Abstract
A homologue of the gene encoding the transcription factor Rim101 (PacC), involved in pH signal transduction in fungi, was identified in the pathogenic basidiomycete Ustilago maydis. The gene (RIM101) encodes a protein of 827 amino acid residues, which shows highest similarity to PacC proteins from Fusarium oxysporum and Aspergillus niger. The gene had the capacity to restore protease activity to rim101 mutants from Yarrowia lipolytica, confirming its homologous function, and was expressed at both acid and neutral pH. Null Δrim101 mutants were not affected in the in vitro pH-induced dimorphic transition, their growth rate, resistance to hypertonic sorbitol or KCl stress, and pathogenicity. However, similar to pacC (rim101) mutants in other fungi, they displayed a pleiotropic phenotype with alterations in morphogenesis, impairment in protease secretion, and increased sensitivity to Na+ and Li+ ions. Other phenotypic characteristics not previously reported in fungal pacC (rim101) mutants (morphological changes, increased sensitivity to lytic enzymes, and augmented polysaccharide secretion) were also observed in U. maydis mutants. All these modifications were alleviated by transformation with the wild-type gene, confirming that all were the result of mutation in RIM101. These data indicate that the Pal/Rim pathway is functional in U. maydis (and probably in other basidiomycetes) and plays complex roles in pH-sensing phenomena, as occurs in ascomycetes and deuteromycetes.
Ustilago maydis, the basidiomycete responsible for corn smut disease, displays a complex life cycle that requires the plant host for completion. In its haploid nonpathogenic phase the fungus grows like yeast. Mating of compatible yeasts (sporidia) gives rise to the formation of a pathogenic dikaryotic mycelium that may invade the host. Abundant mycelial growth and karyogamy occur in the plant, which develops galls or tumors full of dark teliospores (see reference 3). Germination of teliospores produces basidiospores that bud to reinitiate the life cycle (for reviews see references 1, 7, and 27). Dimorphic transition of U. maydis in vitro was reported to occur in haploid or diploid strains as a response to the external pH (38). At neutral pH the fungus grows as a homogeneous population of budding yeast, whereas at acid pH it develops the mycelial form.
As occurs with different physical and chemical stimuli, pH of the medium affects the physiological behavior of fungi in many ways. Nevertheless, specific responses may occur as a result of a change in external pH. The most studied responses of fungi to changes in pH include secretion of different enzymes such as proteases and phosphatases, permease induction, and alterations in the production of some secondary metabolites, etc. (reviewed in references 15, 32, and 33). Studies on the mechanism of the regulation of protein secretion by the external pH led to the discovery in Aspergillus nidulans of a specialized signal transduction pathway responsive to pH (8). This pathway, named Pal/Rim, involves the action of different proteins that convey the pH stimulus to a key zinc finger transcription factor, PacC (Rim101), which activates transcription of genes induced at alkaline pH and represses genes induced at acid pH (8, 15, 26, 44). Further studies proved the wide distribution of similar (although not identical) pathways in other ascomycetes and deuteromycetes (9, 13, 25, 34, 43). These studies have demonstrated that the function of PacC/Rim101 is more complex than simply regulating the transcription of pH-dependent specific enzymes. Additional processes in which PacC/Rim101 plays a regulatory role include meiosis, morphogenesis, and salt stress tolerance (9, 23, 36, 43; reviewed in references 32 and 33).
Interestingly, the Pal/Rim pathway has also been reported to play a role in Candida albicans dimorphism. This fungus grows in the mycelial form at neutral pH, and as pH decreases, a yeast-like morphology is developed (reviewed in reference 30). Mutation of pacC/RIM101 blocks mycelium formation at neutral pH (34) and reduces pathogenesis (12). In contrast, it does not seem to affect the pH-dependent dimorphic transition in Yarrowia lipolytica, whose RIM101 mutants still grow in the mycelial form in serum medium (19).
Considering that the in vitro dimorphic transition of U. maydis is regulated by pH, we investigated whether the Pal/Rim pathway was involved in the process. Accordingly, we have proceeded to the isolation and deletion of the RIM101/pacC homologue gene (RIM101). The data obtained show that the Pal/Rim pathway is operative in this basidiomycete, where, as in other fungi, it controls a series of cellular functions but is not involved in dimorphism.
MATERIALS AND METHODS
Strains and growth conditions.
The following microbial strains were used in this study: Ustilago maydis wild-type strains FB1 (a1b1) and FB2 (a2b2) (2); U. maydis mutants BMA1 and BMA7 (a1b1Δrim101::hyg) and BMA2 and BMA4 (a2b2 Δrim101::hyg), isolated in this work; U. maydis complemented strains PGM1 and PGM2 (a2b2 Δrim101::hyg/pUpacC23) and AAP1(a2b2 Δrim101::hyg/pUpacC22), also isolated in this work; Yarrowia lipolytica FL3ΔR (A rim101-1113 ura3 lys11) and E121 (A lys11) (24); Y. lipolytica strain ETA1 (A rim101-1113 lys11/pUpacC44), isolated in this work; and Escherichia coli DH5α, which was used as a host for plasmid amplification.
U. maydis and Y. lipolytica strains were maintained at −70°C in 50% (vol/vol) glycerol. U. maydis cells were transferred to liquid complex medium (CM) (21) and shaken at 28°C for 2 to 3 days. Y. lipolytica was propagated in yeast extract-peptone-glucose medium (5). These cultures were used as inocula for all subsequent experiments. Yeast or mycelial growth of U. maydis in liquid minimal medium (MM) (21) was obtained as earlier reported (38). Liquid media were supplemented when necessary with hygromycin (100 to 350 μg/ml) and/or 6 μM carboxin. For U. maydis transformant selection, protoplasts were plated on CM (see above) containing 1 M sorbitol plus the appropriate selective agent(s). Skim milk agar plates were used for alkaline protease induction (31). Plate mating assays were carried out as described in reference 2.
Growth rate of fungal strains was measured in liquid medium. Cells (106 per ml) were inoculated into MM or CM (150 ml in 500-ml Erlenmeyer flasks), and shaken (180 rpm) in a water bath at 28°C. At intervals, aliquots were withdrawn and cell numbers were measured with a Neubauer cell counting chamber. The effect of different stress conditions on cell growth was measured in plates of solid MM of pH 3 or 7 (46).
PCR conditions and plasmid constructions.
Amplifications described were performed using U. maydis DNA, and the primers and annealing temperatures are described in Table 1. After an initial denaturing temperature of 94°C for 4 min, 30 cycles of the following program were applied: 1 min at 94°C, 1 min at the annealing temperature (Table 1), and extension time (calculated as 1 min per kb of the expected amplification fragment) at 72°C. This program was followed by 5 min at 72°C for final extension.
TABLE 1.
Oligonucleotides used for PCR
| Primer | 5′-3′ sequence | Tma (°C) | Primer sense |
|---|---|---|---|
| JRH-G181 | CGCGAAGCTTGCCGCAACGTCCGCCGGCA | 65 | Antisense |
| JRH-G182 | GGGGGAATTCCAAATAGTAACGACAAACA | 65 | Sense |
| JRH-G173 | GGGGATATCATTCCTTTGCCCTCGGACGAGT | 65 | Antisense |
| JRH-G174 | GCGCGACACGTATGAAAAAGCCTGAACTCACC | 65 | Sense |
| 1892 | AA(G/A)(C/A)GIGA(C/T)CA(C/T)AT(C/T/A)ACI(A/T)(C/G)C | 50 | Sense |
| 1894 | (C/G)(G/A)TG(T/C)TT(T/C)TTIA(G/A)(G/A)TC(C/T)TGIGG | 50 | Antisense |
| 2075 | CTTGCGACGAGAAGG | 55 | Antisense |
| 2078 | TCTGGGCATTCTTGG | 55 | Sense |
| 2833 | CCCTCCAACACCTCCTCT | 65 | Antisense |
| G-654 | CGCGCGGTACCATGAACCACCTCTCGC | 63 | Sense |
| G-655 | GCGCGGTACCGCAACGTCCGCCGGCA | 63 | Antisense |
Tm, melting temperature.
A 120-bp fragment containing part of the zinc finger coding region of the RIM101 gene was amplified by PCR, using the degenerate oligonucleotides 1892 and 1894 (Table 1). This fragment was cloned in the pCR2.1 plasmid (Invitrogen) to generate pUznf16.
A partial fragment of the RIM101 gene of approximately 3,000 bp was obtained by amplification with oligonucleotides JRH-G182 and JRH-G181 (Table 1) and cloned in pUC19 using EcoRI and HindIII sites included in the primers to generate pUpacC.
Plasmid pUpacH was constructed as follows: plasmid pUpacC was digested with StuI and BamHI, eliminating a 1,289-bp fragment that contained the region encoding the three RIM101 zinc fingers. The BamHI site was filled with the Klenow fragment (Invitrogen) and an EcoRV-EcoRV fragment from pHyg1 (see below) containing the E. coli Hph gene open reading frame (ORF), which confers hygromycin resistance, was cloned in frame at this site. This construction was confirmed by sequencing.
Plasmid pHyg1 was derived from pCR2.1 (Invitrogen) and contained the ORF of the Hph gene from E. coli obtained by PCR with primers JRH-G174 and JRH-G173 (Table 1).
Plasmid pUpacC4 was derived from pCR2.1 (Invitrogen) and contained a 3,832-bp fragment with the complete RIM101 ORF and 1,175 bp of the promoter region, as well as 172 bp of the terminator, obtained by PCR with oligonucleotides JRH-G182 and 2833.
Plasmids pUpacC22 and pUpacC23 were derived from pCBX122 carrying the carboxin resistance gene (22). pUpacC22 contained a 3,000-bp EcoRI fragment from pUpacC (promoter and partial ORF of RIM101), whereas pUpacC23 contained a 3,832-bp fragment from pUpacC4 (1,175 bp of promoter region, RIM101 ORF, and 172 bp of terminator).
The plasmid used to transform Y. lipolytica with the U. maydis gene encoding Rim101 (pUpacC44) was constructed through a series of intermediates. Y. lipolytica plasmid pINA404 (5) was digested with BamHI to excise the lacZ ORF regulated by the Y. lipolytica Xpr2 gene promoter and terminator. Next, an 1,809-bp fragment containing a truncated RIM101 ORF, without the promoter region (amplified with oligonucleotides G-654 and G-655 [both containing a KpnI site; see Table 1]), was cloned at the KpnI site to generate plasmid pRpacC. The following step involved the ligation of an EcoRI-SalI fragment (ca. 3,000 bp) from pRpacC containing the RIM101 fragment, plus the Xpr2 gene promoter and terminator, into the same sites of pUC18, to generate plasmid pYpacC. Finally pYpacC was digested with EcoRI and HindIII, and the resulting fragment containing the truncated RIM101 gene ORF plus the Xpr2 gene promoter and terminator was cloned in plasmid pINA444 (18), which carries the Y. lipolytica URA3 gene and a Y. lipolytica autonomously replicating sequence.
DNA procedures.
DNA was obtained as described in reference 20. Transformation of U. maydis protoplasts was performed by standard methods (see reference 45). Transformed cells were recovered and further grown on CM-sorbitol plates (see above) containing 350-μg/ml hygromycin B or 6 μM carboxin as described previously (4, 22, 45). Y. lipolytica was transformed by the lithium acetate/LiCl method (5), and transformants were recovered in yeast nitrogen broth medium without amino acids (Difco) supplemented with 1 mM lysine, but not uracil.
Standard procedures were followed for molecular cloning. Southern analyses were made with DNA digested with HincII, using as a probe a 1,289-bp StuI-BamHI RIM101 fragment. Probes were labeled with [α-32P]dCTP by random priming with the Rediprime II random prime labeling system (Amersham Biosciences). DNA was sequenced with a Big Dye Terminator v3.1 cycle sequencing kit according to the manufacturer's recommendations (Applied Biosystems). Universal and sequence-specific primers (Sigma Genosys) were used. Analyses of sequences were performed using DNASTAR software (DNASTAR, Madison, WI) and the BLAST program at the National Center for Biotechnology Information. Specific analyses of the RIM101 promoter sequence were done in the BLAST program found at http://www.motif.genome.ad.jp. Sequences in the U. maydis genome were searched at http://mips.gsf.de/genre/proj/ustilago.
RNA procedures.
RNA was prepared by the Trizol technique (Invitrogen) using the manufacturer's suggested conditions. Northern blot analyses were performed with 25 μg total RNA, using ethidium bromide-stained rRNAs as a loading control. Hybridizations were made at 68°C in Church's buffer [5 mM EDTA, 0.25 M Na2HPO4(7H2O), 1% casein hydrolysate, 7% sodium dodecyl sulfate, 0.17% H3PO4] for 12 h, and the membrane was washed twice for 10 min at 68°C with 0.05% sodium dodecyl sulfate (39). As a probe we used a 1,289-bp StuI-BamHI RIM101 fragment labeled as described above.
Pathogenicity assays in maize plants and teliospore isolation and germination.
Assays of pathogenicity of U. maydis strains in maize cv. Cacahuazintle plants were made essentially as previously described (28). Plants were inoculated with 0.1-ml aliquots of suspensions containing equal numbers of sexually compatible yeast cells (105, 104, 103, or 102 cells/aliquot). Plants were incubated in a greenhouse, and disease symptoms (development of chlorosis, anthocyanin production, and tumor formation) were recorded normally after 15 days. Tumors were excised and treated as described previously (10, 28). After this, teliospores were diluted to 1,000/ml, and 0.1-ml aliquots were distributed on plates of solid CM. After 12 to 18 h of incubation at 28°C, teliospores had germinated to produce sporidia.
Miscellaneous procedures.
Proteolysis in solid medium was measured by the development of clear halos on skim milk agar plates.
The polysaccharide secreted to the culture medium was measured at different times of growth in cultures inoculated with 1 × 106 cells/ml. Cells were eliminated by centrifugation, and ice-cold ethanol (50% final concentration) was added to the cell-free medium. After 12 h at 0°C, the mixture was centrifuged at 3,000 rpm for 5 min, and the precipitated material was dried at ambient temperature and weighed. Its content of neutral sugars was measured by the anthrone procedure (16), using glucose as a standard.
Cell wall sensitivity to Trichoderma lysing enzymes (Sigma catalog no. L1412) was determined by protoplast formation. Cells grown in CM were recovered at the end of the log phase by low-speed centrifugation, washed with distilled water twice by centrifugation, and finally resuspended in SCS (1 M sorbitol, 20 mM sodium citrate, pH 5.8). To 1-ml aliquots of SCS containing 108 cells, 50 mg of enzyme was added, and every 20 min 10 μl of the mixture was mounted on a glass slide and observed under the microscope. Photographs of different fields were taken, and the ratio of protoplasts to whole cells was determined.
Cells were observed with a Leica DMRE model microscope using phase contrast and photographed with a Spot camera (Diagnostic Instruments). In some experiments cells were stained with a solution containing 0.05 mg/ml of Calcofluor white, thoroughly washed by centrifugation, and observed by epifluorescence. Cell measurements were made with the Image-ProPlus 4.0 program of Media Cybernetics. We measured 30 cells randomly selected from fungal cultures. Frequency of septation was measured per 30 μm of cell length. Statistical significance of data was analyzed by χ2 determination. Experiments where numerical data were obtained were repeated at least three times with two replicates per experiment.
Nucleotide sequence accession number.
The gene sequence has been registered at the EMBL Nucleotide Sequence Database with the accession no. AJ748125.
RESULTS
Isolation of RIM101 gene.
To analyze whether U. maydis possessed a homologue of the RIM101/pacC gene, we used two degenerate primers (1892 and 1894, Table 1), designed on the basis of the amino acid sequences of the most conserved regions (second and third zinc fingers of the coding region) from several pacC/RIM101 homologues, to amplify a putative fragment by PCR. A single product of 172 bp was obtained, whose DNA sequence revealed an ORF with the same consensus of the zinc finger regions as that of other reported PacC/Rim101 proteins (not shown). Using the information on the U. maydis genome sequence available from the Center for Genome Research (Cambridge, MA), we designed two specific oligonucleotides (JRH-G182 and 2883 [Table 1]) to amplify the complete ORF and promoter region of the gene homologue of pacC/RIM101. The amplified fragment of 3,832 bp was cloned in pUC19 and sequenced (pUpacC4). It was found to contain a 5′ region of 1,175 bp, plus a single 2,484-bp ORF with no consensus introns, encoding a protein of 827 amino acid residues with a molecular mass of 88.5 kDa, and including a 3′ fragment of 172 bp. This sequence has been registered at the EMBL Nucleotide Sequence Database with the accession no. AJ748125. It is identical to ORF um10426 from the U. maydis genomic bank (http://mips.gsf.de/genre/proj/ustilago). In comparison with the GenBank-deposited sequences from several organisms the best BLAST similarity scores were obtained with PacC proteins of Fusarium oxysporum and Aspergillus niger. Homology was very high at the zinc finger zone and low at the C-terminal domain (ca. 51 and 22% and 51 and 21%, respectively). In the 5′ region we identified two adjacent putative A. nidulans PacC consensus binding sites at positions −614 to −598 from the ATG start codon.
Deletion of the RIM101 gene and isolation of null mutants.
Deletion of the RIM101 gene was obtained using plasmid pUpacH. The plasmid was linearized with HindIII and used to transform U. maydis FB2 and FB1 wild-type strains, to obtain RIM101 replacement by the Hph gene through double-crossover homologous recombination. Under these conditions the Hph ORF stays under the control of the RIM101 gene promoter. This strategy made it possible to analyze later on the transcriptional regulation of RIM101 by means of hygromycin resistance (see below). Hygromycin-resistant clones were isolated, and homologous recombination was confirmed by PCR with primers 2075 and 2078 (Table 1). These should amplify a 1,662-bp fragment from null mutants (Fig. 1A, lanes 1 to 3) and a 1,925-bp product from wild-type strains (Fig. 1A, lane 4). Since two EcoRI sites were present in the PCR products (Fig. 1D and 1E), these were digested with EcoRI, giving rise to the expected different products in the wild-type and mutant strains (Fig. 1B, lanes 2 and 4 for mutant and lane 6 for wild-type strains). Gene disruption was also confirmed by Southern analysis (Fig. 1C). For this we used the 1,289-bp StuI-BamHI fragment of the RIM101 ORF as a probe, present only in wild-type strains (Fig. 1D). Two of the transformants obtained from each FB1 and FB2 strain (see Materials and Methods) where homologous gene replacement had occurred were selected for further studies. Strain BMA2 (a2b2 Δrim101::hyg) was used in most of the further analyses.
FIG. 1.
Confirmation analyses of RIM101 mutation. A. DNA was subjected to PCR with primers 2075 and 2078. Lanes 1 to 3, Δrim101 mutants (BMA1, BMA2, and BMA4, respectively); lane 4, FB2 wild-type strain; lane 5, plasmid pUpacH; lane 6, plasmid pUpacC4. B. EcoRI digestion of PCR products. Lane 2, BMA1; lane 4, BMA2; lane 6, FB2; lane 8, pUpacH; lane 10, pUpacC4. Lanes 1, 3, 5, 7, and 9, controls with no restriction enzyme added to BMA1, BMA2, FB2, pUpacH, and pUpacC4, respectively. C. Southern blot hybridization of HincII-digested DNA with the 1.2-kb StuI-BamHI RIM101-specific probe. Lane 1, FB1; lane 2, BMA1; lane 3, BMA7; lane 4, FB2; lane 5, BMA2; lane 6, BMA4. Note absence of hybridization in mutants compared to wild-type strains. Arrows show positions of the molecular size standards. D. Restriction map of RIM101 locus of the wild-type strain showing the primer (solid arrow) and probe (solid line) positions. E. Restriction map of RIM101 locus of the mutant allele used in the experiment showing the position of the primers (solid arrows) and the Hph gene (large open arrow).
Analysis of RIM101 gene expression.
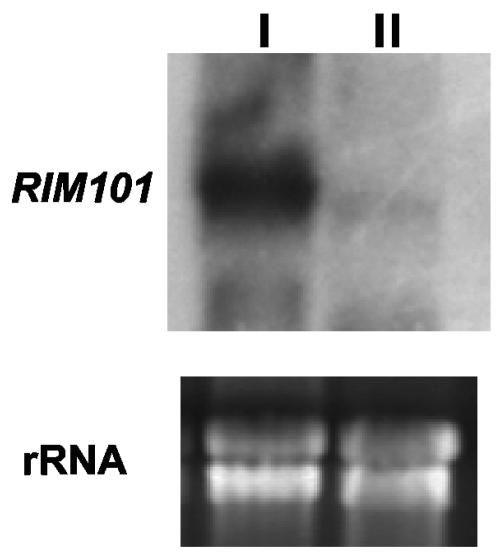
As indicated above, expression of the E. coli Hph gene, and therefore hygromycin resistance in our Δrim101 mutants, was under the control of the RIM101 promoter (see Materials and Methods). This construction allowed the determination of RIM101 expression through the hygromycin resistance of the mutants. We observed that these strains, in contrast to wild type, were resistant to hygromycin at concentrations over 150 μg per ml under acid or neutral conditions in different media, providing evidence that RIM101 was expressed at both acid and neutral pH (not shown). Quantitative determination of expression of the RIM101 gene was done by Northern blotting. The results obtained (Fig. 2) showed that indeed the transcript was observed at both pH values of growth, although expression was much higher at neutral pH.
FIG. 2.
Determination of RIM101 expression. Shown are results of Northern analysis of RIM101 gene expression in strain FB2 grown for 24 h in MM, pH 7 (lane I) or pH 3 (lane II), using the 1.2-kb StuI-BamHI RIM101-specific probe. rRNAs stained with ethidium bromide are shown as a loading control.
Phenotypic analysis of Δrim101 mutants. (i) Growth characteristics.
The growth rates of BMA2 and BMA4 (mutants) and FB2 (wild-type) strains in liquid CM or MM at pH 7.0 did not display any significant difference (data not shown). Further phenotypic characterization included the use of several stress conditions on solid MM at different cellular concentrations. These included salt stress by addition of 0.6 M KCl, 1 M NaCl, or 0.05 M LiCl at pH 7.0 or 3.0; hyperosmotic stress with 1 M sorbitol; growth at temperature above optimum (32 to 33°C); and incubation in media with different concentrations of Calcofluor white (300 to 500 μg/ml). No significant differences in growth and colonial morphology between the different Δrim101 mutants and the wild-type strain were observed under most conditions (not shown). However, exposure to salt stress elicited differential reactions for the mutants and the wild type. Addition of 0.05 M LiCl or 1 M NaCl strongly inhibited the growth of the mutants, in contrast to wild-type strains (Fig. 3). Salt sensitivity occurred only at pH 7 (Fig. 3A and C). KCl had no inhibitory effects at either pH (not shown).
FIG. 3.
Effect of salt stress on U. maydis growth. Different U. maydis cell numbers (107, 106, 105, or 104 cells/aliquot) were inoculated on plates of solid MM and incubated for 96 h. Panels A and C, MM, pH 7; panels B and D, MM, pH 3. Panels A and B, 0.05 M LiCl added to the medium; panels C and D, 1 M NaCl added to the medium. Rows 1, FB2; rows 2, BMA1 (Δrim101); rows 3, BMA2 (Δrim101); rows 4, PGM1 (Δrim101::hyg/pUpacC23).
(ii) Dimorphic transition and cellular morphology.
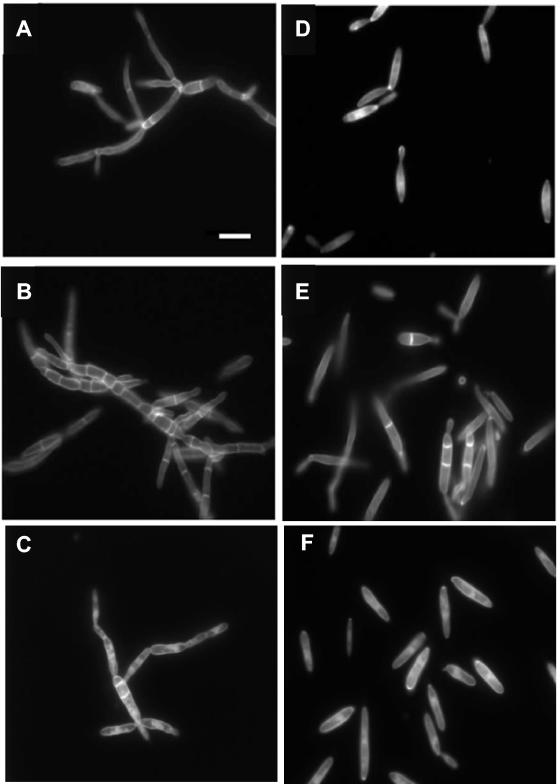
We observed that the several Δrim101 mutants analyzed were not affected in their dimorphic capacity (Fig. 4). Nevertheless, mycelial morphology and the distribution of septa were different from those of the wild-type strain (Fig. 4A and B). Mycelia of Δrim101 mutant BMA2 displayed a significantly higher relative number of septa than did the wild-type FB2 strain (Table 2). Also mutant yeast cells were significantly longer (Table 2), and some of them presented septa (Fig. 4D and E). In contrast, the width of yeast cells was indistinguishable in the mutant (2.83 ± 0.38 μm) and wild-type (2.96 ± 0.38 μm) strains (χ2 value of 1).
FIG. 4.
Cell morphology of U. maydis wild-type and mutant strains. Strains were grown in MM at pH 3 (A, B, and C) or pH 7 (D, E, and F) and stained with Calcofluor white. A and D, FB2 (wild type); B and E, BMA2 (Δrim101); C and F, PGM1 (Δrim101::hyg/pUpacC23). Bar, 10 μm.
TABLE 2.
Several phenotypic characteristics of wild-type, Δrim101 mutant, and complemented strains of U. maydisa
| Strain | Yeast form length (μm) | Septa in myceliumb | Sensitivity to lytic enzymesc |
|---|---|---|---|
| FB2 (wild type) | 12.7 ± 1.30 | 2.40 ± 0.99 | 54.16 ± 4.79 |
| BMA2 (Δrim101) | 17.2 ± 3.50 | 5.00 ± 0.79 | 93.25 ± 0.95 |
| PGM1 (Δrim101/pUpacC23) | 10.51 ± 2.11 | 1.92 ± 0.66 | 39.60 ± 3.60 |
Differences were found to be significant (see text).
Per 30 μm of length.
As percentage of protoplasts formed after 1 h of treatment with Trichoderma lytic enzymes.
(iii) Cell wall sensitivity to lytic enzymes and polysaccharide secretion.
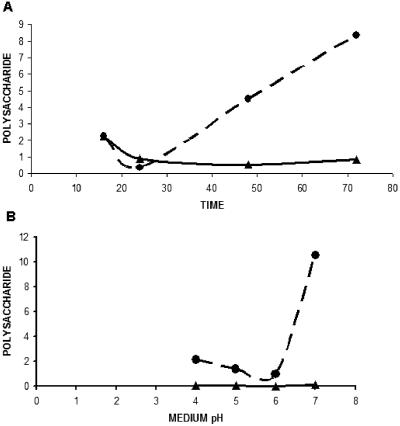
When performing transformation experiments with the standard protocol, we observed that protoplast formation occurred faster in the Δrim101 mutants than in the wild-type strains. Accordingly, we proceeded to perform quantitative experiments and verified that cell wall sensitivity to lytic enzymes was higher in the mutants than in wild-type cells (Table 2). Perhaps related to cell wall alterations, we observed that, when mutant strains were grown in liquid CM at pH 7, the culture medium became highly viscous after a few days. It was presumed that viscosity was related to secretion of a polysaccharide to the medium. Accordingly, the culture medium was treated with cold ethanol, and neutral sugars were measured in the precipitate formed. It was observed that the BMA2 Δrim101 mutant secreted about 8 times more polysaccharide than did the FB2 strain after 48 h and 9.5 times more after 72 h of incubation (Fig. 5A). Interestingly, polysaccharide secretion was pH dependent (Fig. 5B).
FIG. 5.
Polysaccharide secretion by wild-type and mutant strains of U. maydis. A. Strains FB2 and BMA2 were grown in pH 7 CM. At different times, the polysaccharide was precipitated with ethanol, and the content of neutral sugars was measured. B. Cells were grown for 48 h in MM of different pH values. Triangles and solid lines, strain FB2; circles and broken lines, strain BMA2. Polysaccharide data are expressed in mg/ml.
(iv) Mating.
We analyzed mating of mutant strains BMA2 (a2b2 Δrim101::hyg) and BMA7 (a1b1 Δrim101::hyg) and wild-type strains FB2 (a2b2) and FB1 (a1b1) in all the different possible combinations. It was observed that all mixtures of sexually compatible strains developed a positive Fuz reaction, independently of their RIM101 gene status (not shown).
(v) Virulence and development in maize plants.
Virulence of Δrim101 strains was analyzed by inoculation of mixtures of sexually compatible mutants (BMA2 and BMA7) or wild-type strains (FB1 and FB2) in maize seedlings. No qualitative or quantitative differences in virulence symptoms were observed between mutant and wild-type mixtures (Table 3). To analyze whether the normal life cycle of the mutant strains was being completed in the maize plants, teliospores were recovered from the galls formed in the infected plants. Teliospores from mutants or wild-type strains showed no morphological difference, and when inoculated on solid complete medium, they displayed no differences in their germination capacity (not shown), suggesting that they had no defects in meiosis. It has been reported previously that Saccharomyces cerevisiae rim101 mutants have alterations in meiosis (43).
TABLE 3.
Effect of RIM101 deletion on virulence of U. maydis on maize seedlings
| No. of cells inoculated | Plants with galls (%)a
|
|
|---|---|---|
| FB2 × FB1 (wild type) | BMA2 × BMA7 (Δrim101) | |
| 105 | 100 | 100 |
| 104 | 72 | 84 |
| 103 | 50 | 54 |
| 102 | 50 | 31 |
Six-day-old Zea mays seedlings (25 per condition) were inoculated with different cell densities of mixtures of mating-compatible U. maydis strains. After 15 days, disease symptoms were noted.
(vi) Protease secretion.
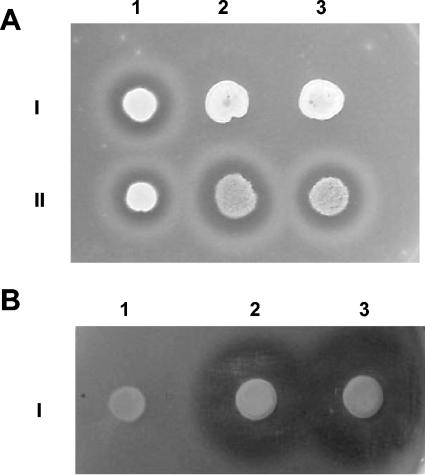
Secretion of alkaline proteases was analyzed in agar plates containing skim milk as the sole nitrogen source at pH 6.8 or 8.0. After 5 days, wild-type strains developed clear halos of proteolysis on the plates, whereas none of the Δrim101 mutants analyzed showed this capacity (Fig. 6A).
FIG. 6.
Proteolytic activities of different U. maydis and Y. lipolytica strains. Strains were inoculated over skim milk plates and incubated for 48 h. A. U. maydis. I/1, FB2 wild-type strain; II/1, FB1 wild-type strain; I/2, BMA2 Δrim101 mutant; I/3, BMA4 Δrim101 mutant; II/2, PGM1 complemented strain; II/3, AAP1 complemented strain. B. Y. lipolytica. I/1, ΔFL3 mutant; I/2, ETA1 complemented strain; I/3, E121 wild-type strain.
Complementation of the RIM101 mutation.
The BMA2 strain was transformed with pUpacC22 or pUpacC23 carrying respectively the truncated or the complete versions of the RIM101 ORF (see Materials and Methods). Carboxin-resistant clones were recovered, and their resistance to carboxin and hygromycin was confirmed. For further studies, we selected one strain obtained by transformation with pUpacC22 (AAP1) and two (PGM1 and PGM2) obtained after transformation with pUpacC23. AAP1 and PGM1 transformants recovered the capacity to secrete the alkaline protease comparable to FB2 (Fig. 6A). Other phenotypic characteristics of the wild-type strains that had been altered by RIM101 disruption were also recovered in strain PGM1. Accordingly, its sensitivity to Trichoderma lytic enzymes was reduced (Table 2). Also polysaccharide secretion was restrained: the PGM1 strain produced 0.500 ± 0.03 mg/ml and PGM2 produced 0.630 ± 0.05 mg/ml, after 48 h of growth in CM, pH 7, compared to 0.567 ± 0.04 mg/ml synthesized by the wild-type strain FB2. Tolerance to Li+ and Na+ salts was also recovered in PGM1 (Fig. 3), and the cell shape of the complemented strain PGM1 was very similar to that of the wild type at either pH 3 or pH 7 (Table 2; Fig. 4C and F).
Complementation of a null rim101 Y. lipolytica mutant with U. maydis RIM101.
Y. lipolytica strain FL3 (A rim101-1113 ura3 lys11) was transformed with the autonomous replication plasmid pUpacC44 bearing the truncated form of the U. maydis RIM101 ORF under the control of the promoter and terminator of the Y. lipolytica Xpr2 gene. Transformants were isolated in uracil-free medium, and their uracil prototrophy was confirmed by transfer to fresh plates of the same medium. One hundred transformants were inoculated on skim milk plates at pH 6.8, and we observed that about 70% of the isolates recovered alkaline protease activity (not shown). Figure 6B shows proteolysis in skim milk plates by one transformant (strain ETA1) compared to Y. lipolytica wild-type strain E121 and contrasting with FL3ΔR (rim101 mutant). Five strains recovered after transformation with the empty plasmid pINA444 carrying Ura3, but not the RIM101 gene, recovered uracil prototrophy but not protease activity (not shown).
DISCUSSION
Signaling by pH in fungi is an important mechanism for their survival in nature. At least three mechanisms have been described to fulfill this function. One of the best-characterized pH-transduction systems is the Pal/Rim pathway (reviewed in references 32 and 33). A mechanism responsive to alkaline pH dependent on calcineurin that acts by activation of a large number of genes through the transcriptional factor Crz1p has been described elsewhere for yeast (41, 42; see reviews in references 32 and 33). Finally a mechanism depending on the MDS3 gene was recently described for C. albicans. MDS3 encodes a polypeptide possessing a kelch-like interaction domain. Homozygous mds3/mds3 C. albicans mutants were affected in the expression of several genes related to mycelial growth, failed to form mycelium at alkaline pH in some media, and displayed reduced virulence in a model system (14). Apparently this protein acts in parallel with Rim101p (14; reviewed in reference 33).
In the present study we analyzed whether the Pal/Rim pathway could be involved in the control of dimorphism of U. maydis, which depends on external pH (38). Accordingly, we proceeded to determine if a homologue of the RIM101/pacC gene was present in U. maydis (all previously described homologues belong to ascomycetes and deuteromycetes) and, if so, to isolate the encoding gene. The homologue of U. maydis RIM101/pacC (named RIM101) identified in this study encodes a protein with highest similarity to PacC proteins from F. oxysporum and A. niger. As for other reported homologues, maximal similarity is concentrated at the zinc finger-encoding regions and reduced at the C termini of the proteins (24).
The expression of RIM101 measured by Northern blotting showed much higher values at neutral pH. Hygromycin resistance of disruptants (a qualitative assay in strains where the hygromycin resistance gene was under the control of the RIM101 promoter) was found also at both pH 3 and pH 7. It is known that, in different fungi, such as C. albicans (13), Y. lipolytica (24), A. niger (26), and Sclerotinia sclerotiorum (36), PacC/Rim101 mRNA is present at all physiological pH values, but PacC/Rim101 stimulates its own transcription only at neutral or alkaline pH. If the two putative PacC/Rim101 binding boxes detected in the promoter region of U. maydis RIM101 are functional, this might explain the difference in expression observed at both pH values.
U. maydis RIM101 was identified as the homologue of pacC/RIM101 by two pieces of evidence: (i) from a structural point of view, by the sequence homology of Rim101p to other Pal/Rim101 proteins, and (ii) from a functional point of view, by its capacity to complement the alkaline protease activity in a Y. lipolytica Δrim101 mutant. Activity of PacC/Rim101 as a transcription factor in other systems (e.g., references 25, 26, and 44) requires its proteolytic processing. In case processing might be different in U. maydis and Y. lipolytica, a partial version of the RIM101 ORF that included the zinc finger coding region was used for transformation. This version (first demonstrated to be functional for the complementation of the alkaline protease secretion in U. maydis Δrim101 mutants) proved to be effective in restoring the proteolytic activity in the Y. lipolytica Δrim101 mutant.
Disruption of RIM101 by double homologous recombination did not affect the growth rate of U. maydis mutants at acid or neutral pH, suggesting that, as in other systems (13, 25, 26, 36), the gene is dispensable for growth. Also, mutants were not affected in their dimorphic capacity, a result suggesting that, in contrast to C. albicans, in which RIM101 is required for pH-dependent filamentation (34), the Pal/Rim pathway is not involved in the pH signal transduction required for dimorphism in U. maydis. Another contrast between U. maydis and C. albicans mutants regards virulence. It has been reported previously that C. albicans RIM101 is involved in persistence of infection (11); in contrast Δrim101 mutants of U. maydis were as virulent in maize as were wild-type strains. Apparently this behavior depends on the fungal species. Fusarium oxysporum pacC mutants are more virulent than the wild-type strains, suggesting that PacC behaves as a negative regulator of virulence (9). In contrast, S. sclerotiorum pacC mutants display reduced virulence (35).
Nevertheless, as occurs with other fungi (9, 23; reviewed in references 32 and 33), U. maydis mutants deficient in the Rim101-encoding gene displayed a pleiotropic phenotype. Δrim101 mutants were abnormal regarding protein secretion, cell morphogenesis, cell wall strength, polysaccharide secretion, and Na+ and Li+ stress tolerance. All these alterations were alleviated by transformation with a plasmid carrying the wild-type version of the RIM101 gene. These results are evidence that the RIM101 mutation was directly responsible for all those phenotypic alterations.
One of the initially observed characteristics of fungal mutants deficient in genes involved in the Pal/Rim pathway was their impaired capacity to secrete different enzymes in response to changes in pH (reviewed in references 32 and 33). In agreement with this behavior, U. maydisΔrim101 mutants were deficient in extracellular protease activity. They were also altered in morphogenesis: mutant yeast cells were larger, and occasionally septated, a rare event in wild-type yeast cells. Frequency of septation in mutant mycelial cells grown at acid pH was increased. These alterations suggest that Rim101 might be involved in morphogenesis and the regulation of the cell cycle. In agreement with this hypothesis, it has been reported that Rim101/PacC is involved in different morphogenetic phenomena in other fungi. Accordingly, evidence exists that that the gene product plays a role in the regulation of sporulation and invasive growth of S. cerevisiae (25, 43), efficient conidiation of A. nidulans (44), and sclerotial development in S. sclerotiorum (35). Morphogenetic alterations of U. maydis occurring in the mycelium of mutants deficient in RIM101 growing at pH 3 suggest that the gene product has an active function at acid pH. The observations that A. nidulans pacC mutants display reduced conidiation and abnormal morphology at acid pH (44) and that chlamydospore formation in medium at pH 5.5 decreases in rim13 and rim101 mutants of C. albicans (29) agree with our results and reinforce this hypothesis. For S. cerevisiae it was reported (25) that the processed (and presumptively active) form of Rim101 protein was present at both alkaline and acid pH.
In regard to other phenotypic alterations of Δrim101 mutants, we may hypothesize that their increased sensitivity to lytic enzymes (an indication of alterations in the structure of the cell wall) and their increased secretion of polysaccharide are related phenomena. In C. albicans, PacC/Rim101 has been related to the regulation of Phr1 and Phr2 surface proteins. These are retained in the plasmalemma through a glycosylphosphatidylinositol anchor and possess a transglycosidase activity that may play a role in the linking of β-1,6 and β-1,3 glucans in the cell wall (17, 40). A similar role for other members of the same transglycosidase family in different fungi has been suggested (for a discussion see reference 37). A plausible hypothesis is that a similar mechanism operates in U. maydis and that mutation of RIM101 alters the correct functioning of putative transglycosidases. As a result, a polysaccharide involved in cell wall growth would be secreted (at least partially) to the medium, instead of being bound to the wall, thus resulting in its abnormal structure and organization.
Another phenotypic alteration brought about by the RIM101 mutation was an elevated susceptibility to Na+ and Li+, but not K+, ions. A similar phenomenon was reported for pacC/rim101 mutants of F. oxysporum and S. cerevisiae. This sensitivity appears to be due to the possible alteration in the expression of a P-type Na+-ATPase, Ena1 (9, 23). Interestingly, in S. cerevisiae a plasma membrane-bound ENA1-type Na+-ATPase was up-regulated by the calcineurin/Crz1 pathway cited above (41). In U. maydis two novel P-type ATPases that mediate K+ or Na+ uptake have been isolated, but these are not related to Ena1 ATPases (6). Also the observation that their transcripts accumulated at acid pH (6) makes improbable their regulation by Rim101, since according to our results sensitivity to salt stress occurred only at alkaline pH. The observation that high concentrations of sorbitol did not affect U. maydis growth is evidence (as suggested in reference 9) of a specific sensitivity to salt and not to hyperosmotic stress.
Originally, the role of the Pal/Rim pathway was considered to be specific to ascomycetes and deuteromycetes and involved in the expression of some selected pH-regulated exoenzymes. Further data have revealed that the pathway plays unsuspected important roles in other cellular functions (9, 23, 35). The results described here are evidence that the pathway also exists in basidiomycetes, having roles in a series of similar complex phenomena with several heretofore-unobserved phenotypic consequences. In this sense it is relevant that an in silico analysis of the U. maydis genome revealed the existence of putative homologues of genes palA/RIM20, palB/RIM13, and palI/RIM9.
Acknowledgments
This work was partially supported by CONACYT, México. The doctoral studies of E.T.A.-C. were supported by a CONACYT fellowship.
We thank Flora Banuett for providing us U. maydis strains FB1 and FB2, Claude Gaillardin for Y. lipolytica strains FL3ΔR and E121, Scott Gold for valuable comments on this work, Peter Schrier for the partial RIM101 sequences provided, and Claudia León for her valuable technical support.
REFERENCES
- 1.Banuett, F. 1995. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genet. 29:179-208. [DOI] [PubMed] [Google Scholar]
- 2.Banuett, F., and I. Herskowitz. 1989. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banuett, F., and I. Herskowitz. 1996. Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122:2965-2976. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, K. J., S. E. Gold, and J. W. Kronstad. 1993. Identification and complementation of a mutation to a constitutive filamentous growth in Ustilago maydis. Mol. Plant-Microbe Interact. 6:274-283. [DOI] [PubMed] [Google Scholar]
- 5.Barth, G., and C. Gaillardin. 1996. Yarrowia lipolytica, p. 313-388. In K. Wolf (ed.), Non-conventional yeasts in biotechnology. A handbook. Springer-Verlag, Berlin, Germany.
- 6.Benito, B., B. Garciadeblás, P. Schreier, and A. Rodríguez-Navarro. 2004. Novel P-type ATPases mediate high-affinity potassium or sodium uptake in fungi. Eukaryot. Cell 3:359-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bölker, M. 2001. Ustilago maydis—a valuable model system for the study of fungal dimorphism and virulence. Microbiology 147:1395-1401. [DOI] [PubMed] [Google Scholar]
- 8.Caddick, M. X., A. G. Brownlee, and H. N. Arst, Jr. 1986. Regulation of gene expression by the pH of the growth medium in Aspergillus nidulans. Mol. Gen. Genet. 203:346-353. [DOI] [PubMed] [Google Scholar]
- 9.Caracuel, Z., C. Casanova, M. I. G. Roncero, A. Di Pietro, and J. Ramos. 2003. pH response transcription factor PacC controls salt stress tolerance and expression of the P-type Na+-ATPase Ena1 in Fusarium oxysporum. Eukaryot. Cell 2:1246-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chávez-Ontiveros, J., A. D. Martinez-Espinoza, and J. Ruiz-Herrera. 2000. Double chitin synthetase mutants from the corn smut fungus Ustilago maydis. New Phytol. 146:335-341. [DOI] [PubMed] [Google Scholar]
- 11.Davis, D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 44:1-7. [DOI] [PubMed] [Google Scholar]
- 12.Davis, D., J. E. Edwards, Jr., A. P. Mitchell, and A. S. Ibrahim. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, D., R. B. Wilson, and A. P. Mitchell. 2000. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, D. A., V. M. Bruno, L. Loza, S. G. Filler, and A. P. Mitchell. 2002. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison, S. H. 2000. pH regulation of gene expression in fungi. Fungal Genet. Biol. 29:61-71. [DOI] [PubMed] [Google Scholar]
- 16.Dimler, R. J., W. C. Schaeffer, C. S. Wise, and C. E. Rist. 1952. Quantitative paper chromatography of D-glucose and its oligosaccharides. Anal. Chem. 24:1411-1414. [Google Scholar]
- 17.Fonzi, W. A. 1999. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of β-1,3 and β-1,6 glucans. J. Bacteriol. 181:7070-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fournier, P., L. Guyaneux, M. Chasles, and C. Gaillardin. 1991. Scarcity of ARS sequences isolated in a morphogenesis mutant of the yeast Yarrowia lipolytica. Yeast 7:25-36. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Lopez, C. I., R. Szabo, S. Blanchin-Roland, and C. Gaillardin. 2002. Genetic control of extracellular protease synthesis in the yeast Yarrowia lipolytica. Genetics 160:417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 21.Holliday, R. 1961. The genetics of Ustilago maydis. Genet. Res. 2:204-230. [Google Scholar]
- 22.Keon, J. P. R., G. A. White, and J. A. Hargreaves. 1991. Isolation, characterization and sequence of a gene conferring resistance to the systemic fungicide carboxin from the maize smut pathogen, Ustilago maydis. Curr. Genet. 19:475-481. [DOI] [PubMed] [Google Scholar]
- 23.Lamb, T. M., and A. P. Mitchell. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes nrg1 and smp1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert, M., S. Blanchin-Roland, F. Le Leuedec, A. Lépingle, and C. Gaillardin. 1997. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol. Cell. Biol. 17:3966-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, W., and A. P. Mitchell. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacCabe, A. P., P. T. W. Johanes, V. D. Homberg, J. Tilburn, H. N. Arst, Jr., and J. Visser. 1996. Identification, cloning and analysis of the Aspergillus niger gene pacC, a wide domain regulatory gene responsive to ambient pH. Mol. Gen. Genet. 250:367-374. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Espinoza, A. D., M. D. Garcia-Pedrajas, and S. E. Gold. 2002. The ustilaginales as plant pests and model systems. Fungal Genet. Biol. 35:1-20. [DOI] [PubMed] [Google Scholar]
- 28.Martínez-Espinoza, A. D., C. León, G. Elizarraraz, and J. Ruiz-Herrera. 1997. Monomorphic nonpathogenic mutants of Ustilago maydis. Phytopathology 87:259-265. [DOI] [PubMed] [Google Scholar]
- 29.Nobile, C. J., V. M. Bruno, M. L. Richard, D. A. Davis, and A. P. Mitchell. 2003. Genetic control of chlamydospore formation in Candida albicans. Microbiology 149:3629-3637. [DOI] [PubMed] [Google Scholar]
- 30.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Bailliere Tindall, London, United Kingdom.
- 31.Ogrydziak, D. M., and R. K. Mortimer. 1977. Genetics of extracellular protease production in Saccharomycopsis lipolytica. Genetics 87:621-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peñalva, M. A., and H. N. Arst, Jr. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peñalva, M. A., and H. N. Arst, Jr. 2004. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 58:425-451. [DOI] [PubMed] [Google Scholar]
- 34.Ramón, A. M., A. Porta, and W. A. Fonzi. 1999. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 181:7524-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rollins, J. A. 2003. The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant-Microbe Interact. 16:785-795. [DOI] [PubMed] [Google Scholar]
- 36.Rollins, J. A., and M. B. Dickman. 2001. pH signaling in Sclerotinia sclerotiorum: identification of a pacC/RIM1 homolog. Appl. Environ. Microbiol. 67:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Herrera, J., M. V. Elorza, P. E. Alvarez, and R. Sentandreu. 2004. Biosynthesis of the fungal cell wall, p. 41-49. In G. San Blas and R. A. Calderone (ed.), Pathogenic fungi: structural biology and taxonomy. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
- 38.Ruiz-Herrera, J., C. León, L. Guevara-Olvera, and A. Cárabez-Trejo. 1995. Yeast-mycelial dimorphism of haploid and diploid strains of Ustilago maydis. Microbiology 141:695-703. [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Saporito-Irwin, S. M., C. E. Birse, P. S. Sypherd, and W. A. Fonzi. 1995. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15:601-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serrano, R., A. Ruiz, D. Bernal, J. R. Chambers, and J. Arino. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46:1319-1333. [DOI] [PubMed] [Google Scholar]
- 42.Stathopoulos, A. M., and M. S. Cyert. 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11:3432-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su, S. S. Y., and A. P. Mitchell. 1993. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 21:3789-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tilburn, J., S. Sarkar, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Peñalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, J., D. Walden, and S. A. Leong. 1988. Gene transfer system for the phytopathogenic fungus Ustilago maydis. Proc. Natl. Acad. Sci. USA 85:865-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xoconostle-Cázares, B., C. A. Spetch, P. W. Robbins, Y. Liu, C. León, and J. Ruiz-Herrera. 1997. Umchs5, a gene coding for a class IV chitin synthase in Ustilago maydis. Fungal Genet. Biol. 22:199-208. [DOI] [PubMed] [Google Scholar]