Abstract
Background
Facioscapulohumeral dystrophy (FSHD) is a myopathy characterized by the loss of repressive epigenetic features affecting the D4Z4 locus (4q35). The assessment of DNA methylation at two regions (DUX4-PAS and DR1) of D4Z4 locus proved to be an effective method to detect epigenetic signatures compatible with FSHD. The present study aims at validating the employment of this method into clinical practice and improving the protocol by refining the classification thresholds of 4qA/4qA patients. To this purpose, 218 subjects with clinical suspicion of FSHD collected in 2022–2023 were analyzed. Each participant underwent in parallel the traditional FSHD molecular testing (D4Z4 sizing) and the proposed methylation assay. The results provided by both analyses were compared to evaluate the concordance and calculate the performance metrics of the methylation test.
Results
Among the 218 subjects, the 4q variant type distribution was 54% 4qA/4qA, 43% 4qA/4qB and 3% 4qB/4qB. The methylation analysis was performed only on carriers of at least one 4qA allele. After refining the classification threshold, the test reached the following performance metrics: sensitivity = 0.90, specificity = 1.00 and accuracy = 0.93. These results confirmed the effectiveness of the methylation assay in identifying patients with genetic signature compatible with FSHD1 and FSHD2 based on their DUX4-PAS and DR1 profile, respectively. The methylation data were also evaluated with respect to the clinical information.
Conclusions
The study confirmed the ability of the method to accurately identify methylation profiles compatible with FSHD genetic signatures considering the 4q genotype. Moreover, the test allows the detection of hypomethylated profiles in asymptomatic patients, suggesting its potential application in identifying preclinical conditions in patients with positive family history and FSHD genetic signatures. Furthermore, the present work emphasizes the importance of interpreting methylation profiles considering the patients’ clinical data.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-024-01747-2.
Keywords: Neuromuscular diseases, FSHD, FSHD signature, FSHD diagnosis, Epigenetic biomarker, DNA methylation, D4Z4
Introduction
Facioscapulohumeral dystrophy (FSHD) is an autosomal dominant muscle disorder caused by genetic and epigenetic alterations at the D4Z4 locus (chromosome 4q35). This macrosatellite region comprises a variable number of Repeated Units (RU), with the last unit containing a complete copy of the DUX4 gene. The D4Z4 locus contains 11-150RU and is transcriptionally repressed in healthy subjects [1, 2].
Two pathogenic alterations have been proposed as the main triggering events leading to disease manifestation. The first one is the reduction of the D4Z4 array to 1-10RU, which is responsible for FSHD1 [3]. The second genetic alteration is usually associated with the FSHD2 form, and it is related to the presence of genetic variants within epigenetic regulators of the locus, namely SMCHD1 (18p11.32), LRIF1 (1p13.3) and DNMT3B (20q11.21) genes [4–9]. Some FSHD2 patients can also show the epigenetic alteration related to the disease (i.e., a global reduction of D4Z4 methylation levels) without being able to detect a pathogenic variant in the known FSHD2 genes. Generally, FSHD2 patients show a borderline to short normal-sized D4Z4 array (8-20RU) [1]. Notably, the critical condition for the establishment of the disease in both FSHD1 and FSHD2 forms is the presence of the 4qA permissive allele containing the Polyadenylation Signal (PAS) for DUX4 mRNA stabilization [10]. Altogether, these molecular alterations lead to the expression of DUX4 gene, which is toxic for skeletal muscle cells [11, 12].
However, the diagnosis of disease is challenged by incomplete penetrance and the clinical heterogeneity, especially in carriers of borderline D4Z4 alleles (in the range of 8–10 RU). A study performed on 208 Dutch healthy subjects reported the presence of D4Z4 alleles of 8-9RU (independently of the haplotype) in the 3% of general population [13]. Importantly, a subsequent study conducted on 801 healthy individuals (560 from Italy and 241 from Brazil) confirmed such result, pointing out that only 1–2% of general population carried a contracted D4Z4 allele with a permissive 4qA haplotype [14].
The gold standard for molecular diagnosis of FSHD consists of pulsed-field gel electrophoresis (PFGE) and Southern Blotting followed by P13-E11 probe hybridization, which enable the assessment of D4Z4 array size [15]. In presence of borderline alleles of 8-20RU, the FSHD2 genes sequencing is performed mainly by direct sequencing or next generation sequencing (NGS) techniques to assess the presence of detrimental variants within SMCHD1, LRIF1 and DNMT3B genes [1].
However, the D4Z4 sizing analyses are labor-intensive and time-consuming, delaying the response and, consequently, the diagnosis of FSHD. Furthermore, the complexity of this diagnostic approach makes it inaccessible to most developing countries and expensive for healthcare systems, even in industrialized countries. Thus, the availability of rapid, cost-effective methods that can provide timely support to FSHD diagnosis is highly desirable.
In a previous study, we developed a method for rapidly identifying subjects with genetic signatures compatible with FSHD through the assessment of DNA methylation levels related to the D4Z4 locus, which is a well-known hallmark of disease [16–20]. In particular, the study relied on the investigation of two regions within the D4Z4 locus, namely DUX4-PAS (10 CpGs, providing methylation status related to the most distal part of the D4Z4 array) and DR1 (29 CpGs, indicative of to the global methylation of the D4Z4 array), which are informative for FSHD1 and FSHD2, respectively. Specifically, a Machine Learning (ML) pipeline facilitated the identification of the most informative CpG sites (DUX4-PAS_CpG6, DUX4-PAS_CpG3, DR1_CpG1, DR1_CpG22), whose methylation levels are able to detect FSHD subjects [21].
Given these premises, the present study aimed at refining the protocol by using specific thresholds for 4qA/4qA and 4qA/4qB genotypes. This approach can improve the sensitivity in case of patients with D4Z4 reduced alleles (DRA) and 4qA/4qA genotype, where the detection of DUX4-PAS methylation levels may be inaccurate due to the presence of a normal-sized D4Z4 allele together with a reduced one. To this purpose, 218 subjects enrolled from 2022 to 2023 were involved in a prospective study. Each participant underwent in parallel the traditional FSHD molecular testing (D4Z4 sizing) and the proposed methylation assay.
Materials and methods
Study cohort and molecular analysis
This prospective study was conducted on a cohort composed of 218 subjects who accessed the laboratory of genomic medicine of Italian Union for the fight against muscular dystrophies (UILDM) at Santa Lucia Foundation IRCCS between 2022 and 2023. As an Italian reference center for the diagnosis of FSHD, our study cohort included patients with the clinical suspicion of FSHD who required genetic confirmation, relatives of FSHD patients undergoing segregation analysis and patients evaluated to exclude the presence of FSHD genetic signature.
The patients were recruited by expert neurologists in collaboration with the UILDM in the following medical centers: IRCCS Santa Lucia Foundation (Rome), IRCCS “Agostino Gemelli” University Hospital (Rome), IRCCS Neuromed Institute (Pozzilli), NeMO Clinical Center for Neuromuscular Diseases (Brescia), Clinical Center for Neuromuscular Disorders University Hospital of Pisa, NeMO Clinical Center for Neuromuscular Diseases (Milano), Neurorehabilitation Unit, SBMC Department, Rome Sapienza University Polo Pontino/ICOT (Latina), “Sant’Andrea” Hospital (Rome), “Sandro Pertini” Hospital (Rome), Multiple Sclerosis Center Binaghi Hospital (Cagliari), IRCCS “San Martino,” University Hospital (Genoa), “Santa Maria” Hospital (Terni), Molinette Hospital (Turin), “San Filippo Neri” Hospital (Rome), A.O.R.N. Cardarelli Hospital (Naples), “San Camillo Forlanini” Hospital (Rome), IRCCS “San Raffaele” Hospital (Milan), “Gaetano Martino” University Hospital (Messina), University Hospital of Campania “Luigi Vanvitelli” (Naples).
All participants provided signed informed consent for research and publication at the time of recruitment. The study was approved by the Ethics Committee of Santa Lucia Foundation (CE/2022_020 approved on June 1, 2022) and complied with the Declaration of Helsinki.
Each participant underwent methylation analysis, D4Z4 sizing and sequencing of FSHD2-associated genes (SMCHD1, LRIF1, DNMT3B). Methylation analysis was performed in blind, without prior information on 4q genotype, D4Z4 sizing or FSHD2 gene results. Subsequently, the data obtained from methylation analysis were compared with those of the traditional molecular tests to assess the concordance between FSHD predictions based on methylation levels and the presence of genetic signatures (reduced D4Z4 allele or pathogenic variants in FSHD2 genes).
All molecular analyses were performed on a fresh blood sample (35 ml) obtained from each patient. The DNA was, in parallel, extracted from peripheral blood mononuclear cells by manual and automatized techniques according to the manufacturer’s instructions. In particular, manual extraction was employed to obtain the high-molecular weight DNA embedded into agarose plugs required for D4Z4 sizing, whereas the DNA extracted by automatized method (MagPurix Blood DNA Extraction Kit and MagPurix Automatic Extraction System, Zinexts, Taiwan, R.O.C.) was utilized for the sequencing of the FSHD2 genes and for the methylation analysis.
The D4Z4 sizing was performed by means of PFGE and Southern blotting followed by hybridization with P13-E11 probe as previously described [15]. The presence of FSHD2-associated variants was assessed by whole exome sequencing (WES) on an Illumina Next-Seq550 system and related kit.
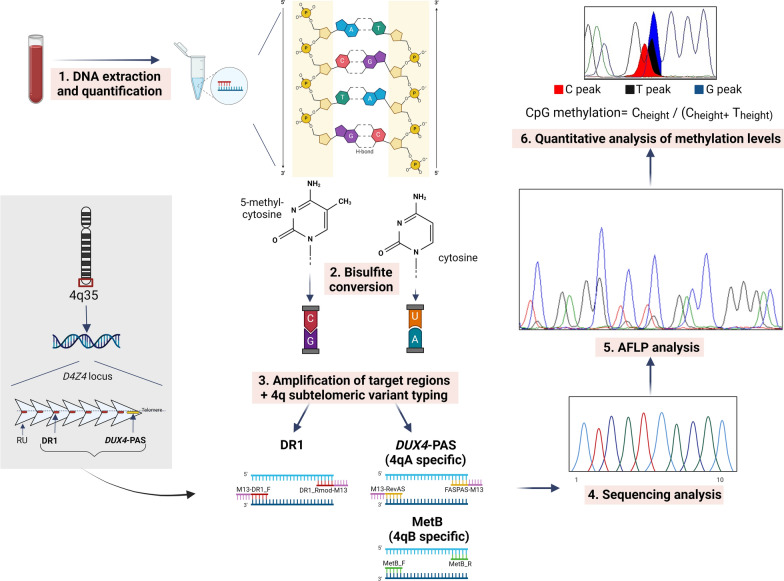
The assessment of methylation levels was performed as described in Caputo et al. [21, 22]. Importantly, the workflow allowed typing the 4q subtelomeric variant using 4qA- and 4qB-allele specific amplification by means of targeted primers reported by Caputo et al. [21]. Only patients with at least one 4qA allele were analyzed by the proposed methylation assay, since carriers of the permissive allele. The DUX4-PAS and DR1 regions were amplified by traditional PCR with specific primers and analyzed by the amplification fragments length polymorphisms (AFLP) protocol previously described [21]. In particular, this step was essential for the quantitative evaluation of methylation levels of all CpG sites contained in the two regions of interest. As reported in Caputo et al. 2022, the methylation levels of four CpG sites (DUX4-PAS_CpG6, DUX4-PAS_CpG3, DR1_CpG1, DR1_CpG22) were evaluated to differentiate between subjects positive for a genetic signature compatible with FSHD and negative subjects. A reduction in methylation levels, compatible with FSHD, was observed considering specific thresholds and following the order of relevance (1) DUX4-PAS_CpG6 ≤ 0.78; (2) DUX4-PAS_CpG3 ≤ 0.34; (3) DR1_CpG1 ≤ 0.53; (4) DR1_CpG22 ≤ 0.99 [21]. The workflow for methylation analysis is illustrated in Fig. 1.
Fig. 1.
Methylation analysis workflow. The protocol employed for 4q subtelomeric variant typing and the assessment of methylation levels at DUX4-PAS and DR1 regions of D4Z4 locus (AFLP = amplification fragments length polymorphisms). Figure adapted from Megalizzi et al. [22] (Created with BioRender.com)
Validation of the classification tool
Methylation data and results from FSHD1/FSHD2 molecular analysis were compared to evaluate the concordance between the results provided by methylation analysis and the data obtained from the gold-standard FSHD testing.
The reliability of the tool was thereby evaluated by calculating the performance metrics. The FSHD classification was considered Concordant-Positive (CP) for patients with 1–10 D4Z4 RU (FSHD1), patients with 11-20RU and likely pathogenic/pathogenic variants in FSHD2 genes (FSHD2), and compound FSHD1 + FSHD2 forms. The non-FSHD classification was deemed to be concordant in the absence of FSHD1 or FSHD2 genetic signatures (namely, Concordant-Negative, CN). In case of methylation profiles not compatible with the presence or absence of FSHD genetic signature, the samples were considered Non-Concordant-Negative (NCN) and Non-Concordant-Positive (NCP), respectively (Table 1).
Table 1.
Criteria for the assignment of the concordance between the molecular results
| Methylation analysis | FSHD1/2 Genetic testing |
Resulting label |
|---|---|---|
| ↓ | + | Concordant-positive (CP) |
| ↓ | − | Non-concordant-positive (NCP) |
| ↑ | − | Concordant-negative (CN) |
| ↑ | + | Non-concordant-negative (NCN) |
The symbol ↓ stands for hypomethylation (with respect to the thresholds employed by the tool), whereas ↑ stands for hypermethylation (with respect to the thresholds employed by the tool). + refers to the positivity to the FSHD1 or FSHD2 test, whereas – is used to indicate negativity to the test
Therefore, the number of concordant (CP, CN) and non-concordant predictions (NCP, NCN) was plotted in a confusion matrix. Firstly, performance metrics were calculated without considering the 4q genotype of the subjects. Successively, metrics were calculated separately for subjects with 4qA/4qA and 4qA/4qB genotypes. This step was crucial as the presence of two 4qA alleles (4qA/4qA) in patients with genetic signature compatible with FSHD may result in overestimated methylation levels in the DUX4-PAS region due to the presence of one 4qA allele > 20RU and a reduced allele (see Additional file 1).
Results
The enrolled patients had an average age of 48.1 ± 18.5 years and a 58:42 M:F ratio.
Initially, the methylation analysis workflow determined the 4q genotype for each patient. As a result, 42.6% of the patients showed a 4qA/4qB genotype, 54.6% were 4qA/4qA and the remaining 2.8% displayed a non-permissive 4qB/4qB subtelomeric configuration. All subjects carrying a 4qB/4qB genotype (6 out of 218) displayed a non-pathogenic D4Z4 allele of > 20RU.
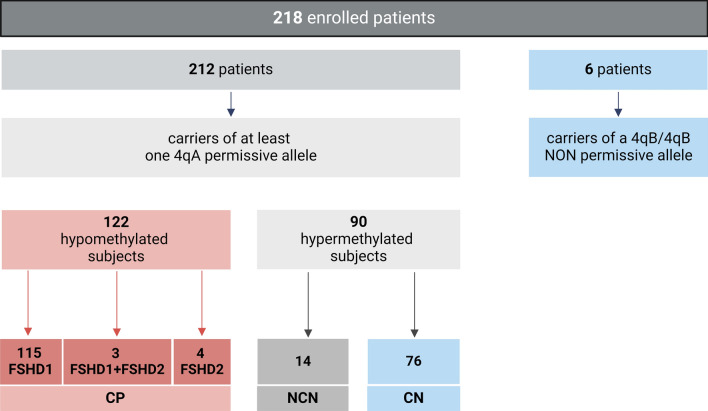
Only subjects carrying at least one 4qA allele (212 out of 218, 97.2%) underwent methylation levels assessment. As a result, 112 patients were classified as positive for a genetic signature of FSHD, consistently with the presence of reduced methylation levels (i.e., hypo-methylation status) compatible with the disease. The remaining 100 subjects were predicted as negative for a genetic signature of FSHD and showed high methylation levels (i.e., hyper-methylation status).
The D4Z4 allele sizing performed on the study cohort revealed that 133 out of 218 patients (61%) carried a 1-10RU permissive 4qA allele whereas the remaining 85 subjects (39%) did not show a D4Z4 reduction (RU > 10). Specifically, the group of subjects carrying ≤ 10RU included 15 patients with 1-3RU, 69 with 4-6RU and 49 with 7-10RU. In the group of subjects carrying > 10RU carriers, 12 out of 85 (14.1%) showed a 4qA allele of 11–20 RU, whereas the remaining 73 (85.9%) had a D4Z4 array > 20RU.
The WES analysis pointed out that seven out of 218 subjects (3%) were positive for the presence of likely pathogenic/pathogenic variants in SMCHD1 gene. In detail, four of them carried a D4Z4 allele in the range of 11-20RU, whereas the remaining three had a 4qA of 9-10RU.
Among the 212 patients carrying at least one 4qA, the methylation data were compared with the results of D4Z4 allele sizing and FSHD2 variants sequencing. For the 112 hypomethylated subjects (52.8%), the data were completely concordant with the presence of 1–10 D4Z4 RU (FSHD1), 11-20RU combined with detrimental variants in FSHD2 genes (FSHD2) and compound FSHD1 + FSHD2 forms. As expected, FSHD2 patients showed significantly lower methylation levels compared to FSHD1 patients, particularly in the DR1 region (p < 0.05) (see Additional file 2). These correctly classified patients were thereby considered as CP. 107 out of the 112 patients predicted as positive carried a contracted 4qA ≤ 10RU (including three patients with a 9-10RU allele combined with a SMCHD1 variant), whereas the remaining five patients had a 4qA allele of 11-20RU. Among them, four patients carried detrimental variants in SMCHD1 gene and one patient showed a variant in the EZH2 gene recently described as a candidate gene for the disease [23].
Additionally, out of 212 patients with at least one 4qA allele, 100 (47.2%) were predicted negative for a genetic signature of FSHD based on D4Z4 methylation levels. Specifically, 76 of the 100 subjects classified as negative were in agreement with the results of traditional FSHD testing (CN), except for 2 subjects carrying a borderline reduced 4qA allele of 8RU.
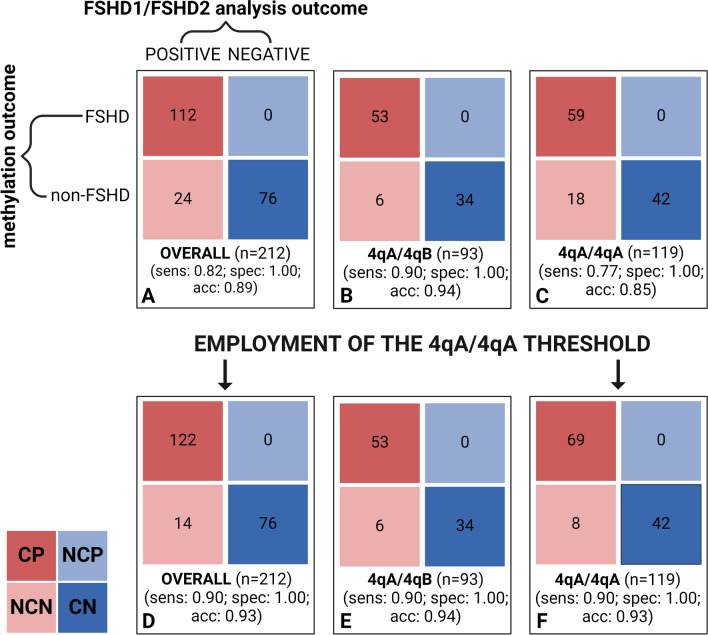
The remaining 24 subjects predicted as negative for a genetic signature of FSHD had slightly elevated methylation levels, although they carried a contracted 4qA allele < 10RU, thus, resulting as NCN. Overall, the model applied on the study cohort achieved the following performance metrics: sensitivity = 0.82, specificity = 1.00 and accuracy = 0.89.
The effectiveness of the tool was further assessed by splitting the study cohort based on the 4q genotype, which included 119 individuals with a 4qA/4qA and 93 with a 4qA/4qB genotype. For patients with a 4qA/4qB genotype, the classification model achieved excellent metrics, with a sensitivity = 0.90, specificity = 1.00 and accuracy = 0.94. In contrast, for patients with a 4qA/4qA genotype, the model showed lower performance metrics (sensitivity = 0.77, specificity = 1.00 and accuracy = 0.85). The lower performance metrics observed in patients with 4qA/4qA genotype highlighted the need for refining the methylation levels thresholds, considering the possible overestimation of DUX4-PAS methylation levels in FSHD1 patients with two 4qA alleles.
To this purpose, the methylation levels of NCN patients with a 4qA/4qA genotype were plotted to observe their distribution compared to 4qA/4qA CN. Specifically, the cohort was filtered to include only subjects with a 4qA/4qA genotype and a DUX4-PAS_CpG6 methylation level > 0.796. This specific filtering criterion was chosen to isolate patients with borderline methylation levels based on the evidence that DUX4-PAS_CpG6 is critical for accurately predicting FSHD, as supported by previous studies [16, 21]. After filtering, the resulting sub-group included 64 subjects (22 positive for FSHD genetic signatures and 42 negative for FSHD genetic signatures). The methylation levels of CpG6 and CpG3 of the DUX4-PAS were therefore used to refine the thresholds for FSHD1 classification, reflecting their relevance as determined by the ML model for subjects’ classification [21]. By observing methylation data distribution, the lowest methylation value at a DUX4-PAS-related CpG site in CN subjects was used to establish a threshold. This threshold was set to prevent overlapping methylation values between genetic positive and negative individuals carrying a 4qA/4qA genotype. The established thresholds were 0.8493 for the CpG6 and 0.2222 for the CpG3, respectively (Fig. 2), and were employed following the previously reported order of relevance.
Fig. 2.
Scatter Plot highlighting the distribution of methylation values of 4qA/4qA subjects for the refinement of DUX4-PAS threshold. Group 1 shows the CN subjects, whereas the Group 2 contains the NCN subjects. Methylation levels are grouped according to the status (y-axis) and visualized into different colors: blue for CN (Group 1) and red for NCN (Group 2). The data are displayed as a collection of shapes plotted with respect to methylation values on x-axis (A for CpG6; B for CpG3). Rounds, crosses, and triangles represent the RU number of the subjects whose methylation levels are plotted. The dashed lines indicate the threshold identified for each CpG site (yellow-highlighted), chosen according to methylation levels of the CN subject displaying the lowest value within each CpG (Created with OrangeDataMining.com)
The application of the new thresholds on the NCN subjects with a 4qA/4qA genotype (n = 18) allowed to correctly predict 10 patients (out of 18) as positive for a genetic signature of FSHD1. After refining methylation thresholds, the re-calculated patients’ classification metrics were improved (sensitivity = 0.90, specificity = 1.00 and accuracy = 0.93). These results were confirmed and validated in an independent set of patients (Additional File 3). The comprehensive performance metrics achieved by the tool on the studied cohort before and after threshold refining are summarized in Fig. 3.
Fig. 3.
Confusion matrix employed to calculate the performance metrics of the ML model. The confusion matrix shows the number of concordant positive (CP), non-concordant-positive (NCP), concordant negative (CN), and non-concordant-negative (NCN). The number of predictions considered for assessing the metrics is indicated at the bottom of each confusion matrix. The performance metrics were calculated with respect to the overall cohort (A, D) as well as by splitting for 4qA/4qA patients (C, F) and 4qA/4qB ones (B, E). Moreover, sensitivity (sens.), specificity (spec.) and accuracy (acc.) are showed for each box (Created with BioRender.com)
The results obtained on the study cohort are summarized in Fig. 4.
Fig. 4.
Flowchart summarizing the results obtained by the employment of D4Z4 methylation assay in the study cohort of 218 patients. CP = concordant-positive; NCN = non-concordant-negative; CN = concordant-negative
Discussion
The epigenetic de-repression of D4Z4 locus in presence of a permissive 4qA allele has been proposed as one of the crucial events for developing FSHD [17, 24]. This alteration can be due to the reduction of D4Z4 locus to ≤ 10RU or to the presence of detrimental variants within genes (SMCHD1, LRIF1, DNMT3B) involved in the epigenetic regulation of the locus. The investigation of the D4Z4 epigenetic profile in patients with a clinical suspicion of FSHD showed to be helpful in supporting FSHD diagnosis. On this subject, the availability of robust and reliable methods for the evaluation of D4Z4 methylation levels is paramount, and the workflow employed in the present study proved to be a valuable tool (Fig. 1). The DNA methylation assay consists of an accessible and rapid (72 h) laboratory protocol, representing thereby a valid method for the prioritization of FSHD patients. Noteworthy, the implementation of such a method fulfills the need for affordable and rapid tests, especially considering the possible difficulties arising from the lack of adequate resources and laboratory equipment in developing countries. Another advantage of the test lies in the combined analysis of DUX4-PAS and DR1 regions which allows the identification of both FSHD1 and FSHD2 patients based on the reduced methylation levels locally observed in these regions [8, 18].
In this context, the present study aimed at validating such a workflow for prioritizing subjects to be tested for FSHD1/FSHD2. In addition, the study aimed to improve the protocol by classifying patients according to their 4q genotype. The proposed workflow was performed on a cohort of 218 patients accessing the laboratory of genomic medicine UILDM at Santa Lucia Foundation IRCCS between 2022 and 2023.
The workflow for assessing methylation was applied only on subjects carrying at least one 4qA allele (namely 119 4qA/4qA and 93 4qA/4qB). Subsequently, the results of methylation analysis were cross-referenced with the outcomes from D4Z4 sizing and FSHD2 genes sequencing to calculate the performance metrics of the assay in predicting genetic signature of FSHD. Moreover, the refinement of the 4qA/4qA thresholds significantly narrowed the gray zone between CP and CN patients.
The employment of such thresholds led to the improvement of the methylation assay in predicting the presence of FSHD genetic signature, considering the 4q genotype (Fig. 3). The refined threshold was additionally validated on an independent cohort of subjects available at the laboratory (Additional File 3), confirming the performance metrics obtained in the present study (Fig. 3D–F). The decisional workflow to detect FSHD based on methylation thresholds is illustrated in Fig. 5.
Fig. 5.
Decisional workflow showing the thresholds of CpGs methylation levels for FSHD prediction and considering the 4q genotype. (Created with BioRender.com)
As a result, the test achieved a positive predictive value of 100%, as all 122 FSHD predictions were concordant with the presence of a genetic signature of FSHD1 or FSHD2 (115 FSHD1, 3 FSHD1+2, 4 FSHD2).
Interestingly, the hypomethylated subjects included not only affected patients but also asymptomatic subjects with positive family history and FSHD-related genetic signatures, indicating that these subjects should be monitored for a possible manifestation of disease.
Conversely, 90 subjects were predicted as negative for FSHD genetic signatures. Notably, 76 of them were CN, whereas the remaining 14 carried a 4qA ≤ 10RU. In CN group, two subjects showed very high methylation levels not consistent with FSHD. In detail, one subject was an asymptomatic young male with an 8RU allele, and a positive family history for FSHD suggesting a need for a clinical follow-up to exclude possible manifestation of disease later in life. The other patient was a male carrying an 8RU allele as well, he had a negative family history and displayed a phenotype not entirely consistent with FSHD, suggesting a potential differential diagnosis. Importantly, these results should also be interpreted considering the known variable penetrance for borderline [8–10] D4Z4 alleles and the occurrence of contracted D4Z4 alleles in 3% of general population independently from the 4q haplotype [14]. The 14 subjects predicted as negative for a genetic signature of FSHD, displayed slightly higher methylation levels than the thresholds defined for predicting FSHD. These patients were found to carry a D4Z4 allele ≤ 10RU and were considered NCN subjects.
The methylation data were also evaluated with respect to the clinical information provided by the medical centers. Specifically, these data were available for 190 patients out of 218. Five of them did not carry any 4qA permissive allele and therefore were excluded from FSHD diagnosis. The methylation levels of the remaining 185 subjects were therefore compared with the presence of genetic signatures (FSHD1/FSHD2 signatures) and with the clinical status (affected, unaffected) (Table 2).
Table 2.
Comparison between methylation levels, FSHD1/FSHD2 genetic signatures and clinical status
| Genetic signatures | Clinical status | Final output | |
|---|---|---|---|
| Hypomethylated patients (61%) | + | + | FSHD1/FSHD2 patients (88%) |
| + | − | Asymptomatic subjects (12%) | |
| Hypermethylated patients (39%) | − | Not applicable | Non-FSHD subjects (79%) |
| + | − | Asymptomatic subjects (7%) | |
| + | + | Cases with inconclusive results (14%) |
Patients are grouped according to D4Z4 methylation status (hypo-/hypermethylated). For the column “Genetic signatures” the + symbol stands for positivity to the corresponding analysis whereas the — symbol stands for negativity. For the column “Clinical status,” the + symbol stands for FSHD-affected status, and the — symbol stands for FSHD-unaffected status. The description of the clinical status was considered “not applicable” in case of patients with a possible differential diagnosis or unaffected relatives of FSHD patients (negative to the methylation analysis and FSHD1/FSHD2 genetic testing)
This analysis highlighted the ability of methylation assay in primarily discriminating FSHD-affected subjects from non-FSHD ones, except for 14% cases with inconclusive results who showed discordant methylation values compared to positive genetic and clinical features. These patients showed an M:F ratio of 60:40 and showed an average age of 53.5 ± 11.8 years. In more detail, one patient (ID94) was reported as a sporadic case of FSHD and exhibited a mild phenotype (FSHD score = 1). Conversely, seven patients (ID8, ID29, ID56, ID70, ID89, ID114, and ID136) with a positive family history for FSHD were reported as affected, most displaying mild muscular involvement (FSHD score ≥ 1), except for two patients who showed moderate signs of disease (FSHD score ≥ 5). Another patient (ID23) without information on family history for the disease displayed a severe phenotype (FSHD score = 11) and a D4Z4 of 5RU, despite the borderline methylation levels. Unfortunately, no clinical data were available for the patient ID116, who has negative family history for FSHD and displayed a borderline D4Z4 size of 9RU. The reduced penetrance and variable expressivity typically observed within the FSHD families can contribute to explain discordant methylation profiles characterized by borderline levels as well as the presence of possible double-trouble conditions that may affect the clinical phenotype. The clinical and molecular details of peculiar cases are summarized in Table 3.
Table 3.
Clinical and molecular details of misclassified patients
| Patient ID | Sex | Age | D4Z4 RU | 4q genotype | DUX4-PAS CpG_6 | DUX4-PAS CpG_3 | DR1 CpG_1 | DR1 CpG_22 | Family history | Clinical status | Age of onset | FSHD score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID8 | M | 42 | 6 | A/A | 0.8557 | 0.3275 | 0.7317 | 1.0000 | Yes | Affected | NA | 1 |
| ID23 | M | 67 | 5 | A/A | 0.8630 | 0.3157 | 0.6194 | 0.8810 | NA | Affected | 55 | 11 |
| ID29 | M | 56 | 4 | A/A | 0.8931 | 0.2884 | 0.5916 | 0.9121 | Yes | Affected | 42 | 5 |
| ID56 | F | 46 | 6 | A/B | 0.8903 | 0.2006 | 0.6121 | 0.9044 | Yes | Affected | NA | 1 |
| ID70 | F | 51 | 5 | A/B | 0.8395 | 0.2854 | 0.6358 | - | Yes | Affected | NA | 1 |
| ID89 | M | 51 | 6 | A/B | 0.8542 | 0.1350 | 0.5884 | 0.8676 | Yes | Affected | NA | 2 |
| ID94 | M | 47 | 7 | A/A | 0.8974 | 0.2378 | 0.6980 | 0.9562 | No | Affected | 35 | 1 |
| ID114 | F | 71 | 9 | A/A | 0.8580 | 0.7938 | 0.6304 | 0.8591 | Yes | Affected | 30 | 10 |
| ID116 | M | 68 | 9 | A/B | 0.8928 | 0.3054 | 0.7280 | 1.0000 | No | Affected | NA | NA |
| ID136 | F | 36 | 7 | A/A | 0.8504 | 0.2736 | - | 0.9025 | Yes | Affected | NA | 1 |
NA: not available. FSHD score refers to Lamperti et al. 2010 [25]
In conclusion, our laboratory experience showed that the introduction of D4Z4 methylation assessment was able to provide clinicians with preliminary evidence for the presence of FSHD molecular signatures to be confirmed by traditional testing or to address the patient toward a clinical re-evaluation and a differential diagnosis. Of note, the test was able to detect differential methylation profiles in asymptomatic subjects, suggesting its potential use to identify possible preclinical status to be monitored in patients with positive family history and presence of genetic signatures compatible with FSHD.
This study validated the ability of the model to accurately identify methylation levels indicative of FSHD genetic signatures considering the 4q genotype. In this regard, this work is in line with other research studies presenting protocols based on other techniques [11, 19, 20, 26, 27]. The employment of long-read sequencing technologies hold great promise, especially for solving complex cases [1]. A number of reports showed its possible application to test for methylation levels and, simultaneously, detect 4q haplotype and D4Z4 size [27–29]. However, this method still has some limitations, mainly related to the generation of limited valid reads for the 4q35 region, the need of validation studies on large-scale sample cohorts, the relatively high costs and the requirement of bioinformatics knowledge and resources that are not always available and accessible in the diagnostic centers.
Notably, although the methylation profile helps in the diagnosis of FSHD, it should always be interpreted considering the clinical evidence of patients. Finally, this study not only highlights the potential of methylation assay as a rapid and cost-effective test in developing countries but also promotes the development of tools that support and streamline the FSHD diagnostic workflow.
Supplementary Information
Acknowledgements
The study was supported by Ministry of Health (Ricerca Corrente) to C.C. and by the PRIN project 2022 PNRR (Prot. P20229XKFC) to E.G.
Author contributions
Concept and design were done by C.S., D.M., R.C. and E.G. Acquisition, analysis, and interpretation of molecular data were done by C.S., D.M., G.T., E.P.P., L.C. and R.C. Recruitment of samples and clinical characterization were done by G.T, M.M., S.Z., G.P., C.S., S.B., E.T., B.R., F.T., G.G., B.R., F.C., F.G., E.C., V.G., M.G., L.T., E.F., R.M., E.M.P., A.P., M.P., A.F., F.G., M.S., P.M., M.G., M.A.M., C.C., E.M., L.P., L.P., R.P., C.R., A.P., S.C.P., V.S., L.V., T.E.M., G.R., G.S., M.F. and E.R. Drafting of the manuscript was done by C.S., D.M and R.C. Revision of the manuscript was done by C.S., D.M., G.T., L.C., G.T, M.M., S.Z., G.P., C.S., S.B., E.T., B.R., F.T., G.G., B.R., F.C., F.G., E.C., V.G., M.G., L.T., E.F., R.M., E.M.P., A.P., M.P., A.F., F.G., M.S., P.M., M.G., M.A.M., C.C., E.M., L.P., L.P., R.P., C.R., A.P., S.C.P., V.S., L.V., T.E.M., G.R., G.S., M.F., E.R. and R.C. Final proofreading was done by C.S. and E.G.
Funding
Not applicable.
Declarations
Ethics approval and consent to participate
All participants provided signed informed consent for research and publication at the time of recruitment. The study was approved by the Ethics Committee of Santa Lucia Foundation (CE/2022_020 approved on June 1, 2022) and complied with the Declaration of Helsinki.
Consent for publication
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article (and its additional files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Claudia Strafella and Domenica Megalizzi shared the first authorship.
References
- 1.Giardina E, Camaño P, Burton-Jones S, Ravenscroft G, Henning F, Magdinier F, et al. Best practice guidelines on genetic diagnostics of facioscapulohumeral muscular dystrophy: update of the 2012 guidelines. Clin Genet. 2024 [DOI] [PMC free article] [PubMed]
- 2.Caputo V, Megalizzi D, Fabrizio C, Termine A, Colantoni L, Caltagirone C, et al. Update on the molecular aspects and methods underlying the complex architecture of FSHD. Cells. 2022;11(17):2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmers RJLF, Wohlgemuth M, van der Gaag KJ, van der Vliet PJ, van Teijlingen CMM, de Knijff P, et al. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81(5):884–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemmers RJLF, Tawil R, Petek LM, Balog J, Block GJ, Santen GWE, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44(12):1370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Boogaard ML, Lemmers RJLF, Balog J, Wohlgemuth M, Auranen M, Mitsuhashi S, et al. Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am J Hum Genet. 2016;98(5):1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamanaka K, Šikrová D, Mitsuhashi S, Masuda H, Sekiguchi Y, Sugiyama A, et al. Homozygous nonsense variant in LRIF1 associated with facioscapulohumeral muscular dystrophy. Neurology. 2020;94(23):e2441–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitsuhashi S, Boyden SE, Estrella EA, Jones TI, Rahimov F, Yu TW, et al. Exome sequencing identifies a novel SMCHD1 mutation in facioscapulohumeral muscular dystrophy 2. Neuromuscul Disord NMD. 2013;23(12):975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen M, Rost S, El Hajj N, Ferbert A, Deschauer M, Walter MC, et al. Diagnostic approach for FSHD revisited: SMCHD1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1. Eur J Hum Genet EJHG. 2015;23(6):808–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strafella C, Caputo V, Galota RM, Campoli G, Bax C, Colantoni L, et al. The variability of SMCHD1 gene in FSHD patients: evidence of new mutations. Hum Mol Genet. 2019;28(23):3912–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mocciaro E, Runfola V, Ghezzi P, Pannese M, Gabellini D. DUX4 role in normal physiology and in FSHD muscular dystrophy. Cells. 2021;10(12):3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones TI, King OD, Himeda CL, Homma S, Chen JCJ, Beermann ML, et al. Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin Epigenetics. 2015;7(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Himeda CL, Jones PL. The genetics and epigenetics of facioscapulohumeral muscular dystrophy. Annu Rev Genomics Hum Genet. 2019;31(20):265–91. [DOI] [PubMed] [Google Scholar]
- 13.van Overveld PG, Lemmers RJ, Deidda G, Sandkuijl L, Padberg GW, Frants RR, et al. Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum Mol Genet. 2000;9(19):2879–84. [DOI] [PubMed] [Google Scholar]
- 14.Scionti I, Greco F, Ricci G, Govi M, Arashiro P, Vercelli L, et al. Large-scale population analysis challenges the current criteria for the molecular diagnosis of fascioscapulohumeral muscular dystrophy. Am J Hum Genet. 2012;90(4):628–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zampatti S, Colantoni L, Strafella C, Galota RM, Caputo V, Campoli G, et al. Facioscapulohumeral muscular dystrophy (FSHD) molecular diagnosis: from traditional technology to the NGS era. Neurogenetics. 2019;20(2):57–64. [DOI] [PubMed] [Google Scholar]
- 16.Calandra P, Cascino I, Lemmers RJLF, Galluzzi G, Teveroni E, Monforte M, et al. Allele-specific DNA hypomethylation characterises FSHD1 and FSHD2. J Med Genet. 2016;53(5):348–55. [DOI] [PubMed] [Google Scholar]
- 17.Huichalaf C, Micheloni S, Ferri G, Caccia R, Gabellini D. DNA methylation analysis of the macrosatellite repeat associated with FSHD muscular dystrophy at single nucleotide level. PLoS ONE. 2014;9(12): e115278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartweck LM, Anderson LJ, Lemmers RJ, Dandapat A, Toso EA, Dalton JC, et al. A focal domain of extreme demethylation within D4Z4 in FSHD2. Neurology. 2013;80(4):392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould T, Jones TI, Jones PL. Precise epigenetic analysis using targeted bisulfite genomic sequencing distinguishes FSHD1, FSHD2, and healthy subjects. Diagn Basel Switz. 2021;11(8):1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erdmann H, Scharf F, Gehling S, Benet-Pagès A, Jakubiczka S, Becker K, et al. Methylation of the 4q35 D4Z4 repeat defines disease status in facioscapulohumeral muscular dystrophy. Brain J Neurol. 2023;146(4):1388–402. [DOI] [PubMed] [Google Scholar]
- 21.Caputo V, Megalizzi D, Fabrizio C, Termine A, Colantoni L, Bax C, et al. D4Z4 methylation levels combined with a machine learning pipeline highlight single CpG sites as discriminating biomarkers for FSHD patients. Cells. 2022;11(24):4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Megalizzi D, Trastulli G, Caputo V, Colantoni L, Caltagirone C, Strafella C, et al. Epigenetic profiling of the D4Z4 locus: optimization of the protocol for studying DNA methylation at single CpG site level. Electrophoresis. 2023;44(19–20):1588–94. [DOI] [PubMed] [Google Scholar]
- 23.Strafella C, Caputo V, Bortolani S, Torchia E, Megalizzi D, Trastulli G, et al. Whole exome sequencing highlights rare variants in CTCF, DNMT1, DNMT3A, EZH2 and SUV39H1 as associated with FSHD. Front Genet. 2023;14:1235589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Greef JC, Wohlgemuth M, Chan OA, Hansson KB, Smeets D, Frants RR, et al. Hypomethylation is restricted to the D4Z4 repeat array in phenotypic FSHD. Neurology. 2007;69(10):1018–26. [DOI] [PubMed] [Google Scholar]
- 25.Lamperti C, Fabbri G, Vercelli L, D’Amico R, Frusciante R, Bonifazi E, et al. A standardized clinical evaluation of patients affected by facioscapulohumeral muscular dystrophy: the FSHD clinical score. Muscle Nerve. 2010;42(2):213–7. [DOI] [PubMed] [Google Scholar]
- 26.Roche S, Dion C, Broucqsault N, Laberthonnière C, Gaillard MC, Robin JD, et al. Methylation hotspots evidenced by deep sequencing in patients with facioscapulohumeral dystrophy and mosaicism. Neurol Genet. 2019;5(6): e372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiramuki Y, Kure Y, Saito Y, Ogawa M, Ishikawa K, Mori-Yoshimura M, et al. Simultaneous measurement of the size and methylation of chromosome 4qA-D4Z4 repeats in facioscapulohumeral muscular dystrophy by long-read sequencing. J Transl Med. 2022;20(1):517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butterfield RJ, Dunn DM, Duval B, Moldt S, Weiss RB. Deciphering D4Z4 CpG methylation gradients in fascioscapulohumeral muscular dystrophy using nanopore sequencing. Genome Res. 2023;33(9):1439–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang M, Zhang Q, Jiao J, Shi J, Xu Y, Zhang C, et al. Comprehensive genetic analysis of facioscapulohumeral muscular dystrophy by nanopore long-read whole-genome sequencing. J Transl Med. 2024;22(1):451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its additional files).