Abstract
Age-related macular degeneration (AMD), a complex neurodegenerative disease, is a leading cause of visual impairment worldwide with a strong genetic component. Genetic studies have identified several loci, but few causal genes with functional characterization. Here we highlight multiple lines of evidence which show a causal role in AMD for SLC16A8, which encodes MCT3, a retinal pigment epithelium (RPE) specific lactate transporter. First, in an unbiased, genome-wide analysis of rare coding variants we show multiple SLC16A8 rare variants are associated with AMD risk, corroborating previous borderline significant reports from AMD rare variant studies. Second, we report a novel SLC16A8 mutation in a three-generation family with early onset macular degeneration. Finally, mis-expression in multiple model organisms shows functional and anatomic retinal consequences. This study highlights the important role for SLC16A8 and lactate regulation towards outer retina/RPE health and highlights a potential new therapeutic opportunity for the treatment of AMD.
Subject terms: Genome-wide association studies, Genetics
Introduction
Age-related macular degeneration (AMD) is a complex, neurodegenerative disease. It is a leading cause of visual impairment worldwide projected to affect 288 million people globally by 20501. A progressive disease, AMD is classified under early, intermediate, and advanced stages leading to the loss of retinal pigment epithelium (RPE) and photoreceptors (PR) located in the macular region responsible for color vision and central visual acuity. Two subtypes for advanced AMD have been described as macular neovascularization (MNV) and geographic atrophy (GA), both of which can develop from early/intermediate stages over time. There is a significant genetic component estimated to account for 47% of variability in disease risk2–6.
While the investigation of multiple AMD risk loci has led to successful identification of causal genes, others have not been as thoroughly interrogated. One approach to identify the causal gene at a locus has been to use rare protein altering variant analyses in combination with functional characterization in laboratory models to pinpoint key molecular pathways for new insights and potential future therapies. Using this approach, we describe multiple lines of evidence which all point to impairment of SLC16A8, which encodes MCT3, a retinal pigment epithelium (RPE)7 specific lactate transporter as a causal contributor to AMD risk burden. The locus containing this gene has been seen in common variant AMD GWAS5,6, however co-localization studies for the lead GWAS SNP have not been able to identify the causal gene at the locus8,9. Furthermore, a rare, splice donor variant in SLC16A8 has been seen in a patient with adult vitelliform macular dystrophy (AVMD)10 and achieved borderline exome-wide significance in a candidate gene, chip-based study of AMD rare variants (RVs)6. Here we show the aforementioned rare splice donor variant achieves genome-wide significance in a meta-analysis, and we broaden the association at SLC16A8 to include several missense variants in a genome-wide RV analysis. Additionally, we report a novel SLC16A8 mutation in a three-generation family with AVMD. Finally, with functional follow-up in Drosophila and mice, we characterize the effect of Slc16a8 reduction to show both functional and anatomic retinal changes.
Results
Rare coding variants are associated with advanced AMD
We utilized whole genome sequencing to perform a genome wide RV association study for AMD susceptibility comparing advanced AMD cases (n = 6055; 60.9% MNV, 39.1% GA) to controls (n = 8294), the largest WGS study of AMD to date (Table 1). We obtained controls of European ancestry and >50 years of age from participants in non-ophthalmic clinical trials that were sequenced and processed with the same informatics pipeline to minimize batch effects that may be introduced via differing sequencing technologies and bioinformatics processing pipelines (Supplementary Table 1). Due to the use of individuals with other, non-ophthalmic diseases as controls, we used a genotype-on-phenotype reverse regression to identify potential association signals originating from the control population11,12.
Table 1.
Population demographics of whole genome sequenced advanced AMD cases and controls
| Population Demographics | |||
|---|---|---|---|
| Covariates | Case | Control | P |
| Age, mean (sd) | 76.3 (8.1) | 61.9 (7.7) | <2E-16 |
| Female, n (%) | 3600 (59.4%) | 4878 (58.8%) | 0.45 |
| Male, n (%) | 2456 (40.5%) | 3416 (41.2%) | 0.45 |
| CNV, n (%) | 3689 (60.9%) | – | – |
| GA, n (%) | 2366 (39.1%) | – | – |
| Total (n) | 6055 | 8294 | – |
Access to WGS data on this scale allowed us to explore the role of RVs to AMD. On a per gene basis, we binned variants with a minor allele frequency <1% in two ways. First, we focused on both high and moderate impact variants predicted for either a loss or reduction in protein function (nonsense, and missense with a damaging PolyPhen score ≥ 0.5, see methods) (Table 2, Supplementary Fig. 1 and Supplementary Data 1). To test this, we used the SKAT-O algorithm using the R package SKAT. The top three hits were complement factor I (CFI; P = 3.13 × 10−14; OR = 4.57), complement factor H (CFH; P = 2.14 × 10−6; OR = 2.05), and solute carrier family 16 member 8 (SLC16A8; P = 1.50 × 10−5; OR = 1.78). CFH and CFI were the only genes in the analysis that exceeded the Bonferroni corrected p value of 2.64 × 10−6. Notably, all three are found within loci identified previously via GWAS from Fritsche et al.6 (Table 2 and Supplementary Fig. 1). Second, in a more restrictive test, we focused only on predicted high impact loss of function variants (nonsense, see methods). The top genetic hit from this loss of function RV burden test was for SLC16A8 (P = 6.91×10−4; OR = 1.71) (Table 2 and Supplementary Fig. 1).
Table 2.
Missense and nonsense protein altering rare variant burden tests predict multiple genes associated with advanced AMD
| Gene | Chr. | Variants (N) | Carriers (N) | Freq. Cases | Freq. Controls | OR | P |
|---|---|---|---|---|---|---|---|
| Missense and/or Nonsense RV burden tests (P 10E-04) | |||||||
| CFI | 4 | 40 | 218 | 0.0261 | 0.0072 | 4.57 | 3.13E-14 |
| CFH | 1 | 39 | 322 | 0.0269 | 0.0192 | 2.05 | 2.14E-06 |
| SLC16A8 | 22 | 25 | 440 | 0.0398 | 0.0240 | 1.78 | 1.50E-05 |
| CFHR5 | 1 | 26 | 292 | 0.0119 | 0.0265 | 0.48 | 5.58E-05 |
| AAED1 | 9 | 12 | 80 | 0.0084 | 0.0035 | 3.28 | 1.67E-04 |
| RGS7 | 1 | 7 | 44 | 0.0046 | 0.0019 | 4.25 | 2.87E-04 |
| MAP1S | 19 | 24 | 511 | 0.0322 | 0.0381 | 0.63 | 3.12E-04 |
| OSGIN2 | 8 | 9 | 29 | 0.0020 | 0.0020 | 6.02 | 3.74E-04 |
| Nonsense RV burden tests (P < 10E-04) | |||||||
| SLC16A8 | 22 | 5 | 301 | 0.0284 | 0.0156 | 1.71 | 6.91E-04 |
| DNAH3 | 16 | 26 | 67 | 0.0036 | 0.0054 | 0.32 | 8.53E-04 |
While Fritsche et al reported rare (<1% MAF) functional variation, it was not a sequencing study and variants had to be pre-selected for inclusion on the array6. Using this method, authors identified CFH, CFI, TIMP3 and SLC16A8 passing a relaxed threshold for RV significance of P = 7×10−5 in their RV burden test. For SLC16A8, RV analysis tested 9 SNPs (P = 3.1×10−6; Fritsche et al Supplementary Dataset 4). The RV association was driven by the signal (P = 9.1×10−6; OR = 1.5) for one predicted truncation (rs77968014, c.214+1 G > C), whereas the other 8 SNPs showed little difference in counts of cases (0.8% with a RV) and controls (0.7%)6.
The result for rs77968014 does not meet the conventional threshold for either genome- or exome-wide significance in Fritsche et al.6. As such, we sought to understand the strength of replication in our dataset, especially as both datasets share common AMD case patient DNA (AREDS1). When removing AREDS1 patients with SNPs tested in Fritsche et al (common data between these studies), we replicate the association at rs77968014 (N = 6039; P = 0.01; OR = 1.50). Seeking further confirmation, we investigated rs77968014 using PheWAS in the FinnGen dataset (r10.finngen.fi) showing a directionally consistent association for “Age-related macular degeneration (whether dry or wet)” (MAF = 0.004 in AMD cases; P = 0.008; OR = 1.44). A meta-analysis for rs77968014 between the three cohorts (Fritsche et al, this study not including AREDS1, and FinnGen) reaches genome-wide significance (P = 2.58 × 10−8).
This study utilized WGS to identify SNPs at any allele frequency. As such, we obtained a clearer picture of the frequency of rare alleles which affect AMD risk compared to chip-based methods. In addition to rs77968014, we found 43 additional rare protein altering SLC16A8 variants in this study population. We tested if, as in Fritsche et al, rs77968014 was driving the burden test association. We removed rs77968014 from inclusion in the burden test for SLC16A8 and tested the remaining 43 SNPs in a separate burden test while conditioning on rs77968014 (in addition to age, sex and genomic ancestry as done previously). The association with AMD remains with a similar difference in case and control frequencies as in Fritsche et al. for SLC16A8 predicted protein altering variants other than rs77968014 (Freq=0.9% in cases; 0.8% in controls; P = 0.04; OR = 1.44). This result is consistent with the hypothesis that multiple SLC16A8 rare coding variants in addition to rs77969014 independently contribute to AMD risk.
SLC16A8 variants were spread across multiple cytoplasmic, extracellular, C- and N-terminal domains. We classified SLC16A8 variants in our AMD cohort according to ACMG/AMP sequence variant interpretation guidelines (Supplementary Data 2)13. We found rs77968014 was pathogenic, located at the 5′ end of intron 3 (of 5 total). We identified other likely pathogenic variants: rs777137024 (Val236fs, c.706delG), rs775208331 (splice donor, c.1198+1 G > C, 5’ end of intron 5) and rs757440454 (splice acceptor, c.1199-2 A > C, 3′ end of intron 5). All missense variants included in the RV burden test had a PolyPhen score ≥0.5. Additionally, we classified the 37 SLC16A8 missense variants with AlphaMissense14. We found 10 variants to be likely pathogenic (score >0.74), one ambiguous (0.53) and the remainder likely benign (<0.3). All 10 likely pathogenic variants map to helical or cytoplasmic domains.
Thus, our findings around SLC16A8 rare variation (1) conclusively ties rs77968014 to AMD risk (P = 2.58 × 10−8), and (2) suggests that not only protein truncation caused by rs77968014, but also overall impairment of SLC16A8 can impact risk for advanced AMD.
Rare SLC16A8 variants are associated with semi-dominant macular degeneration
Further evidence for the causal role in macular degeneration is seen in familial studies of adult vitelliform macular dystrophy (AVMD), part of a wide array of macular disorders which are characterized by the distinct shape and egg-yolk like color of a solitary foveal lesion, but have considerable overlap with AMD15. AVMD onset is typically around 40–50 years of age and tends to be sporadic in nature, but has been canonically associated with mutations in BEST1 (a calcium-activated chloride channel located at the basal side of RPE) or PRPH2 (a photoreceptor specific tetraspanin essential for photoreceptor outer segment phagocytosis by RPE). Patients with mutations in either gene, and with vitelliform lesions in general, exhibit RPE defects due to impaired phagocytosis15,16.
Genetic predisposition to AVMD was expanded to SLC16A8 in a previous report of an AVMD patient homozygous for rs77968014, the same splice donor variant originally seen in Fritsche et al.6,10, and to our knowledge the only report of SLC16A8 variation in AVMD. We further searched for rare SLC16A8 variants in patients with AVMD but without plausible pathogenic variants in canonical disease genes BEST1 and PRPH2.
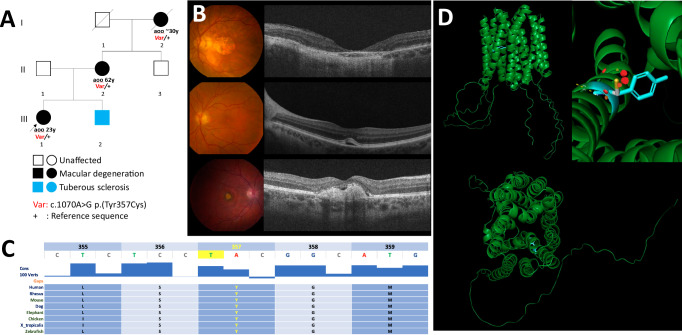
We identified a proband experiencing sudden onset of central visual loss OD at 23 years of age (individual III-1, Fig. 1A). Genome sequencing unveiled a heterozygous variant c.1070 A > G p.(Tyr357Cys) located in exon 5 of SLC16A8. This variant was considered a high-suspicion VUS according to ACMG/AMP sequence variant interpretation guidelines. Her mother (II-2) had vitelliform macular lesion at the age of 62 years and her grandmother (I-2) had macular degeneration first detected around 30 years of age (Fig. 1A). Aspects of the retinal phenotype for this family of European descent are highlighted in Fig. 1B. The top panel shows left eye color fundus photography and optical coherence tomography (OCT) for the grandmother at 94 years of age. Findings include a well circumscribed geographic area of retinal and RPE atrophy in the central macula. A small number of drusen are present temporally. Best-corrected visual acuity (BCVA) at this visit was 20/800 (logMAR 1.6); the right eye was affected to a lesser degree and maintained 20/100 acuity (logMAR 0.7). The middle panel depicts the fundus findings for the proband’s mother at 62 years of age. A vitelliform foveomacular lesion was noted with subRPE hyporeflective changing. Visual acuity was measured at 20/20 in both eyes (logMAR 0.0) and the right eye was not affected. The bottom panel shows the proband’s right eye on presentation at the age of 24 years. The proband presentation was due to acute reduction in right eye visual acuity a few months prior to her visit. The main findings of the assessment were a reduction in BCVA to 20/125 (logMAR 0.8) in the right eye while the acuity was maintained at 20/25 (logMAR 0.1) in the left eye. Right eye fundus exam and imaging were consistent with foveal subretinal fibrotic changes and an RPE tear. At the time of assessment, choroidal neovascularization was suspected. The left eye had a foveal vitelliform lesion with superior pigmentation.
Fig. 1. A family affected by vitelliform macular dystrophy.
A Pedigree. Aoo, Age of onset. All three affected harbors the c.1070 A > G p.(Tyr357Cys) variant in SLC16A8. B Fundus images and OCT of proband (top, 35 yrs), mother (middle, 62 yrs) and grandmother (bottom, 94 yrs). C Tyr357 and neighboring residues are well conserved among vertebrates. Image adapted from the UCSC Genome Browser http://genome.ucsc.edu42. D Structural modeling of MCT3 encoded by SLC16A8 as predicted by AlphaFold (AlphaFold DB version 2022-11-01 created with the AlphaFold Monomer v2.0 pipeline) and visualized in PyMOL. The Tyrosine 357 residue is highlighted in blue. The protein is displayed with its 12 transmembrane domains forming a pore seeing from the cytoplasm view (bottom). Top right panel illustrates the substitution of Tyr357 (blue) by cysteine (pink and yellow stick). The red round fragments represent disruption of the protein structure as predicted by PyMOL.
The p.(Tyr357Cys) variant segregated with disease in the family (Fig. 1A). This variant is absent in gnomADv4 and affects a highly conserved residue (Fig. 1C). It was predicted to be deleterious by multiple in-silico prediction tools including AlphaMissense (score 0.76). To gain further insights into the potential structural implications of this variant, we conducted protein structure modeling using PyMOL based on AlphaFold protein structure prediction. The variant is located in helical transmembrane domain 10 (of a total of 12) that form the channel pore (Fig. 1D). The model suggests that the substitution of the aromatic ring of Tyr357 by a highly reactive sulfur group of the cysteine orienting towards the channel pore may reduce the ability to transport lactate (Fig. 1E).
A previous study utilizing single cell nuclear RNA sequencing in non-AMD controls observed SLC16A8 expression to be highly specific for RPE cells in the human eye7. Given the strong human genetics evidence around SLC16A8 and its RPE specific expression pattern, we sought to further characterize these findings with a functional screening strategy in multiple animal models starting with Drosophila melanogaster.
The Drosophila SLC16A8 orthologue sln maintains retinal homeostasis
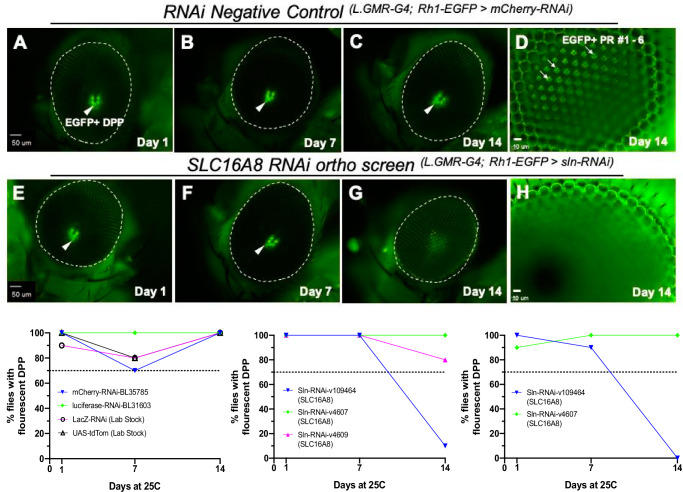
Drosophila melanogaster provides multiple benefits for functional characterization, including (1) the availability of genome-wide libraries of transgenic lines for tissue-specific RNAi knockdowns, (2) cell-specific fluorescent labeling for phenotypic readouts, and (3) fast characterization of the progressive loss of retinal homeostasis. We identified the Drosophila ortholog of SLC16A8, Silnoon (sln) using the DRSC Integrative Ortholog Prediction Tool (DIOPT)17 (Fig. 2 and Supplementary Table 2) and performed functional assessment using available Drosophila RNAi lines18,19 (Supplementary Table 2, methods). We specifically asked whether perturbation of sln in the whole Drosophila eye (L.GMR-GAL4, ninaE-EGFP > UAS-RNAi) (methods) would influence retinal homeostasis, using fluorescent deep pseudopupil (DPP) imaging as a readout20,21. Observation of these ‘pseudopupil’ virtual images depends on normal ommatidial structure and arrangement, and loss of photoreceptor numbers or integrity will be reflected in the loss of the DPP projections20,21. We scored for DPP phenotypes based on the shape and/or intensities of EGFP expression patterns of labeled photoreceptors at days 1, 7, and 14 after induction of dsRNA expression (Fig. 2). A set of negative control RNAi lines targeting genes not normally expressed in Drosophila were also screened. A threshold was set based on the negative control line for intact DPP (70% for UAS-mCherryRNAi); and any gene knockdown resulting in an average DPP loss below this threshold was considered as a phenotype worthwhile investigating towards novel insights into AMD pathobiology (Fig. 2 and Supplementary Table 2; methods).
Fig. 2. Loss of Drosophila photoreceptor EGFP expression patterning in sln RNAi-knockdown eyes by Day 14.
Rhodopsin 1 EGFP labeling of photoreceptor rhabdomeres visualized by fluorescent deep pseudopupil (DPP) and optic neutralization imaging of the cornea (ONC) at days 1, 7, and 14 for negative controls (A–C) Low magnification DPP (4×) and (D) high magnification ONC (40×) imaging of RNAi knockdowns for mCherry (RNAi-BL35785). E–G RNAi knockdown for sln (VDRC#: 109464). D, H Only a subset of EGFP + PR are in focus under the microscope due to the curvature of the compound eye. Bottom panel graphs showing percentage of flies with intact fluorescent DPP. In all negative controls tested (left graph), 70–100% of flies had intact DPP by day 14 depending on the RNAi line and therefore a 70% threshold was set for phenotypes. Two separate knockdown experiments using silnoon (sln) RNAi-VDRC#109464 lines revealed 10% and 0% of flies had intact DPP by day 14 (Bottom: middle and right graphs). For each RNAi line tested n = 10 for each days 1, 7, and 14 timepoints. Scale bars: A–C, E–G = 50 µm; D and H = 10 µm.
The knockdown of Drosophila SLC16A8 ortholog sln (VDRC-109464) showed retinal degeneration over time. On average, only 5% of flies expressing the sln targeting RNAi line VRDC-109464 had intact DPP by day 14 (reproduced in two separate assays; n = 10 for each line and timepoint). Two other sln RNAi lines expressing identical dsRNA constructs (VDRC-4607 and VDRC-4609) were also screened, but showed no DPP loss at any timepoint tested. This was consistent with inefficient sln knockdowns for these two specific lines reported previously22 (Fig. 2 and Supplementary Table 2; and methods). Previous reports identified sln23 as a monocarboxylate transporter in Drosophila and an ortholog to the human proton-coupled monocarboxylate lactate transporter SLC16A8 (DIOPT). Multiple other orthologs of AMD risk genes identified in our genome-wide RV analysis were screened using this approach (such as CFH; Table 2), but revealed limited to no DPP loss in the samples tested (Supplementary Table 2). Based on these intriguing results for sln in Drosophila, we sought to further characterize changes in retinal homeostasis and function after Slc16a8 loss of function in mammalian models.
Slc16a8 knockout mouse models show reduced retinal function by electroretinogram measurements
Mice have an RPE specific expression pattern for Slc16a8, similar to humans10. We examined the functional role of Slc16a8 in a mouse model of retinal degeneration. Constant light exposure (CLE) in rodents has been widely used as an injury model towards better understanding the interplay of genetics and environment that may progressively lead to the loss of retinal homeostasis2,4,24,25. To create a loss of function variant, we used CRISPR to delete exons 1-6 of Slc16a8 on the C57BL/6 J mouse background strain. We then utilized CLE to examine the functional consequences of Slc16a8 loss in combination with light-induced damage (Fig. 3; methods). We utilized electroretinogram (ERG) recordings as a measure of overall retinal function and health at baseline and after CLE. For this study, full-field ERGs (150 cd*s/m2) were measured at baseline and after 7 days of CLE in the same mouse cohorts to determine retinal rod photoreceptor (PR), bipolar cell (BC) and RPE cell function as represented by a-, b-, and c-wave amplitudes, respectively26 (see methods). These non-invasive measurements of retinal activity provide direct longitudinal readouts for comparison between genotypes in the same mouse cohorts over time after CLE for cell types known to be damaged or lost in AMD patients.
Fig. 3. Schematic of constant light exposure (CLE) experimental design for retinal damage in mouse models.
At baseline, measurements were taken using optical coherence tomography (OCT) and electroretinogram (ERG) recordings. Mice were placed in double-housed light boxes at Day 0 and exposed to constant light exposure (CLE) at 100 K lux. On the evening of Day 6, were dark adapted for day 7 ERG and OCT measurements.
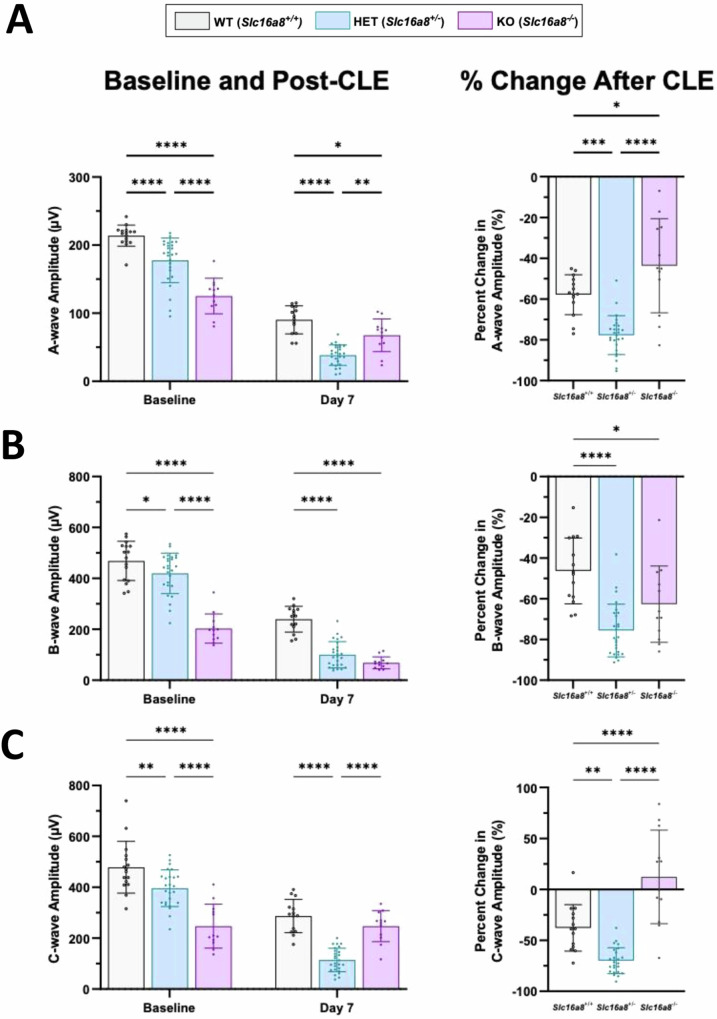
At baseline, eyes from Slc16a8 heterozygote (Het) and homozygote (KO) mice had decreased a-, b-, and c-wave amplitudes compared to WT mice, and significant differences in a-, b-, and c-wave amplitudes were also observed between Slc16a8 KO and Het mice (all P < 0.0001) (Fig. 4A–C). Loss of Slc16a8 thus led to gene dose-dependent visual impairment. CLE reduced a-, b-, and c- wave amplitudes in eyes from Slc16a8 WT mice when compared to baseline measurements, confirming model-induced functional deficits in control mice. After 7 days of CLE, eyes from Slc16a8 KO mice had decreased a-wave (P < 0.05) and b-wave (P < 0.0001) amplitudes, but no significant difference in c-wave amplitudes, when compared to Slc16a8 WT mice post light exposure (Fig. 4A–C). Eyes from Slc16a8 Het mice had decreased a-, b-, and c-wave amplitudes (all P < 0.0001) when compared to Slc16a8 WT eyes following 7 days CLE (Fig. 4A–C).
Fig. 4. ERG measurements before and after CLE-induced retinal damage.
A-, b-, and c-wave (A–C; left panels) ERG amplitudes (µV) were measured at baseline and Day 7 for Slc16a8-WT, Het, and KO mice. Data is plotted as peak amplitudes (Mean ± SD), all n ≥ 12 eyes. A–C; right panels: Percent change following CLE are shown for A-, b- and c-waves (Mean ± SD). A For a-wave amplitudes (µV) at baseline: Slc16a8 WT (213.87 ± 15.5), Het (177.63 ± 32.8), and KO (125.15 ± 26.14). A-wave baseline all P < 0.0001. A-wave amplitudes (µV) day 7: WT (90.30 ± 20.6), Het (38.39 ± 15.1), and KO (67.56 ± 23.96). At day 7 for WT vs. Het (P < 0.0001), WT vs. KO (P = 0.0474), and Het vs. KO (P = 0.0018). A-wave % change WT (-57.84 ± 9.8), Het (-77.70 ± 9.5), and KO (-43.68 ± 23.0). A-wave % change WT vs. Het (P < 0.005), WT vs. KO (P < 0.05), and Het vs. KO (P < 0.0001). B B-wave baseline amplitudes (µV): WT (468.66 ± 77.3), Het (419.65 ± 79.1), and KO (202.97 ± 57.5). B-waves baseline: WT vs. Het (P = 0.0443), WT vs. KO (P < 0.0001), and Het vs. KO (P < 0.0001). Day 7 B-wave amplitudes (µV): WT (239.78 ± 50.67), Het (100.15 ± 51.53), and KO (68.60 ± 23.33). Day 7 b-waves: WT vs. Het and WT vs. KO (both P < 0.0001). B-wave % change WT (-46.3 ± 16.2), Het (-75.60 ± 13.0), and KO (-62.67 ± 18.7). B-wave % change WT vs Het (P < 0.0001) and WT vs. KO (P < 0.05). C C-wave amplitudes (µV) at baseline: WT (478.57 ± 101.43), Het (396.01 ± 72.0), and KO (247.3 ± 86.10). C-wave baseline: WT vs. Het (P < 0.01), WT vs. KO (P < 0.0001), and Het vs. KO (P < 0.0001). Day 7 C-wave amplitudes (µV): WT (286.79 ± 65.24), Het (114.41 ± 46.0), and KO (247.24 ± 61.2). Day 7 WT vs. Het (P < 0.0001), WT vs. KO (P < 0.0001). C-wave % change WT (-37.80 ± 22.8), Het (-70.0 ± 12.64), and KO (12.26 ± 45.87). C-wave % change WT vs. Het (P < 0.01), WT vs. KO (P < 0.0001), and Het vs. KO (P < 0.0001). For a, b, and c-wave analysis: (A–C; left panels) Mixed-effects analysis: a-wave: F(2, 49) = 25.61, P < 0.0001, b-wave: F(2, 49) = 19.02, P < 0.0001, c-wave: F(2, 99) = 31.41, P < 0.0001); multiple comparisons are between groups within timepoints, Sidak post-hoc., (A-C; right. panels) One-way ANOVA (a-wave: F(2, 49) = 27.13, P < 0.0001, b-wave: F(2, 49) = 16.82, P < 0.0001, c-wave: F(2, 48) = 39.14, P < 0.0001), Sidak post-hoc, % Change computed as (Day 7–Baseline)/Baseline *100.
Moreover, we also compared the percent change in amplitude from baseline and after 7 days CLE as a measure of longitudinal change between genotypes over time. In eyes from Slc16a8 WT mice there were reductions in a-, b-, and c-wave amplitudes following CLE in comparison to baseline (Fig. 4A–C, right panel). Moreover, eyes from Slc16a8 Het mice also had reductions in a-, b-, and c-wave amplitudes following CLE in comparison to baseline (Fig. 4A–C, right panel). However, eyes from Slc16a8 KO mice had reduced a- and b-wave amplitudes with no change in c-wave amplitudes following CLE (Fig. 4A–C, right panel). When comparing between genotypes as a measure of percentage change over time, eyes from Slc16a8 Hets had significant reductions in percent change for a-wave amplitudes (µV) vs Slc16a8 WT and KO (Fig. 4A, right panel; P < 0.001 for both). Meanwhile, eyes from Slc16a8 Hets had significant percent change reductions in b-wave amplitudes vs Slc16a8 WT (P < 0.0001) with no significant differences in comparison to Slc16a8 KO (Fig. 4B, right panel). In addition, eyes from Slc16a8 Hets had more significant reductions in c-wave amplitudes in comparison to Slc16a8 WT and KO over time following CLE (Fig. 4C, right panel; P < 0.01 and P < 0.0001, respectively). Together, these findings reveal a Slc16a8 dose dependent loss of photoreceptor, bipolar cell, and RPE cell function at baseline as well as significantly increased sensitivity to CLE in the eyes from Slc16a8 Het mice compared to both Slc16a8 WT and KO groups.
Measurements of total retinal thickness by OCT
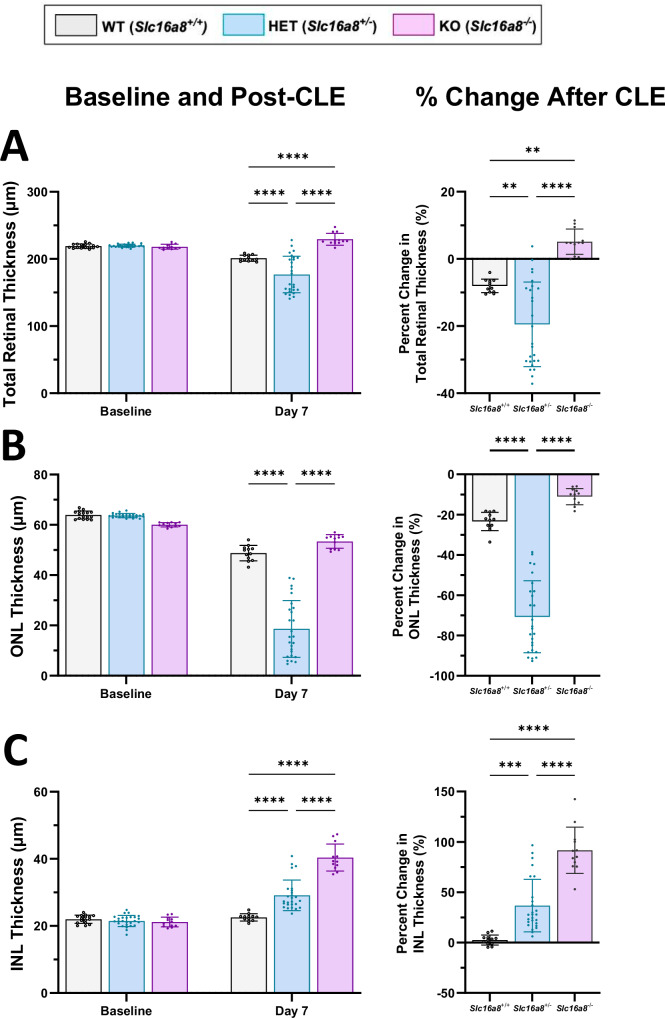
OCT is an established, non-invasive method to measure retinal thickness and determine retinal integrity. In this study, OCT measurements were acquired at multiple timepoints for total retina, outer nuclear layer (ONL), and inner nuclear layer (INL) thickness of the same mice used for ERG measurements in this study27. At baseline, no differences in total retinal layer thickness were observed between genotypes of 10-week old mice (Fig. 5A). Following 7 days CLE, retinas from Slc16a8 Het (P < 0.0001) and WT (P < 0.0001) showed reduced total thickness, while Slc16a8 KO retina total thickness increased compared to both Het and WT (P < 0.0001 for both; Fig. 5A). Similar findings were observed when total retinal thickness in Slc16a8 Het eyes were measured as a percent change in comparison to WT (P < 0.005) or KO (P < 0.0001) (Fig. 5A).
Fig. 5. OCT measurements of total, ONL, and INL retinal thickness before and after CLE-induced damage.
Total retinal, ONL and INL thickness were measured (µM) (A-C; left panels) by OCT at baseline and Day 7 of CLE. All data shown as mean ± SD, all n ≥ 12 eyes. Percent change following CLE are shown for Total, ONL and INL thickness in right panels (Mean ± SD). A For total retina thickness (µm) at baseline: Slc16a8 WT (219.11 ± 3.19), Het (219.86 ± 2.24), and KO (218.18 ± 3.6). For total retinal thickness (µm) at day 7: WT (201.11 ± 4.5), Het (176.91 ± 27.3), and KO (229.32 ± 8.9). For total retinal thickness at day 7 all P < 0.0001. For % change total retinal thickness (µm) WT (-8.07 ± 2.0), Het (-19.50 ± 12.59), and KO (5.11 ± 3.77). For % change total retinal thickness (µm) WT vs. Het (P < 0.01), WT vs. KO (p val<0.01), and Het vs. KO (p val<0.0001). B For ONL thickness at baseline: WT (63.93 ± 1.57), Het (63.93 ± 0.82), and KO (60.03 ± 0.85). For ONL thickness (µm) at day 7: WT (48.79 ± 3.06), Het (18.61 ± 11.3), and KO (53.40 ± 2.73). For ONL thickness (µm) at day 7: WT vs Het and Het vs. KO (p val<0.0001). For % change ONL thickness (µm) ONL WT (-23.39 ± 4.54), Het (-70.72 ± 17.84), and KO (-11.07 ± 4.03). C For INL thickness (µm) at baseline: WT (21.98 ± 1.22), Het (21.48 ± 1.65), and KO (21.15 ± 1.45). For INL thickness (µm) at day 7: WT (22.55 ± 1.05), Het (29.13 ± 4.56), and KO (40.38 ± 4.03). For INL thickness all p val<0.0001. For % change INL thickness (µm) WT (2.60 ± 5.02), Het (36.86 ± 26.11), and KO (91.70 ± 23.0). For % change INL thickness (µm) WT vs. Het (p val<0.001), WT vs. KO p val<0.0001), Het vs. KO (p val<0.0001). For total, ONL, and INL thickness comparisons: (A–C; left panels). Mixed-effects analysis (interaction between genotype and timepoint as follows TRT: F(2, 98) = 29.97, P < 0.0001, ONL: F(2, 98) = 102.9, P < 0.0001, INL: F(2, 98) = 67.21, P < 0.0001), multiple comparisons reported are between groups within timepoints, Sidak post-hoc., (A–C; right.panels) One-way ANOVA (TRT: F(2, 47) = 28.75, P < 0.0001, ONL: F(2, 47) = 102.9, P < 0.0001, INL: F(2, 47) = 49.78, P < 0.0001), Sidak post-hoc % Change computed as (Day 7 – Baseline)/Baseline *100.
OCT segmentation analyses were carried out to pinpoint the effects of CLE on ONL/INL thickness. At baseline, no differences in ONL/INL retinal layer thickness were observed between genotypes at 10 weeks old (Fig. 5B, C). Following 7 days CLE, the ONL thickness of Slc16a8 Hets was significantly reduced in comparison to WT or KO and when measured as percentage change (Fig. 5B; P < 0.0001 for all). Meanwhile, the INL thickness of Slc16a8 KO (P < 0.0001) and Het (P < 0.0001) retinas significantly increased in comparison to WT (Fig. 5C). Similar results were observed when INL thickness in Slc16a8 Het and KO eyes were measured as a percent change in comparison to WT (P < 0.001 and P < 0.0001, respectively; Fig. 5C). These findings suggest that loss of one Slc16a8 allele is sufficient to sensitize the mouse retina to CLE, leading to the loss of cells in the ONL and edema of the INL, both involved in AMD pathology. It remains unclear why ONL thickness was not reduced in animals with complete loss of Slc16a8, but previous studies using Slc16a8 KO mice show complete loss of Slc16a8 leads to compensatory increase in MCT1, a related family member which imports lactate from photoreceptors found on the apical side of the RPE28,29. Further studies are needed to explore the impact of such compensation on retinal structure and function.
Slc16a8 promotes retinal integrity in the mouse
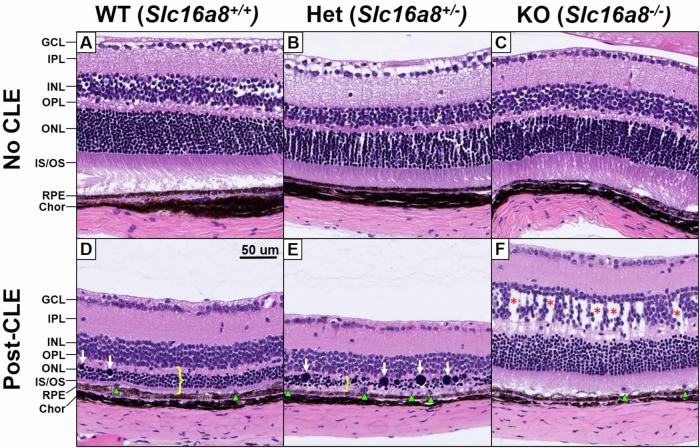
To better understand the changes in thickness of retinal layers observed by OCT, we used hematoxylin/eosin (H&E) histological staining of formalin-fixed, paraffin-embedded eyes on sagittal sections at the level of the optic disc. All slides were evaluated by a pathologist blinded to genotype and treatment group. CLE pathology is typically inconsistent and patchy across the retina, with large regions entirely spared, thus only regions with the strongest pathological features for each slide were evaluated, focusing on the region 0.5–1.0 mm from the optic disc when possible. At baseline, 10-week old mice had normal retinal architecture and cell numbers for all genotypes (Slc16a8 WT, Het, and KO) (Fig. 6A–C). After 7 days of CLE, typical features of CLE were observed in the retinas of WT animals (n = 6), including thinning of the ONL and PR layers, swollen and hyperchromatic ONL nuclei, displaced PR nuclei across the external limiting membrane into the PR layer, subretinal phagocytic cells, and less prominently, dysmorphic RPE cells demonstrating decreased pigmentation and multilayering. These ONL and PR features were more severe in the Slc16a8 Het retinas (n = 14), but no additional or unique features were observed. In contrast, all of the Slc16a8 KO retinas (n = 6) showed thickening in the INL and OPL, which was not present in any of the WT or Het retinas by H&E histology. In addition, the pathologic features in the ONL, PR, and RPE observed in WT and Het retinas were dramatically reduced in the KO retinas (Fig. 6D–F). Overall, these features are consistent with the OCT analysis, and demonstrate more severe pathology in Hets and alternative pathology (INL/OPL edema) in the KOs.
Fig. 6. Histologic evaluation of Slc16a8 loss of function mouse retinae at baseline and post-CLE.
Representative sections of hematoxylin and eosin (H&E) stained retina, photographed at the areas with greatest pathology (approximately 0.5–1 mm from the optic disc). At baseline, 10-week-old non-CLE samples demonstrate normal cellularity and architecture across all retinal layers in Slc16a8 WT, Het, and, KO genotypes (A–C). Total retinal thinning following CLE was exacerbated in Slc16a8 Het retinas compared to WT, attributable entirely to degeneration of the photoreceptors, manifesting as thinning of the OPL, ONL, and OS/IS layers (D, E; yellow bracket). This was accompanied by and proportional to swollen and hyperchromatic photoreceptor nuclei (white arrows) subretinal phagocytic cells (green arrowheads), and dysmorphic RPE cells. Edema of the INL was present in all Slc16a8 KO retinas after CLE (F; red asterisks), but not observed in any WT or Het retinas. GCL ganglion cell layer, INL/ONL inner/outer nuclear layers, IPL/OPL inner/outer plexiform layers, IS/OS inner/outer segmented layers, RPE retinal pigment epithelium, Chor choroid.
Discussion
Here, we report data from multiple lines of evidence showing a causal role for SLC16A8 in macular degeneration. First, the genetic association at SLC16A8 was broadened to include additional variants in addition to the two identified in the Fritsche et al.6 study and we show genome-wide statistical significance for rs77968014, a rare splice donor variant in SLC16A8. Second, Drosophila knockdown showed SLC16A8 ortholog sln mediated monocarboxylate transport deficiencies can lead to changes in retinal homeostasis. Third, ERG and OCT measures reveal that absence of Slc16a8 leads to dose dependent loss of retinal function over time, and, in combination with CLE damage, reveal reduced ONL cellularity and increased INL edema. Future work should evaluate these structural and functional changes at various timepoints after CLE to determine if there is any retinal recovery or more retinal decay as time progresses. These observations provide new insights into SLC16A8 function using longitudinal measures in a mammalian model before and after CLE damage. Additionally, the RPE specific expression pattern for SLC16A8 further highlights the potential importance of this gene for retinal health with potential therapeutic implications for the treatment of AMD8.
Two recent studies, from Acar et al.30 and Kwong et al.31, have used WGS in a similar manner to identify AMD risk genes, containing data from ~2400 AMD cases each (approximately 40% of the cases used in this study). Acar et al contains a subset of AMD patient DNA used in this current study from clinical trials for geographic atrophy secondary to AMD (CHROMA, SPECTRI and PROXIMA) and neovascular AMD (HARBOR). Kwong et al, contains a different subset of AMD patient DNA (AREDS1). All three studies identify associations at CFH and CFI with AMD risk. Interestingly, Kwong et al also reports SLC16A8, but suggest this finding may be a false positive due to possible differences in sequencing depth, variant calling algorithms or population structure. The results from the current study, which does not have these differences between cases and controls, shows the SLC16A8 result from Kwong et al was not a false positive, and that missense and nonsense variation in SLC16A8 are significant contributors to AMD risk. Kwong et al also identified rare variation in ORMDL2 as associated with AMD risk, which did not replicate here (P > 0.4).
We see a genome-wide specific P value for SLC16A8 splice donor variant rs77968014 in a meta-analysis. A recent study recently characterized the effect of this variant using SLC16A8 loss of function human iPSC-RPE derived from the fibroblasts of a homozygous rs77968014 carrier with AFVD10. Analysis revealed rs77968014 leads to the loss of SLC16A8 functionality and deficits in lactate transport10. The model organism findings in this current study show that absence of Slc16a8 and sln directly results in decreased retinal health, which, based on the iRPE data described above, can at least be partially attributed to impaired lactate transport; other SLC16 transporters have been shown to transport other monocarboxylates such as pyruvate32.
Additional evidence implicating SLC16A8 in macular degeneration is derived from a three-generation family sharing the c.1070 A > G variant, which is considered a variant of uncertain significance according to the current ACMG/AMP sequence variant interpretation guidelines13. Functional studies and phenotyping of additional individuals harboring this variant would be needed to understand its role in AVMD. The retinal phenotype in this family includes findings of retinal drusen and macular circumscribed geographic atrophy with severe loss of vision by the 9th decade of life, unilateral vitelliform deposits, and subretinal fibrosis with RPE tear and suspected neovascularization. The variable expressivity of the phenotype in this family is consistent with the suggested autosomal dominant inheritance. The age of onset supports the diagnosis of an AFVD and some of the clinical manifestations are similar to findings in AMD.
SLC16A8 encodes monocarboxylate transporter 3 (MCT3), localized to the basal side of the RPE. Together with MCT1 (encoded by SLC16A1), a lactate transporter found at the apical side of the RPE, it transports lactate from the outer retina through the RPE and into the choroid10. Both MCT1 and MCT3 are proton-linked transporters of lactate and the direction of transport is determined by the concentration of protons and lactate across the plasma membrane32. As such, impairment of MCT3, at the basal side of the RPE facing the choroid, would increase lactate concentration in the RPE33. This in turn alters the gradient needed to import lactate into the RPE from the photoreceptors where it would accumulate, and indeed this has been observed previously by Daniele et al in Slc16a8-/- mice28. However, the authors observed healthy photoreceptor cells and normal ultrastructure after 36 weeks. The current study also observes normal retinal structure without CLE, however, after CLE Slc16a8 Het retinas had more severe ONL and PR features than WT. This result shows cell loss and damage caused by aberrant Slc16a8 is only seen over time under environmental stress, similar to what an AMD patient would experience over a lifetime.
In addition to the data on SLC16A8 we have presented here, the current WGS study has broadened knowledge around rare variation CFH and CFI. We see greater than 5% of AMD patients have a rare, predicted protein altering variant in either CFH or CFI, prevalence divided equally between the two (Table 2). This is over 5X higher than estimated in AMD cases by the SNP array used in Fritsche et al.
Here, we highlight multiple lines of evidence which show a causal role for SLC16A8 in AMD. First, in the largest genome-wide, hypothesis independent RV burden analysis for AMD risk in humans we see multiple SLC16A8 rare variants contributing to AMD risk. Second, we describe a novel SLC16A8 variant in a family with AVMD. Finally, we present functional characterization in two model organism systems. This study highlights the important role for SLC16A8 and lactate regulation towards outer retina/RPE health and highlights a potential new therapeutic opportunity for the treatment of AMD.
Methods
Study design and populations
We performed a rare-variant gene-burden study using DNA derived from blood samples obtained for age-related macular degeneration patients from clinical trials for ranibizumab (NCT00061594/NCT00056836 [DAWN], NCT00891735 [HARBOR], NCT02960828 [SAVE], FU and PALMER), ranibizumab Port Delivery System (PDS) (NCT03677934 [ARCHWAY], NCT02510794 [LADDER]), faricimab (NCT03823300 [LUCERNE], NCT03823287 [TENAYA]), lampalizumab (NCT02247479 [CHROMA], NCT01229215 [MAHALO], NCT02479386 [PROXIMA A], NCT02399072 [PROXIMA B], NCT02247531 [SPECTRI], NCT02288559 [LAMPA Dosing Study]), and the National Eye Institute (NEI) studies (NCT00000145 [AREDS1], NCT00345176 [AREDS2]) (Supplementary Table 1). These study populations were selected for inclusion on the basis of available phenotypic information and DNA availability for WGS. All AMD patients and non-AMD controls in this study were aged 50 years or older and were of European ancestry, which were determined by comparison with samples from the International Haplotype Map (HapMap) Project. Family members presented in Fig. 1 were evaluated at NEI (NCT02077894).
Samples and data for controls in the study population were obtained from clinical trial studies of asthma, autoimmune disease, Crohn’s disease, chronic obstructive pulmonary disease, cancer, inflammatory bowel disease, interstitial lung disease, idiopathic pulmonary fibrosis, rheumatoid arthritis, systemic lupus erythematosus and ulcerative colitis.
Study population ethics statements
All research in this study was conducted in accordance with the Declaration of Helsinki. The Roche/Genentech data used in this study were generated from clinical trial participants who signed informed consent forms approved by the ethics committee or IRB responsible for the country or site where the trial’s participants donated samples for research. Informed consent included use of these data for genetics research. Before execution of the study, an internal Genentech team of informed consent form experts reviewed the forms from all the studies to ensure appropriate use of the samples.
For both the AREDS and AREDS2, institutional review board approval was obtained at each site and written informed consent was obtained from all participants. The research was conducted under the tenets of the Declaration of Helsinki and, for the AREDS2, complied with the Health Insurance Portability and Accountability Act.
A population used in this study consisted of patients enrolled in the FinnGen Biobank cohort. Patients and control subjects in FinnGen provided informed consent for biobank research, based on the Finnish Biobank Act. Alternatively, separate research cohorts, collected prior the Finnish Biobank Act came into effect (in September 2013) and start of FinnGen (August 2017), were collected based on study-specific consents and later transferred to the Finnish biobanks after approval by Fimea (Finnish Medicines Agency), the National Supervisory Authority for Welfare and Health. Recruitment protocols followed the biobank protocols approved by Fimea. The Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS) statement number for the FinnGen study is Nr HUS/990/2017.
The FinnGen study is approved by Finnish Institute for Health and Welfare (permit numbers: THL/2031/6.02.00/2017, THL/1101/5.05.00/2017, THL/341/6.02.00/2018, THL/2222/6.02.00/2018, THL/283/6.02.00/2019, THL/1721/5.05.00/2019, THL/1524/5.05.00/2020, and THL/2364/14.02/2020), Digital and population data service agency (permit numbers: VRK43431/2017-3, VRK/6909/2018-3, VRK/4415/2019-3), the Social Insurance Institution (permit numbers: KELA 58/522/2017, KELA 131/522/2018, KELA 70/522/2019, KELA 98/522/2019, KELA 138/522/2019, KELA 2/522/2020, KELA 16/522/2020, Findata THL/2364/14.02/2020 and Statistics Finland (permit numbers: TK-53-1041-17 and TK/143/07.03.00/2020 (earlier TK-53-90-20).
The Biobank Access Decisions for FinnGen samples and data utilized in FinnGen Data Freeze 7 include: THL Biobank BB2017_55, BB2017_111, BB2018_19, BB_2018_34, BB_2018_67, BB2018_71, BB2019_7, BB2019_8, BB2019_26, BB2020_1, Finnish Red Cross Blood Service Biobank 7.12.2017, Helsinki Biobank HUS/359/2017, Auria Biobank AB17-5154 and amendment #1 (August 17 2020), Biobank Borealis of Northern Finland_2017_1013, Biobank of Eastern Finland 1186/2018 and amendment 22 § /2020, Finnish Clinical Biobank Tampere MH0004 and amendments (21.02.2020 & 06.10.2020), Central Finland Biobank 1-2017, and Terveystalo Biobank STB 2018001.
DNA analysis
The WGS data for the study population was generated to a read depth of 30X using the HiSeq platform (Illumina X10, San Diego, CA, USA) processed using the Burrows-Wheeler Aligner (BWA) / Genome Analysis Toolkit (GATK) best practices pipeline. WGS short reads were mapped to hg38 / GRCh38 (GCA_000001405.15), including alternate assemblies, using BWA version 0.7.9a-r786 to generate BAM files. All WGS data was subject to quality control and checked for concordance with SNP fingerprint data collected before sequencing. After filtering for genotypes with a GATK genotype quality greater than 90, samples with heterozygote concordance with SNP chip data of less than 75% were removed. Sample contamination was determined with VerifyBamID software, and samples with a freemix parameter of more than 0.03 were excluded. Joint variant calling was done using the GATK best practices joint genotyping pipeline to generate a single variant call format (VCF) file. The called variants were then processed using ASDPEx to filter out spurious variant calls in the alternate regions.
Quality control—patient DNA samples
Prior to genotype QC, 15,438 samples were eligible for inclusion. Samples were excluded if the call rate was less than 90% (n = 5). Identity-by-descent analysis was used to detect and filter out relatedness in the dataset. Samples were excluded if PI_HAT was 0.4 or higher (n = 102). Samples were removed if they showed excess heterozygosity with more than three SDs of the mean (n = 222). Ancestry was assigned using an ancestry threshold of 0.7 obtained from the supervised analysis with ADMIXTURE 1.334 using samples from phase 3 of the 1000 Genomes Project as reference samples labeled by their superpopulation. Due to sample sizes, this analysis was restricted to those with EUR ancestry and the remaining ancestry groups were removed (n = 760). This resulted in cohorts comprised of controls (n = 8294) and advanced AMD cases (n = 6055) diagnosed with geographic atrophy (GA; n = 2366) and/or macular neovascularization (MNV; n = 3689) in the study population.
Quality control—genotype
Sample genotypes were set to missing if the Genotype Quality score was less than 20 and SNPs were removed if the missingness was higher than 5%. SNPs were filtered out if the significance level for the Hardy-Weinberg equilibrium test was less than 5×10-8. The allele depth balance test was performed to test for equal allele depth at heterozygote carriers using a binomial test; SNPs were excluded if the p value was less than 1×10-5.
Statistical analysis—WGS
The SKAT-O algorithm using the R package SKAT35 (https://CRAN.R-project.org/package=SKAT) was used to assess the cumulative effect of rare variants (MAF < 1%). Tests conducted in the analysis were two-sided. Variants were annotated using SnpEff and SnpSift annotation softwares36 as well as the Ensembl Variant Effect Predictor37. Variants were included in the gene burden analysis if they were classified as missense variants with a PolyPhen38 score that was predicted to be damaging (>=0.5) or if they were high impact variants (stop gain, stop loss, frameshift, start loss, and canonical splicing). The rare variant gene burden test was adjusted for age, sex and genetic ancestry. In addition, METAL software was used to perform a meta-analysis of summary statistics for 34 AMD susceptibility loci from the International AMD Genomics Consortium (IAMDGC) with summary statistics from this study6.
Since samples from other disease areas were used as controls, we used a genotype-on-phenotype reverse regression11 to test for association with the non-AMD controls. We calculated the posterior probability (PP) that a gene burden score is also associated with each individual disease area included in the control cohort; genes passing the 0.6 PP threshold in any of the control cohorts disease areas were flagged and excluded from the results. The SLC16A8 variant enrichment in AMD cohort was analyzed using Fisher’s exact test (two tailed) in Supplementary Data 2.
Variant classification
Variant classification was performed according to the ACMG/AMP variant interpretation framework with the following modifications13. PVS1 was applied to frameshift and canonical splicing variants, by downgrading c.102_109del p.(Phe35fs) for the presence of Methionine at a.a. 47, and c.1449_1459del p.(Ser483fs) for being located at the last exon and C-terminal to the basolateral sorting signal at a.a. 451-48439. PM2 was applied to variants with gnomAD v3.1.2 popmax AF < 0.0005, the threshold was 22 folds less than the maximum credible population allele frequency threshold determined based on the prevalence (~20%) of late-stage AMD among people greater than 95 years old40, the maximum allelic contribution by the SLC16A8 c.214+1 G > C (2.8%) in our AMD cohort, and penetrance of 50% (20%*2.8%/50% = 0.0112). The maximum credible population allele frequency threshold at a penetrance of 10% was 0.056 (20%*2.8%/10%), thus gnomAD v3.1.2 popmax AF of 0.05 was used for BS1 cut-off. As the only statistically-significantly enriched variant in our AMD cohort was c.214+1 G > C with gnomAD popmax AF of 0.0096, variants with gnomAD popmax AF > 0.01 and not enriched in AMD cohort (OR of allele count < 1.2) were assigned BS1_P. PP3/BP4 was applied for missense variants based on AlphaMissense prediction14.
Drosophila orthologs
Subsets of human genes were prioritized and a list of corresponding orthologs were generated using the DRSC Integrative Ortholog Prediction Tool (DIOPT) which uses a ranking system of low, moderate, and high orthologs for each gene queried. Screenings focused on high ranked orthologs for all genes. Due to the replication of SLC16A8 with Fritsche et al.6 study, one of the top-scoring moderately ranked orthologs (sln) was prioritized for screening at the time of DIOPT queries. All other human genes with only low or moderately ranked Drosophila orthologs were not included in fly screens. Genes with no readily available Drosophila RNAi stocks were also excluded from functional screens (methods; see Supplementary Table 2).
Drosophila stocks
Flies crosses were initially cultured on standard cornmeal food at 18 °C to minimize potential GAL4 mediated developmental eye defects. Then male offspring were transferred to 25 °C at 65% humidity with a 12 h light/dark cycle post-eclosion for RNAi screenings using a fluorescent microscope at days 1, 7, and/or 14. An eye-specific conditional Gal4-UAS expression system was used to conditionally express UAS-linked transgenes for RNAi screenings of candidate genes and controls. Furthermore, an EGFP reporter under the control of the rhodopsin 1 promoter (ninaE-EGFP) was used to specifically label photoreceptors #1-6 towards fluorescent imaging of fly screens.
The following fly lines were obtained from the Bloomington Drosophila Stock Center (BDSC)18: L.GMR-GAL4 (8605), ninaE-EGFP (8601), rsg7RNAi (28574), haspRNAi (65101), luciferaseRNAi (31603), mCherryRNAi (35785).
The following fly lines were obtained from the Vienna Drosophila RNAi Center (VDRC)19: slnRNAi (109464, 4607, and 4609), rsg7RNAi (101733), hasp RNAi (101250).
The following fly lines were obtained from the Jasper lab stock: UAS-tdTomato and LacZRNAi.
Drosophila sample preparation
A small drop of clear nail polish was applied on either standard microscope slides when using the 4X objective. Alternatively, nail polish was applied on small 35 mm diameter petri dishes mounted on top of microscope slides (to hold in place for microscope stage) for use with a 40X water immersion objective. The 4X objective was used for fluorescent deep pseudopupil (DPP) visualization and the 40X water immersion objective for optic neutralization of the cornea (ONC) visualization techniques21. Each CO2 anesthetized fly was then placed carefully on top of each drop of nail polish in sets of 5–10 male flies per slide/petri dish for phenotypic readouts and then discarded after screenings. The heads were carefully arranged with forceps to ensure eyes were perpendicular to the plane of the objectives. Females were not used for screenings. An Olympus BX61 fluorescent upright with low mag 4X objective (UPlanSApo) and high mag 40X water dipping objective (LUMPlanFL-N) were used with standard FITC and Texas Red filters21. Only ~10–20 photoreceptor units in the approximate center of view, rather than in the peripheral regions, can be directly visualized due to curvature of the fly compound eye after imaging with 40X immersion objective for ONC visualization. All Drosophila images were acquired and pseudocolored using SlideBook 6 software and processed for brightness, contrast, and labeled using Microsoft Powerpoint 2021 and Adobe Photoshop CC 2019 Software. Drosophila deep pseudopupil graphic images were prepared using GraphPad Prism Software v9.
Drosophila screening
For all crosses (n = 10 flies per timepoint, per fly line) offspring were screened for EGFP expression patterns of photoreceptors and ommatidia morphology at days 1, 7, and, 14 with a fluorescent low mag 4X objective by DPP. If a phenotype was detected, then a more low-throughput, fluorescent 40X immersion objective was used to further validate eye phenotypes directly by ONC. To account for potential phenotypic variability, multiple negative control screens were carried out (L.GMR-GAL4; ninaE-EGFP crossed to LacZRNAi, luciferaseRNAi, mCherryRNAi, or UAS-tdTom) and a mean loss of DPP below 70% (n = 10 flies per line, per timepoint) was then determined to be a plausible phenotype for each RNAi line screened. For each gene screened, attempts were made to prioritize one VDRC and one BDSC RNAi line if available. Parents were rotated into new tubes with food every 3-4 days at 18 C. Then male offspring were collected for aging at 25 C and imaging was prioritized for days 1 and 14. For negative controls or lines with DPP loss below the 70% threshold screens were also acquired at day 7 by microscopy.
Mouse Models
Animal experiments were conducted in accordance with protocols approved by the Genentech Institutional Animal Care and Use Committee and with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. CRISPR mediated deletion of Slc16a8 exons 1-6 (4298 bp deletion) was outsourced and carried out by Taconic Inc. The founder Slc16a8 KO line was backcrossed to the original C57BL/6 J WT littermates used for CRISPR deletions to generate Slc16a8 Hets, and then these Slc16a8 Het littermates were bred to each other to eventually generate 10-week old Slc16a8 WT (n = 8; 4 male and 4 female), Het (n = 13; 10 male and 3 female), and KOs (n = 6; 3 male and 3 female) for subsequent ERG, OCT, and histological analyses. All animals were housed in pathogen-free animal facility at Genentech with 12/12 h of light/dark cycle prior to the start of constant light exposure.
Randomization and blinding
Randomization and blinding was used whenever possible. During CLE, mice were randomly assigned to a light box in the room, regardless of gender or genotype. For ERG and OCT measurements, mice were assessed in random order and the operator did not have knowledge of the mouse’s gender or genotype. For data analysis, ERGs and OCTs were marked in random order and the scientist had no knowledge of the gender or genotype associated with a given ERG waveform or OCT image.
Data exclusions
One WT mouse was excluded from analysis because they died inadvertently between baseline and day 7. Additionally, one eye from a mouse from the heterozygous group at baseline had a waveform in which the c-wave could not be confidently marked and thus was excluded from analysis. Finally, one mouse from the WT group at day 7 obtained cataracts before the OCT measurements so these datapoints were excluded.
Constant light exposure (CLE)
See Natoli et al. for more information regarding CLE methods24. Briefly, age matched Slc16a8 -WT, Het, and KO animals were placed 2 per plastic box with free access to food and water. The mice were exposed to constant (24 hr) 100 K lux natural white LEDs for 7 days and pupils were dilated twice per day with 1% atropine sulfate. Immediately following 7 days of constant light exposure, mice were dark-adapted overnight in preparation for subsequent functional measurements.
Electroretinogram recordings (ERGs)
The Celeris ERG system (Diagnosys) was used to perform full-field scotopic ERGs at baseline and after CLE. A-, b-, and c-wave amplitudes were measured in order to determine photoreceptor, bipolar, and RPE cell function, respectively. Slc16a8 + /+ (WT, n = 8), Slc16a8 + /− (HET, n = 13), and Slc16a8−/− (KO, n = 6) mice were dark-adapted overnight and then anesthetized with an intraperitoneal (IP) injection of a Ketamine (75–80 mg/kg) and Xylazine (7.5–15 mg/kg) in 200-300 uL of pharmaceutical grade saline. Mouse body temperature was maintained at 37 °C using a homeothermic plate connected to its control unit and pupils were dilated with 1% tropicamide under dark conditions. Mice were placed on a platform and a reference electrode inserted subcutaneously through the forehead and a ground electrode inserted at the base of the tail. ERGs from both eyes were recorded simultaneously. Gonak hypermellose ophthalmic demulcent solution was placed on the cornea to establish an electrical contact between the cornea and the electrode, and to protect eyes from drying during the experiment. Electrodes with light stimulator simultaneously placed on both eyes. In scotopic conditions, retinas were stimulated with a flash intensity of 150 cd.s/m2. Recorded signals were bandpass-filtered at 0.15–1000 Hz and sampled at 2 kHz. Espion v6.59.9 software (Diagnosys) was used to record the waveforms of each response. Responses to 3 flashes of light stimulation were averaged. All of the recorded waveforms were then analyzed using custom Matlab software (MathWorks) and the amplitudes were marked as follows: a-wave amplitude measured from the baseline to the trough of the a-wave while b-wave amplitude from the trough of the a-wave to the peak of the b-wave. C-wave amplitude was recorded from the trough following the b-wave to the c-wave peak. Data was then analyzed using GraphPad (GraphPad Prism Software v10, Boston, MA). For comparisons between groups (genotypes) within timepoints (baseline and day 7) a mixed-effects analysis with a Sidak post-hoc was used. For percent change comparisons a one-way ANOVA with a Sidak post-hoc was used.
Optical coherence tomography (OCT)
Immediately following ERG recordings, retinal morphology and thickness were examined by spectral domain optical coherence tomography (SD-OCT) scans using a Bioptigen Envisu R machine (Leica Microsystems, IL, USA). Mice were still anesthetized from ERG recordings (by intraperitoneal injection of ketamine (75-80 mg/kg body weight) and xylazine (15 mg/kg body weight)). Pupils were dilated with drops of Tropicamide Ophthalmic Solution USP 1% (Akorn). Drops of Systane lubricant eye drop (Alcon) were applied bilaterally to prevent corneal dehydration during procedure. Mouse body temperature was again maintained at 37 C using a homeothermic plate. Total retina thickness was defined as the width from the nerve fiber layer to the RPE/choroid layer on the cross-sectional images with an algorithm. Data were exported to the Matlab software (MathWorks) and analyzed. Data was then analyzed using GraphPad (GraphPad Prism Software v10, Boston, MA). For comparisons between groups (genotypes) within timepoints (baseline and day 7) a mixed-effects analysis with a Sidak post-hoc was used. For percent change comparisons a one-way ANOVA with a Sidak post-hoc was used.
H&E histology
Mice were euthanized via C02 overdose. Formalin-fixed, paraffin-embedded (FFPE) mouse eyes were sectioned in the sagittal plane at 4 um thickness and stained with hematoxylin and eosin (H&E). Slides were examined by a pathologist blinded to the treatment groups. Also see Weber et al Scientific Reports 2022 for more methods information41.
Supplementary information
Acknowledgements
This study was funded by Roche/Genentech except as follows. AREDS and AREDS2 studies were funded by the Intramural Research Program at NEI and supported by Intramural Research Program funds and contracts (contract HHS-N-260-2005-00007-C; ADB contract N01- EY-5-0007) from the National Eye Institute, National Institutes of Health, Department of Health and Human Services, Bethesda, MD. Funds were generously contributed to these contracts by the following National Institutes of Health institutes: Office of Dietary Supplements; National Center for Complementary and Alternative Medicine; National Institute on Aging; National Heart, Lung, and Blood Institute; National Institute of Neurological Disorders and Stroke. The familial study was funded by the Intramural Research Program of the National Eye Institute (NIH). The funding for the SAVE study (NCT02960828) was obtained through a research grant from Genentech, Inc.
Author contributions
NN, BHG, AS, HJ, BLY contributed to the design of this study. NN, AS, BLY contributed to the human genetics sections. NN, KCB, HJ contributed to the Drosophila melanogaster sections. JRLdeC, EU, RAR, WMZ, BG contributed to the familial genetics sections. SVE, CCW, DB, AF, JDP, EYC contributed clinical and genetic data from patient cohorts. NN, BHG, TT, GJK, YM, AS, JWH, SYC, HJ contributed to murine model sections. MIMcC reviewed the manuscript and provided valuable feedback. All authors approved the submission of this manuscript.
Data availability
Summary statistics for the complete set of genome-wide rare variant burden tests are listed by gene and found in Supplementary Data 1. Data for all rare, predicted protein altering variants in SLC16A8 are found in Supplementary Data 2.
Competing interests
N.N., B.H.G., A.S., T.T., G.J.K., K.C.B., Y.M., A.S., S.V.E., MIMcC, J.W.H., S.Y.C., H.J., B.L.Y. are (or were at the time of contribution) employees of Genentech/Roche with stock and stock options in Roche. N.N., H.J., B.L.Y. are listed inventors on the Genentech Inc. patent application WO2023212273A, published November 2, 2023 covering the human genetics, Drosophila melanogaster and murine model portions of the text. All other authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Navid Nouri, Bailey Hannon Gussler, Amy Stockwell.
Contributor Information
Heinrich Jasper, Email: jasperh@gene.com.
Brian L. Yaspan, Email: yaspan.brian@gene.com
Supplementary information
The online version contains supplementary material available at 10.1038/s41525-024-00442-8.
References
- 1.Wong, W. L. et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob. Health2, e106–e116 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Lambert, N. G. et al. Risk factors and biomarkers of age-related macular degeneration. Prog. Retin Eye Res.54, 64–102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Lookeren Campagne, M., LeCouter, J., Yaspan, B. L. & Ye, W. Mechanisms of age-related macular degeneration and therapeutic opportunities. J. Pathol.232, 151–164 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Gorin, M. B. Genetic insights into age-related macular degeneration: controversies addressing risk, causality, and therapeutics. Mol. Asp. Med.33, 467–486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fritsche, L. G. et al. Seven new loci associated with age-related macular degeneration. Nat. Genet.45, 433–439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fritsche, L. G. et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet.48, 134–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orozco, L. D. et al. Integration of eQTL and a single-cell atlas in the human eye identifies causal genes for age-related macular degeneration. Cell Rep.42, 112298 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Orozco, L. D. et al. Integration of eQTL and a single-cell atlas in the human eye identifies causal genes for age-related macular degeneration. Cell Rep.30, 1246–1259.e1246 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Ratnapriya, R. et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet.51, 606–610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klipfel, L. et al. A splice variant in SLC16A8 gene leads to lactate transport deficit in human ips cell-derived retinal pigment epithelial cells. Cells10, 179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tom, J. et al. Enabling genome-wide association testing with multiple diseases and no healthy controls. Gene684, 118–123 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Chang, D. et al. A whole genome sequencing study of moderate to severe asthma identifies a lung function locus associated with asthma risk. Sci. Rep.12, 5574 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med.17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng, J. et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science381, eadg7492 (2023). [DOI] [PubMed] [Google Scholar]
- 15.Iovino, C. et al. Vitelliform maculopathy: Diverse etiologies originating from one common pathway. Surv. Ophthalmol.68, 361–379 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Tebbe, L. et al. Prph2 disease mutations lead to structural and functional defects in the RPE. FASEB J.36, e22284 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, Y. et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinforma.12, 357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkins, L. A. et al. The transgenic RNAi Project at Harvard Medical School: Resources and validation. Genetics201, 843–852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietzl, G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature448, 151–156 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Huang, Y., Xie, J. & Wang, T. A fluorescence-based genetic screen to study retinal degeneration in drosophila. PLoS One10, e0144925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pichaud, F. & Desplan, C. A new visualization approach for identifying mutations that affect differentiation and organization of the Drosophila ommatidia. Development128, 815–826 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Silva, B. et al. Glia fuel neurons with locally synthesized ketone bodies to sustain memory under starvation. Nat. Metab.4, 213–224 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang, C., Lee, G. & Chung, J. LKB1 induces apical trafficking of Silnoon, a monocarboxylate transporter, in Drosophila melanogaster. J. Cell Biol.183, 11–17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natoli, R. et al. A model of progressive photo-oxidative degeneration and inflammation in the pigmented C57BL/6J mouse retina. Exp. eye Res.147, 114–127 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Katschke, K. J. Jr et al. Classical and alternative complement activation on photoreceptor outer segments drives monocyte-dependent retinal atrophy. Sci. Rep.8, 7348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, J., Peachey, N. S. & Marmorstein, A. D. Light-evoked responses of the mouse retinal pigment epithelium. J. Neurophysiol.91, 1134–1142 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer, M. D. et al. Noninvasive, in vivo assessment of mouse retinal structure using optical coherence tomography. PLoS One4, e7507 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniele, L. L., Sauer, B., Gallagher, S. M., Pugh, E. N. Jr & Philp, N. J. Altered visual function in monocarboxylate transporter 3 (Slc16a8) knockout mice. Am. J. Physiol. Cell Physiol.295, C451–C457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chidlow, G., Wood, J. P., Graham, M. & Osborne, N. N. Expression of monocarboxylate transporters in rat ocular tissues. Am. J. Physiol. Cell Physiol.288, C416–C428 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Acar, I. E. et al. Whole genome sequencing identifies novel common and low-frequency variants associated with age-related macular degeneration. Investig. Ophthalmol. Vis. Sci.64, 24 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwong, A. et al. Whole genome sequencing of 4,787 individuals identifies gene-based rare variants in age-related macular degeneration. Hum. Mol. Genet.33, 374–385 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halestrap, A. P. & Wilson, M. C. The monocarboxylate transporter family–role and regulation. IUBMB Life64, 109–119 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Leveillard, T. & Sahel, J. A. Metabolic and redox signaling in the retina. Cell Mol. Life Sci.74, 3649–3665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, M. C. et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet.89, 82–93 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (Austin)6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol.17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castorino, J. J. et al. Basolateral sorting signals regulating tissue-specific polarity of heteromeric monocarboxylate transporters in epithelia. Traffic12, 483–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rein, D. B. et al. Prevalence of age-related macular degeneration in the US in 2019. JAMA Ophthalmol.140, 1202–1208 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber, M. et al. Ocular phenotypes in a mouse model of impaired glucocerebrosidase activity. Sci. Rep.11, 6079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nassar, L. R. et al. The UCSC Genome Browser database: 2023 update. Nucleic Acids Res.51, D1188–D1195 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics for the complete set of genome-wide rare variant burden tests are listed by gene and found in Supplementary Data 1. Data for all rare, predicted protein altering variants in SLC16A8 are found in Supplementary Data 2.