Abstract
Candida parapsilosis is responsible for ca. 15% of Candida infections and is of particular concern in neonates and surgical intensive care patients. The related species Candida albicans has recently been shown to possess a functional mating pathway. To analyze the analogous pathway in C. parapsilosis, we carried out a genome sequence survey of the type strain. We identified ca. 3,900 genes, with an average amino acid identity of 59% with C. albicans. Of these, 23 are predicted to be predominantly involved in mating. We identified a genomic locus homologous to the MTLa mating type locus of C. albicans, but the C. parapsilosis type strain has at least two internal stop codons in the MTLa1 open reading frame, and two predicted introns are not spliced. These stop codons were present in MTLa1 of all eight C. parapsilosis isolates tested. Furthermore, we found that all isolates of C. parapsilosis tested appear to contain only the MTLa idiomorph at the presumptive mating locus, unlike C. albicans and C. dubliniensis. MTLα sequences are present but at a different chromosomal location. It is therefore likely that all (or at least the majority) of C. parapsilosis isolates have a mating pathway that is either defective or substantially different from that of C. albicans.
Candida species are an important cause of nosocomial infection and rank fourth in bloodstream infections in the United States (8). Invasive candidiasis has an attributable mortality of up to 38%. Although Candida albicans has in the past been the most common causative organism, other Candida species are becoming increasingly more prevalent. A worldwide study of bloodstream infections from 1997 to 1999 showed that at least 45% of yeast infections were caused by Candida species other than C. albicans (37). The most common emerging species are C. glabrata (16%), C. parapsilosis (15%), and C. tropicalis (10%) (11, 35, 36). C. parapsilosis is now the second most commonly isolated Candida species from blood cultures in Europe, Canada, and Latin America and in some European hospitals even outranks C. albicans (19, 34, 37). It is responsible for 24% of Candida infections in children less than 1 year old (29, 36, 40). Unlike other Candida species, C. parapsilosis has been found on the hands of health care workers, resulting in subsequent nosocomial infection associated with handling central venous catheters (20, 25, 41). C. parapsilosis is a particular problem since it tend to grow as biofilms on implanted medical devices, conferring almost total resistance to antifungal drugs (2). The ability to grow as a biofilm is directly related to clinically significant disease, which is a unique attribute of C. parapsilosis (44).
C. parapsilosis strains have historically been categorized as group I, II, or III on the basis of molecular fingerprinting (10, 21, 26). Group I strains (which include the type strain CLIB214, synonymous with ATCC 22019) are predominant in clinical isolates. Analysis of levels of heterozygosity and of mitochondrial genome architecture (39) supports the hypothesis that the three groups represent three different species, and recently group II and group III isolates have been renamed as the new species C. orthopsilosis and C. metapsilosis (46). All three species are distantly related to C. albicans, relative to C. dubliniensis and C. tropicalis. Group I isolates form the basis of the present study.
There is some debate regarding the ploidy of C. parapsilosis isolates. The type strain, however, is highly likely to be diploid, since it is heterozygous for the MET locus (49), and fluorescence-activated cell sorting analysis shows that the DNA content is similar to that of C. albicans (7). Heterozygosity levels between C. parapsilosis isolates are less than 1 base in 1,000 (10), but analysis of those that exist indicates that other isolates are not haploid; they are either diploid or aneuploid (10, 46).
Until recently, it was assumed that C. albicans and related yeasts had no sexual cycle. It is now known that C. albicans MTLa homozygous strains can mate with MTLα homozygotes (14, 31). Mating within C. dubliniensis species and between C. dubliniensis and C. albicans has also been reported (38). Mating is facilitated by white/opaque phenotype switching (27, 32). The distantly related yeast C. glabrata also contains many of the genes required for mating and is capable of undergoing mating type switching (3, 5, 45, 51). Mating in C. albicans and C. dubliniensis is facilitated by growth on skin, suggesting it may play a role in virulence (22, 38). We therefore decided to investigate whether a similar pathway existed in C. parapsilosis.
We carried out a genome sequence survey of C. parapsilosis, producing >10 million bases and identifying 3,900 genes. Many mating-related genes are present, and we show here that the MTLa idiomorph has a similar structure to that of C. albicans. However, the MTLa1 open reading frame (ORF) is defective in all isolates tested.
MATERIALS AND METHODS
Strains and media.
The type strain of C. parapsilosis (CLIB214) was obtained from the Collection de Levure d'Intérêt Biotechnologique, Thiverval Grignon, France. All other C. parapsilosis isolates were obtained from Frank Odds, Aberdeen, United Kingdom (Table 1) (46). The strains were routinely maintained on yeast extract-peptone-dextrose (YPD) medium. The oligonucleotide (oligo) primers used are shown in Table 2.
TABLE 1.
C. parapsilosis strains used in this study
| Isolatea | Geographic origin | Source | MTLa1 sequencedb |
|---|---|---|---|
| CLIB214 (type strain) | Puerto Rico | Feces | Yes |
| 711701 | Aberdeen, United Kingdom | Unknown | Yes |
| 73/107 | London, United Kingdom | Mouth | Yes |
| J931058 | Belgium | Nail | Yes |
| J931845 | Japan | Unknown | Yes |
| 81/042 | London, United Kingdom | Ear | Yes |
| 81/041 | Mayo Clinic, Rochester, Minn. | Vagina | |
| 81/040 | London, United Kingdom | Toe space | |
| 73/037 | Leeds, United Kingdom | Vagina | |
| 81/253 | London, United Kingdom | Nail | Yes |
| 74/046 | Leeds, United Kingdom | Aortic valve | |
| 90-137 | San Jose, Calif. | Orbital tissue | Yes |
All isolates, apart from the type strain, were obtained from Frank Odds, Aberdeen, United Kingdom, and are described by Tavanti et al. (47).
Where indicated, a 200-bp fragment of the MTLa1 region was amplified, and the DNA sequence was determined.
TABLE 2.
Oligonucleotides used in this study
| Oligo | Sequence |
|---|---|
| CP4 | 5′-CATTTCCGTAAAGCCTCTCAAAGTAGTCTTTCAAC-3′ |
| CP2 | 5′-CCAAATTCTAGCATACGTTGATCGTAAAGGCCGTCC-3′ |
| CP6 | 5′-CAGATTAGCACTGTCAACTCTTCACTCACAAATGAC-3′ |
| CP3 | 5′-CGCACACCTTGTACATCATAGGAATAGATTGTGG-3′ |
| 3190_1 | 5′-TACTAGGAGTGTTGACTGATGAAATACGCGGGACA-3′ |
| CCT7_1 | 5′-CGATGACATTGCAGTTGTGGCACATTTCTCCAATG-3′ |
| MPR1 | 5′-GAGCTTCTTGTAAGAATTTTTCTCGTTA-3′ |
| MTX1 | 5′-ATCCTGTCAATCAATGTAACAC-3′ |
| MTX2 | 5′-GCAACTCAAGGATGTACTACT-3′ |
| MTX3 | 5′-ATGAGTTCCGAGTATTATAAAAGC-3′ |
| MTX4 | 5′-TGCTATAAGTTCCCTTTGTCG-3′ |
| MTX5 | 5′-GGACTCGGTCAATTACCCAAG-3′ |
| MTX6 | 5′-TACCTCTCATTCTTTTGTTCGCG-3′ |
| MAR3 | 5′-GCTTTCGATAAAACCTGACCAC-3′ |
| MAR4 | 5′-CTGATATTGACACATTGGTTG-3′ |
| MAR5 | 5′-ATTAATTCTTTTATTGCATTTAGAT-3′ |
| MAR6 | 5′-GTATGATTGTGTGTATTGTTTC-3′ |
Genome sequence survey.
High-molecular-weight DNA was prepared from C. parapsilosis CLIB214 and used to generate a plasmid-based random genomic library with an average insert size of ca. 5 kb (AGOWA [Germany]). Sequences were obtained from both ends of the insert for 8,160 clones. For submission of high-quality reads to GenBank, base calling was carried out by using PHRED (error rate cutoff of 0.05 and a PHRED score of 15) and vector clipping using CROSS_MATCH and a dedicated Perl script. Mitochondrial, rRNA, and tRNA elements were removed by BLASTN searches of a database containing the equivalent sequences from Saccharomyces cerevisiae and C. albicans and the mitochondrial genome from C. parapsilosis (33). Any read with a significant expect value (E-value) of <1e-3 was excluded from subsequent analysis. Contigs were assembled by using PHRAP. Approximately 100 high-quality reads with a significant match (E < 1e-10) to Pseudomonas sequences and no or much weaker match to fungal sequences were also removed, since these were assumed to be derived from a low-level contamination of the genomic library. PCR analysis of two of these sequences confirmed that they were not present in the C. parapsilosis genome. The sequence reads have been deposited in GenBank with accession numbers from CZ278593 to CZ293207.
Putative identities were assigned to many of the C. parapsilosis sequences by using a comparison to a recent annotation of the S. cerevisiae genome containing 5,616 ORFs (18). The S. cerevisiae protein with the strongest BLASTX score was considered to be the ortholog, providing that the E-value for this hit was <1e-10 and more than 1e10 lower than the E-value for the second-best hit. For each ortholog the cellular function in S. cerevisiae was determined by using the “cellular role” categories of the Yeast Proteome Database of the Incyte Bioknowledge Library (12).
Comparisons with C. albicans proteins were determined by using a haploid gene set of 7,677 ORFs (including small and overlapping ORFs) predicted from assembly 19 (16). Classification of C. albicans proteins into functional categories was performed by using the same criteria as for C. parapsilosis. Sequences that did not match the C. albicans genome with BLASTX scores of <1e-3 were compared to the nonredundant database.
For the calculation of average sequence identity, only orthologs that are reciprocal best hits were used. This corresponded to 3,547 genes compared between C. parapsilosis and C. albicans and 5,511 genes compared between C. dubliniensis and C. albicans. The C. dubliniensis gene sequences were obtained from the Sanger Institute (assembly of 1 May 2004 [http://www.sanger.ac.uk/Projects/C_dubliniensis/]).
The C. tropicalis MTL sequences were obtained from assembly 1 at the MIT sequencing project (http://www.broad.mit.edu). We did not analyze the ORFs in detail since there appear to be several frameshifts and possible sequencing errors in the current assembly.
Isolation of MTLa
locus. The sequence of the entire insert from clones cp45f11 (from CpPIKa to Cp19.3202) and cp12c03 (from Cp19.3192 to CpGap1; see Fig. 1) was determined by AGOWA. Oligos CP4 (designed from the CpPIKa ortholog) and CP2 (designed from the CpGAP1 ortholog) were used with Expand Long-Template DNA polymerase (Roche) to amplify a region of 6.3 kb between the two inserts. The PCR product was purified by using a QiaQuick kit (Qiagen) according to the manufacturer's instructions, and the sequence was determined directly by GATC (Germany). The sequence of the PCR product was combined with the overlapping sequence of the cp45f11 and cp12c03 inserts to give a 15.7-kb sequence that includes the entire predicted CpMTLa idiomorph. The sequence has been deposited with GenBank (accession number AY961981). For restriction analysis of the MTL locus, fragments of 14 to 21 kb were amplified by PCR by using Expand Long-Template DNA polymerase (Roche) and oligos 3190_1 (designed from within the Cp19.3190 ORF), CP6 (designed from sequence 900 bp upstream from the start of CpGAP1), CP3 (designed from within CpRCY1), and CCT7_1 (designed from within the CpCCT7 ortholog). High-molecular-weight genomic DNA was prepared by using a Genomic-Tip 20/G kit (Qiagen).
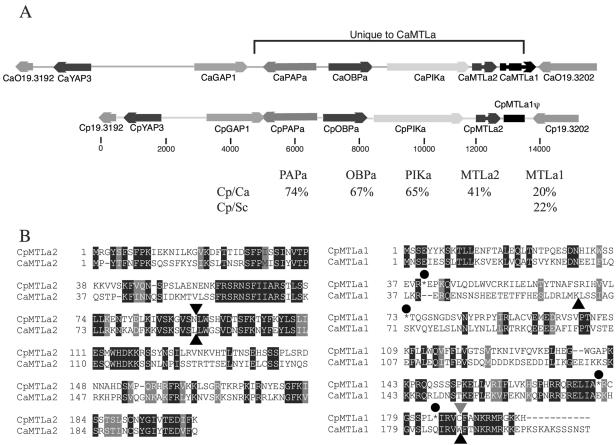
FIG. 1.
Identification of an MTLa idiomorph from C. parapsilosis. (A) The top line shows the structure of the MTLa sequence and surrounding region from C. albicans. The genes are drawn approximately to scale. The region indicated by a bracket is unique to MTLa, and the remaining sequences surround both MTLa and MTLα idiomorphs. The line below illustrates the corresponding structure in the orthologous CpMTLa idiomorph that was sequenced in full. The C. albicans genome annotation places an ORF (Orf19.3194) between CaYAP3 and CaGAP1. However, it is not conserved and is likely to be spurious and so is omitted in this diagram. Orf19.3192 corresponds to the N terminus of the Sti1 protein in S. cerevisiae. The sequence of the Cp19.3202 and Cp19.3192 orthologs at the ends of the fragment is not complete. Distances are shown in base pairs. The percent identity between proteins from C. parapsilosis (Cp) and C. albicans (Ca) or S. cerevisiae (Sc) is shown below. (B) An alignment of the MTLa2 proteins from C. parapsilosis (CpMTLa2) and C. albicans (CaMTLa2) was generated by using CLUSTAL W and is shown on the left-hand side. The intron locations are indicated with black triangles. The right-hand side shows an alignment between the C. albicans MTLa1 protein (CaMTLa1) and the hypothetical protein translation of the pseudogene from C. parapsilosis (CpMTLa1). Introns in the C. albicans sequence are indicated with black triangles, and a putative intron in C. parapsilosis is indicated with a gray triangle. Black circles highlight four stop codons in the C. parapsilosis sequence.
To confirm the mutations in MTLa1, a 200-bp fragment of the ORF was amplified from eight C. parapsilosis isolates by using oligos MPR1 and MTX6. The PCR product was purified as described above, and the sequence was determined directly by GATC.
Southern blot analysis.
Digested genomic DNA was separated by electrophoresis on 1% agarose gels, transferred to positively charged nylon membranes (Roche Diagnostics), and treated according to the manufacturer's instructions. Probes for CaPAPα and CaMTLα1 were amplified from C. albicans SC5314 genomic DNA by using MAR3/MAR4 and MAR5/MAR6 primer combinations. The CpMTLa1 probe was amplified from C. parapsilosis CLIB214 genomic DNA by using the primers MPR1 and MTX6. All probes were labeled with [32P]dCTP by using a High Prime DNA Labeling Kit (Roche Diagnostics). For low-stringency conditions, hybridizations were carried out overnight at 60°C in PerfectHyb (Sigma). The blots were washed twice for 30 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 60°C (6). High-stringency hybridizations were carried out at 68°C, and the blots were washed once for 5 min at room temperature in 2× SSC-0.1% SDS and twice for 20 min at 68°C in 0.5× SSC-0.1% SDS. The blots were then exposed to X-ray film.
Analysis of RNA splicing.
C. parapsilosis CLIB214 was cultured overnight in YPD medium, diluted to an A600 of 0.2 in 50 ml of fresh YPD medium, and incubated at 30°C for 4 h. Total RNA was isolated from 5 ml of culture by using a Ribopure kit (Ambion). DNase I treatment was carried out during the purification process. To generate cDNA, 3 μg of RNA and 291 ng of oligo dT-15 primer in a final volume of 15 μl was incubated at 70°C for 10 min and then chilled on ice. A 45-μl cocktail containing 0.6 mM deoxynucleoside triphosphates, 120 U of RNasin RNase inhibitor (Promega), 600 U of Moloney murine leukemia virus reverse transcriptase (Promega), and 1× Promega reverse transcription-PCR (RT-PCR) buffer was then added. The mixture was incubated at 37°C for 1 h, followed by 95°C for 2 min, and the resulting cDNA was held at 4°C. For each PCR, 5 μl of cDNA was used as a template in a 50-μl volume containing 0.25 mM deoxynucleoside triphosphates, 1× reaction buffer, 1.5 mM MgCl2, 25 pmol of each primer, and 2.5 U of Taq DNA polymerase (Invitrogen). RNA that was not reverse transcribed was used as a control to test for the presence of genomic DNA. The same PCR amplification was also performed with genomic DNA as a template. The MTLa2 region was amplified by using oligos MTX1 and MTX2, and the MTLa1 region was amplified by using oligos MTX3, MTX4, MTX5, and MTX6. The MTX1/MTX2 and MTX3/MTX6 RT-PCR products were combined from five reactions, purified as described above, and the DNA sequence was determined directly by GATC.
RESULTS
Genome sequence survey of C. parapsilosis.
We constructed a plasmid library with random genomic inserts of ca. 5 kb and sequenced both ends of ca. 8,000 clones. After removal of mtDNA and rRNA, 10.1 Mb of high-quality primary sequence data was obtained, corresponding to ca. 0.8× coverage of the haploid genome size. The sequences were assembled into 5,374 contigs. Putative ORFs were identified by using BLASTX searches of the C. albicans assembly 19 ORFs. We identified partial sequence of 3,898 orthologs in C. parapsilosis, corresponding to ca. 60% of the 6,419 nonoverlapping ORFs predicted in the haploid C. albicans genome (16). We also identified orthologs of seven additional ORFs found in other fungal genomes but which are not present in the C. albicans genome sequence (Table 3). The average protein sequence identity between C. parapsilosis and C. albicans is 59%. In contrast, the average identity between C. dubliniensis and C. albicans is 82% (based on a comparison of 5,511 C. albicans genes).
TABLE 3.
Some C. parapsilosis genes not present in C. albicans
| Gene namea | Organism | GenBank accession no. |
|---|---|---|
| DEHA0B13200g (capsid protein, virus L-A) | Debaryomyces hansenii | CAG85523 |
| MG10213.4 (Gentisate 1,2-dioxygenase) | Magnaporthe grisea | EAA48150 |
| DEHA0E11440g (LMBR1-like membrane protein) | Debaryomyces hansenii | XP_459779 |
| FG04776.1 (monooxygenase) | Gibberella zeae | EAA75735 |
| SPAC11D3.03c | Schizosaccharomyces pombe | NP_592800 |
| DEHA0E15411g (esterase) | Debaryomyces hansenii | XP_459950 |
| KLLA0A12045g | Kluyveromyces lactis | XP_451524 |
The name of the closest ortholog is shown and, where possible, a function is inferred from sequences in other organisms and is shown in parentheses.
Since the cellular function of the majority of C. albicans genes is not yet determined, we inferred gene function in C. parapsilosis from the known functions of the S. cerevisiae orthologs. The “cellular role” divisions of the Yeast Proteome Database were used to categorize a total of 2,650 C. parapsilosis genes with identifiable S. cerevisiae orthologs. A similar analysis was performed with 7,677 ORFs from C. albicans derived from the haploid gene set (16; see also Table S5 in the supplemental material). We identified orthologs for 47% of the S. cerevisiae genes in C. parapsilosis and 51% in C. albicans. The category “conjugation with cellular fusion” included 94 genes from C. parapsilosis, several with known functions in mating. Although many have roles in other cellular processes, we extracted a list of 19 that function predominantly in mating (Table 4). These were augmented with orthologs of genes shown to be mating specific in C. albicans by Tsong et al. (48). The genes were placed into three classes: a-specific, α-specific, and genes more generally involved in mating. Although some may have several roles (such as Ram1 and Ram2, which are required for farnesylation of a-factor and Ras proteins in S. cerevisiae), others (such as Ste2, the α-factor receptor in S. cerevisiae) have no known function outside mating.
TABLE 4.
Some C. parapsilosis genes with potential roles in mating
| Gene type and namea | Function |
|---|---|
| α-Specific genes | |
| KEX1 | Processing of killer toxin and α-factor |
| STE13 | α-Factor processing |
| SAG1 | Agglutination of α-factor |
| a-Specific genes | |
| RAM1 | Farnesylation of a-factor |
| RAM2* | Farnesylation of a-factor |
| STE23 | a-Factor processing |
| STE6* | Component of a-factor secretory pathway |
| AXL1 | Maturation and cleavage of a-factor |
| STE2 | α-Factor receptor |
| SST2 | GAP protein, desensitisation to α-factor |
| RBT1* | Predicted glycosylated cell wall protein |
| Orf19.4688*† | Unknown function |
| Orf19.2825*† | Unknown function |
| MTLa2 | Regulator of a-specific mating |
| General mating genes | |
| FAR1 | Cyclin-dependent kinase inhibitor, mediates cell cycle arrest in response to pheromone |
| AKR1 | Negative regulation of pheromone response pathway, required for endocytosis of pheromone receptors |
| PLP2 | Positive regulation of transcription from polymerase II promoter by pheromones |
| FAR11 | G1 arrest in response to mating pheromones |
| LRG1 | Rho GTPase, involved in mating and sporulation |
| GPA1 | G protein alpha subunit, coupled to mating factor receptors |
| STE4 | G protein beta subunit, coupled to mating factor receptors |
| FIG4 | Polyphosphoinositide phosphatase, required for efficient mating |
| KAR4 | Karyogamy, functions in mating and meiosis |
*, Genes described as a-specific in C. albicans by Tsong et al. (48); †, original designations were as follows: Orf19.4688 = Orf6.1768 and Orf19.2825 = Orf6.1251.
Identification of a putative C. parapsilsosis MTL locus.
In C. albicans, mating type is determined by the genes residing at the MTL locus (13, 48). There are two idiomorphs. MTLa contains the genes PAPa [poly(A) polymerase], OBPa (oxysterol-binding protein), PIKa (phosphatidyl inositol kinase), MTLa2 (a HMG-box protein), and MTLa1 (a homeodomain protein) (Fig. 1). MTLα contains three genes related to those in the MTLa locus (PAPα, OBPα, and PIKα), plus MTLα2 (a homeodomain protein) and MTLα1 (an α domain protein). The genes surrounding the MTL idiomorphs are the same on both chromosomes. Under certain conditions, cells with two copies of MTLa (a/a; or hemizygous for MTLa) can mate with cells with two copies of MTLα (α/α; or hemizygous for MTLα). While analyzing the data from the sequence survey of C. parapsilosis, we noted that several reads matched many of the genes within and surrounding the MTLa locus. The sequence reads matching MTLa1, however, contained stop codons in the predicted ORF. To confirm this observation, we determined the sequence of the entire region orthologous to C. albicans MTLa (Fig. 1). This region contains orthologs of all of the genes found in the C. albicans MTLa locus, and the order and transcriptional orientations of the genes are conserved between the two species.
The C. parapsilosis MTLa1 locus is a pseudogene.
Although the majority of the genes in the C. parapsilosis MTLa idiomorph are intact, CpMTLa1 appears to be a pseudogene, because there are four stop codons in the region corresponding to CaMTLa1 (Fig. 1B). Since C. parapsilosis CLIB214 is most likely diploid (7, 49), we considered the possibility that the genome might contain two copies of the MTLa idiomorph, one with a defective and one with a functional MTLa1 sequence. We therefore amplified a portion of the MTLa1 region from genomic DNA, derived from the conserved homeodomain and encompassing the last two stop codons. The sequence of the PCR product was determined directly. If two alleles are present we should obtain a mixed DNA sequence. However, only one sequence was detected, identical to the CpMTLa sequence (Fig. 1). We also amplified this fragment from seven other C. parapsilosis isolates (Table 1). All contained the same sequence, with no evidence of heterozygozity.
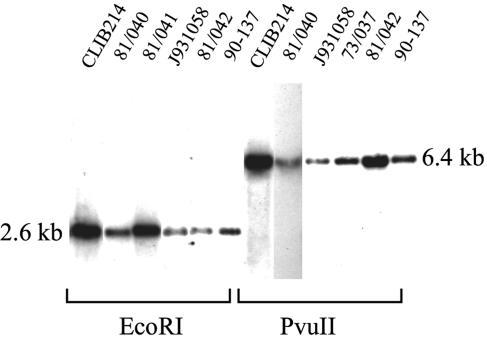
To confirm there is no copy of CpMTLa1 elsewhere in the genome, we performed a Southern blot, using the same 200-bp CpMTLa1 fragment as a probe. As shown in Fig. 2, only one fragment of the predicted size is detected under low-stringency conditions in the genomes of all C. parapsilosis isolates tested. Since C. albicans probes hybridize to C. parapsilosis DNA under these conditions (Fig. 4), it is highly likely that we would detect any related CpMTLa1 sequence present. Identical results were obtained at high stringencies (not shown).
FIG. 2.
CpMTLa1 is detected in a single location in the C. parapsilosis genome. Genomic DNA was isolated from the C. parapsilosis strains indicated, digested with EcoRI or PvuII, and transferred to nylon membranes. The blots were probed at low stringency with a 200-bp fragment derived from the homeodomain region of CpMtla1p. The fragments detected are consistent with those predicted from the sequence of the MTL locus.
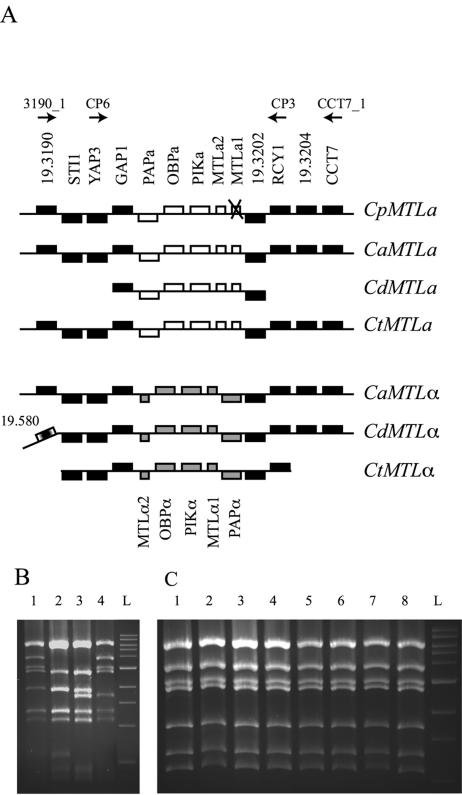
FIG. 4.
Amplification of the C. parapsilosis MTL locus. (A) The structure of the region surrounding CpMTLa was extrapolated from overlapping clones and is compared to the gene order around the MTL loci in other Candida species. The extent of the available contigs is indicated. The unique MTLa genes are shown in white, the MTLα genes in gray, and the shared surrounding genes in black. The order upstream of the STI1 gene in C. dubliniensis MTLα differs from the other sequences. The CpMTLa1 pseudogene is indicated with an “X.” The C. tropicalis fragments contain many putative sequencing errors that are not marked. The location of the oligo primers used in PCR amplification reactions are shown above. The region is not drawn to scale. (B) The MTL region from C. parapsilosis CLIB214 was amplified by using oligo primers noted and digested with EcoRI. Lane 1, primers 3190_1 and CCT7_1 (∼21 kb); lane 2, primers 3190_1 and CP3 (∼16.2 kb); lane 3, primers CP6 and CP3 (∼14.5 kb); lane 4, primers CP6 and CCT7_1 (∼20 kb); L, 1-kb ladder. (C) The MTL locus from eight C. parapsilosis isolates was amplified by using primers CP6 and CCT7_1 and digested with EcoRI. Lane 1, CLIB 214; lane 2, 81/041; lane 3, 81/042; lane 4, 81/040; lane 5, 90/137; lane 6, 74/046; lane 7, 73/037; lane 8, J931058. L, 1-kb ladder.
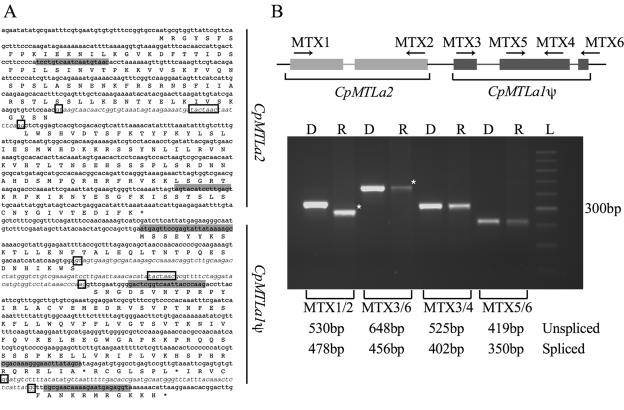
The structure of CpMTLa1 as shown in Fig. 1 has one intron at a position equivalent to intron 2 of CaMTLa1, although the predicted 5′ splice site is GG, and there is no TACTAAC box (Fig. 3). The gene also has four internal stop codons. We considered another possible structure for this gene, one that incorporated an additional intron at a position similar to, but not identical to, intron 1 of the C. albicans gene (Fig. 1 and 3). The possible intron has GT and AG splice sites and a TACTAAC lariat site and skips over two of the four predicted stop codons, but it also drastically shortens the predicted C. parapsilosis protein compared to C. albicans and other a1 proteins. To determine whether any splicing occurs, we compared the sizes of products obtained by RT-PCR from an RNA template to those generated by direct amplification of the genomic DNA. Primers were chosen that amplify the entire MTLa1 region, including the two possible introns (Fig. 3). As a control, we also amplified the MTLa2 region, which is predicted to contain an intron in both C. albicans and C. parapsilosis. As shown in Fig. 3, the RT-PCR product from CpMTLa2 is indeed smaller than the product amplified from genomic DNA. Sequence analysis confirmed that the predicted intron in MTLa2 is correctly spliced. To our knowledge, this is the first experimental demonstration of splicing of MTLa2 in Candida species. The RT-PCR products from CpMTLa1, however, are identical in size to those derived from genomic DNA. The sequence of the largest RT-PCR product was determined, confirming that it is generated from the CpMTLa1 sequence and that no splicing occurs. We are thus unsure whether CpMTLa1 is a pseudogene of a single-intron gene that has gained four stop codons or a pseudogene of a two-intron gene that has gained two stop codons, but in either case it is undoubtedly a pseudogene.
FIG. 3.
The CpMTLa1 sequence is not spliced. (A) Predicted translation of the CpMTLa2 and CpMTLa1 region is shown. The positions of putative introns are indicated in italics, and the oligos used in PCR amplifications are shaded in gray. Putative splice sites and TACTAAC sites are boxed. (B) Proposed structure of the MTLa2 and MTLa1 region and location of oligos used in PCR. Putative exons are shown as rectangles, and introns as lines. The oligo combinations were used in a PCR amplification using genomic DNA (D) and cDNA (R) as templates. The size of predicted spliced and unspliced products are indicated below the gel. Products highlighted with an asterisk were sequenced. L, 100-bp ladder.
Structure of the C. parapsilosis MTL locus.
Our genome sequencing data yielded several random genomic clones that originated from MTLa, but none that matched MTLα. To look for MTLα sequences in this and other isolates, we first used overlapping clone data to extrapolate the structure of the region surrounding the CpMTL locus (Fig. 4). The order of the surrounding genes is identical to the MTL loci in C. albicans (16). We used four primer combinations to amplify the region surrounding the MTL locus from C. parapsilosis CLIB214, yielding fragments varying in size from 14.2 to more than 21 kb (Fig. 4). Primer CP6 lies within the region for which the entire sequence was determined (Fig. 1), and the remaining primers were derived from the sequences of overlapping clones. If two idiomorphs are present we should amplify two fragments. However, as shown in Fig. 4B, restriction analysis shows that only one fragment is present. The restriction pattern is also identical for eight isolates amplified by using the primer pair CP6 and CCT7_1 (Fig. 4C), and for 14 isolates amplified by using CP6 and CP3 (not shown), indicating that these PCR products also contain MTLa idiomorphs.
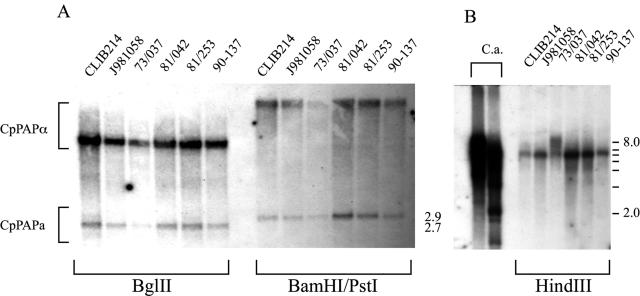
To look for evidence of CpMTLα sequences elsewhere in the genome, we performed low-stringency Southern blot analysis. We first used an 856-bp fragment derived from CaPAPα, which is found in the MTLα idiomorph of C. albicans (13), C. dubliniensis (38), and C. tropicalis (http://www.broad.mit.edu). We reasoned that at low stringency, this probe should hybridize to the related gene CpPAPa, which we have shown is present in the CpMTLa idiomorph, and to any ortholog of PAPα present. As shown in Fig. 5, CaPAPα hybridizes to DNA fragments of 2.7 kb (BglII digest) and 2.9 kb (BamHI/PstI digest). These correspond to the predicted size of CpPAPa, which we confirmed in other blots (not shown). However, it hybridizes at higher intensities to a larger fragment in both digests (>8 kb), suggesting the C. parapsilosis genome does contain an ortholog of CaPAPα. A 145-bp fragment derived from the conserved domain of CaMTLα1 also hybridized to sequences in C. parapsilosis (Fig. 5B). Both probes detected large fragments in all of the digests tested (not shown), and we were unable to determine whether the sequences are contiguous. It is therefore likely that C. parapsilosis does contain MTLα sequences, but the PCR results suggest that they are not in the same chromosomal context as CpMTLa, even though the order of the genes flanking CpMTLa is very similar to those flanking the MTL loci of C. albicans, C. dubliniensis, and C. tropicalis (Fig. 4).
FIG. 5.
Identification of CpMTLα sequences. Genomic DNA was isolated from the C. parapsilosis strains indicated, digested, and transferred to nylon membranes. Hybridizations were carried out at low stringencies. (A) An 865-bp fragment of CaPAPα detects two fragments in C. parapsilosis genomic DNA. One (2.7 kb with BglII or 2.9 kb with BamHI/PstI) corresponds to the CpPAPa sequence in the CpMTLa idiomorph. The other fragment (>8 kb in both digests) most likely corresponds to a CpPAPα ortholog. (B) A 145-bp fragment derived from CaMTLα1 hybridizes to a 7-kb HinDIII fragment in genomic DNA from C. parapsilosis isolates. C.a., C. albicans.
DISCUSSION
The results of the genome sequence survey of C. parapsilosis yielded partial DNA sequence information for more than 3,900 potential ORFs. The amino acid sequence identity between C. parapsilosis proteins and C. albicans proteins and between C. dubliniensis proteins and C. albicans proteins both ranged from ca. 20 to 100%. The most similar sequences included ribosomal proteins, actin, and polyubiquitin (see Table S6 in the supplemental material). Proteins with <25% conservation between C. parapsilosis and C. albicans are mostly uncharacterized, but include AXL1, a putative a-pheromone maturase (see Table S7 in the supplemental material) (1). The average sequence identity between C. parapsilosis and C. albicans proteins, however, is 59%, and between C. dubliniensis and C. albicans it is 82%.
To identify genes that may be unique to C. parapsilosis, we extracted sequences with no similarity to the C. albicans genome and looked for orthologs in other species. We identified seven genes all present in other fungal genomes (Table 3). One is strongly similar to the major coat protein of the S. cerevisiae L-A virus, suggesting that C. parapsilosis may harbor double-stranded RNA viruses or that its genome contains genes derived from such a virus. In other yeasts these viruses encode toxins, some of which have anti-Candida activities (42, 43, 50). It will therefore be interesting to look for similar activities in C. parapsilosis.
Disruption of the mating locus in C. parapsilsosis.
In C. albicans, the ability to mate is closely associated with white-opaque phenotype switching. In heterozygous diploid cells containing both a and α idiomorphs, a1 and α2 proteins heterodimerize to generate a negative regulator. As a result, white-opaque switching is repressed and mating-specific functions are switched off (27, 31, 32, 48). White-opaque switching occurs in C. albicans cells that have only one functional MTL idiomorph because they are homozygous or hemizygous (24, 27). In opaque cells that contain only the MTLa idiomorph, the a2 protein is a positive regulator of mating functions. Similarly, α1 positively regulates mating in opaque cells containing only the MTLα idiomorph. The closely related species C. dubliniensis also undergoes white-opaque switching and mates in a similar fashion (38). Phenotypic switching does occur in C. parapsilosis (9, 23, 28), but we have not observed anything directly comparable to white-opaque phenotypes. We have, however, shown that the type strain of C. parapsilosis carries a defective MTLa1 allele, with up to four internal stop codons in the presumptive ORF.
The final two stop codons in CpMTLa1 from C. parapsilosis CLIB214 are also present in seven other randomly selected isolates, and PCR amplification of genomic DNA and Southern blot analysis indicates that there is no functional copy of MTLa1 elsewhere. It is therefore extremely unlikely that C. parapsilosis can generate a heterodimeric a1/α2 protein. The MTLa2 gene, however, is transcribed and spliced, and so should be capable of inducing a-specific mating type functions.
If mating occurs in C. parapsilosis in a manner that is in any way similar to that in C. albicans or C. dubliniensis, we would expect to find an MTLα idiomorph in an analogous position to MTLa. Our sequence survey resulted in 0.8× coverage of the genome, with several matches to MTLa-specific genes such as PIKa, MTLa1, and MTLa2. No similarities to MTLα genes were detected. To look for a second idiomorph, we first compared the structures of the MTL loci from related Candida species. In C. albicans, the region unique to MTLa is 8,742 bp, and the unique MTLα sequence is 8,861 bp (13). The equivalent unique regions in C. dublinienis are 8,728 bp (CdMTLa) and 8,845 bp (CdMTLα) (http://www.sanger.ac.uk/Projects/C_dubliniensis/), and in C. tropicalis they are 8,473 bp (CtMTLa) and 10,337 bp (CtMTLα) (http://www.broad.mit.edu). The gene order within the equivalent idiomorphs is identical (Fig. 4). It is not possible to determine the order of all of the genes surrounding the MTL loci, since the assemblies for C. dubliniensis and C. tropicalis are not complete. However, the synteny is very similar for all of the loci for which sequence is currently available (Fig. 4), although there does appear to be a break upstream from STI1 in C. dubliniensis MTLα. We therefore amplified regions surrounding CpMTL by using primers derived from the sequence of the unique genes that lie well outside the borders of the MTL locus in related Candida species (Fig. 4A). We amplified only one DNA fragment, corresponding to CpMTLa, in all isolates tested (Fig. 4B and C). Since there is very little sequence variability in C. parapsilosis isolates (46) and we used four primer combinations, it is highly unlikely that our failure to amplify an MTLα-containing fragment was caused by polymorphisms in the primer sequences. It therefore appears that the chromosomal location that corresponds to MTL in other Candida species contains only MTLa in C. parapsilosis. However, our Southern blot analysis does suggest that there are MTLα-like sequences somewhere in the C. parapsilosis genome, since we identified fragments that hybridize to CaPAPα and CaMTLα1 (Fig. 5). We conclude that MTLα in C. parapsilosis has been rearranged somehow, breaking its colinearity with the genomic context of CpMTLa and the MTL loci in other Candida species. It is difficult to envisage how C. parapsilosis strains could become mating competent through homozygozity or hemizygozity at the MTL genes, when these genes are not at allelic locations in the genome. It is also unlikely that the MTLα sequences represent a silent mating cassette involved in mating type switching, since our analysis has shown that this mechanism evolved relatively recently and is restricted to the Saccharomyces sensu stricto group and their close relatives (5). The most likely scenario is that the MTLα locus has degenerated after rearrangement and has become inactivated, although further sequence analysis is required to support this hypothesis. Despite the overall similarities, however, it is very unlikely that C. parapsilosis can mate in the same way as C. albicans (15) or C. dubliniensis (38).
Despite the defects in the MTL sequences, we did identify many genes that are predominantly required for mating in C. albicans and S. cerevisiae (Table 4). These include orthologs of STE13, required for processing of α-factor (17), and STE2, the α-factor receptor in S. cerevisiae (4). We also identified several genes that are probably a-specific, including orthologs of STE6, RAM2, Orf19.4688, Orf19.2825, and RBT1 whose expression in C. albicans requires both MTLa2 and exposure to α-factor (48). STE6 is known to be required for mating in C. albicans (30). The fact that many components of the pathway are apparently intact suggests that C. parapsilosis has only recently undergone a rearrangement at the MTL locus. However, this contrasts with the observation that the MTLa1 sequence contains four stop codons, which were presumably acquired in four separate mutational events.
Supplementary Material
Acknowledgments
This study was supported by Science Foundation Ireland.
We are grateful to Jozef Nosek for providing access to the C. parapsilosis mitochondrial genome prior to publication and to Frank Odds for providing the strain collection.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adames, N., K. Blundell, M. N. Ashby, and C. Boone. 1995. Role of yeast insulin-degrading enzyme homologs in propheromone processing and bud site selection. Science 270:464-467. [DOI] [PubMed] [Google Scholar]
- 2.Baillie, G. S., and L. J. Douglas. 1998. Effect of growth rate on resistance of Candida albicans biofilms to antifungal agents. Antimicrob. Agents Chemother. 42:1900-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brockert, P. J., S. A. Lachke, T. Srikantha, C. Pujol, R. Galask, and D. R. Soll. 2003. Phenotypic switching and mating type switching of Candida glabrata at sites of colonization. Infect. Immun. 71:7109-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkholder, A. C., and L. H. Hartwell. 1985. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 13:8463-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, G., C. Kenny, A. Fagan, C. Kurischko, C. Gaillardin, and K. H. Wolfe. 2004. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. USA 101:1632-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, Y., Y. L. Qiu, P. Kuhlman, and J. D. Palmer. 1998. Explosive invasion of plant mitochondria by a group I intron. Proc. Natl. Acad. Sci. USA 95:14244-14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dvorak, J. A., W. L. Whelan, J. P. McDaniel, C. C. Gibson, and K. J. Kwon-Chung. 1987. Flow cytometric analysis of the DNA synthetic cycle of Candida species. Infect. Immun. 55:1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 9.Enger, L., S. Joly, C. Pujol, P. Simonson, M. Pfaller, and D. R. Soll. 2001. Cloning and characterization of a complex DNA fingerprinting probe for Candida parapsilosis. J. Clin. Microbiol. 39:658-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fundyga, R. E., R. J. Kuykendall, W. Lee-Yang, and T. J. Lott. 2004. Evidence for aneuploidy and recombination in the human commensal yeast Candida parapsilosis. Infect. Genet. Evol. 4:37-43. [DOI] [PubMed] [Google Scholar]
- 11.Hajjeh, R. A., A. N. Sofair, L. H. Harrison, G. M. Lyon, B. A. Arthington-Skaggs, S. A. Mirza, M. Phelan, J. Morgan, W. Lee-Yang, M. A. Ciblak, L. E. Benjamin, L. T. Sanza, S. Huie, S. F. Yeo, M. E. Brandt, and D. W. Warnock. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 42:1519-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodges, P. E., A. H. McKee, B. P. Davis, W. E. Payne, and J. I. Garrels. 1999. The Yeast Proteome Database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 27:69-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271-1275. [DOI] [PubMed] [Google Scholar]
- 14.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, A. 2003. The biology of mating in Candida albicans. Nat. Rev. Microbiol. 1:106-116. [DOI] [PubMed] [Google Scholar]
- 16.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 11:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julius, D., L. Blair, A. Brake, G. Sprague, and J. Thorner. 1983. Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell 32:839-852. [DOI] [PubMed] [Google Scholar]
- 18.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 19.Krcmery, V., Jr., G. Kovacicova, et al. 2000. Longitudinal 10-year prospective survey of fungaemia in Slovak Republic: trends in etiology in 310 episodes. Diagn. Microbiol. Infect. Dis. 36:7-11. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn, D. M., P. K. Mukherjee, T. A. Clark, C. Pujol, J. Chandra, R. A. Hajjeh, D. W. Warnock, D. R. Soll, and M. A. Ghannoum. 2004. Candida parapsilosis characterization in an outbreak setting. Emerg. Infect. Dis. 10:1074-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 22.Lachke, S. A., S. R. Lockhart, K. J. Daniels, and D. R. Soll. 2003. Skin facilitates Candida albicans mating. Infect. Immun. 71:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laffey, S. F., and G. Butler. 2005. Biofilm formation is associated with phenotypic switching in Candida parapsilosis. Microbiology, 151:1073-1081. [DOI] [PubMed] [Google Scholar]
- 24.Legrand, M., P. Lephart, A. Forche, F. M. Mueller, T. Walsh, P. T. Magee, and B. B. Magee. 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol. Microbiol. 52:1451-1462. [DOI] [PubMed] [Google Scholar]
- 25.Levin, A. S., S. F. Costa, N. S. Mussi, M. Basso, S. I. Sinto, C. Machado, D. C. Geiger, M. C. Villares, A. Z. Schreiber, A. A. Barone, and M. L. Branchini. 1998. Candida parapsilosis fungemia associated with implantable and semi-implantable central venous catheters and the hands of healthcare workers. Diagn. Microbiol. Infect. Dis. 30:243-249. [DOI] [PubMed] [Google Scholar]
- 26.Lin, D., L. C. Wu, M. G. Rinaldi, and P. F. Lehmann. 1995. Three distinct genotypes within Candida parapsilosis from clinical sources. J. Clin. Microbiol. 33:1815-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lockhart, S. R., C. Pujol, K. J. Daniels, M. G. Miller, A. D. Johnson, M. A. Pfaller, and D. R. Soll. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lott, T. J., R. J. Kuykendall, S. F. Welbel, A. Pramanik, and B. A. Lasker. 1993. Genomic heterogeneity in the yeast Candida parapsilosis. Curr. Genet. 23:463-467. [DOI] [PubMed] [Google Scholar]
- 29.Lupetti, A., A. Tavanti, P. Davini, E. Ghelardi, V. Corsini, I. Merusi, A. Boldrini, M. Campa, and S. Senesi. 2002. Horizontal transmission of Candida parapsilosis candidemia in a neonatal intensive care unit. J. Clin. Microbiol. 40:2363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magee, B. B., M. Legrand, A. M. Alarco, M. Raymond, and P. T. Magee. 2002. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol. 46:1345-1351. [DOI] [PubMed] [Google Scholar]
- 31.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 32.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 33.Nosek, J., M. Novotna, Z. Hlavatovicova, D. W. Ussery, J. Fajkus, and L. Tomaska. 2004. Complete DNA sequence of the linear mitochondrial genome of the pathogenic yeast Candida parapsilosis. Mol. Genet. Genomics. [DOI] [PubMed]
- 34.Pagano, L., A. Antinori, A. Ammassari, L. Mele, A. Nosari, L. Melillo, B. Martino, M. Sanguinetti, F. Equitani, F. Nobile, M. Carotenuto, E. Morra, G. Morace, and G. Leone. 1999. Retrospective study of candidemia in patients with hematological malignancies: clinical features, risk factors, and outcome of 76 episodes. Eur. J. Hematol. 63:77-85. [DOI] [PubMed] [Google Scholar]
- 35.Pfaller, M. A., and D. J. Diekema. 2004. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin. Microbiol. Infect. 10(Suppl. 1):11-23. [DOI] [PubMed] [Google Scholar]
- 36.Pfaller, M. A., D. J. Diekema, R. N. Jones, S. A. Messer, and R. J. Hollis. 2002. Trends in antifungal susceptibility of Candida spp. isolated from pediatric and adult patients with bloodstream infections: SENTRY Antimicrobial Surveillance Program, 1997 to 2000. J. Clin. Microbiol. 40:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaller, M. A., D. J. Diekema, R. N. Jones, H. S. Sader, A. C. Fluit, R. J. Hollis, and S. A. Messer. 2001. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and in vitro susceptibilities to fluconazole, ravuconazole, and voriconazole of isolates collected from 1997 through 1999 in the SENTRY antimicrobial surveillance program. J. Clin. Microbiol. 39:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pujol, C., K. J. Daniels, S. R. Lockhart, T. Srikantha, J. B. Radke, J. Geiger, and D. R. Soll. 2004. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot. Cell 3:1015-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rycovska, A., M. Valach, L. Tomaska, M. Bolotin-Fukuhara, and J. Nosek. 2004. Linear versus circular mitochondrial genomes: intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology 150:1571-1580. [DOI] [PubMed] [Google Scholar]
- 40.Saiman, L., E. Ludington, J. D. Dawson, J. E. Patterson, S. Rangel-Frausto, R. T. Wiblin, H. M. Blumberg, M. Pfaller, M. Rinaldi, J. E. Edwards, R. P. Wenzel, and W. Jarvis. 2001. Risk factors for Candida species colonization of neonatal intensive care unit patients. Pediatr. Infect. Dis. J. 20:1119-1124. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez, V., J. A. Vazquez, D. Barth-Jones, L. Dembry, J. D. Sobel, and M. J. Zervos. 1993. Nosocomial acquisition of Candida parapsilosis: an epidemiologic study. Am. J. Med. 94:577-582. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt, M. J., and F. Neuhausen. 1994. Killer toxin-secreting double-stranded RNA mycoviruses in the yeasts Hanseniaspora uvarum and Zygosaccharomyces bailii. J. Virol. 68:1765-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitt, M. J., O. Poravou, K. Trenz, and K. Rehfeldt. 1997. Unique double-stranded RNAs responsible for the anti-Candida activity of the yeast Hanseniaspora uvarum. J. Virol. 71:8852-8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin, J. H., S. J. Kee, M. G. Shin, S. H. Kim, D. H. Shin, S. K. Lee, S. P. Suh, and D. W. Ryang. 2002. Biofilm production by isolates of Candida species recovered from nonneutropenic patients: comparison of bloodstream isolates with isolates from other sources. J. Clin. Microbiol. 40:1244-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srikantha, T., S. A. Lachke, and D. R. Soll. 2003. Three mating type-like loci in Candida glabrata. Eukaryot. Cell 2:328-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tavanti, A., A. D. Davidson, N. A. Gow, M. C. Maiden, and F. C. Odds. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43:284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavanti, A., A. D. Davidson, N. A. R. Gow, M. C. J. Martin, and F. C. Odds. Candida orthopsilosis and Candida metapsilosis, spp. nov., to replace Candida parapsilosis groups II and III. J. Clin. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 48.Tsong, A. E., M. G. Miller, R. M. Raisner, and A. D. Johnson. 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115:389-399. [DOI] [PubMed] [Google Scholar]
- 49.Whelan, W. L., and K. J. Kwon-Chung. 1988. Auxotrophic heterozygosities and the ploidy of Candida parapsilosis and Candida krusei. J. Med. Vet. Mycol. 26:163-171. [PubMed] [Google Scholar]
- 50.Wickner, R. B. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol. Rev. 60:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, S., M. A. Fares, W. Zimmermann, G. Butler, and K. H. Wolfe. 2003. Evidence from comparative genomics for a complete sexual cycle in the ‘asexual’ pathogenic yeast Candida glabrata. Genome Biol. 4:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.