Abstract
The class II and class I chitin synthases of the filamentous fungus Aspergillus nidulans are encoded by chsA and chsC, respectively. Previously, we presented several lines of evidence suggesting that ChsA and ChsC have overlapping functions in maintaining cell wall integrity. In order to determine the functions of these chitin synthases, we employed electron and fluorescence microscopy and investigated in detail the cell wall of a ΔchsA ΔchsC double mutant (ΔAC mutant) along with the localization of ChsA and ChsC. In the lateral cell wall of the ΔAC mutant, electron-transparent regions were thickened. Septa of the ΔAC mutant were aberrantly thick and had a large pore. Some septa were located abnormally close to adjacent septa. A functional hemagglutinin (HA)-tagged ChsA (HA-ChsA) and a functional FLAG-tagged ChsC (FLAG-ChsC) were each localized to a subset of septation sites. Comparison with the localization pattern of actin, which is known to localize at forming septa, suggested that ChsA and ChsC transiently exist at the septation sites during and shortly after septum formation. Double staining of HA-ChsA and FLAG-ChsC indicated that their localizations were not identical but partly overlapped at the septation sites. Fluorescence of FLAG-ChsC, but not of HA-ChsA, was also observed at hyphal tips. These data indicate that ChsA and ChsC share overlapping roles in septum formation.
Chitin, a polymer of β-1,4-linked N-acetylglucosamine, is one of the major cell wall constituents in many filamentous fungi. Chitin synthases are membrane proteins that catalyze the polymerization of N-acetylglucosamine. To date, many chitin synthase genes have been identified from various fungi, and the polypeptides deduced from these genes have been divided into six classes (32, 37). Studies in the budding yeast Saccharomyces cerevisiae have revealed the localization and specific functions of its three chitin synthases (Chs1, Chs2, and Chs3) (5, 34). In this organism, chitin is concentrated mostly in septa. Chs3, a class IV chitin synthase, is required for chitin ring formation at the base of emerging buds and for chitin synthesis in the lateral cell wall during vegetative growth. Chs3 is found at the plasma membrane and in an intracellular compartment called the chitosome, and its correct localization requires the functioning of several proteins (5, 32, 34, 37). Chs2, a class II enzyme, synthesizes chitin in primary septa. Chs2 appears at a late stage of mitosis and is localized to the septation site; it is degraded immediately after septum formation (7, 36). Chs1, a class I enzyme, repairs the weakened cell walls of daughter cells after their separation from mother cells; this separation is executed by the activity of a chitinase that digests the primary septa. Chs1 exists at a constant level throughout the cell cycle and is reported to be present on the plasma membrane and chitosomes (60), although its precise localization has not been reported.
In our previous work, we identified five chitin synthase genes, chsA, chsB, chsC, chsD, and csmA, that encode chitin synthases of classes II, III, I, IV, and V, respectively, in Aspergillus nidulans (12, 29, 30, 58). A search through the A. nidulans genome database (http://www-genome.wi.mit.edu/annotation/fungi/aspergillus/) identified eight chitin synthase genes, including chsA-chsD and csmA. Elimination or reduction of the expression of chsB or csmA caused obvious defects in hyphal growth and asexual development (3, 18, 21, 43, 58). Analogous observations have also been reported in other filamentous fungi (2, 23, 25, 31, 59). Deletion mutants of chsA, chsC, or chsD exhibited no clear morphological differences from the wild-type strain (29, 30, 58). Several combinations of double mutants have also been constructed in A. nidulans (9, 13, 18, 20, 21, 29). Among these, ΔchsA ΔchsC double mutants (ΔAC mutants) showed remarkable synergistic defects in hyphal growth as well as in sexual and asexual development in comparison with the single mutants (13). Although the growth rate of the ΔAC mutants under ordinary conditions was similar to that of the wild-type strain, the growing edge of their colonies was irregular and their hyphae were occasionally lysed. The growth rate was reduced by chitin-binding dyes, salts (1.2 M NaCl or KCl), or a detergent (0.005% sodium dodecyl sulfate [SDS]). Conidial production by the ΔAC mutants was less than 0.01% of that by the wild-type strain due to the reduced number and altered morphology of conidiophores. The wild-type conidiophore consists of a stalk, a vesicle, two tiers of sterigmata (metulae and phialides), and conidia (1). Although the ΔAC mutants formed normal conidiophore vesicles at the tips of conidiophore stalks, they produced chains of sterigmata and occasionally reproduced conidiophores upon vesicles, which are termed secondary conidiophores (13).
In this study, we investigated the cell wall ultrastructure of the ΔAC mutant by electron microscopy and identified defects in both the lateral wall construction and septum formation. We constructed strains that expressed epitope-tagged ChsA and/or ChsC. These fusion proteins were functional, and either of them was found at septation sites when their subcellular localization was examined. Double staining of these fusion proteins suggested that their localization at the septation sites partially overlapped. ChsC, but not ChsA, was found at hyphal tips. These results suggest that ChsA and ChsC have overlapping functions in proper septum formation in A. nidulans.
MATERIALS AND METHODS
Strains and growth conditions.
The A. nidulans strains used in this study are listed in Table 1. Strains with the same genotype (e.g., AC-7 and AC-8) showed indistinguishable phenotypes. Standard genetic manipulation techniques were used for A. nidulans (24, 33, 35). Strains were cultured at 37°C unless otherwise mentioned. Minimal medium (MMG) was prepared as described previously by Rowlands and Turner (38), except that glucose was added to 2% in MMG. Y2G medium (0.5% Bacto yeast extract, 2% d-glucose, 0.1% [vol/vol] trace elements solution [38]) was also used. To support the growth of pyrG89 strains, we supplemented media with 5 mM uridine and 10 mM uracil. For the growth of argB2, biA1, and pyroA4 mutants, 0.20 mg/ml arginine, 0.02 μg/ml biotin, and 0.50 μg/ml pyridoxin, respectively, were added in MMG. Solid media were prepared by adding 1.5% agar. Although ΔAC mutants produced few conidiophores on MMG or Y2G plates, they produced a much larger number of conidiophores on plates supplemented with a moderate amount of osmotic stabilizers, such as 0.6 M sorbitol or sucrose (M. Ichinomiya et al., unpublished data). Those conidiophores containing a small number of conidia were scraped from plates and used for liquid cultivation.
TABLE 1.
A. nidulans strains used in this study
| Strain | Genotypeb | Source |
|---|---|---|
| FGSC A26 | biA1 | FGSCa |
| ABPU/AU | biA1 pyrG89 wA3 argB2 pyroA4 [pSS1][pP1] | 29 |
| A-4 | biA1 pyrG89 wA3 argB2 pyroA4 ΔchsA::argB | 29 |
| C2-11/A-16 | biA1 pyrG89 wA3 argB2 pyroA4 ΔchsC::pyr-4 [pSS1] | 13 |
| AC-7, -8 | biA1 pyrG89 wA3 argB2 pyroA4 ΔchsA::argB ΔchsC::pyr-4 | 13 |
| ABPUS14 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 | This study |
| ///-8, -10 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 [pUCPYR1][pUSC][pSS1] | 56 |
| ABPUS////-7 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 [pUCPYR1][pUSC][pSS1][pUCPYROA] | This study |
| AΔPG/sC-9 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG [pUSC] | 56 |
| AΔPG/sC/A-1 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG [pUSC][pSS1] | 56 |
| AΔPG/sC/A/P-1, -12 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG [pUSC][pSS1] [pUCPYROA] | This study |
| CΔsC/PG/A-1 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsC::sC [pUCPYR1][pSS1] | 56 |
| ΔACa4 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC | 56 |
| ΔAC/A-4 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC [pSS1] | 56 |
| ΔAC/pHBS-4, -5, -6 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC chsA(p)::chsA::argB | This study |
| ΔAC/pHBSBXHA-7, -8 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC chsA(p)::HA-chsA::argB | This study |
| AΔPG/sC/pHBSBXHA-2, -3, -9 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG chsA(p)::HA-chsA::argB [pUSC] | This study |
| CΔsC/PG/A/FLAGC-1, -2, -4 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsC::sC chsC(p)::FLAG-chsC::pyroA [pUCPYR1][pSS1] | This study |
| ΔAC/A/P-2, -5, -8 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC [pSS1][pUCPYROA] | This study |
| ΔAC/FLAGC-2, -4 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC chsC(p)::FLAG-chsC::pyroA | This study |
| ΔAC/A/FLAGC-2, -5 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC chsC(p)::FLAG-chsC::pyroA [pSS1] | This study |
| ΔAC/ChsC-1, -3 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC chsC(p)::chsC::pyroA | This study |
| ΔAC/A/ChsC-8, -9, -13 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC chsC(p)::chsC::pyroA [pSS1] | This study |
| ΔAC/HAA/FLAGC-1, -2 | biA1 pyrG89 wA3 argB2 sC114 pyroA4 ΔchsA::pyrG ΔchsC::sC chsA(p)::HA-chsA::argB chsC(p)::FLAG-chsC::pyroA | This study |
Fungal Genetics Stock Center, Kansas City, Kans.
The plasmids in brackets were digested with a restriction enzyme that has the sole recognition site on the auxotrophic marker. The linearized plasmid was used for transformation.
Transmission electron microscopy.
Fixation, embedding, and examination of samples with transmission electron microscopy were carried out as described previously (18). Mycelia were fixed in 5% glutaraldehyde diluted in 0.1 M phosphate buffer (pH 7.0) and in 1% buffered osmium tetroxide (pH 7.0). Specimens were embedded in epoxy resin. Samples were prepared from the wild-type strain and the chs deletion mutants grown on MMG plates for 96 h.
Fluorescence microscopy.
Conidia were spread on a thin MMG plate and incubated for 12 h. A small piece of agar containing germlings was subjected to fixation and staining as described previously by Harris et al. (16), except for the concentration of reagents used for the staining. Germlings of the wild-type strain, ΔchsA mutant (ΔA mutant), or ΔchsC mutant (ΔC mutant) were stained with a solution containing 500 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) (Wako) and 12 μg/ml calcofluor white (CFW) (Fluorescent Brightener 28, F-6259; Sigma). For ΔAC mutants, 6 μg/ml CFW was used. Germlings were observed using an Olympus BX52. Images were taken with an ORCA-ER charge-coupled-device camera (HAMAMATSU) and analyzed by AQUACOSMOS software (HAMAMATSU).
Counting of septa.
Conidia of each strain were inoculated onto a thin MMG plate and grown for 72 h. A small piece of agar, on which the colony edge was advancing, was removed and mounted on a slide glass. Septa were visualized by using 0.01% CFW. The number of septa formed in 200 μm of a hypha was counted. Apical parts of hyphae containing the three septa closest to the hyphal tip or containing foot cells of conidiophores were not subjected to measurement. A total of 60 hyphae from three colonies of each strain were examined, and the average and standard deviations were calculated.
DNA manipulation and construction of plasmids.
Standard techniques were used for DNA manipulation and construction of plasmids (39). PCR was performed using an Expand High Fidelity PCR system (Roche Diagnostics) or KOD-Plus (TOYOBO). DNA sequences were determined using ABI PRISM BigDye Terminator, version 3.0, Cycle Sequencing Ready Reaction kit and ABI PRISM 310 genetic analyzer (Applied Biosystems).
pHBS, which contains the chsA gene and the argB gene, was constructed by inserting the 1.7-kb BamHI-SphI fragment of pSS1 into the HindIII site on the multiple-cloning site of pchsA (29). A 0.2-kb hemagglutinin (HA)-encoding sequence was PCR amplified from pHA6 (47) using primers 6×HA.Sma.F (5′-GGCCC GGGTA CCCAT ACGAT G-3′ [italics indicate the SmaI site]) and 6×HA.Sma.R (5′-CCCCC GGGGC TAGCG TAATC TG-3′ [italics indicate the SmaI site]). The fragment was digested with SmaI and inserted into the SmaI site on the chsA-encoding region of pHBS, yielding pHBSBXHA. p3XFLAG-myc-CMV-26 expression vector (E6401; Sigma) contains a sequence encoding three repeats of the FLAG epitope. The FLAG-encoding sequence was PCR amplified from p3XFLAG-myc-CMV-26 using primers FLAG-F (5′-GGAAT TCAGA ATTAA CCATG GACTA C-3′ [italics indicate the EcoRI site]) and FLAG-R (5′-GAAGA TCTCG CAAGC TTGTC ATCGT CATC-3′ [italics indicate the BglII site]). The PCR products and p3XFLAG-myc-CMV-26 were digested with BglII and EcoRI and ligated, yielding p6XFLAG. A sequence encoding six repeats of the FLAG epitope was PCR amplified using primers 6× FF (5′-CCGCT CGAGT GAACC GTCAG AATTA ACC-3′ [italics indicate the XhoI site]) and 6×FR (5′-CCGCT CGAGA TATCA GATCT CGCAA G-3′ [italics indicate the XhoI site]). The fragment was digested with XhoI and inserted into the XhoI site of pchsC, yielding pchsC-X-6XFLAG. The pyroA-encoding sequence was PCR amplified from FGSC A26 total DNA using primers pyroA1s (5′-GGCTG CAGAA GTGCG CG-3′ [italics indicate the PstI site]) and pyroA2684as (5′-CCGGA TCCAG GAGTA TACG-3′ [italics indicate the BamHI site]). The PCR products were digested with BamHI and PstI and ligated into the BamHI and PstI sites of pUC118, yielding pUCPYROA. A 2.7-kb BamHI-PstI fragment of pUCPYROA was inserted into the KpnI site of pchsC-X-6XFLAG and pchsC, yielding pC6XF-pyroA and pCpyroA, respectively. By sequencing, it was confirmed that for every cloned PCR product, no mutation was introduced into the sequence.
Construction of A. nidulans strains by transformation.
All strains constructed in this study are derived from ABPUS14. Plasmids used in transformation were pHBS, pSS1, pHBSBXHA, pUCPYROA, pCpyroA, and pC6XF-pyroA.
ΔACa4 was transformed by pHBS [containing chsA(p)::chsA], pHBSBXHA [containing chsA(p)::HA-chsA], or pSS1 (containing only the argB marker). A single copy of the plasmids was integrated into the genomic argB locus, which was confirmed by Southern analysis probed with a 1.1-kb fragment of pSS1. pHBS was linearized by digestion with HindIII and used for transformation. Total DNAs prepared from transformants were digested with XbaI and subjected to Southern analysis. A 3.0-kb band was detected in the wild-type strain. In some transformants, 2.5- and 12.1-kb bands were detected. Three of them were designated ΔAC/pHBS-4, ΔAC/pHBS-5, and ΔAC/pHBS-6. pHBSBXHA was linearized by digestion with HindIII and used for transformation. Total DNAs prepared from transformants were digested with XbaI and subjected to Southern analysis. In some transformants, 2.5- and 12.3-kb bands were detected. Two of them were designated ΔAC/pHBSBXHA-7 and ΔAC/pHBSBXHA-8. pSS1 was linearized by digestion with BglII and used for transformation. Two transformants that presented 2.5- and 5.4-kb bands were designated ΔAC/A-4 and ΔAC/A-6.
ΔAC/A/P-2, ΔAC/A/P-5, and ΔAC/A/P-8 were constructed by transforming ΔAC/A-4 with pUCPYROA. Integration of pUCPYROA into the genomic pyroA locus was confirmed by detection of 4.5- and 10.7-kb bands instead of the 9.4-kb wild-type pyroA band in Southern hybridization probed with a 1.2-kb EcoRV fragment of pUCPYROA against total DNAs digested with HindIII.
ΔAC/ChsC-1 and ΔAC/ChsC-3 were constructed by transforming ΔACa4 with pCpyroA [containing chsC(p)::chsC] linearized by digestion with MluI. Integration of pCpyroA into the genomic pyroA locus was confirmed by detection of 6.5- and 10.7-kb bands instead of the 9.4-kb wild-type pyroA band in Southern hybridization probed with a 1.2-kb EcoRV fragment of pUCPYROA against total DNAs digested with HindIII. ΔAC/A/ChsC-8, ΔAC/A/ChsC-9, and ΔAC/A/ChsC-13 were constructed by transforming ΔAC/ChsC-1 with pSS1 as described above.
ΔAC/FLAGC-2 and ΔAC/FLAGC-4 were constructed by transforming ΔACa4 with pCX6F-pyroA [containing chsC(p)::FLAG-chsC] linearized by digestion with MluI. Integration of pC6XF-pyroA into the genomic pyroA locus was confirmed by detection of 6.5- and 10.7-kb bands instead of the 9.4-kb wild-type pyroA band in Southern hybridization probed with a 1.2-kb EcoRV fragment of pUCpyroA against total DNAs digested with HindIII. ΔAC/A/FLAGC-2 and ΔAC/A/FLAGC-5 were constructed by transforming ΔAC/ChsC-1 with pSS1 as described above.
ΔAC/HAA/FLAGC-1 and ΔAC/HAA/FLAGC-2, strains that express both HA-tagged ChsA (HA-ChsA) and FLAG-tagged ChsC (FLAG-ChsC), were constructed from ΔAC/pHBSBXHA-7 by transforming with pC6XF-pyroA as described above.
ABPUS////-7 was obtained by transforming ///-10 with pUCPYROA as described above.
AΔPG/sC/A/P-1 and AΔPG/sC/A/P-12 were obtained by transforming AΔPG/sC/A-1 with pUCPYROA as described above. AΔPG/sC/pHBSBXHA-2, AΔPG/sC/pHBSBXHA-3, and AΔPG/sC/pHBSBXHA-9 were obtained by transforming AΔPG/sC-9 with pHBSBXHA as described above.
CΔsC/PG/A/FLAGC-1, CΔsC/PG/A/FLAGC-2, and CΔsC/PG/A/FLAGC-4 were obtained by transforming with pC6XF-pyroA as described above.
Examination of production of conidia.
Conidia were point inoculated at the center of MMG plates, which were incubated for 72 h. The conidia produced by a colony were scraped thoroughly and suspended in H2O containing 0.01% Tween 20. The number of conidia was counted using a hemocytometer. Colony diameter was also measured. Data were taken from five colonies for each strain to determine the average and standard deviation of conidia number and colony diameter.
Northern analysis.
Total RNA isolation and Northern analysis were performed as described previously (20). Total RNAs were prepared from mycelia cultured for 20 h in 50 ml of MMG. To detect chsA mRNA, a 1.25-kb ClaI fragment from pAM was used.
Preparation of A. nidulans cell lysate and Western analysis.
Approximately 5 × 107 conidia were inoculated into 50 ml of MMG and were cultured for 20 h. Preparation of cell lysates and fractionation by centrifugation were performed as described previously (47). Proteins were extracted in an extraction buffer (25 mM Tris-HCl [pH 7.3], 137 mM NaCl, 5 μl/ml protease inhibitor cocktail [P 8215; Sigma]) or another buffer (10 mM Tris-HCl [pH 8.2], 0.8 M sucrose, 5 μl/ml protease inhibitor cocktail). Protein content was determined using Bio-Rad protein assay dye reagent and bovine serum albumin as a standard. Samples containing 10 to 50 μg protein were boiled in 1× sample buffer (58 mM Tris-HCl [pH 6.8], 5% glycerol, 2% SDS, 1.55% dithiothreitol, 0.002% bromophenol blue) for 3 min and electrophoresed on 6% or 7.5% SDS-polyacrylamide gel. Proteins were transferred to Hybond ECL nitrocellurose membrane (Amersham Biosciences). HA-ChsA was detected using a mouse anti-HA monoclonal antibody, HA.11 (BAbCO), at a 1:1,000 dilution and an anti-mouse immunoglobulin G (IgG), horseradish peroxidase-linked antibody (Cell Signaling Technology) at a 1:5,000 dilution. FLAG-ChsC was detected using a mouse anti-FLAG M2 monoclonal antibody (F 3165; Sigma) at a 1:1,000 dilution. Signals were detected using ECL Western blotting detection system (Amersham Bioscience).
Indirect immunofluorescence microscopy.
Indirect immunofluorescence was performed as described previously by Harris et al. (16) or Momany (26) with the following modifications. Conidia (1 × 106 to 5 × 106) were inoculated into 10 ml of MMG and incubated for 12 to 14 or 20 h. Cell wall digestion was performed with a digestion solution (20 mg/ml Yatalase [TaKaRa], 5 mM EGTA [pH 7], 10 mg/ml egg white [Sigma]) for 15 to 30 min at 30°C or in another solution (30 mg/ml Yatalase, 3 mg/ml lysing enzyme [L 2265; Sigma], and 10 mg/ml egg white in phosphate-buffered saline]) for 10 min at 37°C. Cell wall digestion of AC-7 (a ΔAC mutant) was performed with a digestion solution (10 mg/ml Yatalase [TaKaRa], 5 mM EGTA [pH 7], 10 mg/ml egg white [Sigma]) for 15 min at 30°C. To detect α-tubulin and actin, a mouse anti-α-tubulin monoclonal antibody, DM 1A (1:250) (T 9026; Sigma), and a rabbit anti-actin antibody (1:250) (A 2066; Sigma), respectively, were used. To detect HA-ChsA, a mouse anti-HA monoclonal antibody (1:1,000; BAbCO) was used. To detect FLAG-ChsC, a mouse anti-FLAG M2 monoclonal antibody (1:800) (F 3165; Sigma) or a rabbit anti-FLAG polyclonal antibody (1:800) (F 7425; Sigma) was used. As secondary antibodies, an anti-mouse IgG fluorescein isothiocyanate-conjugated antibody from goat (1:100) (F 0257; Sigma) and an anti-rabbit IgG Cy3-conjugated antibody from sheep (1:400) (C2306; Sigma) were used. Germlings were observed using an Olympus BX52. Images were taken with an ORCA-ER charge-coupled-device camera (HAMAMATSU) and analyzed by AQUACOSMOS software (HAMAMATSU).
RESULTS
Cell wall and septal ultrastructures of the chsA chsC double mutant.
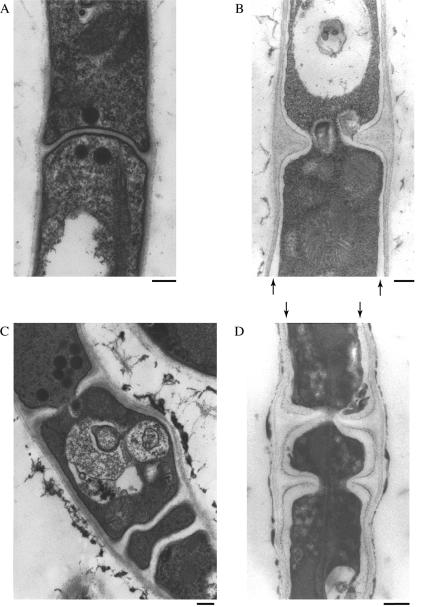
Since our previous results suggested a cell wall defect in the ΔAC mutants, we examined the cell wall structure of the ΔAC mutant in detail by transmission electron microscopy (Fig. 1). In the wild-type strain, the lateral cell wall was observed as a layer of moderate electron density and uniform thickness (Fig. 1A). Longitudinal sectioning of hyphae revealed septa of uniform thickness. When hyphae were sectioned at the center of septa, we observed a central pore (data not shown), an observation consistent with previous reports by other researchers (27). ΔchsA or ΔchsC single mutants (ΔA or ΔC mutants) did not display different appearances in their cell wall and septa in comparison with those of the wild-type strain (data not shown). In contrast, the cell wall of the ΔAC mutant exhibited several marked abnormalities (Fig. 1B to D). The lateral cell wall of the ΔAC mutant was heterogeneous, and we often observed electron-transparent regions immediately outside the plasma membrane (Fig. 1B and D, arrows) which were never observed in the wild-type strain or the single mutants. The septa of the ΔAC mutant were thicker than those of the wild-type strain, and septal pores were abnormally large. In addition, some septa were formed less than 5 μm apart. In the ΔA and ΔC mutants as well as in the wild-type strain, septa were formed at a sufficient distance from one another, and multiple septa formed in the vicinity were never observed under the conditions employed in this study. These observations indicate that either chsA or chsC is required for the construction of normal lateral walls and septa and for proper septum placement.
FIG. 1.
Transmission electron microscopy of chs mutant hyphae. Panels are (A) ABPU/AU (wild type) and (B to D) AC-8 (ΔAC mutant). Strains were grown on MMG for 4 days. Arrows in panels B and D indicate an electron-transparent layer. Bars, 250 nm.
Aberrant septum placement and uneven nuclear distribution in the chsA chsC double mutant.
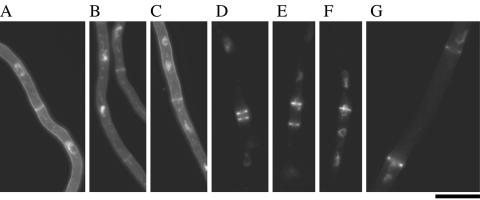
It has been suggested that mitotic nuclei have some roles in determining the position of septum formation in A. nidulans (53). We examined the positions of septa and nuclei in the wild-type strain and the chs mutants (Fig. 2). In the wild-type strain, a septum was positioned at a distance from the neighboring septa (Fig. 2A and data not shown). Subapical regions of wild-type hyphae contained 5.1 ± 1.5 septa per 200 μm (average ± standard deviation). Nuclei were evenly distributed, and each hyphal compartment contained at least two nuclei. Septation and nuclear distribution in the ΔA and ΔC mutants were similar to those in the wild-type strain (Fig. 2B and C). In the ΔAC mutant, closely placed septa were readily observed (Fig. 2D and E). The number of septa formed in subapical regions of the ΔAC hyphae was 8.2 ± 2.9 septa per 200 μm. Some subapical regions contained 16 septa per 200 μm. These results suggest an elevated frequency of septum formation in the ΔAC mutant. Apart from septum placement, nuclear distribution was also aberrant in the ΔAC mutant. Some nuclei were found at septation sites and appeared as though they had been trapped while penetrating the septum pores, or a new septum was formed in the middle of a nucleus (Fig. 2E to G). The ΔAC mutant possessed anucleate compartments: of 112 compartments examined, 21 were anucleate, whereas the wild-type strain had no anucleate compartments (n = 118). The length of anucleate compartments exceeded 25 μm in one case (Fig. 2G). Since microtubule organization is known to be crucial for proper nuclear distribution (10, 44, 54), we examined the localization of α-tubulin in the wild-type strain and in the ΔAC mutant (Fig. 3). In the wild-type hyphae, arrays of cytoplasmic microtubules were observed, except in regions around septa (Fig. 3A and B). Such arrays were also observed in the ΔAC mutant (Fig. 3C and D), although their structure was not as straight as that of the arrays in the wild-type strain.
FIG. 2.
Distribution of nuclei and septa in chs mutant hyphae. ABPU/AU (wild type, A), A-4 (ΔA mutant, B), C2-11/A-16 (ΔC mutant, C), and AC-7 (ΔAC mutant, D to G) were grown on MMG for 12 h. Hyphae were stained with CFW and DAPI to visualize septa and nuclei, respectively. Bar, 10 μm.
FIG. 3.
Microtubule organization in the wild-type strain and the ΔAC mutant. ABPU/AU (A and B) and AC-7 (C and D) grown on MMG for 13 h were subjected to indirect immunofluorescence microscopy using anti-actin antibody (α-actin) and anti-α-tubulin antibody (α-tubulin). Hyphae were also stained with DAPI and CFW. The arrowhead in panel D indicates the septum with a microtubule passing through. Bars, 10 μm.
A functional HA-tagged ChsA localizes to septa.
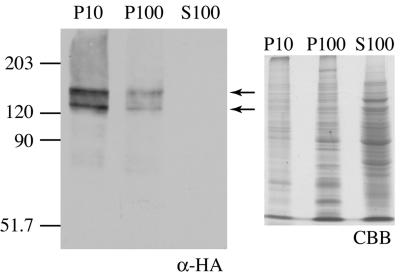
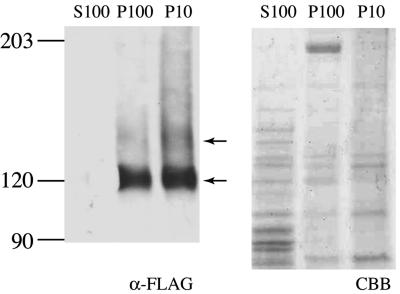
In order to obtain clues for identifying the roles of ChsA in vivo, we constructed strains that express the HA epitope-tagged ChsA (HA-ChsA) under the chsA promoter. Based on the fact that Chs2, an S. cerevisiae ortholog of ChsA, is functional without the N-terminal 200 amino acids (11), six copies of the HA epitope were inserted between proline 103 and arginine 104 of ChsA. To determine whether HA-ChsA was functional, HA-ChsA was expressed in a ΔAC mutant at the genomic argB locus and was tested for its ability to rescue conidiation defects in the ΔAC mutants. Southern analysis confirmed that two transformants, ΔAC/pHBSBXHA-7 and ΔAC/pHBSBXHA-8, had a single copy of chsA(p)::HA-chsA at the argB locus (data not shown). Strains that express wild-type ChsA at the genomic argB locus (ΔAC/pHBS-4, ΔAC/pHBS-5, and ΔAC/pHBS-6) were also constructed as controls. As previously reported, the ΔAC mutant produced very few conidia, less than 0.01% of the number produced by the wild-type strain (Table 2) (13). Conidial production by the HA-chsA-expressing ΔAC mutant (ΔAC/pHBSBXHA-7) was approximately 60% of that by the wild-type or the ΔC mutant strain (CΔsC/PG/A-1); however, it was virtually the same as that by the chsA-expressing ΔAC mutant (ΔAC/pHBS-4) (Table 2), indicating that HA-ChsA is as functional as the wild-type ChsA. Northern analysis showed that the mRNA levels of ectopically expressed HA-chsA and wild-type chsA were not higher than that of the ΔC mutant (data not shown), confirming that a single copy of HA-chsA or wild-type chsA is expressed under the chsA promoter. We examined the HA-ChsA protein level in liquid-cultured mycelia by Western analysis using anti-HA monoclonal antibody. Cell extracts were prepared from mycelia that had been cultured in minimal medium for 20 h; the extracts were then fractionated by serial centrifugation (Fig. 4). The predicted molecular mass of HA-ChsA was 121 kDa. Several bands of higher molecular mass (two of these were approximately 123 and 146 kDa) were detected in the P10 and P100 fractions but not in the soluble fraction (S100) (Fig. 4). Similar results were obtained when the cultivation was carried out for 13 h in minimal medium or in rich medium (data not shown).
TABLE 2.
Conidial formation by the wild-type strain and mutants
| Strain | Relevant genotype | Colony diameter (mm) | No. of conidia/colony (%)a |
|---|---|---|---|
| ///-10 | Wild type | 32.5 ± 0.5 | (1.22 ± 0.25) × 108 (100) |
| CΔsC/PG/A-1 | ΔchsC | 33.4 ± 2.3 | (1.27 ± 2.85) × 108 (104) |
| ΔAC/A-4 | ΔchsA ΔchsC | 32.8 ± 1.4 | (<0.01) |
| ΔAC/pHBS-4 | ΔchsA ΔchsC chsA(p)::chsA | 35.1 ± 1.5 | (8.13 ± 0.96) × 107 (67) |
| ΔAC/pHBSBXHA-7 | ΔchsA ΔchsC chsA(p)::HA-chsA | 35.1 ± 0.8 | (6.97 ± 0.35) × 107 (57) |
| AΔPG/sC/A-4 | ΔchsA | 31.0 ± 1.3 | (3.49 ± 0.39) × 107 (29) |
| AΔPG/sC/pHBSBXHA-2 | ΔchsA chsA(p)::HA-chsA | 31.4 ± 1.1 | (1.37 ± 0.13) × 108 (122) |
| ΔAC/A/FLAGC-5 | ΔchsA ΔchsC chsC(p)::FLAG-chsC | 31.0 ± 1.5 | (2.90 ± 0.35) × 107 (24) |
| ΔAC/A/ChsC-8 | ΔchsA ΔchsC chsC(p)::chsC | 30.7 ± 0.6 | (3.50 ± 0.79) × 107 (29) |
The ratio of conidiation efficiency of each strain to that of the wild-type strain is shown in parentheses. Conidiation efficiency of ΔAC/A-4 was below the measurable level.
FIG. 4.
Western analysis of the ΔAC mutant containing chsA(p)::HA-chsA. Cell extract from ΔAC/pHBSBXHA-7 [a ΔAC mutant containing chsA(p)::HA-chsA] was fractionated by centrifugation and subjected to Western analysis with anti-HA antibody (α-HA). P10, P100, and S100 indicate the 10,000 × g pellet fraction, 100,000 × g pellet fraction, and 100,000 × g supernatant fraction, respectively. Arrows indicate two major bands of 123 kDa and 146 kDa. In order to show the amount of proteins loaded, 20% of the volume of extract used for Western analysis was subjected to SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue (CBB).
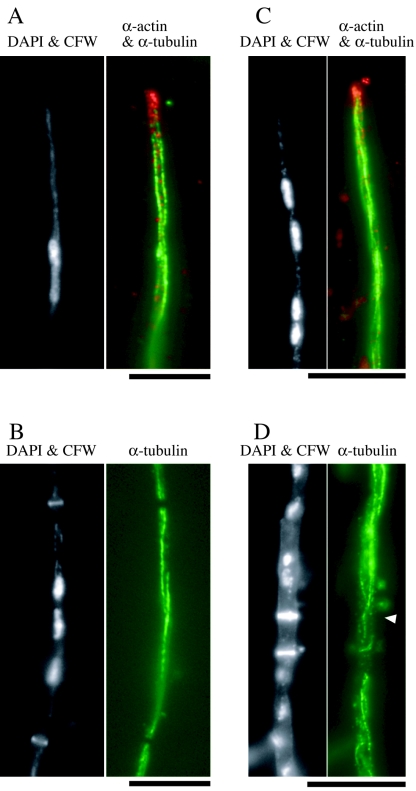
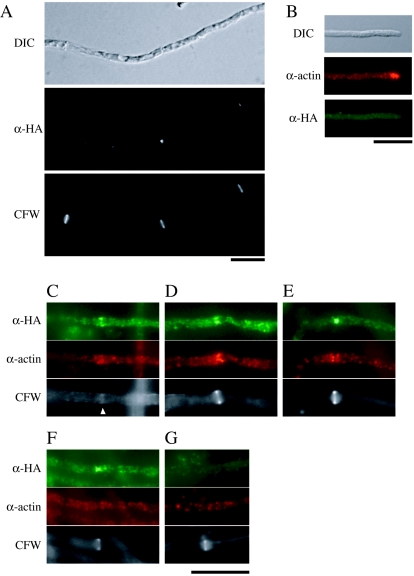
Localization of HA-ChsA in hyphae was observed in the ΔAC mutants expressing HA-ChsA by indirect immunofluorescence (Fig. 5A). Many fluorescent dots were observed in the cytoplasm. Since the amino acid sequence of ChsA has several putative transmembrane domains and since HA-ChsA was not present in the soluble fraction in the Western analysis, these dots would be localized on membranous structures. Fluorescence was intense at a subset of septa (Fig. 5A). However, the majority of septa did not have intense fluorescence. HA-ChsA fluorescence was not concentrated at the hyphal tips (see below and Fig. 5B).
FIG. 5.
Localization of HA-ChsA. (A) ΔAC/pHBSBXHA-7 [ΔAC mutant containing chsA(p)::HA-chsA] was grown for 12 h and subjected to indirect immunofluorescence microscopy using anti-HA antibody (α-HA). Cells were also stained with CFW. A DIC image is also shown. (B to G) AΔPG/sC/pHBSBXHA-2 [ΔA mutant containing chsA(p)::HA-chsA] was double stained with anti-actin antibody (α-actin) and anti-HA antibody (α-HA). Corresponding DIC or CFW-stained images are shown. The arrowhead in panel C indicates a faintly CFW-stained chitin ring. Bars, 10 μm.
To exclude the possibility that the lack of ChsC affects the expression of HA-chsA or the localization of HA-ChsA, we constructed ΔA mutants expressing HA-chsA under the chsA promoter. Whereas the conidial production by the ΔA mutant was approximately 30% of that by the wild-type strain under this condition, the ΔA mutant expressing HA-chsA produced conidia at the wild-type level (Table 2, compare AΔPG/sC/A-4 with AΔPG/sC/pHBSBXHA-2), again indicating that HA-ChsA is functional. The HA-ChsA protein level, fractionation pattern, and mobility of bands in Western analysis did not differ from those in the absence of ChsC (data not shown). The localization pattern was also indistinguishable from that of the ΔAC mutant expressing HA-ChsA (Fig. 5B to G). These results indicate that the chsC deletion affects neither the expression of HA-ChsA nor the localization of HA-ChsA.
To examine the timing of HA-ChsA localization during septum formation, we compared the localization of HA-ChsA with that of actin, which is known to localize at forming septa (27) (Fig. 5C to G). In the early stages of septation, when obvious CFW stains were not detectable, actin rings were observed (data not shown). At the CFW-stained septa, actin signals were observed as a band (Fig. 5C), as an hourglass shape (Fig. 5D), or as signals on both sides of the septum (Fig. 5E), and septum formation was presumed to occur in that order. When we observed septa with actin signals and CFW stains (n = 47), 46 of the septa were associated with intense HA-ChsA signals. HA-ChsA signals were observed at the periphery of hyphae (Fig. 5C and D) or at the center of septa (Fig. 5E). When we observed septa with HA-ChsA signals and CFW stains (n = 35), actin signals were not intense at 17 of the septa; judging from differential interference contrast (DIC) images (data not shown), CFW stains, and HA-ChsA patterns, those septa seemed to be in the later stages of septation (Fig. 5F). At presumptive septation sites without CFW stains, either the HA-ChsA signal, the actin signal, or both signals were observed (data not shown). The majority of septation sites did not have intense signals of either actin or HA-ChsA (Fig. 5G). These results suggest that ChsA localizes on forming septa and may remain for a short period after the actin disappears.
A functional FLAG-tagged ChsC localizes to septation sites and hyphal tips.
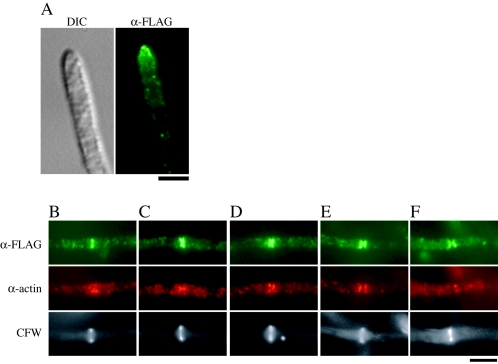
We next constructed strains that express FLAG epitope-tagged ChsC (FLAG-ChsC) under the chsC promoter. Six copies of the FLAG epitope were inserted between serine 56 and proline 57 of ChsC and expressed at the genomic pyroA locus of the ΔAC mutant (ΔACa4) or of the ΔC mutant (CΔsC/PG/A-1). Integration of a single copy of chsC(p)::FLAG-chsC was confirmed by Southern analysis (see Materials and Methods) and Northern analysis (data not shown). Conidial production by the FLAG-ChsC-expressing ΔAC mutant (ΔAC/A/FLAGC-5) was at a level similar to that by the ΔAC mutant expressing wild-type ChsC (ΔAC/A/ChsC-8) or the ΔA mutant (AΔPG/sC/A-4) (Table 2). The conidiophore morphology and colony appearance of the FLAG-ChsC-expressing ΔAC mutant were very similar to those of the ΔA mutant (data not shown). Therefore, we concluded that the functions of ChsC were not disturbed by the insertion of FLAG epitopes into the N-terminal region.
The predicted molecular mass of FLAG-ChsC is approximately 120 kDa. When the intracellular protein level of FLAG-ChsC expressed in the ΔC mutant was determined by Western analysis using anti-FLAG antibody, a rather broad band of approximately 120 kDa was detected in the P10 and P100 fractions but not in the S100 fraction (Fig. 6). In addition, a faint band with lower mobility (approximately 144 kDa) was reproducibly observed accompanying the 120 kDa band. These bands were not detected in the samples prepared from strains that do not express FLAG-ChsC (data not shown).
FIG. 6.
Western analysis of the ΔC mutant containing chsC(p)::FLAG-chsC. Cell extract from CΔsC/PG/A/FLAGC-1 [ΔchsC mutant expressing chsC(p)::FLAG-chsC] was fractionated by centrifugation and subjected to Western analysis using anti-FLAG antibody (α-FLAG). Arrows indicate a broad band of 120 kDa (lower arrow) and 144 kDa (upper arrow). The same volume of extracts used for Western analysis was subjected to SDS-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue (CBB).
To investigate the localization of FLAG-ChsC expressed in the ΔC mutant, we performed indirect immunofluorescence microscopy (Fig. 7). In addition to dots in the cytoplasm, intense fluorescence was observed at the hyphal tips (Fig. 7A) and at some septa (Fig. 7B to F). However, most septa were not accompanied by intense FLAG-ChsC fluorescence (data not shown). To examine the timing of FLAG-ChsC localization to the septum, we performed double staining with anti-FLAG and anti-actin antibodies. Figure 7B to F shows septation sites with various patterns of actin localization. When we observed septa with actin signals and CFW stains (n = 53), 51 of the septa were intensely stained with FLAG-ChsC signals. Signals of FLAG-ChsC were observed as a band (Fig. 7B and C), as an hourglass shape (Fig. 7D and E), or as two plates on the both sides of a septum (Fig. 7F). When we observed septa in the late stages with FLAG-ChsC signals in the appearance of two plates (n = 20), actin signals were not intense at nine of the septa (Fig. 7F). At presumptive septation sites without CFW stains, either the FLAG-ChsC signal, the actin signal, or both signals were observed (data not shown). Thus, it is possible that ChsC localized to forming septa and may remain after actin disappearance.
FIG. 7.
Localization of FLAG-ChsC. CΔsC/PG/A/FLAGC-1 was subjected to indirect immunofluorescence using anti-FLAG antibody (α-FLAG) and anti-actin antibody (α-actin). Signals of FLAG-ChsC were observed at the hyphal tips (A) and a subset of the septation sites (B to F). (B to E) Septa with obvious actin and FLAG-ChsC localization. (F) A septum with only FLAG-ChsC localization. Bar, 4 μm.
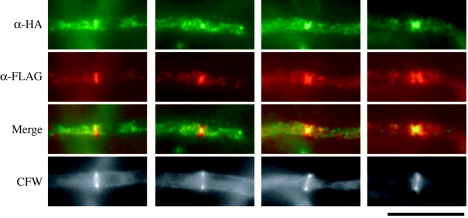
ChsA and ChsC occupy overlapping regions on forming septa.
The results of the double staining with actin suggested that both HA-ChsA and FLAG-ChsC localize to forming septa, although they exhibited distinct fluorescence patterns. We constructed ΔAC mutants expressing both HA-ChsA and FLAG-ChsC (see Materials and Methods) and examined their localization (Fig. 8). At septation sites, intense fluorescent signals of HA-ChsA and FLAG-ChsC partially overlapped. The HA-ChsA signals were restricted to a narrow area of the septation sites, while FLAG-ChsC occupied a broader area. We also found septa with either one of the signals (7% of septa had HA-ChsA signals alone [n = 67], and 35% of septa had FLAG-ChsC signals alone [n = 51]), which included those in the early and late stages of septation (data not shown). In areas other than the septation sites, fluorescent dots of HA-ChsA and FLAG-ChsC scarcely overlapped. These results suggest that ChsA and ChsC occupy overlapping regions on forming septa and might use distinct localization mechanisms.
FIG. 8.
Localization of HA-ChsA and FLAG-ChsC. ΔAC/HAA/FLAGC-2 [ΔchsA ΔchsC mutant containing chsA(p)::HA-chsA and chsC(p)::FLAG-chsC] was subjected to indirect immunofluorescence using anti-HA antibody (α-HA) and anti-FLAG antibody (α-FLAG). Mergers of α-HA and α-FLAG images are shown to indicate colocalization of HA-ChsA and FLAG-ChsC (Merge). Bar, 10 μm.
DISCUSSION
It has been proposed that in filamentous fungi, hyphal septa contain chitin. In fact, chitin is a major component of the septa formed in the hyphae of Neurospora crassa (19). Septum formation in the budding yeast S. cerevisiae requires many proteins, including the class II chitin synthase Chs2, which is required for primary septum formation, and members of well-known protein families, such as actin, formin, and septin (5, 50). In A. nidulans, orthologues of actin, formin, and septin have been identified and were shown to be involved in hyphal septum formation (16, 27, 42, 52). However, there has been no evidence that the class II chitin synthase is involved in septum formation in A. nidulans or in any other filamentous fungi. In this study, we found septation defects in the ΔAC mutants by electron and fluorescence microscopy. Epitope-tagged ChsA and ChsC localized at the sites where septa were forming. These results suggest that ChsA and ChsC are directly involved in septum formation. Recently, our group reported that CsmA, a class V chitin synthase, localizes to hyphal tips and septa (46). The authors of that work suggested that regulation of chitin synthesis at septation sites may be disturbed in deletion mutants of csmA.
The aberrant septa of the ΔAC mutant were intensely stained with CFW, and this suggests a high chitin content in these septa. Therefore, chitin synthase(s) other than ChsA and ChsC would have been involved in the construction of such aberrant septa. The normal thin plate of the primary septum is not observed in the chs2 mutants of S. cerevisiae; instead, aberrantly thick septa are formed by lateral wall thickening, and chs3 has been suggested to be necessary for this process (41). Recently, we constructed ΔAC mutants in which chsD or csmA was deleted (56). These mutants formed septa that were stained with CFW, which suggested that chsD and csmA are dispensable in the formation of abnormal septa in ΔAC mutants. However, four chitin synthase genes remain in the A. nidulans genome, and some of these may be responsible for the process.
An elevated number of septa were formed in the ΔAC mutant, and these septa were not evenly spaced. We frequently observed that some of these were very close to adjacent septa, which is in contrast with the regular distribution in the wild-type strain and in the ΔA and ΔC single mutants. There are two possible mechanisms by which these septation defects can occur. First, the cell wall defect may disturb the function of an apparatus that defines the positioning of septa. Such an apparatus, a septation marker, has been proposed and is supposedly formed at the hyphal tips (22). Second, an uneven distribution of mitotic nuclei may affect septation. Wolkow et al. observed that in the nud mutants that are grown at semirestrictive temperatures, nuclei aggregate in clumps and that septation is induced at the positions of the nuclear aggregates (53). Although no such nuclear aggregates were observed in the ΔAC hyphae, many nuclei were unevenly distributed and had rather distorted shapes. In addition, some nuclei associated with a septum in a manner similar to the “cut” phenotype in certain Schizosaccharomyces pombe cell division mutants (57). This “cut”-like phenotype might be caused by the mistimed activation of septation.
Nuclei were not evenly distributed in the hyphae in the ΔAC mutant. In the A. nidulans wild-type strain, nuclei migrate toward the hyphal tip; subsequently, they are fixed at specific positions (45). We prefer the idea that the fixation process is disturbed, because nuclei did migrate through hyphae and because microtubules were present in the ΔAC mutant, though they were less polarized than in the wild-type strain. ApsA and ApsB are believed to be involved in the regulation of this fixation process (10, 44, 54). Since ApsA was shown to localize at the hyphal cortex (45), it is possible that certain machinery that regulates nuclear positioning and possibly interacts with ApsA and ApsB exists at a similar cortical location. If this is the case, its function and/or localization may be disturbed by the cell wall defects in the ΔAC mutant, which were observed in Fig. 1. Interestingly, a deletion mutant of the nudC gene, which has been identified as necessary for normal nuclear distribution (55), has been shown to have cell wall defects (6). This result, together with ours, supports the hypothesis that the fungal cell wall might provide rigid supports for nuclei to migrate to and stay at specific positions in the hyphae.
When HA-ChsA-expressing strains were subjected to Western analysis, we detected several bands with molecular masses higher than 121 kDa, the predicted molecular mass of HA-ChsA (Fig. 4). It is possible that ChsA is posttranslationally modified. To date, it has been shown that chitin synthases of S. cerevisiae receive posttranslational modifications. Chs3 is glycosylated (40, 48) and phosphorylated (49). Chs1, Chs2, and Chs3 are ubiquitination substrates at the plasma membrane (17). The smeared FLAG-ChsC low-mobility band might be generated by posttranslational modification(s), although it is not as prominent as that of ChsA. Alternatively, these higher-molecular-mass bands might be artifacts. Cos et al. reported that two bands (160 and 230 kDa) were detected in cells expressing HA-tagged Chs3, the predicted size of which was 131 kDa in S. cerevisiae, and they considered the 230-kDa band to be a running artifact (8). A smeared high-molecular-mass band is also observed in the analysis of class III chitin synthase in Wangiella dermatitidis (51).
HA-ChsA and FLAG-ChsC localized to forming septa and partially colocalized there. This suggests that they construct an overlapping fraction of chitin on septa, which would explain the synthetic defect in the septum formation of the ΔAC mutants. However, we could not observe their colocalization at a subset of septa. Since the lack of ChsA or ChsC did not change the localization of the other, and since their colocalization was not observed in the cytoplasmic space, it is possible that their localization and disappearance at septation sites may be independent of each other. Such independence might mean that their localization and disappearance occur at different timing. Colocalization was not observed at the hyphal tips, either; localization of ChsC at the hyphal tips is consistent with its involvement in vegetative growth, which was suggested from the analysis of chsB chsC double mutants (20). The transport and retention mechanisms of chitin synthases to different hyphal regions in filamentous fungi remain to be clarified. Identification of proteins interacting with ChsA or ChsC would help in understanding their transport and retention mechanisms.
Although we identified septation defects in the ΔAC mutant in this study, this mutant also exhibited other defects (13). It is possible that the overlap of functions between ChsA and ChsC is not restricted to septum formation; it may extend to the maintenance of cell wall integrity and to conidiation. Alternatively, the primary defect in the ΔAC mutants might occur in septation during hyphal growth, and other phenotypes might be caused indirectly. Although several temperature-sensitive mutants that failed to form septa (sep mutants) have been isolated and characterized in A. nidulans (4, 14-16, 28, 42), whether they have cell wall defects like the ΔAC mutants has not been examined. Further investigations are required to elucidate how ChsA and ChsC are involved in the maintenance of cell wall integrity and in conidiation.
Acknowledgments
This work was supported by a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and partly by a grant from Bio-oriented Technology Research Advancement Institution.
This work was performed using the facilities of the Biotechnology Research Center of the University of Tokyo.
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aufauvre-Brown, A., E. Mellado, N. A. Gow, and D. W. Holden. 1997. Aspergillus fumigatus chsE: a gene related to CHS3 of Saccharomyces cerevisiae and important for hyphal growth and conidiophore development but not pathogenicity. Fungal Genet. Biol. 21:141-152. [DOI] [PubMed] [Google Scholar]
- 3.Borgia, P. T., N. Iartchouk, P. J. Riggle, K. R. Winter, Y. Koltin, and C. E. Bulawa. 1996. The chsB gene of Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet. Biol. 20:193-203. [DOI] [PubMed] [Google Scholar]
- 4.Bruno, K. S., J. L. Morrell, J. E. Hamer, and C. J. Staiger. 2001. SEPH, a Cdc7p orthologue from Aspergillus nidulans, functions upstream of actin ring formation during cytokinesis. Mol. Microbiol. 42:3-12. [DOI] [PubMed] [Google Scholar]
- 5.Cabib, E., D. H. Roh, M. Schmidt, L. B. Crotti, and A. Varma. 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276:19679-19682. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, Y. H., X. Xiang, A. L. Dawe, and N. R. Morris. 1997. Deletion of nudC, a nuclear migration gene of Aspergillus nidulans, causes morphological and cell wall abnormalities and is lethal. Mol. Biol. Cell 8:1735-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang, J. S., and R. W. Schekman. 1996. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 135:597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cos, T., R. A. Ford, J. A. Trilla, A. Duran, E. Cabib, and C. Roncero. 1998. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur. J. Biochem. 256:419-426. [DOI] [PubMed] [Google Scholar]
- 9.Culp, D. W., C. L. Dodge, Y. Miao, L. Li, D. Sag-Ozkal, and P. T. Borgia. 2000. The chsA gene from Aspergillus nidulans is necessary for maximal conidiation. FEMS Microbiol. Lett. 182:349-353. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, R. 1999. Nuclear movement in filamentous fungi. FEMS Microbiol. Rev. 23:39-68. [DOI] [PubMed] [Google Scholar]
- 11.Ford, R. A., J. A. Shaw, and E. Cabib. 1996. Yeast chitin synthases 1 and 2 consist of a non-homologous and dispensable N-terminal region and of a homologous moiety essential for function. Mol. Gen. Genet. 252:420-428. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara, M., H. Horiuchi, A. Ohta, and M. Takagi. 1997. A novel fungal gene encoding chitin synthase with a myosin motor-like domain. Biochem. Biophys. Res. Commun. 236:75-78. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara, M., M. Ichinomiya, T. Motoyama, H. Horiuchi, A. Ohta, and M. Takagi. 2000. Evidence that the Aspergillus nidulans class I and class II chitin synthase genes, chsC and chsA, share critical roles in hyphal wall integrity and conidiophore development. J. Biochem. (Tokyo) 127:359-366. [DOI] [PubMed] [Google Scholar]
- 14.Harris, S. D., and J. E. Hamer. 1995. sepB: an Aspergillus nidulans gene involved in chromosome segregation and the initiation of cytokinesis. EMBO J. 14:5244-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, S. D., L. Hamer, K. E. Sharpless, and J. E. Hamer. 1997. The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 16:3474-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, S. D., J. L. Morrell, and J. E. Hamer. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitchcock, A. L., K. Auld, S. P. Gygi, and P. A. Silver. 2003. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. USA 100:12735-12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horiuchi, H., M. Fujiwara, S. Yamashita, A. Ohta, and M. Takagi. 1999. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 181:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunsley, D., and G. W. Gooday. 1974. The structure and development of septa in Neurospora crassa. Protoplasma 82:125-146. [DOI] [PubMed] [Google Scholar]
- 20.Ichinomiya, M., H. Horiuchi, and A. Ohta. 2002. Different functions of the class I and class II chitin synthase genes, chsC and chsA, are revealed by the repression of the chsB expression of Aspergillus nidulans. Curr. Genet. 41:51-58. [DOI] [PubMed] [Google Scholar]
- 21.Ichinomiya, M., T. Motoyama, M. Fujiwara, M. Takagi, H. Horiuchi, and A. Ohta. 2002. Repression of chsB expression reveals the functional importance of class IV chitin synthase gene chsD in hyphal growth and conidiation of Aspergillus nidulans. Microbiology 148:1335-1347. [DOI] [PubMed] [Google Scholar]
- 22.Kaminskyj, S. G. 2000. Septum position is marked at the tip of Aspergillus nidulans hyphae. Fungal Genet. Biol. 31:105-113. [DOI] [PubMed] [Google Scholar]
- 23.Madrid, M. P., A. Di Pietro, and M. I. Roncero. 2003. Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47:257-266. [DOI] [PubMed] [Google Scholar]
- 24.May, G. S., C. A. McGoldrick, C. L. Holt, and S. H. Denison. 1992. The bimB3 mutation of Aspergillus nidulans uncouples DNA replication from the completion of mitosis. J. Biol. Chem. 267:15737-15743. [PubMed] [Google Scholar]
- 25.Mellado, E., A. Aufauvre-Brown, N. A. Gow, and D. W. Holden. 1996. The Aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Mol. Microbiol. 20:667-679. [DOI] [PubMed] [Google Scholar]
- 26.Momany, M. 2001. Cell biology of the duplication cycle in fungi, p. 119-125. In N. Talbot (ed.), Molecular and cellular biology of filamentous fungi. Oxford University Press, Oxford, United Kingdom.
- 27.Momany, M., and J. E. Hamer. 1997. Relationship of actin, microtubules, and crosswall synthesis during septation in Aspergillus nidulans. Cell Motil. Cytoskel. 38:373-384. [DOI] [PubMed] [Google Scholar]
- 28.Morris, N. R. 1976. A temperature-sensitive mutant of Aspergillus nidulans reversibly blocked in nuclear division. Exp. Cell Res. 98:204-210. [DOI] [PubMed] [Google Scholar]
- 29.Motoyama, T., M. Fujiwara, N. Kojima, H. Horiuchi, A. Ohta, and M. Takagi. 1996. The Aspergillus nidulans genes chsA and chsD encode chitin synthases which have redundant functions in conidia formation. Mol. Gen. Genet. 251:442-450 (Corrected and republished 253: 520-528, 1997.) [DOI] [PubMed] [Google Scholar]
- 30.Motoyama, T., N. Kojima, H. Horiuchi, A. Ohta, and M. Takagi. 1994. Isolation of a chitin synthase gene (chsC) of Aspergillus nidulans. Biosci. Biotechnol. Biochem. 58:2254-2257. [DOI] [PubMed] [Google Scholar]
- 31.Muller, C., C. M. Hjort, K. Hansen, and J. Nielsen. 2002. Altering the expression of two chitin synthase genes differentially affects the growth and morphology of Aspergillus oryzae. Microbiology 148:4025-4033. [DOI] [PubMed] [Google Scholar]
- 32.Munro, C. A., and N. A. Gow. 2001. Chitin synthesis in human pathogenic fungi. Med. Mycol. 39:41-53. [PubMed] [Google Scholar]
- 33.Oakley, C. E., C. F. Weil, P. L. Kretz, and B. R. Oakley. 1987. Cloning of the riboB locus of Aspergillus nidulans. Gene 53:293-298. [DOI] [PubMed] [Google Scholar]
- 34.Orlean, P. 1997. Biogenesis of yeast wall and surface components. .In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces cerevisiae: cell cycle and cell biology, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 35.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 36.Roh, D. H., B. Bowers, M. Schmidt, and E. Cabib. 2002. The septation apparatus, an autonomous system in budding yeast. Mol. Biol. Cell 13:2747-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roncero, C. 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41:367-378. [DOI] [PubMed] [Google Scholar]
- 38.Rowlands, R. T., and G. Turner. 1973. Nuclear and extranuclear inheritance of oligomycin resistance in Aspergillus nidulans. Mol. Gen. Genet. 126:201-216. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Santos, B., and M. Snyder. 1997. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 136:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt, M., B. Bowers, A. Varma, D. H. Roh, and E. Cabib. 2002. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115:293-302. [DOI] [PubMed] [Google Scholar]
- 42.Sharpless, K. E., and S. D. Harris. 2002. Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13:469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Specht, C. A., Y. Liu, P. W. Robbins, C. E. Bulawa, N. Iartchouk, K. R. Winter, P. J. Riggle, J. C. Rhodes, C. L. Dodge, D. W. Culp, and P. T. Borgia. 1996. The chsD and chsE genes of Aspergillus nidulans and their roles in chitin synthesis. Fungal Genet. Biol. 20:153-167. [DOI] [PubMed] [Google Scholar]
- 44.Suelmann, R., and R. Fischer. 2000. Nuclear migration in fungi—different motors at work. Res. Microbiol. 151:247-254. [DOI] [PubMed] [Google Scholar]
- 45.Suelmann, R., N. Sievers, and R. Fischer. 1997. Nuclear traffic in fungal hyphae: in vivo study of nuclear migration and positioning in Aspergillus nidulans. Mol. Microbiol. 25:757-769. [DOI] [PubMed] [Google Scholar]
- 46.Takeshita, N., A. Ohta, and H. Horiuchi. 2005. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol. Biol. Cell 16:1961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeshita, N., A. Ohta, and H. Horiuchi. 2002. csmA, a gene encoding a class V chitin synthase with a myosin motor-like domain of Aspergillus nidulans, is translated as a single polypeptide and regulated in response to osmotic conditions. Biochem. Biophys. Res. Commun. 298:103-109. [DOI] [PubMed] [Google Scholar]
- 48.Trilla, J. A., A. Duran, and C. Roncero. 1999. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 145:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valdivia, R. H., and R. Schekman. 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 100:10287-10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walther, A., and J. Wendland. 2003. Septation and cytokinesis in fungi. Fungal Genet. Biol. 40:187-196. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Z., and P. J. Szaniszlo. 2000. WdCHS3, a gene that encodes a class III chitin synthase in Wangiella (Exophiala) dermatitidis, is expressed differentially under stress conditions. J. Bacteriol. 182:874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westfall, P. J., and M. Momany. 2002. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolkow, T. D., S. D. Harris, and J. E. Hamer. 1996. Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J. Cell Sci. 109:2179-2188. [DOI] [PubMed] [Google Scholar]
- 54.Xiang, X., and R. Fischer. 2004. Nuclear migration and positioning in filamentous fungi. Fungal Genet. Biol. 41:411-419. [DOI] [PubMed] [Google Scholar]
- 55.Xiang, X., and N. R. Morris. 1999. Hyphal tip growth and nuclear migration. Curr. Opin. Microbiol. 2:636-640. [DOI] [PubMed] [Google Scholar]
- 56.Yamada, E., M. Ichinomiya, A. Ohta, and H. Horiuchi. 2005. The class V chitin synthase gene csmA is crucial for the growth of the chsA chsC double mutant in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 69:87-97. [DOI] [PubMed] [Google Scholar]
- 57.Yanagida, M. 1998. Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol. 8:144-149. [DOI] [PubMed] [Google Scholar]
- 58.Yanai, K., N. Kojima, N. Takaya, H. Horiuchi, A. Ohta, and M. Takagi. 1994. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans. Biosci. Biotechnol. Biochem. 58:1828-1835. [DOI] [PubMed] [Google Scholar]
- 59.Yarden, O., and C. Yanofsky. 1991. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa. Genes Dev. 5:2420-2430. [DOI] [PubMed] [Google Scholar]
- 60.Ziman, M., J. S. Chuang, and R. W. Schekman. 1996. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell 7:1909-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]