G. Z. Hu, and J. O. Nehlin. 1991. Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol. Cell. Biol. 11:4876-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roopchand, D. E., J. M. Lee, S. Shahinian, D. Paquette, H. Bussey, and P. E. Branton. 2001. Toxicity of human adenovirus E4orf4 protein in Saccharomyces cerevisiae results from interactions with the Cdc55 regulatory B subunit of PP2A. Oncogene 20:5279-5290. [DOI] [PubMed] [Google Scholar]
- 24.Rose, M. D., R. Winston, and P. Heiter. 1990. Methods of yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Shu, Y., and R. L. Hallberg. 1995. SCS1, a multicopy suppressor of hsp60-ts mutant alleles, does not encode a mitochondrially targeted protein. Mol. Cell. Biol. 15:5618-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shu, Y., H. Yang, E. Hallberg, and R. Hallberg. 1997. Molecular genetic analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 17:3242-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverstein, A. M., C. A. Barrow, A. J. Davis, and M. C. Mumby. 2002. Actions of PP2A on the MAP kinase pathway and apoptosis are mediated by distinct regulatory subunits. Proc. Natl. Acad. Sci. USA 99:4221-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sneddon, A. A., P. T. Cohen, and M. J. Stark. 1990. Saccharomyces cerevisiae protein phosphatase 2A performs an essential cellular function and is encoded by two genes. EMBO J. 9:4339-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sontag, E., V. Nunbhakdi-Craig, G. Lee, R. Brandt, C. Kamibayashi, J. Kuret, C. L. White III, M. C. Mumby, and G. S. Bloom. 1999. Molecular interactions among protein phosphatase 2A, tau, and microtubules. Implications for the regulation of tau phosphorylation and the development of tauopathies. J. Biol. Chem. 274:25490-25498. [DOI] [PubMed] [Google Scholar]
- 31.Tian, Q., and J. Wang. 2002. Role of serine/threonine protein phosphatase in Alzheimer's disease. Neurosignals 11:262-269. [DOI] [PubMed] [Google Scholar]
- 32.Tolstykh, T., J. Lee, S. Vafai, and J. B. Stock. 2000. Carboxyl methylation regulates phosphoprotein phosphatase 2A by controlling the association of regulatory B subunits. EMBO J. 19:5682-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Zyl, W., W. Huang, A. A. Sneddon, M. Stark, S. Camier, M. Werner, C. Marck, A. Sentenac, and J. R. Broach. 1992. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:4946-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Zyl, W. H., N. Wills, and J. R. Broach. 1989. A general screen for mutant of Saccharomyces cerevisiae deficient in tRNA biosynthesis. Genetics 123:55-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Virshup, D. M. 2000. Protein phosphatase 2A: a panoply of enzymes. Curr. Opin. Cell Biol. 12:180-185. [DOI] [PubMed] [Google Scholar]
- 36.Wei, H., D. G. Ashby, C. S. Moreno, E. Ogris, F. M. Yeong, A. H. Corbett, and D. C. Pallas. 2001. Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J. Biol. Chem. 276:1570-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, J., T. Tolstykh, J. Lee, K. Boyd, J. B. Stock, and J. R. Broach. 2000. Carboxyl methylation of the phosphoprotein phosphatase 2A catalytic subunit promotes its functional association with regulatory subunits in vivo. EMBO J. 19:5672-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, X. X., X. Du, C. S. Moreno, R. E. Green, E. Ogris, Q. Feng, L. Chou, M. J. McQuoid, and D. C. Pallas. 2001. Methylation of the protein phosphatase 2A catalytic subunit is essential for association of Balpha regulatory subunit but not SG2NA, striatin, or polyomavirus middle tumor antigen. Mol. Biol. Cell 12:185-199. [DOI] [PMC free article] [PubMed] [Google Scholar]