Abstract
Objective
The emergence of resistance to colistin, the last resort for treating severe infections caused by Pseudomonas aeruginosa, poses a significant threat to public health. This meta-analysis aimed to investigate the prevalence of colistin resistance in clinical isolates of P. aeruginosa.
Method
A comprehensive search of MEDLINE (PubMed), Web of Science, and Scopus databases was conducted to identify relevant articles published until December 2023. Subsequently, a meta-analysis was performed using Stata software to examine the pooled prevalence of colistin resistance and to conduct subgroup analyses.
Results
A total of 619 studies were included in the meta-analysis, revealing a global prevalence of colistin resistance of 1% among all P. aeruginosa isolates. Furthermore, cystic fibrosis patients exhibited the highest resistance to colistin, with a prevalence of 7% among the examined diseases.
Conclusion
The increase in colistin resistance in P. aeruginosa in recent years from 2% (in the period of 2006–2010) to 5% (in the period of 2020–2023) underscores the need for implementing infection prevention programs, using appropriate treatment regimens, and disseminating comprehensive information on antimicrobial resistance patterns. These measures are crucial for addressing this growing public health concern.
Keywords: Pseudomonas aeruginosa, cystic fibrosis, infection prevention, treatment regimens, public health
Introduction
Pseudomonas aeruginosa is recognized as an opportunistic pathogen and a major causative agent of hospital-acquired infections, including urinary tract infections, pneumonia, bloodstream infections, and surgical site infections (Pang et al., 2019; Sadikot et al., 2005). The development of intrinsic and acquired resistance in P. aeruginosa is attributed to the inappropriate and excessive use of antibiotics, leading to the emergence of antibiotic resistance (El-Mokhtar and Hetta, 2018).
The management of P. aeruginosa infections has always presented challenges. Carbapenems such as imipenem and meropenem were introduced as effective treatments for severe multidrug-resistant (MDR) P. aeruginosa infections. However, the overuse of antibiotics has resulted in the emergence of carbapenem-resistant isolates, posing a significant concern (Wi et al., 2017; Bonyadi et al., 2022). In 2017, the World Health Organization (WHO) identified carbapenem-resistant P. aeruginosa as a priority pathogen necessitating the development of new antibiotics for treatment (Tacconelli et al., 2018).
The increasing rate of infections caused by multidrug-resistant (MDR), extensively drug-resistant (XDR), and particularly carbapenem-resistant P. aeruginosa has led to the resurgence of colistin as a critical last-resort therapeutic option (Wi et al., 2017; Biswas et al., 2012; Al-Orphaly et al., 2021). Despite its potent antimicrobial activity against P. aeruginosa and its designation as a potentially effective drug, the increased utilization of colistin has resulted in the emergence of bacterial strains with reduced susceptibility to this antibiotic class worldwide (Pechorsky et al., 2009; Lee et al., 2012).
Colistin resistance primarily arises through various mechanisms, including enzymatic modification of lipid A, leading to a decrease in the outer membrane’s negative charge and reduced colistin affinity. Resistance to colistin may also stem from chromosomally encoded mutations or plasmid-mediated colistin resistance gene mcr, facilitating horizontal dissemination of resistance (Cannatelli et al., 2013; Paterson and Harris, 2016; Hasman et al., 2015; Arcilla et al., 2016). The prevalence of colistin resistance is typically below 10%, but this rate is steadily increasing in the Mediterranean, Southeast Asia, and certain African countries (Bialvaei and Samadi, 2015). Recent observations indicate that resistance to colistin has emerged in several Enterobacteriaceae species, including Klebsiella pneumoniae, Escherichia coli, and Enterobacter aerogenes. This resistance has been linked to the extensive use of polymyxins for infection control in veterinary medicine (Baron et al., 2016; Al-Kadmy et al., 2020). Given the potential for both horizontal transfer of resistance genes through conjugative plasmids and vertical transfer through chromosomal mutation, the emergence of colistin-resistant isolates poses a significant global health threat, especially considering the importance of colistin as a last-resort treatment option (Liu et al., 2016; Abd El-Baky et al., 2020).
The rise of MDR, XDR, and pan drug-resistant (PDR) P. aeruginosa poses a significant public health challenge, leading to delays in antimicrobial therapy, treatment failures, and increased mortality rates (Abd El-Baky et al., 2020). This situation necessitates urgent attention, as these resistant strains may exhibit resistance to all available antimicrobials or show susceptibility only to colistin or polymyxins, severely limiting treatment options for healthcare providers managing severe infections associated with MDR P. aeruginosa. The emergence of colistin-resistant strains is particularly concerning for patients with critical infections. Consequently, this systematic review and meta-analysis aims to investigate the global prevalence of colistin resistance in P. aeruginosa, thereby enhancing our understanding of antibiotic resistance in this pathogen.
Methods
Search strategy
We conducted a comprehensive search for eligible studies published from 1990 to December 2023 using MEDLINE (PubMed), Web of Science, and Scopus. The search terms included (“Pseudomonas aeruginosa” OR P. aeruginosa) AND (Colisticin OR “Polymyxin E” OR Colimycin OR colistin OR colistimethate). This review was carried out and reported in accordance with current guidelines, and the results were reported following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement (Moher et al., 2009).
Inclusion and exclusion criteria
All original articles that provided data on the total number of clinical P. aeruginosa isolates and the number of colistin-resistant P. aeruginosa isolates were included. Studies were excluded if they met the following criteria: (Pang et al., 2019) did not present P. aeruginosa colistin resistance; (Sadikot et al., 2005) did not clearly report resistance rates (the exact number of primary isolates and the number of resistant isolates are not provided); (El-Mokhtar and Hetta, 2018) conducted antimicrobial susceptibility tests for colistin without specifying the method; (Wi et al., 2017) were written in languages other than English; and (Bonyadi et al., 2022) sourced data from conference abstracts, editorials, case reports, meta-analyses, systematic reviews, narrative reviews, experimental studies on animal models, and articles without full text after contacting the corresponding author.
Data extraction
After consolidating the articles using EndNote X20 Citation Manager Software, duplicate articles were removed before review. The citations were then imported into Rayyan, a citation classification application (Ouzzani et al., 2016). Three reviewers independently screened all titles and abstracts to exclude irrelevant topics. In the subsequent assessment stage, qualified studies were downloaded, and the full text of selected articles was retrieved based on the inclusion and exclusion criteria.
Three reviewers developed a data extraction form and collected data from all qualified studies. The extracted data included the first author’s name, year of publication, year of collection, continent and countries where the study was conducted, sample size (number of P. aeruginosa isolates and number of colistin-resistant isolates), origin of isolates, drug resistance categories, disease, guideline, and susceptibility test methodology (agar dilution, broth microdilution, disk elution, E-test, and disk diffusion).
Quality assessment
The quality of the included studies was evaluated independently by three reviewers using a modified version of the Joanna Briggs Institute (JBI) assessment tool for prevalence studies (Munn et al., 2014). The checklist includes the following questions: Was the sample frame appropriate to address the target population? Were study participants sampled appropriately? Was the sample size adequate? Were the study subjects and the setting described in detail? Was the data analysis conducted with sufficient coverage of the identified sample? Were valid methods used for the identification of the condition? Was the condition measured in a standard, reliable way for all participants? Was there an appropriate statistical analysis? Was the response rate adequate, and if not, was the low response rate managed appropriately? Each item is evaluated as “yes,” “no,” or “unclear.” A “yes” response is assigned a score of 1 point, while responses categorized as “no” or “unclear” receive 0 points. Studies that score 7 or higher are classified as high quality, those with scores between 5 and 6 are considered medium quality, and studies scoring 4 or lower are designated as low quality. In cases of disagreement, a fourth reviewer provided adjudication.
Statistical analysis
We conducted a prevalence meta-analysis using the metaprop package in Stata 17 software. We calculated the pooled prevalence of colistin-resistant P. aeruginosa, along with the associated 95% confidence intervals (CIs), utilizing the Freeman-Tukey double arcsine transformation within a random-effects model.
To identify publication bias, we employed Egger’s test, with a significance threshold set at p < 0.05, indicating the presence of statistically significant bias. Additionally, a trim-and-fill analysis was conducted to address potential bias. Funnel plots were also utilized for a visual assessment of publication bias.
Heterogeneity among studies was measured using the I2 statistic. Specifically, I2 ≤ 25% indicated low heterogeneity, 25% < I2 ≤ 75% indicated moderate heterogeneity, and I2 > 75% indicated high heterogeneity.
Subgroup analyses were conducted based on various factors, including publication year (from 2009 to 2023), collection period (five distinct periods), continent (five continents), country (thirty-two countries), guidelines followed (CLSI and EUCAST), disease type (including Urinary Tract Infections, Pneumonia, Lower Respiratory Tract Infections, Intra-abdominal Infection, COVID-19, cancer, Cystic Fibrosis, Bacteremia, and Bloodstream Infection), method of colistin resistance detection (agar dilution, E-test, disk diffusion, and broth microdilution), different resistance categories (Multidrug Resistance, Extensively Drug Resistant, and Carbapenem Resistance), and sample origin (urine, sputum, endotracheal aspirate, burn wounds, and blood).
Results
Studies selection
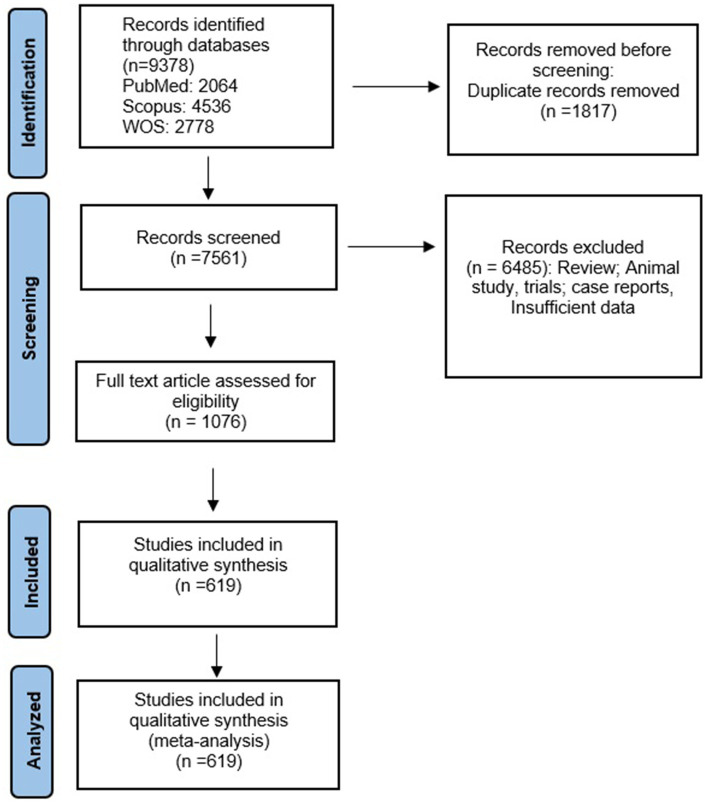
Our initial search yielded a total of 9,378 articles. After removing duplicates, we screened the titles and abstracts of 7,561 articles. From this screening process, 1,076 articles met the inclusion criteria and were selected for a full-text review. After the full-text review, we identified 619 articles that were suitable for analysis (Abavisani et al., 2021; Abd El-Baky et al., 2020; Abdelatti et al., 2023; Abed and Kareem, 2021; Abubakar et al., 2022; Abulzahra and Ismail, 2020; Addis et al., 2021; Agrawal et al., 2013; Aguilar-Rodea et al., 2017; Ahani Azari and Fozouni, 2020; Ahmed et al., 2022; Ahmed et al., 2019; Aiyegoro et al., 2007; Akgül et al., 2021; Akhi et al., 2015; Akram et al., 2022; Al Dawodeyah et al., 2018; Al-Agamy et al., 2012; Al-Bayssari et al., 2021; Al-Kabsi et al., 2011; Al-kaffas et al., 2022; Al-Khudhairy and Al-Shammari, 2020; Al-shimmary, 2018; Al-Zahrani and Al-Ahmadi, 2021; Alam et al., 2021; Alcántar-Curiel et al., 2023; Alexopoulou et al., 2016; Alfouzan et al., 2022; Alfouzan et al., 2018; Alhanout et al., 2009; Ali et al., 2021; Ali et al., 2015; Alkhulaifi and Mohammed, 2023; Alotaibi et al., 2023; Alruways, 2023; Amabile-Cuevas, 2017; Appalaraju et al., 2020; Aprile et al., 2019; Arab et al., 2023; Araújo Lima et al., 2020; Arca-Suárez et al., 2021; Arici et al., 2023; Arif et al., 2022; Armengol et al., 2020; Armengol et al., 2019; Aruhomukama et al., 2019; Asar et al., 2019; Aydemir et al., 2022; Aydın et al., 2018; Azimi et al., 2012; Azimi L. et al., 2016; Azimi S. et al., 2016; Babu and Menon, 2018; Badawy et al., 2023; Badierah et al., 2019; Bae and Stone, 2022; Baek et al., 2020; Bagheri-Nesami et al., 2017; Bahabri et al., 2022; Bahador et al., 2019; Bahçe et al., 2022; Baiomy et al., 2023; Bakht et al., 2022; Balkhair et al., 2023; Bandic-Pavlovic et al., 2020; Banerjee et al., 2024; Bangera et al., 2016; Basu et al., 2013; Bayram et al., 2013; Bazgir et al., 2021; Beirao et al., 2020; Ben Nejma et al., 2018; Berwal et al., 2020; Bian et al., 2022; Blondeau et al., 2023; Bogiel et al., 2022; Bono et al., 2015; Bourgi et al., 2020; Boustanshenas et al., 2023; Brauncajs et al., 2022; Brzozowski et al., 2020; Bunsow et al., 2020; Buzilă et al., 2021; Cabot et al., 2011; Camargo et al., 2023; Candel et al., 2022; Canton et al., 2022; Carvalhaes et al., 2020; Castanheira et al., 2018; Cavallo et al., 2022; Cesur et al., 2012; Çetin et al., 2022; Chang et al., 2023; Chaturvedi et al., 2021; Chaudhary et al., 2020; Chauhan et al., 2022; Chen et al., 2022; Chen et al., 2015; Chen et al., 2023; Chen Q. et al., 2020; Chen X. et al., 2020; Chen et al., 2014; Chew et al., 2019; Chittawatanarat et al., 2014; Chukamnerd et al., 2023; Cillóniz et al., 2016; Cipriano et al., 2007; Çopur Çiçek et al., 2021; Czekajło-Kołodziej et al., 2006; da Costa Júnior et al., 2020; Dadmanesh et al., 2014; Darji and Patel, 2023; Dassner et al., 2017; Datar et al., 2021; de Dios et al., 2016; De Francesco et al., 2013; de Oliveira Santos et al., 2019; De Vecchi et al., 2013; Dehbashi et al., 2018; Del Barrio-Tofino et al., 2017; Del Giacomo et al., 2022; Delgado-Valverde et al., 2020; Delroshan et al., 2023; Depka et al., 2020; Descours et al., 2018; Dharati et al., 2021; Di Carlo et al., 2021; Di Domenico et al., 2017; Dias et al., 2017; Díaz-Cañestro et al., 2018; Diekema et al., 2019; Din et al., 2023; Do Tran et al., 2022; Dogonchi et al., 2018; Dong et al., 2012; Doumith et al., 2022; Durdu et al., 2018; Ebadati et al., 2023; Ece et al., 2014; Eftekhar et al., 2009; Eid et al., 2020; Ejaz, 2022; Ekkelenkamp et al., 2020; El Mekes et al., 2020; El-Sokkary et al., 2021; Eladawy et al., 2021; Elshafie et al., 2007; Emami et al., 2017; Emami et al., 2019; Emami et al., 2020; Ergul et al., 2017a; Ergul et al., 2017b; Escolà-Vergé et al., 2018; Evans et al., 2019; Fakhkhari et al., 2022; Falagas et al., 2017; Fang et al., 2023; Farag et al., 2020; Farhan et al., 2019; Farrell et al., 2013; Farrell et al., 2014a; Farrell et al., 2014b; Fekri Kohan et al., 2020; Feretzakis et al., 2019; Ferjani et al., 2022; Fernández-Olmos et al., 2013; Flamm et al., 2014; Flores-Paredes et al., 2021; Fluge et al., 2001; Ford et al., 2022; Fournier et al., 2021; Fournier et al., 2013; Fraenkel et al., 2023; Franco et al., 2010; Franco et al., 2023; Gaber et al., 2020; Gahlot and Kasana, 2021; Gajdács et al., 2021a; Gajdács et al., 2019; Gajdács et al., 2021b; Galani et al., 2008; Galani et al., 2020; Gales et al., 2012; Gales et al., 2011; Gales et al., 2003; Galindo-Mendez et al., 2023; Gangwar et al., 2021; Gant et al., 2021; García-Castillo et al., 2011; Garcia-Fernandez et al., 2019; Garg et al., 2019; Gaudereto et al., 2020; Ghanem et al., 2023; Ghasemian et al., 2023; Ghasemshahi et al., 2022; Gherardi et al., 2019; Giani et al., 2018; Gilani et al., 2015; Goli et al., 2017; Goli et al., 2016; Golli et al., 2022; Gomez et al., 2023; Gómez-Garcés et al., 2009; Gomila et al., 2013; Gunalan et al., 2021; Guo et al., 2023; Gutierrez-Santana et al., 2022; Guzek et al., 2017; Guzek et al., 2015; Güzel and Gerçeker, 2008; Hackel et al., 2018; Hakeam et al., 2022; Hallit et al., 2020; Hansen et al., 2008; Hao et al., 2023; Hawser et al., 2021; Heidari et al., 2022; Henderson et al., 2018; Herrera-Espejo et al., 2020; Hindler and Humphries, 2013; Hishinuma et al., 2018; Hishinuma et al., 2020; Holger et al., 2023; Holger et al., 2022a; Hong et al., 2015; Hoşbul et al., 2022; Hosseininassab Nodoushan et al., 2017; Howard-Anderson et al., 2022; Hrbacek et al., 2020; Hsueh et al., 2019; Hu et al., 2022; Hu et al., 2023; Hu et al., 2021a; Hu et al., 2021b; Huang et al., 2022; Huband et al., 2016; Huband et al., 2020; Humphries et al., 2023; Ibrahim, 2018; Ioannou et al., 2023; Ismail and Mahmoud, 2018; Izadi Pour Jahromi et al., 2018; Jalali et al., 2021; Jamalifar et al., 2019; Jansen et al., 2016; Japoni et al., 2011; Jauhari et al., 2020; Jayarani et al., 2020; Jazani et al., 2012; Jean et al., 2009; Jean et al., 2016; Jean et al., 2022; Jeong et al., 2024; Jimenez-Guerra et al., 2018; Jones et al., 2013a; Jones et al., 2014; Jones et al., 2013b; Juhász et al., 2017a; Juhasz et al., 2017b; Jung et al., 2019; Kabic et al., 2023; Kakhandki et al., 2020; Kanwar et al., 2023; Karami et al., 2020; Karimzadeh et al., 2017; Karlowsky et al., 2013; Karlowsky et al., 2022; Karlowsky et al., 2019a; Karlowsky et al., 2019b; Karlowsky et al., 2023; Karlowsky et al., 2018a; Karlowsky et al., 2018b; Karlowsky et al., 2021; Karruli et al., 2022; Kashfi et al., 2017; Katchanov et al., 2018; Kaur et al., 2022; Kazmierczak et al., 2018; Kazmierczak et al., 2016; Kc et al., 2019; Keepers et al., 2014; Khajuria et al., 2013; Khan et al., 2017; Khan et al., 2023; Khater, 2022; Khater and AlFaki, 2022; Khorvash et al., 2017; Khoshnood et al., 2019; Khosravi et al., 2017; Khosravi et al., 2016; Kidd et al., 2009; Kim et al., 2022; Kim et al., 2017; Kiratisin et al., 2021; Kiratisin et al., 2023; Kırac et al., 2018; Ko and Stone, 2020; Ko and Stone, 2020; Kohira et al., 2023; Kokkayil et al., 2018; Kombade et al., 2021; Korzekwa et al., 2021; Kothari et al., 2022; Kovacevic et al., 2019; Kragh et al., 2021; Krause et al., 2019; Kresken et al., 2020a; Kresken et al., 2020b; Kristof et al., 2021; Kuo et al., 2021; Kurihara et al., 2022; Lai et al., 2019; Laurentiu et al., 2017; Lee J.-Y. et al., 2011; Lee S.-C. et al., 2011; Lee et al., 2023; Lee et al., 2019; Li et al., 2021; Li et al., 2022; Li et al., 2023; Licata et al., 2020; Lin J. et al., 2019; Lin et al., 2016; Lin Z. et al., 2019; Liu et al., 2018; Liu et al., 2020; Livermore et al., 2017; Llanes et al., 2013; Lob et al., 2023; Lockwood and Lawson, 1973; Longshaw et al., 2020; López Montesinos et al., 2022; Lopez-Causape et al., 2017; Lopez-Causape et al., 2013; Macin and Akyon, 2017; MacKenzie et al., 2004; Mahar et al., 2020; Mahdy, 2022; Mahmoud et al., 2021; Maina et al., 2023; Makharita et al., 2020; Malekzadegan et al., 2019; Mallikarjuna and Dhanashree, 2023; Manno et al., 2005; Mansour et al., 2013; Maraki et al., 2015; Maraki et al., 2012; Marteva-Proevska et al., 2021; Martínez et al., 2012; Mataraci Kara et al., 2020; Matuschek et al., 2018; McCracken et al., 2019; McCracken et al., 2019; Medell et al., 2013; Medell et al., 2012; Mellouli et al., 2021; Memar et al., 2016; Mendes et al., 2013; Meradji et al., 2016; Meradji et al., 2015; Meschiari et al., 2021; Mickymaray, 2019; Mikucionyte et al., 2016; Milczewska et al., 2020; Milojković et al., 2020; Mirbagheri et al., 2015; Mishra et al., 2020; Mobaraki et al., 2018; Mohamed et al., 2021; Mohamed et al., 2022; Mohamed and Youssef, 2011; Mohammadi Barzelighi et al., 2020; Mohammadzadeh et al., 2017; Mohammadzamani et al., 2020; Mohanty et al., 2013; Momenah et al., 2023; Monogue and Nicolau, 2018; Montero M. et al., 2020; Montero et al., 2019; Montero M. M. et al., 2020; Moradi et al., 2021; Morata et al., 2012; Mordi and Erah, 2006; MaI et al., 2005; Morrow et al., 2013; Morteza et al., 2019; Mostafa et al., 2022; Moubareck et al., 2019; Mulet et al., 2021; Mustafa et al., 2016; Naas et al., 2021; Nair et al., 2020; Najafi et al., 2015; Nedeljković et al., 2015; Negm et al., 2023; Negm et al., 2021; Nelson and Rosowsky, 2002; Nguyen et al., 2021; Nichols et al., 2016; Nitescu et al., 2023; Nitz et al., 2021; Nojookambari et al., 2019; Nolan et al., 2021; Nwabuisi and Ologe, 2002; O'Carroll et al., 2004; Odumosu et al., 2012; Olowo-okere et al., 2020; Öncül et al., 2014; Ozumba, 2005; Pani et al., 2022; Papagiannitsis et al., 2017; Paprocka et al., 2021; Park et al., 2017; Parsa et al., 2020; Pasca et al., 2012; Peña et al., 2012; Pérez et al., 2019; Perez et al., 2014; Pérez-Vázquez et al., 2020; Perovic et al., 2023; Petca, 2021; Petca, 2021; Petrova et al., 2016; Peymani et al., 2015; Pfaller et al., 2022; Pfaller et al., 2017; Pfaller et al., 2018; Picão Renata et al., 2009; Pierard and Stone, 2021; Pitt et al., 2003; Pourakbari et al., 2016; Pragsam et al., 2018; Prasanth Manohar et al., 2017; Kokkayil et al., 2018; Priyadarshi et al., 2023; Pujji et al., 2019; Qadeer et al., 2016; Radan et al., 2016; Rafalskiy et al., 2020; Rahimi et al., 2021; Rajenderan et al., 2014; Ramadan et al., 2018; Ramanathan et al., 2017; Rashid et al., 2014; Rattanaumpawan et al., 2013; Raza et al., 2019; Reddy et al., 2014; Rizek et al., 2014; Rodulfo et al., 2019; Rolston et al., 2020; Rosales-Reyes et al., 2020; Rossolini et al., 2008; Rout et al., 2023; Roy Chowdhury et al., 2016; Ruekit et al., 2022; Ruh et al., 2016; Ruiz-Roldan et al., 2018; Sacha et al., 2015; Sader et al., 2020a; Sader et al., 2020b; Sader et al., 2019; Sader et al., 2018a; Sader et al., 2015a; Sader et al., 2017a; Sader et al., 2016; Sader et al., 2017b; Sader et al., 2017c; Sader et al., 2018b; Sader et al., 2017d; Sader et al., 2021a; Sader et al., 2014a; Sader et al., 2014b; Sader et al., 2014c; Sader et al., 2018c; Sader et al., 2018d; Sader et al., 2018e; Sader et al., 2017e; Sader et al., 2015b; Sader et al., 2021b; Saderi et al., 2015; Saffari et al., 2017; Saleem M. et al., 2023; Saleem et al., 2022; Saleem and Bokhari, 2020; Saleem Z. et al., 2023; Salimizand et al., 2023; Samonis et al., 2012; Samonis et al., 2010; Samonis et al., 2012; Sanchez-Lopez et al., 2021; Santella et al., 2021; Santimaleeworagun et al., 2020; Sarwat et al., 2021; Schaumburg et al., 2022; Schechner et al., 2011; Schülin, 2002; Seale et al., 1979; Sefraoui et al., 2014; Seifert et al., 2018; Şen et al., 2016; Sendra et al., 2022; Shahri et al., 2022; Sharan et al., 2016; Shariati et al., 2018; Sharifi et al., 2019; Sharma et al., 2023; Sherchan et al., 2022; Sherchan and Humagain, 2020; Sheth et al., 2012a; Sheth et al., 2012b; Shiralizadeh et al., 2023; Shivshetty et al., 2020; Shortridge et al., 2021a; Shortridge et al., 2021b; Shortridge et al., 2017; Shortridge et al., 2019a; Shortridge et al., 2019b; Shortridge et al., 2019c; Shortridge et al., 2018; Shortridge et al., 2020; Shortridge et al., 2022; Shravani et al., 2023; Sid Ahmed et al., 2020; Sid Ahmed et al., 2019; Sid Ahmed et al., 2021; Sid Ahmed et al., 2021; Sid Ahmed et al., 2023; Simar et al., 2017; Singh-Moodley et al., 2018; Singhal et al., 2019; Sleiman et al., 2023; Soliman et al., 2020; Soni et al., 2023; Stone and Ponce-de-Leon, 2020; Stone et al., 2020; Stracquadanio et al., 2021; Tada et al., 2019; Tada et al., 2013; Tahmasebi et al., 2020a; Tahmasebi et al., 2020b; Tahmasebi et al., 2020c; Takahashi et al., 2021; Taleb et al., 2023; Talebi and Hakemi-Vala, 2019; Tan and Ng, 2006; Tantisiriwat et al., 2022; Tarashi et al., 2016; Taylor et al., 2021; Tchakal-Mesbahi et al., 2021; Tekin et al., 2013; Tenover et al., 2022; Thabet et al., 2022; Thapa et al., 2023; Thelen et al., 2022; Tiengrim et al., 2017; Tohamy et al., 2018; Tsitsopoulos et al., 2016; Tumbarello et al., 2013; Tumbarello et al., 2011; Tuon et al., 2020; Khudair and Mahmood, 2021; Ullah et al., 2019; Ullah et al., 2023; Uskudar Guclu et al., 2021; Uzun et al., 2014; Valenza et al., 2010; Valenza et al., 2008; Valenza et al., 2015; Vamsi et al., 2023; Van An et al., 2023; Van An et al., 2023; van Burgh et al., 2019; van der Heijden et al., 2007; Vasudeva et al., 2016; Vata et al., 2018; Vata et al., 2018; Veeraraghavan et al., 2018; Viasus et al., 2020; Viedma et al., 2012; Vosahlikova et al., 2007; Wadhwa et al., 2016; Walkty et al., 2012; Walkty et al., 2011; Walkty et al., 2009; Walkty et al., 2013; Walkty et al., 2022; Walkty et al., 2017; Walters et al., 2019; Wang et al., 2020; Wang and Wang, 2020; Wattal et al., 2019; Wattal et al., 2014; Wemambu and Joshi, 1983; Wendel et al., 2022; Wi et al., 2017; Wi et al., 2018; Willmann et al., 2013; Wise et al., 2023a; Wise et al., 2023b; Wu et al., 2022; Xi et al., 2022; Yadav et al., 2020; Yayan et al., 2015; Yilmaz et al., 2023; Yilmaz et al., 2017; Yousefi et al., 2013; Zerouali et al., 2016; Zhanel et al., 2019; Zhanel et al., 2013; Zhanel et al., 2011; Zhanel et al., 2010; Zhang et al., 2021; Zhao et al., 2023; Zhu et al., 2021; Zhu et al., 2023; Zorgani et al., 2015; Zubair and Iregbu, 2018; Farzana et al., 2013).
A total of 262 articles investigated colistin resistance in P. aeruginosa using the microbroth dilution method, while 242 articles employed methods other than microbroth dilution. Additionally, 115 articles examined colistin resistance in multidrug-resistant (MDR), extensively drug-resistant (XDR), and carbapenem-resistant (CR) bacteria.
We followed the PRISMA guidelines and presented the article selection process in a flow diagram (Figure 1). Supplementary Table S1 provides a summary of the characteristics and quality assessment of all included studies. Additionally, references to the included studies can be found in Supplementary File 2.
Figure 1.
The study PRISMA flow diagram.
Meta-analysis results
For our meta-analysis, we focused on studies that utilized standard methods such as the broth microdilution and disk elution, which is recommended by the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines for evaluating resistance rates. Out of the total articles reviewed, 262 articles investigated colistin resistance in P. aeruginosa isolates using the microbroth dilution method. The pooled prevalence of colistin resistance among clinical P. aeruginosa isolates was estimated to be 1% (95% CI: 1–2%; I2 = 97.47%; p < 0.001).
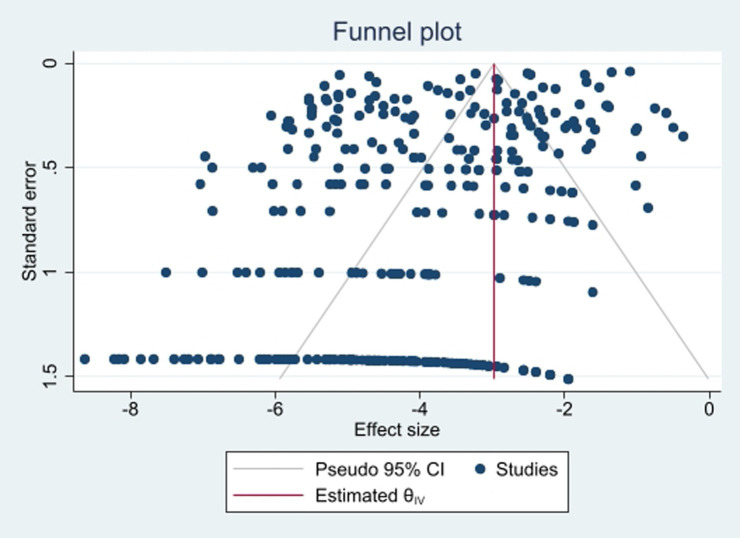
To assess publication bias, we examined a funnel plot (Figure 2) and conducted Egger’s tests, which yielded a p-value of 0.053, indicating no evidence of publication bias. Additionally, the results following the Trim-and-Fill adjustment indicated that the prevalence of colistin resistance remained unchanged.
Figure 2.
Funnel plot of the prevalence of colistin-resistant P. aeruginosa isolates.
Subgroup meta-analysis
Subgroup meta-analyses were conducted based on various factors, including the year of publication, period of sample collection, continent, country, guideline used, disease assessed, origin of samples, different resistance categories, and methods.
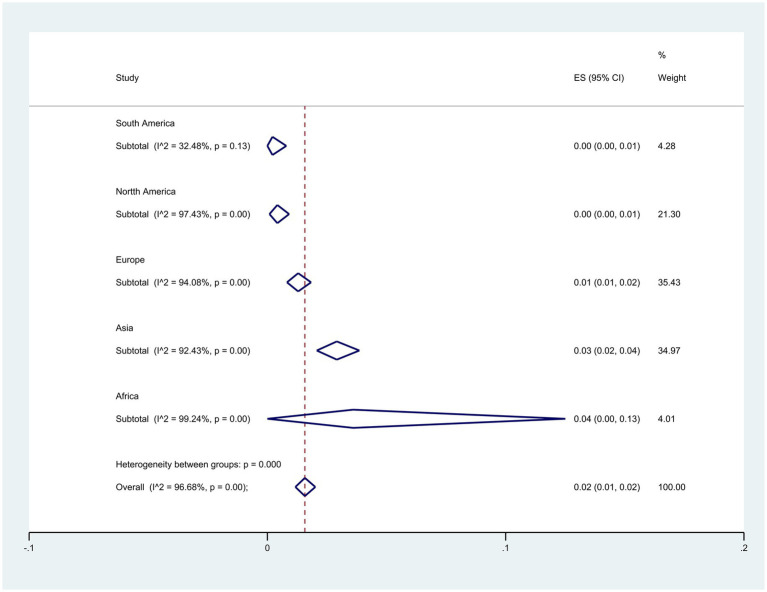
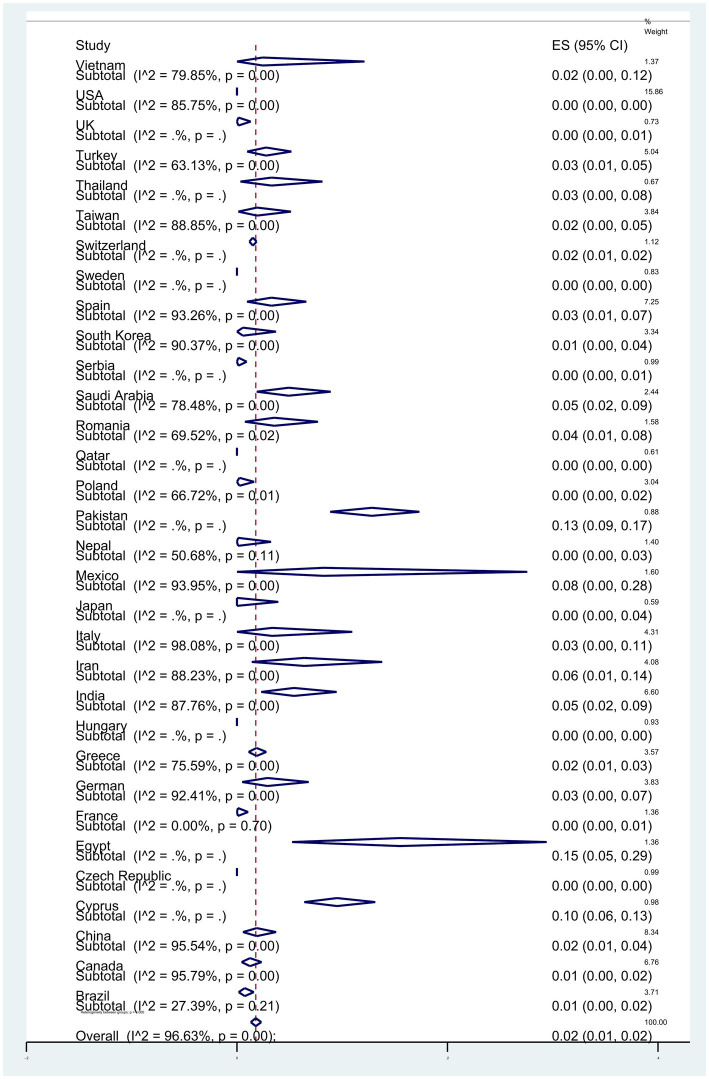
Subgroup meta-analyses based on continents indicated that Africa exhibited the highest resistance rate at 4% (95% CI: 0–13%) (p < 0.001) (Figure 3). The studies encompassed 32 countries, with Egypt 15% (95% CI: 5–29%), and Pakistan 13% (95% CI: 9–17%) had the highest resistance (p < 0.001) (Figure 4).
Figure 3.
Subgroup analysis for continents of colistin-resistant P. aeruginosa isolates.
Figure 4.
Subgroup analysis for countries of the colistin-resistant P. aeruginosa isolates.
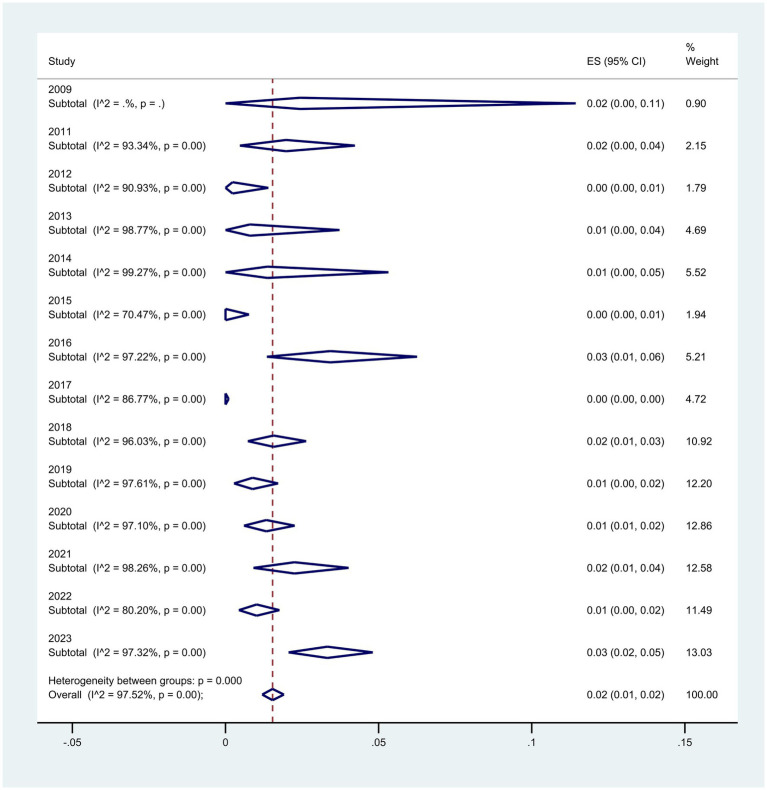
Regarding the subgroup meta-analyses based on the year of article publication, the rate of P. aeruginosa resistance to colistin has increased from 2% (95% CI: 0–11%) in 2009 to 3% (95% CI: 2–5%) in 2023, representing a 1% increase (p < 0.001) (Figure 5).
Figure 5.
Subgroup analysis for year of publication of the colistin-resistant P. aeruginosa isolates.
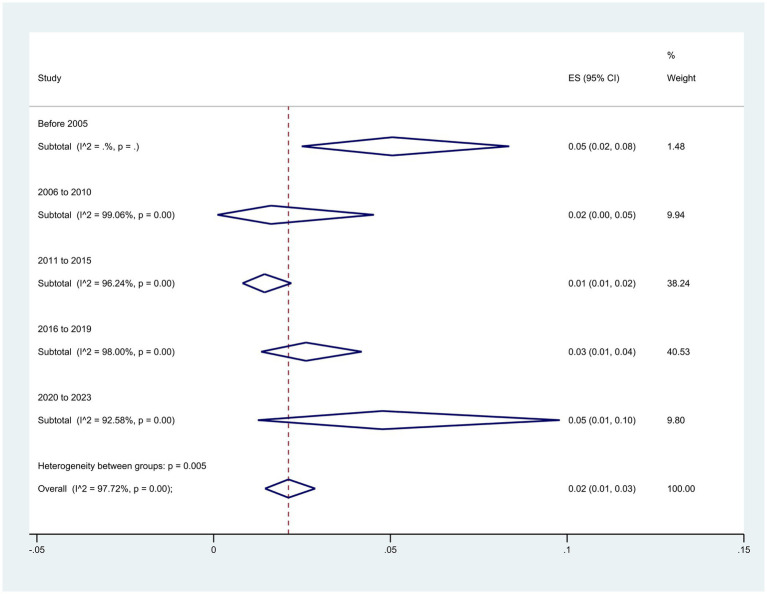
When the sample collection time was divided into five periods, the subgroup meta-analyses revealed an increase in resistance over time from 2% (95% CI: 0–5%) to 5% (95% CI: 0–10%), from 2010–2006 to 2020–2023 (p = 0.005) (Figure 6).
Figure 6.
Subgroup analysis for period of sample collection of the colistin-resistant P. aeruginosa isolates.
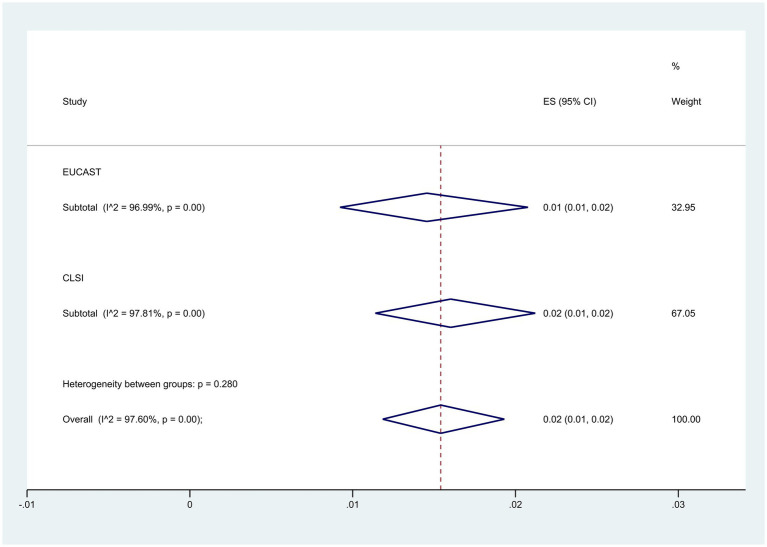
Subgroup analysis based on guidelines demonstrated that the CLSI group had a higher resistance level at 2% (95% CI: 1–2%) compared to 1% (95% CI: 1–2%) in the EUCAST group (p = 0.280) (Figure 7).
Figure 7.
Subgroup meta-analysis for guidelines of colistin-resistant P. aeruginosa isolates.
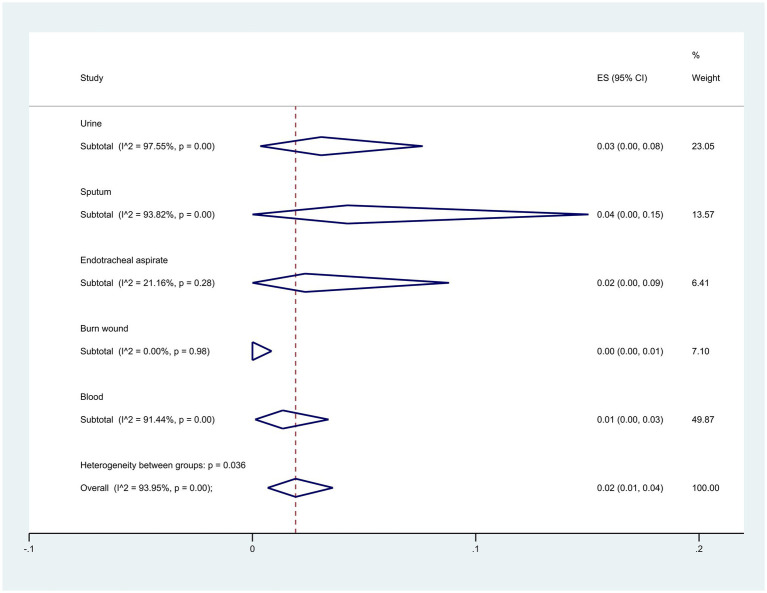
Regarding the origin of samples, sputum samples exhibited the highest resistance at 4% (95% CI: 0–15%), whereas burn wound samples showed the lowest at 0% (95% CI: 0–1%) (p = 0.036) (Figure 8).
Figure 8.
Subgroup analysis for origin of samples of colistin-resistant P. aeruginosa isolates.
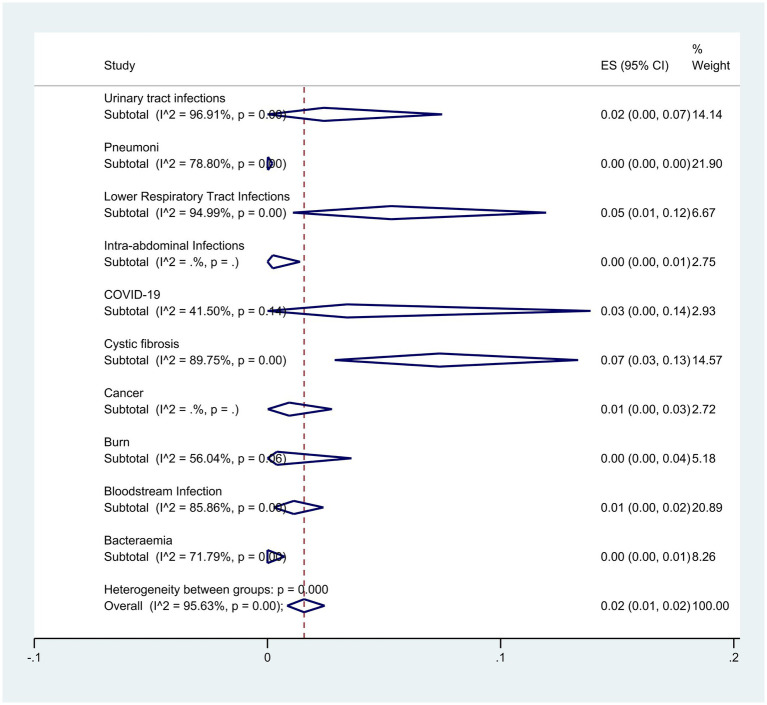
Subgroup analysis based on disease type showed that patients with cystic fibrosis and lower respiratory infection had the highest resistance with rates of 7% (95% CI: 13–3%) and 5% (95% CI: 1–12%) respectively (p < 0.001) (Figure 9).
Figure 9.
Subgroup analysis for infection of colistin-resistant P. aeruginosa isolates.
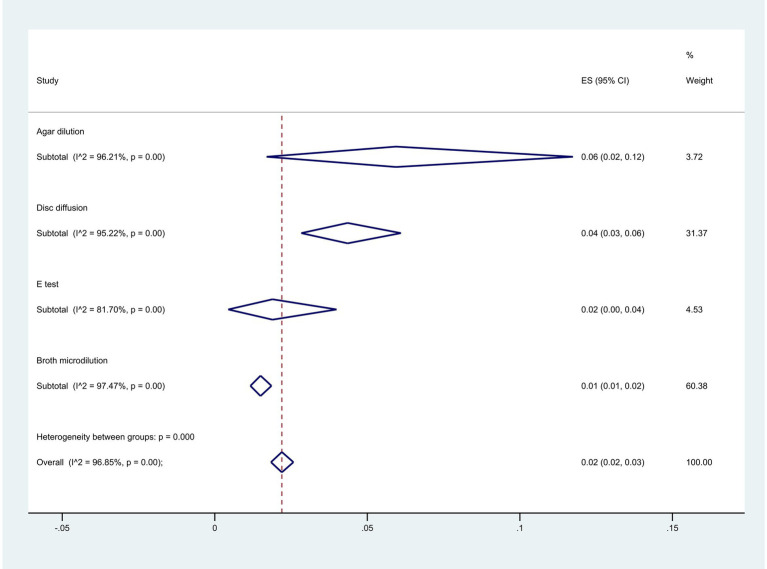
Among the methodologies used, 22 articles employed the agar dilution method, 28 used the E-test method, 192 utilized the disk diffusion method, and 262 opted for the broth microdilution method. The analysis indicated the highest resistance level with the agar dilution method at 6% (95% CI: 2–12%), while the broth microdilution method showed the lowest at 1% (95% CI: 1–2%) (p < 0.001) (Figure 10).
Figure 10.
Subgroup meta-analysis for AST methods of colistin-resistant P. aeruginosa isolates.
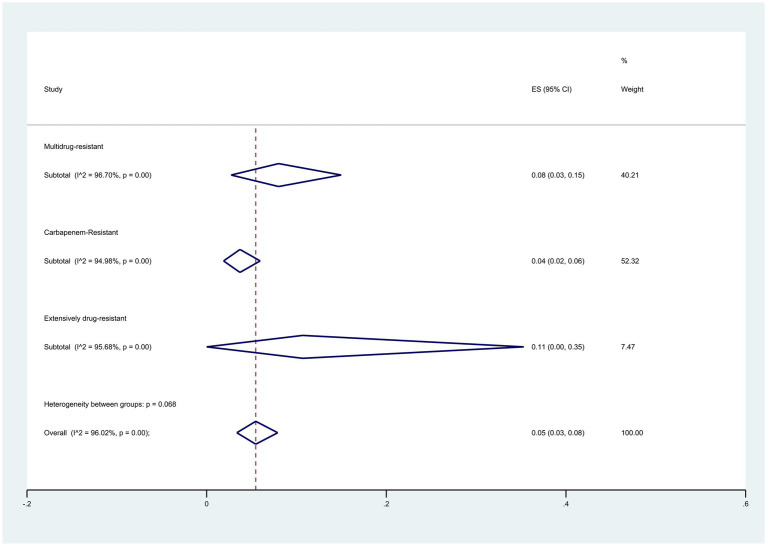
When exploring colistin resistance in isolates with different resistance categories, findings based on the broth microdilution and disk elution method revealed that Extensively Drug-Resistant (XDR) isolates displayed a resistance rate of 11% (95% CI: 0–35%), Multidrug-resistant (MDR) isolates displayed a resistance rate of 8% (95% CI: 3–5%), and Carbapenem-Resistant (CR) isolates displayed a resistance rate of 4% (95% CI: 2–6%) (p = 0.068) (Figure 11).
Figure 11.
Subgroup meta-analysis for different resistance categories of colistin-resistant P. aeruginosa isolates.
Discussion
Antimicrobial resistance poses a significant threat to public health, leading to increased treatment expenses, prolonged hospital stays, and higher mortality rates. Currently, the rise in antibiotic resistance is particularly concerning in Enterobacteriaceae family members and the hospital bacterium P. aeruginosa (Talebi Bezmin Abadi et al., 2019). Additionally, the inappropriate and excessive use of antibiotics in medical and veterinary settings has contributed to the emergence of resistant strains.
P. aeruginosa exhibits various intrinsic and acquired antimicrobial resistance mechanisms, including AmpC cephalosporinases, diverse carbapenemases, and multidrug efflux pumps, resulting in resistance to a wide range of antimicrobial agents (Pang et al., 2019). The emergence and dissemination of MDR and XDR strains of P. aeruginosa, coupled with the limited availability of effective antimicrobial agents against these bacteria, have severely restricted treatment options (del Barrio-Tofiño et al., 2020; Willmann et al., 2015). Numerous studies have indicated that P. aeruginosa is resistant to most beta-lactam antibiotics, quinolones, and aminoglycosides (Pachori et al., 2019). Although carbapenems have been considered one of the primary treatment choices for P. aeruginosa infections, increasing resistance to this antibiotic has imposed limitations on its use (Pang et al., 2019; Balkhair et al., 2019). Despite colistin and tigecycline being commonly viewed as the only available antimicrobial agents for treating XDR P. aeruginosa infections, some strains have developed resistance to these last-line treatment options (Pang et al., 2019; Ibrahim et al., 2021; Cai et al., 2012). The widespread use of colistin has created conditions conducive to the emergence of resistant strains (Bialvaei and Samadi, 2015). The objective of this meta-analysis was to investigate the global prevalence of colistin resistance in P. aeruginosa isolates.
Our findings revealed that the estimated overall prevalence of colistin resistance in clinical isolates of P. aeruginosa was 1%. Several other meta-analyses have examined colistin resistance in different Gram-negative bacteria, finding rates of 6.9% in Iran for Klebsiella pneumoniae (Narimisa et al., 2022) and 4% in Acinetobacter baumannii (Bostanghadiri et al., 2024). While the level of colistin resistance in P. aeruginosa has been lower than in other Gram-negative bacteria, our analysis indicates that this resistance has been increasing in recent years. The recent rise in colistin resistance among P. aeruginosa can be attributed to multiple factors, particularly the overuse and misuse of antibiotics in both clinical and agricultural contexts, which have intensified selective pressure on bacterial populations (Hassen et al., 2022). Colistin, recognized as a last-resort antibiotic for multidrug-resistant infections, has seen increased utilization due to the emergence of resistant pathogens (Sharma et al., 2022), especially during the COVID-19 pandemic. As healthcare systems faced a surge in respiratory infections, colistin was often employed as a last-resort treatment, further heightening selective pressure on bacteria (de Blasio, 2021). This surge in usage, frequently driven by empirical treatment strategies amid uncertainty, has facilitated the emergence and spread of resistant strains (Ghosh et al., 2021). Additionally, the pandemic disrupted routine healthcare practices and antibiotic stewardship programs, creating an environment conducive to the development of resistance (Campbell et al., 2023). The implications of this growing resistance are significant, complicating treatment options for infections caused by multidrug-resistant organisms and presenting considerable challenges for public health.
Our analysis revealed that the highest rates of colistin resistance were observed in Egypt (15%) and Pakistan (13%). The implementation of effective management strategies is essential for the appropriate use of this antibiotic in these regions. This issue may be attributed to the lack of adequate diagnostic tools, as patient management often relies heavily on drug prescriptions, particularly antibiotics, in developing countries. Additionally, the availability of substandard antibiotics sold over the counter further exacerbates the problem, contributing to the rising rates of antimicrobial resistance in these areas (Dadgostar, 2019; Chaw et al., 2018; Chokshi et al., 2019).
Having a comprehensive standard protocol for determining antibiotic sensitivity is critical for several antibiotics. The CLSI recommends utilizing broth microdilution, colistin broth disc elution (CBDE), and the colistin agar test (CAT) for antimicrobial susceptibility testing against colistin. Additionally, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) also advocates for broth microdilution as the preferred method for evaluating susceptibility to colistin (Pancholi et al., 2018; Özhak et al., 2019). The disk diffusion method, a widely employed and cost-effective approach in clinical microbiology laboratories, particularly in developing countries, lacks a standardized protocol for colistin sensitivity testing (Waites et al., 2011). In a study by Irene Galani and colleagues in Greece, two phenotypic methods, E-test and disk diffusion, were compared for measuring colistin resistance in Gram-negative bacilli. The researchers concluded that the disk diffusion method is not suitable or reliable for assessing antimicrobial sensitivity to colistin (Galani et al., 2008). Despite this, numerous articles have utilized non-endorsed methods, such as disk diffusion, to assess resistance. A subgroup meta-analysis focusing on measurement methods consistently found higher resistance rates when alternative methods were used, compared to the standard method. This discrepancy may be attributed to the lack of sensitivity in other methods for detecting and distinguishing resistant strains. Therefore, adherence to established standard guidelines for measurement methods is imperative.
The findings of our study revealed that P. aeruginosa isolated from respiratory samples, particularly in patients with respiratory infections such as cystic fibrosis, exhibited the highest level of resistance to colistin. In a meta-analysis conducted by Bonyadi et al. (2022), the resistance rate of P. aeruginosa isolates from cystic fibrosis patients to colistin was reported as 5%. However, our study demonstrated a resistance rate of 7%, suggesting a potential increase in resistance over the past two years. Notably, our subgroup meta-analysis focused solely on studies where resistance rates were confirmed by established guidelines, which may account for the variance in resistance percentages among cystic fibrosis patients.
Given the challenges in discovering new antibiotics, optimizing the use of existing treatments is crucial. Colistin is considered the last resort for treating extensively drug-resistant (XDR) Gram-negative bacteria (Ozsurekci et al., 2016). To address the increasing rates of antibiotic resistance, it is vital to implement innovative strategies. For instance, the combination of colistin and other antibiotics has demonstrated a synergistic effect against antibiotic-resistant Gram-negative pathogens, potentially curtailing the development of resistance (Ly et al., 2015). Other approaches may include combination therapies utilizing nanoparticles, natural components, and phage-based strategies (Holger et al., 2022b; Yassin et al., 2022; Wang et al., 2022). Additionally, promoting antibiotic stewardship and preventing the misuse and overprescription of colistin, particularly among physicians in developing countries, is essential for maintaining its effectiveness.
The considerable heterogeneity among the studies represents a primary limitation of this research. Nonetheless, through the use of subgroup analysis, we were able to identify sources of heterogeneity and mitigate its impact on the outcomes. Another limitation of this article is the exclusion of non-English studies, which may contain valuable data. The decision to focus solely on English-language research aimed to ensure accurate comprehension of the studies and maintain consistency in data quality and reporting. However, we recognize that this exclusion may compromise the comprehensiveness of our analysis. We encourage future research to incorporate studies in other languages to provide a more comprehensive view of the topic.
Conclusion
Our study indicates that while the overall rate of resistance to colistin in P. aeruginosa is relatively low, there has been a recent upward trend in resistance levels. This underscores the importance of accurate surveillance of resistance rates, particularly in regions with higher prevalence, and the judicious prescription of antibiotics for patients with P. aeruginosa infection. Promoting antibiotic stewardship and preventing the misuse and overprescription of colistin, especially among healthcare professionals in developing countries, is crucial for preserving its efficacy.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by Microbial Biotechnology Research Center (Iran University of Medical Sciences) by a research grant (No. 18205).
Abbreviations
P. aeruginosa, Pseudomonas aeruginosa; PRISMA, Preferred reporting items for systematic reviews and meta-analyses guidelines; CI, Confidence interval; CLSI, The Clinical and Laboratory Standards Institute; EUCAST, European Committee on Antimicrobial Susceptibility Testing.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
NN: Data curation, Formal analysis, Software, Writing – original draft, Writing – review & editing. AK: Software, Validation, Writing – review & editing. LD-Z: Data curation, Methodology, Writing – review & editing. NB: Data curation, Methodology, Writing – original draft, Writing – review & editing. YF: Data curation, Methodology, Writing – review & editing. SS: Data curation, Methodology, Writing – review & editing. AZ: Supervision, Validation, Writing – review & editing. SR: Conceptualization, Supervision, Validation, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1477836/full#supplementary-material
References
- Abavisani M., Goudarzi M., Ghalavand Z., Hajikhani B., Rad Z. R., Rad Z. R., et al. (2021). Evaluation of efflux pumps overexpression and β-lactamase genes among colistin resistant Pseudomonas aeruginosa. Gene Rep. 24:101301. doi: 10.1016/j.genrep.2021.101301 [DOI] [Google Scholar]
- Abd El-Baky R. M., Masoud S. M., Mohamed D. S., Waly N. G., Shafik E. A., Mohareb D. A., et al. (2020). Prevalence and some possible mechanisms of colistin resistance among multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa. Infect. Drug Resist. 13, 323–332. doi: 10.2147/IDR.S238811, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelatti M. A., Abd El-Aziz N. K., El-Naenaeey E.-s. Y., Ammar A. M., Alharbi N. K., Alharthi A., et al. (2023). Antibacterial and anti-efflux activities of cinnamon essential oil against pan and extensive drug-resistant Pseudomonas aeruginosa isolated from human and animal sources. Antibiotics 12:514. doi: 10.3390/antibiotics12101514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abed W. H., Kareem S. M. (2021). Molecular detection of gyrA and mexA genes in Pseudomonas aeruginosa. Mol. Biol. Rep. 48, 7907–7912. doi: 10.1007/s11033-021-06820-0, PMID: [DOI] [PubMed] [Google Scholar]
- Abubakar U., Zulkarnain A. I., Rodríguez-Baño J., Kamarudin N., Elrggal M. E., Elnaem M. H., et al. (2022). Treatments and predictors of mortality for carbapenem-resistant gram-negative bacilli infections in Malaysia: a retrospective cohort study. Trop. Med. Infect. Dis. 7:415. doi: 10.3390/tropicalmed7120415, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abulzahra A. N., Ismail M. I. (2020). Antimicrobial resistance patterns and phenotypic-βlactamases detection of MDR Pseudomonas aeruginosa isolates from wounds and burns of Iraqi patients. Biochem. Cell. Arch. 20. [Google Scholar]
- Addis T., Araya S., Desta K. (2021). Occurrence of multiple, extensive and Pan drug-resistant Pseudomonas aeruginosa and Carbapenemase production from presumptive isolates stored in a biobank at Ethiopian Public Health Institute. Infect. Drug Resist. 14, 3609–3618. doi: 10.2147/IDR.S327652, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A., Kumar D., Goyal A., Goyal S., Singh N., Khandelwal G. (2013). Microbiological profile and their antimicrobial sensitivity pattern in patients of otitis media with ear discharge. Indian J. Otol. 19, 5–8. doi: 10.4103/0971-7749.108149 [DOI] [Google Scholar]
- Aguilar-Rodea P., Zuniga G., Rodríguez-Espino B. A., Olivares Cervantes A. L., Gamiño Arroyo A. E., Moreno-Espinosa S., et al. (2017). Identification of extensive drug resistant Pseudomonas aeruginosa strains: new clone ST1725 and high-risk clone ST233. PLoS One 12:e0172882. doi: 10.1371/journal.pone.0172882, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahani Azari A., Fozouni L. (2020). Incidence of multidrug-resistant, extensively drug-resistant, and Pandrug-resistant Pseudomonas aeruginosa strains isolated from clinical specimens. Infect. Epidemiol. Microbiol. 6, 211–217. doi: 10.29252/iem.6.3.211 [DOI] [Google Scholar]
- Ahmed M. A. S., Hadi H. A., Jarir S. A., Khan F. A., Arbab M. A., Hamid J. M., et al. (2022). Prevalence and microbiological and genetic characteristics of multidrug-resistant Pseudomonas aeruginosa over three years in Qatar. Antimicrobial Stewardship Healthcare Epidemiol. 2:e96. doi: 10.1017/ash.2022.226, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M. S., Hassan A., Jarir S. A., Hadi H. A., Bansal D., Wahab A. A., et al. (2019). Emergence of multidrug-and pandrug-resistant Pseudomonas aeruginosa from five hospitals in Qatar. Infect. Prevent. Pract. 1:100027. doi: 10.1016/j.infpip.2019.100027, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyegoro O., Igbinosa O., Ogunmwonyi I., Odjadjare E., Igbinosa O., Okoh A. (2007). Incidence of urinary tract infections (UTI) among children and adolescents in Ile-Ife, Nigeria. Afr. J. Microbiol. Res. 1, 13–19. [Google Scholar]
- Akgül Ö., Arvas G., KuŞTan A. (2021). Phenotypic and genotypic analysis of the antibiotic resistance profiles of gram negative Bacteria isolated from the blood culture samples. Eastern J. Med. 26, 299–307. doi: 10.5505/ejm.2021.03880 [DOI] [Google Scholar]
- Akhi M. T., Ghotaslou R., Beheshtirouy S., Asgharzadeh M., Pirzadeh T., Asghari B., et al. (2015). Antibiotic susceptibility pattern of aerobic and anaerobic Bacteria isolated from surgical site infection of hospitalized patients. Jundishapur J. Microbiol. 8:e20309. doi: 10.5812/jjm.20309v2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akram W., Raza A., Munir S., Waqas K., Muddassir M., Islam M., et al. (2022). Incidence, characterization and anti-microbial susceptibility pattern of Pseudomonas aeruginosa isolated from clinical subjects. Bacteriology 13, 962–965. [Google Scholar]
- Al Dawodeyah H. Y., Obeidat N., Abu-Qatouseh L. F., Shehabi A. A. (2018). Antimicrobial resistance and putative virulence genes of Pseudomonas aeruginosa isolates from patients with respiratory tract infection. Germs 8, 31–40. doi: 10.18683/germs.2018.1130, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Agamy M. H., Shibl A. M., Tawfik A. F., Elkhizzi N. A., Livermore D. M. (2012). Extended-spectrum and metallo-beta-lactamases among ceftazidime-resistant Pseudomonas aeruginosa in Riyadh, Saudi Arabia. J. Chemother. 24, 97–100. doi: 10.1179/1120009X12Z.00000000015 [DOI] [PubMed] [Google Scholar]
- Alam M. M., Islam M. N., Hossain Hawlader M. D., Ahmed S., Wahab A., Islam M., et al. (2021). Prevalence of multidrug resistance bacterial isolates from infected wound patients in Dhaka, Bangladesh: a cross-sectional study. Int. J. Surg. Open 28, 56–62. doi: 10.1016/j.ijso.2020.12.010 [DOI] [Google Scholar]
- Al-Bayssari C., Dagher T. N., El Hamoui S., Fenianos F., Makdissy N., Rolain J.-M., et al. (2021). Carbapenem and colistin-resistant bacteria in North Lebanon: coexistence of mcr-1 and NDM-4 genes in Escherichia coli. J. Infect. Dev. Countries 15, 934–342. doi: 10.3855/jidc.14176, PMID: [DOI] [PubMed] [Google Scholar]
- Alcántar-Curiel M. D., Huerta-Cedeño M., Jarillo-Quijada M. D., Gayosso-Vázquez C., Fernández-Vázquez J. L., Hernández-Medel M. L., et al. (2023). Gram-negative ESKAPE bacteria bloodstream infections in patients during the COVID-19 pandemic. PeerJ 11:e15007. doi: 10.7717/peerj.15007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou A., Vasilieva L., Agiasotelli D., Siranidi K., Pouriki S., Tsiriga A., et al. (2016). Extensively drug-resistant bacteria are an independent predictive factor of mortality in 130 patients with spontaneous bacterial peritonitis or spontaneous bacteremia. World J. Gastroenterol. 22, 4049–4056. doi: 10.3748/wjg.v22.i15.4049, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfouzan W., Dhar R., Mohsin J., Khamis F., Mokaddas E., Abdullah A., et al. (2022). Evaluation of in vitro activity of ceftolozane/tazobactam and comparators against recent clinical bacterial isolates, and genomics of Pseudomonas aeruginosa, Klebsiella pneumoniae and Escherichia coli isolates that demonstrated resistance to ceftolozane/tazobactam: data from Kuwait and Oman. JAC-Antimicrob. Resist. 4:35. doi: 10.1093/jacamr/dlac035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfouzan W., Dhar R., Nicolau D. P. (2018). In vitro activity of newer and conventional antimicrobial agents, including Fosfomycin and Colistin, against selected gram-negative Bacilli in Kuwait. Pathogens 7:75. doi: 10.3390/pathogens7030075, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhanout K., Brunel J.-M., Raoult D., Rolain J.-M. (2009). In vitro antibacterial activity of aminosterols against multidrug-resistant bacteria from patients with cystic fibrosis. J. Antimicrob. Chemother. 64, 810–814. doi: 10.1093/jac/dkp281, PMID: [DOI] [PubMed] [Google Scholar]
- Ali A., Ahmad K., Rahat S. (2021). Diversity of extended Spectrum β-lactamases among multi drug resistant clinical isolates of Pseudomonas aeruginosa collected fromTertiary care hospitals of Peshawar, Pakistan. Pakistan J. Zool. 53:705. doi: 10.17582/journal.pjz/20190712160705 [DOI] [Google Scholar]
- Ali Z., Mumtaz N., Naz S. A., Jabeen N., Shafique M. (2015). Multi-drug resistant pseudomonas aeruginosa: a threat of nosocomial infections in tertiary care hospitals. J. Pak. Med. Assoc. 65, 12–16, PMID: [PubMed] [Google Scholar]
- Al-Kabsi A. M., Yusof M., Sekaran S. D. (2011). Antimicrobial resistance pattern of clinical isolates of Pseudomonas aeruginosa in the University of Malaya Medical Center, Malaysia. Afr. J. Microbiol. Res. 5, 5266–5272. doi: 10.5897/AJMR11.284 [DOI] [Google Scholar]
- Al-Kadmy I. M., Ibrahim S. A., Al-Saryi N., Aziz S. N., Besinis A., Hetta H. F. (2020). Prevalence of genes involved in colistin resistance in Acinetobacter baumannii: first report from Iraq. Microb. Drug Resist. 26, 616–622. doi: 10.1089/mdr.2019.0243, PMID: [DOI] [PubMed] [Google Scholar]
- Al-kaffas M., Haggag M. G., Soliman S. M., Ghalwash A. A., Alkaffas M. (2022). Genetic identification of Pseudomonas aeruginosa virulence genes associated with keratitis in Egyptian population. J. Pure Appl. Microbiol. 16, 1714–1721. doi: 10.22207/JPAM.16.3.12 [DOI] [Google Scholar]
- Al-Khudhairy M. K., Al-Shammari M. M. M. (2020). Prevalence of metallo-beta-lactamase-producing Pseudomonas aeruginosa isolated from diabetic foot infections in Iraq. New Microbes New Infect. 35:100661. doi: 10.1016/j.nmni.2020.100661, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhulaifi Z. M., Mohammed K. A. (2023). Prevalence and molecular analysis of antibiotic resistance of Pseudomonas aeruginosa isolated from clinical and environmental specimens in Basra, Iraq. Iranian J. Microbiol. 15, 45–54. doi: 10.18502/ijm.v15i1.11917, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Orphaly M., Hadi H. A., Eltayeb F. K., Al-Hail H., Samuel B. G., Sultan A. A., et al. (2021). Epidemiology of multidrug-resistant Pseudomonas aeruginosa in the Middle East and North Africa region. Msphere 6, e00202–e00221. doi: 10.1128/mSphere.00202-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alotaibi B. S., Tantry B. A., Farhana A., Alammar M. A., Shah N. N., Mohammed A. H., et al. (2023). Resistance pattern in mostly gram-negative Bacteria causing urinary tract infections. Infect. Disord. Drug Targets 23:e280922209238. doi: 10.2174/1871526522666220928115043, PMID: [DOI] [PubMed] [Google Scholar]
- Alruways M. W. (2023). Phenotypic and molecular characterization of Carbapenemase-producing gram-negative isolates detected from wound infections. Medical Forum Monthly. [Google Scholar]
- Al-shimmary S. (2018). Improvement rapid molecular detection of Pseudomonas aeruginosa infected some Iraqi patients and It's antimicrobial susceptibility. Res. J. Pharm., Biol. Chem. Sci. 7, 1256–1264. [Google Scholar]
- Al-Zahrani I. A., Al-Ahmadi B. M. (2021). Dissemination of VIM-producing Pseudomonas aeruginosa associated with high-risk clone ST654 in a tertiary and quaternary hospital in Makkah, Saudi Arabia. J. Chemother. 33, 12–20. doi: 10.1080/1120009X.2020.1785741, PMID: [DOI] [PubMed] [Google Scholar]
- Amabile-Cuevas C. F. (2017). Selection of amikacin hyper-resistant Pseudomonas aeruginosa after stepwise exposure to high amikacin concentrations. Microb. Drug Resist. 23, 32–36. doi: 10.1089/mdr.2015.0218, PMID: [DOI] [PubMed] [Google Scholar]
- Appalaraju B., Baveja S., Baliga S., Shenoy S., Bhardwaj R., Kongre V., et al. (2020). In vitro activity of a novel antibacterial agent, levonadifloxacin, against clinical isolates collected in a prospective, multicentre surveillance study in India during 2016-18. J. Antimicrob. Chemother. 75, 600–608. doi: 10.1093/jac/dkz493 [DOI] [PubMed] [Google Scholar]
- Aprile A., Caio C., Gona F., Stefani S., Mezzatesta M. L. (2019). In vitro evidence of the synergistic interaction of ceftopibrole and other antibiotics against multidrug-resistant gram-negative isolates. Diagn. Microbiol. Infect. Dis. 95:114884. doi: 10.1016/j.diagmicrobio.2019.114884, PMID: [DOI] [PubMed] [Google Scholar]
- Arab O., Al-Kayali R., Khouri A., Haj K. S. (2023). Resistance patterns of bacterial pathogens causing lower respiratory tract infections: Aleppo-Syria. Ann. Med. Surg. 85, 2655–2661. doi: 10.1097/MS9.0000000000000778, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo Lima A. V., da Silva S. M., do Nascimento Júnior J. A. A., Correia M. T. S., Luz A. C., Leal-Balbino T. C., et al. (2020). Occurrence and diversity of intra-and Interhospital drug-resistant and biofilm-forming Acinetobacter baumannii and Pseudomonas aeruginosa. Microb. Drug Resist. 26, 802–814. doi: 10.1089/mdr.2019.0214, PMID: [DOI] [PubMed] [Google Scholar]
- Arca-Suárez J., Lasarte-Monterrubio C., Rodiño-Janeiro B.-K., Cabot G., Vázquez-Ucha J. C., Rodríguez-Iglesias M., et al. (2021). Molecular mechanisms driving the in vivo development of OXA-10-mediated resistance to ceftolozane/tazobactam and ceftazidime/avibactam during treatment of XDR Pseudomonas aeruginosa infections. J. Antimicrob. Chemother. 76, 91–100. doi: 10.1093/jac/dkaa396, PMID: [DOI] [PubMed] [Google Scholar]
- Arcilla M. S., van Hattem J. M., Matamoros S., Melles D. C., Penders J., de Jong M. D., et al. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 147–149. doi: 10.1016/S1473-3099(15)00541-1 [DOI] [PubMed] [Google Scholar]
- Arici N., Kansak N., Adaleti R., Aksaray S. (2023). Comparison of broth microdilution and Colistin disk elution methods for the determination of Colistin susceptibility in multidrug resistant Pseudomonas aeruginosa isolates. Mediterr J Infect Microb Antimicrob:2023. doi: 10.4274/mjima.galenos.2023.2023.14 [DOI] [Google Scholar]
- Arif A., Ullah I., Ullah O., Zaman R. (2022). Identification of colistin resistance and its bactericidal activity against uropathogenic gram negative bacteria from Hayatabad medical complex Peshawar. Pakistan J. Med. Sci. 38:5221. doi: 10.12669/pjms.38.4.5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol E., Asuncion T., Vinas M., Sierra J. M. (2020). When combined with Colistin, an otherwise ineffective rifampicin-linezolid combination becomes Active in Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii. Microorganisms 8:86. doi: 10.3390/microorganisms8010086, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengol E., Domenech O., Fuste E., Perez-Guillen I., Borrell J. H., Sierra J. M., et al. (2019). Efficacy of combinations of colistin with other antimicrobials involves membrane fluidity and efflux machinery. Infect Drug Resist. 12, 2031–2038. doi: 10.2147/IDR.S207844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruhomukama D., Najjuka C. F., Kajumbula H., Okee M., Mboowa G., Sserwadda I., et al. (2019). Bla VIM-and Bla OXA-mediated carbapenem resistance among Acinetobacter baumannii and Pseudomonas aeruginosa isolates from the Mulago hospital intensive care unit in Kampala, Uganda. BMC Infect. Dis. 19, 1–8. doi: 10.1186/s12879-019-4510-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asar L., Pfefferle S., Lutgehetmann M., Hoffmann A., Katchanov J., Aepfelbacher M., et al. (2019). Influence of local epidemiology on the performance of common colistin drug susceptibility testing methods. PLoS One 14:e0217468. doi: 10.1371/journal.pone.0217468, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydemir O., Aydemir Y., Şahin E. Ö., Şahin F., Koroglu M., Erdem A. F. (2022). Secondary bacterial infections in patients with coronavirus disease 2019-associated pneumonia. Rev. Assoc. Med. Bras. 68, 142–146. doi: 10.1590/1806-9282.20210745, PMID: [DOI] [PubMed] [Google Scholar]
- Aydın M., Ergönül Ö., Azap A., Bilgin H., Aydın G., Çavuş S., et al. (2018). Rapid emergence of colistin resistance and its impact on fatality among healthcare-associated infections. J. Hosp. Infect. 98, 260–263. doi: 10.1016/j.jhin.2017.11.014, PMID: [DOI] [PubMed] [Google Scholar]
- Azimi S., Kafil H. S., Baghi H. B., Shokrian S., Najaf K., Asgharzadeh M., et al. (2016). Presence of exoY, exoS, exoU and exoT genes, antibiotic resistance and biofilm production among Pseudomonas aeruginosa isolates in Northwest Iran. GMS Hyg Infect. Control. 11:Doc04. doi: 10.3205/dgkh000264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi L., Lari A. R., Alaghehbandan R., Alinejad F., Mohammadpoor M., Rahbar M. (2012). KPC-producer gram negative bacteria among burned infants in Motahari hospital, Tehran: first report from Iran. Ann. Burns Fire Disasters 25, 74–77, PMID: [PMC free article] [PubMed] [Google Scholar]
- Azimi L., Namvar A. E., Lari A. R., Jamali S., Lari A. R. (2016). Comparison of efflux pump involvement in antibiotic resistance among Pseudomonas aeruginosa isolates of burn and non-burn patients. Arch. Pediatric Infect. Dis. 4:e36160. doi: 10.5812/pedinfect.36160 [DOI] [Google Scholar]
- Babu M., Menon V. P. (2018). Prevalence of antimicrobial resistant pathogens in severe sepsis and septic shock patients. J. Young Pharm. 10, 358–361. doi: 10.5530/jyp.2018.10.79 [DOI] [Google Scholar]
- Badawy M. S. E., Elkhatib W. F., Shebl R. I. (2023). Mathematical pharmacodynamic modeling for antimicrobial assessment of ceftazidime/colistin versus gentamicin/meropenem combinations against carbapenem-resistant Pseudomonas aeruginosa biofilm. Ann. Clin. Microbiol. Antimicrob. 22:53. doi: 10.1186/s12941-023-00597-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badierah R. A., Natto Z. S., Nassar M. S., Al-Ghamdi A. A., Jiman-Fatani A. A., Bakhrebah M. A. (2019). The prevalence of antibiotic-resistant bacterial nasal carriage at a Saudi university hospital. Acta Microbiologica Hellenica.
- Bae I. G., Stone G. G. (2022). In vitro activity of ceftazidime-avibactam and comparators against bacterial isolates collected in South Korea as part of the ATLAS global surveillance program (2016-2018). Diagn. Microbiol. Infect. Dis. 102:115553. doi: 10.1016/j.diagmicrobio.2021.115553, PMID: [DOI] [PubMed] [Google Scholar]
- Baek M. S., Chung E. S., Jung D. S., Ko K. S. (2020). Effect of colistin-based antibiotic combinations on the eradication of persister cells in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 75, 917–924. doi: 10.1093/jac/dkz552, PMID: [DOI] [PubMed] [Google Scholar]
- Bagheri-Nesami M., Rezai M. S., Ahangarkani F., Rafiei A., Nikkhah A., Eslami G., et al. (2017). Multidrug and co-resistance patterns of non-fermenting gram-negative bacilli involved in ventilator-associated pneumonia carrying class 1 integron in the north of Iran. Germs 7, 123–131. doi: 10.18683/germs.2017.1117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahabri N. M., Al-Alawi M. M., Qutub M. O., Tashkandi W. A., AlTurki R., Janah S. S., et al. (2022). In-vitro activity of ceftolozane/tazobactam against recent clinical bacterial isolates from two Saudi Arabian hospitals. J. Infect. Public Health 15, 486–490. doi: 10.1016/j.jiph.2022.02.009, PMID: [DOI] [PubMed] [Google Scholar]
- Bahador N., Shoja S., Faridi F., Dozandeh-Mobarrez B., Qeshmi F. I., Javadpour S., et al. (2019). Molecular detection of virulence factors and biofilm formation in Pseudomonas aeruginosa obtained from different clinical specimens in Bandar Abbas. Iranian J. Microbiol. 11, 25–30. doi: 10.18502/ijm.v11i1.701, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahçe Y. G., Acer Ö., Özüdoğru O. (2022). Evaluation of bacterial agents isolated from endotracheal aspirate cultures of Covid-19 general intensive care patients and their antibiotic resistance profiles compared to pre-pandemic conditions. Microb. Pathog. 164:105409. doi: 10.1016/j.micpath.2022.105409, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiomy A. A., Serry F. E., Kadry A. A., Yahya G., Doijad S., Mostafa A., et al. (2023). Genome analysis of Pseudomonas aeruginosa strains from chronically infected patients with high levels of persister formation. Pathogens 12:426. doi: 10.3390/pathogens12030426, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakht M., Alizadeh S. A., Rahimi S., Kazemzadeh Anari R., Rostamani M., Javadi A., et al. (2022). Phenotype and genetic determination of resistance to common disinfectants among biofilm-producing and non-producing Pseudomonas aeruginosa strains from clinical specimens in Iran. BMC Microbiol. 22:124. doi: 10.1186/s12866-022-02524-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkhair A., Al Saadi K., Al A. B. (2023). Epidemiology and mortality outcome of carbapenem-and colistin-resistant Klebsiella pneumoniae, Escherichia coli, Acinetobacter baumannii, and Pseudomonas aeruginosa bloodstream infections. IJID Regions 7, 1–5. doi: 10.1016/j.ijregi.2023.01.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkhair A., Al-Muharrmi Z., Al’adawi B., Al Busaidi I., Taher H., Al-Siyabi T., et al. (2019). Prevalence and 30-day all-cause mortality of carbapenem-and colistin-resistant bacteraemia caused by Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae: description of a decade-long trend. Int. J. Infect. Dis. 85, 10–15. doi: 10.1016/j.ijid.2019.05.004, PMID: [DOI] [PubMed] [Google Scholar]
- Bandic-Pavlovic D., Zah-Bogovic T., Zizek M., Bielen L., Bratic V., Hrabac P., et al. (2020). Gram-negative bacteria as causative agents of ventilator-associated pneumonia and their respective resistance mechanisms. J. Chemother. 32, 344–358. doi: 10.1080/1120009X.2020.1793594, PMID: [DOI] [PubMed] [Google Scholar]
- Banerjee T., Adwityama A., Sharma S., Mishra K., Prusti P., Maitra U. (2024). Comparative evaluation of colistin broth disc elution (CBDE) and broth microdilution (BMD) in clinical isolates of Pseudomonas aeruginosa with special reference to heteroresistance. Indian J. Med. Microbiol. 47:100494. doi: 10.1016/j.ijmmb.2023.100494, PMID: [DOI] [PubMed] [Google Scholar]
- Bangera D., Shenoy S. M., Saldanha D. R. (2016). Clinico-microbiological study of Pseudomonas aeruginosa in wound infections and the detection of metallo-beta-lactamase production. Int. Wound J. 13, 1299–1302. doi: 10.1111/iwj.12519, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S., Hadjadj L., Rolain J.-M., Olaitan A. O. (2016). Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int. J. Antimicrob. Agents 48, 583–591. doi: 10.1016/j.ijantimicag.2016.06.023, PMID: [DOI] [PubMed] [Google Scholar]
- Basu S., Mukherjee S., Samanta A. (2013). Epidemiological study of bacterial microbiology in AECOPD patients of Kolkata, India. Asian J. Pharm. Clin. Res. 6, 112–116. [Google Scholar]
- Bayram Y., Parlak M., Aypak C., Bayram İ. (2013). Three-year review of bacteriological profile and antibiogram of burn wound isolates in Van, Turkey. Int. J. Med. Sci. 10, 19–23. doi: 10.7150/ijms.4723, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazgir Z. N., Ahanjan M., Goli H. R., Gholami M., Ghasemian R., Hashemi-Soteh M. B. (2021). Frequency of blaIMP and blaSPM metallo-β-lactamase genes among carbapenem-resistant pseudomonas aeruginosa clinical isolates in Sari, North of Iran. Recent Advances in Anti-Infective Drug Discovery Formerly Recent Patents on Anti-Infective Drug Discovery 16, 148–156. [DOI] [PubMed] [Google Scholar]
- Beirao E. M., Rodrigues S. D. S., Andrade T. K., Serra F. B., Paula M. D. N., Polis T. J. B., et al. (2020). Activity of ceftolozane-tazobactam and comparators against gram-negative bacilli: results from the study for monitoring antimicrobial resistance trends (SMART – Brazil; 2016-2017). Braz. J. Infect. Dis. 24, 310–321. doi: 10.1016/j.bjid.2020.05.010, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Nejma M., Sioud O., Mastouri M. (2018). Quinolone-resistant clinical strains of Pseudomonas aeruginosa isolated from University Hospital in Tunisia. 3 Biotech 8:1019. doi: 10.1007/s13205-017-1019-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwal A., Shobha K. L., Gupta R., Gupta K., Ashraf A. A. (2020). Non-fermenting gram negative Bacteria as Uropathogens in causing urinary tract infection and its antimicrobial susceptibility pattern at a tertiary Care Centre of South India. J. Pure Appl. Microbiol. 14, 2033–2038. doi: 10.22207/JPAM.14.3.43 [DOI] [Google Scholar]
- Bialvaei A. Z., Samadi K. H. (2015). Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 31, 707–721. doi: 10.1185/03007995.2015.1018989 [DOI] [PubMed] [Google Scholar]
- Bian X., Liu X., Hu F., Feng M., Chen Y., Bergen P. J., et al. (2022). Pharmacokinetic/pharmacodynamic based breakpoints of polymyxin B for bloodstream infections caused by multidrug-resistant gram-negative pathogens. Front. Pharmacol. 12:785893. doi: 10.3389/fphar.2021.785893, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Brunel J.-M., Dubus J.-C., Reynaud-Gaubert M., Rolain J.-M. (2012). Colistin: an update on the antibiotic of the 21st century. Expert Rev. Anti-Infect. Ther. 10, 917–934. doi: 10.1586/eri.12.78, PMID: [DOI] [PubMed] [Google Scholar]
- Blondeau J., Charles M. K., Loo V., Adam H., Gonzalez Del Vecchio M., Ghakis C., et al. (2023). A nested cohort 5-year Canadian surveillance of gram-negative antimicrobial resistance for optimized antimicrobial therapy. Sci. Rep. 13:14142. doi: 10.1038/s41598-023-40012-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogiel T., Depka D., Rzepka M., Mikucka A. (2022). Decoding genetic features and antimicrobial susceptibility of Pseudomonas aeruginosa strains isolated from bloodstream infections. Int. J. Mol. Sci. 23:9208. doi: 10.3390/ijms23169208, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono L., Cavoli G. L., Verde M. S., Sodano C., Tortorici C., Ferrantelli A., et al. (2015). Prevalence of bacterial pathogens and their emerging resistance patterns in patients with renal diseases. Diálisis y Trasplante. 36, 78–82. doi: 10.1016/j.dialis.2015.02.006 [DOI] [Google Scholar]
- Bonyadi P., Saleh N. T., Yamini M., Dehghani M., Amini K. (2022). Prevalence of antibiotic resistance of Pseudomonas aeruginosa in cystic fibrosis infection: a systematic review and meta-analysis. Microb. Pathog. 165:105461. doi: 10.1016/j.micpath.2022.105461, PMID: [DOI] [PubMed] [Google Scholar]
- Bostanghadiri N., Narimisa N., Mirshekar M., Dadgar-Zankbar L., Taki E., Navidifar T., et al. (2024). Prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: a systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 13:24. doi: 10.1186/s13756-024-01376-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgi J., Said J. M., Yaakoub C., Atallah B., Al Akkary N., Sleiman Z., et al. (2020). Bacterial infection profile and predictors among patients admitted to a burn care center: a retrospective study. Burns 46, 1968–1976. doi: 10.1016/j.burns.2020.05.004, PMID: [DOI] [PubMed] [Google Scholar]
- Boustanshenas M., Bakhshi B., Mobasseri P., Kiani P., Abadi F. H. H., Seyfi E., et al. (2023). Genetically diverse, extremely resistant, and Pan-drug resistant Pseudomonas aeruginosa as the Main cause of nosocomial infection among hospitalized patients. Arch. Clin. Infect. Dis. 18:136338. doi: 10.5812/archcid-136338 [DOI] [Google Scholar]
- Brauncajs M., Bielec F., Macieja A., Pastuszak-Lewandoska D. (2022). Carbapenem-resistant gram-negative fermenting and non-fermenting rods isolated from hospital patients in Poland—what are they susceptible to? Biomedicines 10:3049. doi: 10.3390/biomedicines10123049, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski M., Krukowska Ż., Galant K., Jursa-Kulesza J., Kosik-Bogacka D. (2020). Genotypic characterisation and antimicrobial resistance of Pseudomonas aeruginosa strains isolated from patients of different hospitals and medical centres in Poland. BMC Infect. Dis. 20:693. doi: 10.1186/s12879-020-05404-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunsow E., Los-Arcos I., Martin-Gómez M. T., Bello I., Pont T., Berastegui C., et al. (2020). Donor-derived bacterial infections in lung transplant recipients in the era of multidrug resistance. J. Infect. 80, 190–196. doi: 10.1016/j.jinf.2019.12.006 [DOI] [PubMed] [Google Scholar]
- Buzilă E. R., Năstase E. V., Luncă C., Bădescu A., Miftode E., Iancu L. S. (2021). Antibiotic resistance of non-fermenting gram-negative bacilli isolated at a large infectious diseases Hospital in North-Eastern Romania, during an 11-year period. Germs 11, 354–362. doi: 10.18683/germs.2021.1272, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabot G., Ocampo-Sosa A. A., Tubau F., Macia M. D., Rodríguez C., Moya B., et al. (2011). Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob. Agents Chemother. 55, 1906–1911. doi: 10.1128/AAC.01645-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Chai D., Wang R., Liang B., Bai N. (2012). Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67, 1607–1615. doi: 10.1093/jac/dks084 [DOI] [PubMed] [Google Scholar]
- Camargo C. H., Yamada A. Y., Souza A. R., Lima M. J. C., Cunha M. P. V., Ferraro P. S. P., et al. (2023). Genomics and antimicrobial susceptibility of clinical Pseudomonas aeruginosa isolates from hospitals in Brazil. Pathogens 12:918. doi: 10.3390/pathogens12070918, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A., Borek A. J., McLeod M., Tonkin-Crine S., Pouwels K. B., Roope L. S., et al. (2023). Impact of the COVID-19 pandemic on antimicrobial stewardship support for general practices in England: a qualitative interview study. BJGP Open 7:193. doi: 10.3399/BJGPO.2022.0193, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candel F. J., Henriksen A. S., Longshaw C., Yamano Y., Oliver A. (2022). In vitro activity of the novel siderophore cephalosporin, cefiderocol, in gram-negative pathogens in Europe by site of infection. Clin. Microbiol. Infect. 28, 447.e1. e6–447.e6. doi: 10.1016/j.cmi.2021.07.018 [DOI] [PubMed] [Google Scholar]
- Cannatelli A., D'Andrea M. M., Giani T., Di Pilato V., Arena F., Ambretti S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton R., Hamed K., Wiktorowicz T., Redder N., Jemmely N., Quevedo J., et al. (2022). In vitro activity of ceftobiprole and comparator antibiotics against contemporary European isolates (2016–19). JAC-Antimicrobial Resistance 4:dlac030. doi: 10.1093/jacamr/dlac030, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhaes C. G., Shortridge D., Sader H. S., Castanheira M. (2020). Activity of Meropenem-Vaborbactam against bacterial isolates causing pneumonia in patients in U.S. hospitals during 2014 to 2018. ASM J CD 64:e02177. doi: 10.1128/AAC.02177-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira M., Davis A. P., Mendes R. E., Serio A. W., Krause K. M., Flamm R. K. (2018). In vitro activity of plazomicin against gram-negative and gram-positive isolates collected from US hospitals and comparative activities of aminoglycosides against carbapenem-resistant Enterobacteriaceae and isolates carrying carbapenemase genes. Antimicrob. Agents Chemother. 62, 313–318. doi: 10.1128/AAC.00313-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo I., Lesnoni La Parola I., Sivori F., Toma L., Koudriavtseva T., Sperduti I., et al. (2022). Homocysteine and inflammatory cytokines in the clinical assessment of infection in venous leg ulcers. Antibiotics. 11:1268. doi: 10.3390/antibiotics11091268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesur S., Yıldız E., Irmak H., Gülay Z., Arslan U., Özen S., et al. (2012). Metallobeta-lactamase enzymes and antibiotic susceptibilities in strains of Pseudomonas Aeruginosa isolated from intensive care units in Turkey. Turkiye Klinikleri J Med Sci 32, 687–693. doi: 10.5336/medsci.2011-24972 [DOI] [Google Scholar]
- Çetin Y. S., Mollamehmetoğlu S. O., Düzenli U., Turan M., Bozan N. (2022). Treatment of multi-drug resistant microorganisms in chronic suppurative otitis media. B-ENT 18, 44–51. doi: 10.5152/B-ENT.2022.21425 [DOI] [Google Scholar]
- Chang F., Wang X., Huang X., Liu X., Huang L. (2023). Analysis on bacterial distribution and change of drug resistance rate in ICUs across Southwest China from 2018 to 2022. Infect. Drug Resist., 5685–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P., Lamba M., Sharma D., Mamoria V. P. (2021). Bloodstream infections and antibiotic sensitivity pattern in intensive care unit. Trop. Dr. 51, 44–48. doi: 10.1177/0049475520977043 [DOI] [PubMed] [Google Scholar]
- Chaudhary B. R., Malla K. K., Poudel S., Jha B. K. (2020). Study of antibiotic susceptibility among bacterial isolates in neonatal intensive care unit of a tertiary care hospital: a descriptive cross-sectional study. JNMA J. Nepal Med. Assoc. 58, 893–899. doi: 10.31729/jnma.5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S., Kaur N., Saini A. K., Aman S., Chauhan J., Kumar H. (2022). Colistin resistant gram-negative Bacteria isolated from various clinical samples in north Indian tertiary care center.
- Chaw P., Höpner J., Mikolajczyk R. (2018). The knowledge, attitude and practice of health practitioners towards antibiotic prescribing and resistance in developing countries—a systematic review. J. Clin. Pharm. Ther. 43, 606–613. doi: 10.1111/jcpt.12730, PMID: [DOI] [PubMed] [Google Scholar]
- Chen J., Liang Q., Ding S., Xu Y., Hu Y., Chen J., et al. (2023). Ceftazidime/avibactam for the treatment of carbapenem-resistant Pseudomonas aeruginosa infection in lung transplant recipients. Infect. Drug Resist. 16, 2237–2246. doi: 10.2147/IDR.S407515, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Lu W., Zhou D., Zheng G., Liu H., Qian C., et al. (2020). Characterization of two macrolide resistance-related genes in multidrug-resistant Pseudomonas aeruginosa isolates. Pol. J. Microbiol. 69, 349–356. doi: 10.33073/pjm-2020-038, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Mai H., Lopes B., Wen F., Patil S. (2022). Novel Pseudomonas aeruginosa strains co-Harbouring Bla (NDM-1) Metallo beta-lactamase and mcr-1 isolated from immunocompromised Paediatric patients. Infect Drug Resist. 15, 2929–2936. doi: 10.2147/IDR.S368566, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sun M., Wang M., Lu Y., Yan Z. (2014). Dissemination of IMP-6-producing Pseudomonas aeruginosa ST244 in multiple cities in China. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1181–1187. doi: 10.1007/s10096-014-2063-5, PMID: [DOI] [PubMed] [Google Scholar]
- Chen H., Wang Z., Li H., Wang Q., Zhao C., He W., et al. (2015). In vitro analysis of activities of 16 antimicrobial agents against gram-negative bacteria from six teaching hospitals in China. Jpn. J. Infect. Dis. 68, 263–267. doi: 10.7883/yoken.JJID.2014.202 [DOI] [PubMed] [Google Scholar]
- Chen X., Xu J., Zhu Q., Ren Y., Zhao L. (2020). Polymyxin B resistance rates in carbapenem-resistant Pseudomonas aeruginosa isolates and a comparison between Etest® and broth microdilution methods of antimicrobial susceptibility testing. Exp. Ther. Med. 20, 762–769. doi: 10.3892/etm.2020.8777, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew K. L., Octavia S., Ng O. T., Marimuthu K., Venkatachalam I., Cheng B., et al. (2019). Challenge of drug resistance in Pseudomonas aeruginosa: clonal spread of NDM-1-positive ST-308 within a tertiary hospital. J. Antimicrob. Chemother. 74, 2220–2224. doi: 10.1093/jac/dkz169, PMID: [DOI] [PubMed] [Google Scholar]
- Chittawatanarat K., Jaipakdee W., Chotirosniramit N., Chandacham K., Jirapongcharoenlap T. (2014). Microbiology, resistance patterns, and risk factors of mortality in ventilator-associated bacterial pneumonia in a northern Thai tertiary-care university based general surgical intensive care unit. Infect. Drug Resist. 7, 203–210. doi: 10.2147/IDR.S67267, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokshi A., Sifri Z., Cennimo D., Horng H. (2019). Global contributors to antibiotic resistance. J. Global Infect. Dis. 11, 36–42. doi: 10.4103/jgid.jgid_110_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukamnerd A., Pomwised R., Chusri S., Singkhamanan K., Chumtong S., Jeenkeawpiam K., et al. (2023). Antimicrobial susceptibility and molecular features of colonizing isolates of Pseudomonas aeruginosa and the report of a novel sequence type (ST) 3910 from Thailand. Antibiotics 12:165. doi: 10.3390/antibiotics12010165, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillóniz C., Gabarrús A., Ferrer M., de la Bellacasa J. P., Rinaudo M., Mensa J., et al. (2016). Community-acquired pneumonia due to multidrug-and non–multidrug-resistant Pseudomonas aeruginosa. Chest 150, 415–425. doi: 10.1016/j.chest.2016.03.042, PMID: [DOI] [PubMed] [Google Scholar]
- Cipriano R., Vieira V. V., Fonseca E. L., Rangel K., Freitas F. S., Vicente A. C. P. (2007). Coexistence of epidemic colistin-only-sensitive clones of Pseudomonas aeruginosa, including the Bla SPM clone, spread in hospitals in a Brazilian Amazon city. Microb. Drug Resist. 13, 142–146. doi: 10.1089/mdr.2007.708, PMID: [DOI] [PubMed] [Google Scholar]
- Çopur Çiçek A., Ertürk A., Ejder N., Rakici E., Kostakoğlu U., Esen Yıldız İ., et al. (2021). Screening of antimicrobial resistance genes and epidemiological features in hospital and community-associated carbapenem-resistant pseudomonas aeruginosa infections. Infect. Drug Resist. 14, 1517–1526. doi: 10.2147/IDR.S299742, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czekajło-Kołodziej U., Giedrys-Kalemba S., Medrala D. (2006). Phenotypic and genotypic characteristics of Pseudomonas aeruginosa strains isolated from hospitals in the north-west region of Poland. Pol. J. Microbiol. 55, 103–112, PMID: [PubMed] [Google Scholar]
- da Costa Júnior S. D., da Silva W. R. C., da Silva A. M. C. M., Maciel M. A. V., Cavalcanti I. M. F. (2020). Synergistic effect between usnic acid and polymyxin B against resistant clinical isolates of Pseudomonas aeruginosa. Evid. Based Complement. Alternat. Med. 2020:9852145. doi: 10.1155/2020/9852145, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadgostar P. (2019). Antimicrobial resistance: implications and costs. Infect. Drug Resist. 12, 3903–3910. doi: 10.2147/IDR.S234610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadmanesh M., Pilehvarzadeh M., Eramabadi M., Eramabadi P., Moghadam M. B., Mashayekhi F. (2014). Community acquired Pseudomonas aeroginosa urinary tract infections in children hospitalized in a Baqiatallah hospital, Tehran, Iran: virulence profile and antibiotic resistance properties. Biosci. Biotechnol. Res. Asia 11, 417–426. doi: 10.13005/bbra/1290 [DOI] [Google Scholar]
- Darji S. M., Patel N. (2023). Central line associated blood stream infection: microbiological profile and its antimicrobial susceptibility pattern at tertiary care Centre. J. Pure Appl. Microbiol. 17, 911–918. doi: 10.22207/JPAM.17.2.18 [DOI] [Google Scholar]
- Dassner A. M., Sutherland C., Girotto J., Nicolau D. P. (2017). In vitro activity of ceftolozane/tazobactam alone or with an aminoglycoside against multi-drug-resistant Pseudomonas aeruginosa from pediatric cystic fibrosis patients. Infect. Dis. Ther. 6, 129–136. doi: 10.1007/s40121-016-0141-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar R., Coello Pelegrin A., Orenga S., Chalansonnet V., Mirande C., Dombrecht J., et al. (2021). Phenotypic and genomic variability of serial Peri-Lung transplantation Pseudomonas aeruginosa isolates from cystic fibrosis patients. Front. Microbiol. 12:604555. doi: 10.3389/fmicb.2021.604555, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Blasio B. F. (2021). Infectious disease threats: Antibiotic resistance, COVID-19, and future pandemics. Meeting the challenges of existential threats through educational innovation. London: Routledge. [Google Scholar]
- de Dios C. J., Del Campo R., Royuela A., Solé A., Máiz L., Olveira C., et al. (2016). Bronchopulmonary infection-colonization patterns in Spanish cystic fibrosis patients: results from a national multicenter study. J. Cyst. Fibros. 15, 357–365. doi: 10.1016/j.jcf.2015.09.004 [DOI] [PubMed] [Google Scholar]
- De Francesco M. A., Ravizzola G., Peroni L., Bonfanti C., Manca N. (2013). Prevalence of multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa in an Italian hospital. J. Infect. Public Health 6, 179–185. doi: 10.1016/j.jiph.2012.11.006, PMID: [DOI] [PubMed] [Google Scholar]
- de Oliveira Santos I. C., de Andrade N. F. P., da Conceição Neto O. C., da Costa B. S., de Andrade M. E., Rocha-de-Souza C. M., et al. (2019). Epidemiology and antibiotic resistance trends in clinical isolates of Pseudomonas aeruginosa from Rio de janeiro-Brazil: importance of mutational mechanisms over the years (1995–2015). Infect. Genet. Evol. 73, 411–415. doi: 10.1016/j.meegid.2019.05.015 [DOI] [PubMed] [Google Scholar]
- De Vecchi E., Sitia S., Romano C. L., Ricci C., Mattina R., Drago L. (2013). Aetiology and antibiotic resistance patterns of urinary tract infections in the elderly: a 6-month study. J. Med. Microbiol. 62, 859–863. doi: 10.1099/jmm.0.056945-0 [DOI] [PubMed] [Google Scholar]
- Dehbashi S., Tahmasebi H., Arabestani M. R. (2018). Association between Beta-lactam antibiotic resistance and virulence factors in AmpC producing clinical strains of P. aeruginosa. Osong. Public Health Res. Perspect. 9, 325–333. doi: 10.24171/j.phrp.2018.9.6.06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Barrio-Tofino E., Lopez-Causape C., Cabot G., Rivera A., Benito N., Segura C., et al. (2017). Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob. Agents Chemother. 61:1589. doi: 10.1128/AAC.01589-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Barrio-Tofiño E., López-Causapé C., Oliver A. (2020). Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents 56:106196. doi: 10.1016/j.ijantimicag.2020.106196 [DOI] [PubMed] [Google Scholar]
- Del Giacomo P., Raffaelli F., Losito A. R., Fiori B., Tumbarello M. (2022). XDR-Pseudomonas aeruginosa outside the ICU: is there still place for colistin? Antibiotics. 11:193. doi: 10.3390/antibiotics11020193, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Valverde M., Conejo M. D. C., Serrano L., Fernandez-Cuenca F., Pascual A. (2020). Activity of cefiderocol against high-risk clones of multidrug-resistant Enterobacterales, Acinetobacter baumannii, Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 75, 1840–1849. doi: 10.1093/jac/dkaa117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delroshan N., Ghandehari F., Mirzaei R., Hoveida L. (2023). Molecular typing of multidrug-resistant Pseudomonas aeruginosa isolates obtained from hospitalized burn patients by rep-PCR. Infect. Epidemiol. Microbiol. 9, 99–106. doi: 10.61186/iem.9.2.99 [DOI] [Google Scholar]
- Depka D., Mikucka A., Bogiel T., Gospodarek-Komkowska E. (2020). Comparison of the recommended colistin susceptibility testing methods with colistin gradient strips and semi-automated method for antimicrobial-resistant non-fermenting rods. J. Microbiol. Methods 172:105905. doi: 10.1016/j.mimet.2020.105905, PMID: [DOI] [PubMed] [Google Scholar]
- Descours G., Desmurs L., Hoang T. L. T., Ibranosyan M., Baume M., Ranc A.-G., et al. (2018). Evaluation of the accelerate Pheno™ system for rapid identification and antimicrobial susceptibility testing of gram-negative bacteria in bloodstream infections. Eur. J. Clin. Microbiol. Infect. Dis. 37, 1573–1583. doi: 10.1007/s10096-018-3287-6, PMID: [DOI] [PubMed] [Google Scholar]
- Dharati S., Atit S., Lata P., Jayshri P., Urvashi L., Hiral S. (2021). Microbiological profile and Antibiogram of Uropathogens isolated at a tertiary care hospital. J. Krishna Inst. Med. Sci. 10. [Google Scholar]
- Di Carlo P., Serra N., Lo Sauro S., Carelli V. M., Giarratana M., Signorello J. C., et al. (2021). Epidemiology and pattern of resistance of gram-negative bacteria isolated from blood samples in hospitalized patients: a single center retrospective analysis from southern Italy. Antibiotics 10:1402. doi: 10.3390/antibiotics10111402, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Domenico E. G., Farulla I., Prignano G., Gallo M. T., Vespaziani M., Cavallo I., et al. (2017). Biofilm is a major virulence determinant in bacterial colonization of chronic skin ulcers independently from the multidrug resistant phenotype. Int. J. Mol. Sci. 18:1077. doi: 10.3390/ijms18051077, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias V. C., Resende J. A., Bastos A. N., De Andrade Bastos L. Q., De Andrade Bastos V. Q., Bastos R. V., et al. (2017). Epidemiological, physiological, and molecular characteristics of a Brazilian collection of Carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Microb. Drug Resist. 23, 852–863. doi: 10.1089/mdr.2016.0219 [DOI] [PubMed] [Google Scholar]
- Díaz-Cañestro M., Periañez L., Mulet X., Martin-Pena M. L., Fraile-Ribot P. A., Ayestarán I., et al. (2018). Ceftolozane/tazobactam for the treatment of multidrug resistant Pseudomonas aeruginosa: experience from the Balearic Islands. Eur. J. Clin. Microbiol. Infect. Dis. 37, 2191–2200. doi: 10.1007/s10096-018-3361-0, PMID: [DOI] [PubMed] [Google Scholar]
- Diekema D. J., Hsueh P.-R., Mendes R. E., Pfaller M. A., Rolston K. V., Sader H. S., et al. (2019). The microbiology of bloodstream infection: 20-year trends from the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 63, 00355–00319. doi: 10.1128/AAC.00355-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din M., Awan M. A., Ur Rahman S., Ali M., Aslam M. (2023). Co-existence of blaIMP, blaNDM-1, and blaSHV, genes of Pseudomonas aeruginosa isolated from Quetta: antimicrobial resistance and clinical significance. Pakistan J. Med. Sci. 39:1507. doi: 10.12669/pjms.39.5.7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do Tran H., Nguyen Y. T. B., Tran T. T., Le T. T. T., Nguyen H. T. T., Nguyen C. M., et al. (2022). Community-acquired pneumonia-causing bacteria and antibiotic resistance rate among Vietnamese patients: a cross-sectional study. Medicine 101:e30458. doi: 10.1097/MD.0000000000030458, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogonchi A. A., Ghaemi E. A., Ardebili A., Yazdansetad S., Pournajaf A. (2018). Metallo-beta-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: a potential threat to clinical therapeutics. Ci Ji Yi Xue Za Zhi. 30, 90–96. doi: 10.4103/tcmj.tcmj_101_17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S.-X., Wang J.-T., Chang S.-C. (2012). Activities of doripenem against nosocomial bacteremic drug-resistant gram-negative bacteria in a medical center in Taiwan. J. Microbiol. Immunol. Infect. 45, 459–464. doi: 10.1016/j.jmii.2012.08.022 [DOI] [PubMed] [Google Scholar]
- Doumith M., Alhassinah S., Alswaji A., Alzayer M., Alrashidi E., Okdah L., et al. (2022). Genomic characterization of carbapenem-non-susceptible Pseudomonas aeruginosa clinical isolates from Saudi Arabia revealed a global dissemination of GES-5-producing ST235 and VIM-2-producing ST233 sub-lineages. Front. Microbiol. 12:765113. doi: 10.3389/fmicb.2021.765113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durdu B., Kritsotakis E. I., Lee A. C., Torun P., Hakyemez I. N., Gultepe B., et al. (2018). Temporal trends and patterns in antimicrobial-resistant gram-negative bacteria implicated in intensive care unit-acquired infections: a cohort-based surveillance study in Istanbul, Turkey. J. Global Antimicrob. Resist. 14, 190–196. doi: 10.1016/j.jgar.2018.04.015, PMID: [DOI] [PubMed] [Google Scholar]
- Ebadati A., Oshaghi M., Saeedi S., Parsa P., Mahabadi V. P., Karimi M., et al. (2023). Mechanism and antibacterial synergies of poly(Dabco-BBAC) nanoparticles against multi-drug resistant Pseudomonas aeruginosa isolates from human burns. Bioorg. Chem. 140:106718. doi: 10.1016/j.bioorg.2023.106718, PMID: [DOI] [PubMed] [Google Scholar]
- Ece G., Samlioglu P., Atalay S., Kose S. (2014). Evaluation of the in vitro colistin susceptibility of Pseudomonas aeruginosa and Acinetobacter baumannii strains at a tertiary care Centre in Western Turkey. Infez. Med. 22, 36–40, PMID: [PubMed] [Google Scholar]
- Eftekhar F., Hosseinkhan N., Asgharzadeh A., Tabatabaei A. (2009) Genetic profiling of Pseudomonas aeruginosa isolates from Iranian patients with cystic fibrosis using RAPD-PCR and PFGE. Iranian Journal of Basic Medical Sciences.
- Eid D., Sayed O. M., Hozayen W. G., Azmy A. F. (2020). Battling biofilm forming nosocomial pathogens using chitosan and Pluronic F127. J. Pure Appl. Microbiol. 14, 1893–1903. doi: 10.22207/JPAM.14.3.28 [DOI] [Google Scholar]
- Ejaz H. (2022). Molecular characterization and antibiogram of the carbapenemase gene variants in clinical strains of Pseudomonas aeruginosa. Mol. Biol. Rep. 49, 10531–10539. doi: 10.1007/s11033-022-07930-z [DOI] [PubMed] [Google Scholar]
- Ekkelenkamp M. B., Cantón R., Díez-Aguilar M., Tunney M. M., Gilpin D. F., Bernardini F., et al. (2020). Susceptibility of Pseudomonas aeruginosa recovered from cystic fibrosis patients to murepavadin and 13 comparator antibiotics. Antimicrob. Agents Chemother. 64, 01541–01519. doi: 10.1128/AAC.01541-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mekes A., Zahlane K., Ait Said L., Tadlaoui Ouafi A., Barakate M. (2020). The clinical and epidemiological risk factors of infections due to multi-drug resistant bacteria in an adult intensive care unit of University Hospital Center in Marrakesh-Morocco. J. Infect. Public Health 13, 637–643. doi: 10.1016/j.jiph.2019.08.012 [DOI] [PubMed] [Google Scholar]
- Eladawy M., El-Mowafy M., El-Sokkary M. M., Barwa R. (2021). Antimicrobial resistance and virulence characteristics in ERIC-PCR typed biofilm forming isolates of P. aeruginosa. Microb. Pathog. 158:105042. doi: 10.1016/j.micpath.2021.105042, PMID: [DOI] [PubMed] [Google Scholar]
- El-Mokhtar M. A., Hetta H. F. (2018). Ambulance vehicles as a source of multidrug-resistant infections: a multicenter study in Assiut City, Egypt. Infect. Drug Resist. 11, 587–594. doi: 10.2147/IDR.S151783, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshafie S. S., Wahab A. A., Al J. I. (2007). Antimicrobial resistance of bacterial strains isolated from respiratory tract of cystic fibrosis patients with CFTR I1234V mutation. J. Pediatr. Infect. Dis. 2, 039–043. doi: 10.1055/s-0035-1557010 [DOI] [Google Scholar]
- El-Sokkary R., Uysal S., Erdem H., Kullar R., Pekok A. U., Amer F., et al. (2021). Profiles of multidrug-resistant organisms among patients with bacteremia in intensive care units: An international ID-IRI survey. Eur. J. Clin. Microbiol. Infect. Dis. 40, 2323–2334. doi: 10.1007/s10096-021-04288-1, PMID: [DOI] [PubMed] [Google Scholar]
- Emami A., Kazempour A., Pirbonyeh N., Keshavarzi A., Zardosht M. (2017). Hospitalization length survey and relation with distribution of LasA protease and type III secretion system encoding-genes in multi-drug resistant Pseudomonas aeruginosa isolates from burn wounds in southwest of Iran. Gene Rep. 9, 81–85. doi: 10.1016/j.genrep.2017.09.006 [DOI] [Google Scholar]
- Emami A., Keshavarzi A., Pirbonyeh N., Behbahani M. R. (2019). Identification of different faces of Pseudomonas aeruginosa isolates in burn patients by genetic fingerprinting. Gene Rep. 15:100377. doi: 10.1016/j.genrep.2019.100377 [DOI] [Google Scholar]
- Emami A., Pirbonyeh N., Keshavarzi A., Bazargani A., Hassanpour S., Javanmardi F. (2020). Evaluating the saliva of burn ICU patients for resistant Infections Harbor Metallo-beta-lactamase genes. J. Burn Care Res. 41, 647–651. doi: 10.1093/jbcr/iraa007, PMID: [DOI] [PubMed] [Google Scholar]
- Ergul A. B., Cetin S., Altintop Y. A., Bozdemir S. E., Ozcan A., Altug U., et al. (2017a). Evaluation of microorganisms causing ventilator-associated pneumonia in a pediatric intensive care unit. Eurasian J. Med. 49, 87–91. doi: 10.5152/eurasianjmed.2017.16262, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergul A. B., Isik H., Altintop Y. A., Torun Y. A. (2017b). A retrospective evaluation of blood cultures in a pediatric intensive care unit: a three year evaluation. Turk Pediatri Ars. 52, 154–161. doi: 10.5152/TurkPediatriArs.2017.5451, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolà-Vergé L., Pigrau C., Los-Arcos I., Arévalo Á., Viñado B., Campany D., et al. (2018). Ceftolozane/tazobactam for the treatment of XDR Pseudomonas aeruginosa infections. Infection 46, 461–468. doi: 10.1007/s15010-018-1133-5 [DOI] [PubMed] [Google Scholar]
- Evans S. R., Tran T. T. T., Hujer A. M., Hill C. B., Hujer K. M., Mediavilla J. R., et al. (2019). Rapid molecular diagnostics to Inform empiric use of ceftazidime/avibactam and Ceftolozane/Tazobactam against Pseudomonas aeruginosa: PRIMERS IV. Clin. Infect. Dis. 68, 1823–1830. doi: 10.1093/cid/ciy801, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhkhari P., Tajeddin E., Azimirad M., Salmanzadeh-Ahrabi S., Abdi-Ali A., Nikmanesh B., et al. (2022). Involvement of Pseudomonas aeruginosa in the occurrence of community and hospital acquired diarrhea, and its virulence diversity among the stool and the environmental samples. Int. J. Environ. Health Res. 32, 61–71. doi: 10.1080/09603123.2020.1726300, PMID: [DOI] [PubMed] [Google Scholar]
- Falagas M. E., Skalidis T., Vardakas K. Z., Legakis N. J., Hellenic Cefiderocol Study G (2017). Activity of cefiderocol (S-649266) against carbapenem-resistant gram-negative bacteria collected from inpatients in Greek hospitals. J. Antimicrob. Chemother. 72, 1704–1708. doi: 10.1093/jac/dkx049 [DOI] [PubMed] [Google Scholar]
- Fang Y., Wang N., Wu Z., Zhu Y., Ma Y., Li Y., et al. (2023). An XDR pseudomonas aeruginosa ST463 strain with an IncP-2 plasmid containing a novel transposon Tn 6485f encoding Bla IMP-45 and bla AFM-1 and a second plasmid with two copies of Bla KPC-2. Microbiol. Spectr. 11, e04462–e04422. doi: 10.1128/spectrum.04462-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag A. M., Tawfick M. M., Abozeed M. Y., Shaban E. A., Abo-Shadi M. A. (2020). Microbiological profile of ventilator-associated pneumonia among intensive care unit patients in tertiary Egyptian hospitals. J. Infect. Dev. Ctries. 14, 153–161. doi: 10.3855/jidc.12012 [DOI] [PubMed] [Google Scholar]
- Farhan S. M., Ibrahim R. A., Mahran K. M., Hetta H. F., Abd El-Baky R. M. (2019). Antimicrobial resistance pattern and molecular genetic distribution of metallo-β-lactamases producing Pseudomonas aeruginosa isolated from hospitals in Minia, Egypt. Infect. Drug Resist. 12, 2125–2133. doi: 10.2147/IDR.S198373, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell D. J., Flamm R. K., Sader H. S., Jones R. N. (2013). Antimicrobial activity of ceftolozane-tazobactam tested against Enterobacteriaceae and Pseudomonas aeruginosa with various resistance patterns isolated in U.S. hospitals (2011-2012). Antimicrob. Agents Chemother. 57, 6305–6310. doi: 10.1128/AAC.01802-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell D. J., Flamm R. K., Sader H. S., Jones R. N. (2014a). Ceftobiprole activity against over 60,000 clinical bacterial pathogens isolated in Europe, Turkey, and Israel from 2005 to 2010. Antimicrob. Agents Chemother. 58, 3882–3888. doi: 10.1128/AAC.02465-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell D. J., Sader H. S., Flamm R. K., Jones R. N. (2014b). Ceftolozane/tazobactam activity tested against gram-negative bacterial isolates from hospitalised patients with pneumonia in US and European medical centres (2012). Int. J. Antimicrob. Agents 43, 533–539. doi: 10.1016/j.ijantimicag.2014.01.032, PMID: [DOI] [PubMed] [Google Scholar]
- Farzana R., Shamsuzzaman S., Mamun K. Z. (2013). Isolation and molecular characterization of New Delhi metallo-beta-lactamase-1 producing superbug in Bangladesh. J. Infect. Dev. Countries 7, 161–168. doi: 10.3855/jidc.2493 [DOI] [PubMed] [Google Scholar]
- Fekri Kohan S., Asadpour L., Houshmand E. (2020). The study of frequency of SIM and AmpC genes in clinical isolates of Pseudomonas aeruginosa in Gilan, Iran. Infect. Epidemiol. Microbiol 6, 117–125. doi: 10.29252/iem.6.2.117 [DOI] [Google Scholar]
- Feretzakis G., Loupelis E., Sakagianni A., Skarmoutsou N., Michelidou S., Velentza A., et al. (2019). A 2-year single-Centre audit on antibiotic resistance of Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae strains from an intensive care unit and other wards in a general public hospital in Greece. Antibiotics 8:62. doi: 10.3390/antibiotics8020062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferjani S., Maamar E., Ferjani A., Kanzari L., Boubaker I. B. B. (2022). Evaluation of three carbapenemase-phenotypic detection methods and emergence of diverse VIM and GES variants among Pseudomonas aeruginosa isolates in Tunisia. Antibiotics 11:858. doi: 10.3390/antibiotics11070858, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Olmos A., García-Castillo M., Alba J. M., Morosini M. I., Lamas A., Romero B., et al. (2013). Population structure and antimicrobial susceptibility of both nonpersistent and persistent Pseudomonas aeruginosa isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 51, 2761–2765. doi: 10.1128/JCM.00802-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm R. K., Farrell D. J., Sader H. S., Jones R. N. (2014). Ceftazidime/avibactam activity tested against gram-negative bacteria isolated from bloodstream, pneumonia, intra-abdominal and urinary tract infections in US medical centres (2012). J. Antimicrob. Chemother. 69, 1589–1598. doi: 10.1093/jac/dku025, PMID: [DOI] [PubMed] [Google Scholar]
- Flores-Paredes W., Luque N., Albornoz R., Rojas N., Espinoza M., Pons M. J., et al. (2021). Evolution of antimicrobial resistance levels of ESKAPE microorganisms in a Peruvian IV-level hospital. Infect Chemother. 53, 449–462. doi: 10.3947/ic.2021.0015, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluge G., Ojeniyi B., Høiby N., Digranes A., Ciofu O., Hunstad E., et al. (2001). Typing of Pseudomonas aeruginosa strains in Norwegian cystic fibrosis patients. Clin. Microbiol. Infect. 7, 238–243. doi: 10.1046/j.1469-0691.2001.00247.x [DOI] [PubMed] [Google Scholar]
- Ford M. B., Mende K., Kaiser S. J., Beckius M. L., Lu D., Stam J., et al. (2022). Clinical characteristics and resistance patterns of Pseudomonas aeruginosa isolated from combat casualties. Mil. Med. 187, 426–434. doi: 10.1093/milmed/usab259, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier D., Carriere R., Bour M., Grisot E., Triponney P., Muller C., et al. (2021). Mechanisms of resistance to Ceftolozane/Tazobactam in Pseudomonas aeruginosa: results of the GERPA multicenter study. Antimicrob. Agents Chemother. 65:20. doi: 10.1128/AAC.01117-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier D., Richardot C., Müller E., Robert-Nicoud M., Llanes C., Plésiat P., et al. (2013). Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 68, 1772–1780. doi: 10.1093/jac/dkt098, PMID: [DOI] [PubMed] [Google Scholar]
- Fraenkel C. J., Starlander G., Tano E., Sutterlin S., Melhus A. (2023). The first Swedish outbreak with VIM-2-producing Pseudomonas aeruginosa, occurring between 2006 and 2007, was probably due to contaminated hospital sinks. Microorganisms 11:974. doi: 10.3390/microorganisms11040974, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M. R. G., Caiaffa-Filho H. H., Burattini M. N., Rossi F. (2010). Metallo-beta-lactamases among imipenem-resistant Pseudomonas aeruginosa in a Brazilian university hospital. Clinics 65, 825–829. doi: 10.1590/S1807-59322010000900002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R., de Oliveira Santos I. C., Mora M. F. M., López P. V. A., Alvarez V. E. T., Arce F. H. O., et al. (2023). Genotypic characterization and clonal relatedness of metallo-β-lactamase-producing non-fermentative gram negative bacteria in the first 5 years of their circulation in Paraguay (2011-2015). Braz. J. Microbiol. 54, 179–190. doi: 10.1007/s42770-022-00888-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber S. N., Hemeda E. E. M., Elsayeh H.-A. S., Wahed W. Y. A., Khalil M. A. F., Ibrahim E. G. (2020). Propolis extract: a possible antiseptic Oral care against multidrug-resistant non-fermenting Bacteria isolated from non-ventilator hospital-acquired pneumonia. J. Pure Appl. Microbiol. 14, 123–131. doi: 10.22207/JPAM.14.1.13 [DOI] [Google Scholar]
- Gahlot T., Kasana D. (2021). A cross-sectional study of etiological and sensitivity profiling of meningitis in under-five children. Int J Mycobacteriol. 10, 149–154. doi: 10.4103/ijmy.ijmy_61_21, PMID: [DOI] [PubMed] [Google Scholar]
- Gajdács M., Barath Z., Karpati K., Szabo D., Usai D., Zanetti S., et al. (2021a). No correlation between biofilm formation, virulence factors, and antibiotic resistance in Pseudomonas aeruginosa: results from a laboratory-based in vitro study. Antibiotics (Basel) 10:1134. doi: 10.3390/antibiotics10091134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdács M., Burián K., Terhes G. (2019). Resistance levels and epidemiology of non-fermenting gram-negative bacteria in urinary tract infections of inpatients and outpatients (RENFUTI): a 10-year epidemiological snapshot. Antibiotics 8:143. doi: 10.3390/antibiotics8030143, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdács M., Kárpáti K., Stájer A., Zanetti S., Donadu M. G. (2021b). Insights on carbapenem-resistant Pseudomonas aeruginosa. Acta Biologica Szegediensis. 65, 105–112. doi: 10.14232/abs.2021.1.105-112 [DOI] [Google Scholar]
- Galani I., Kontopidou F., Souli M., Rekatsina P.-D., Koratzanis E., Deliolanis J., et al. (2008). Colistin susceptibility testing by Etest and disk diffusion methods. Int. J. Antimicrob. Agents 31, 434–439. doi: 10.1016/j.ijantimicag.2008.01.011, PMID: [DOI] [PubMed] [Google Scholar]
- Galani I., Papoutsaki V., Karantani I., Karaiskos I., Galani L., Adamou P., et al. (2020). In vitro activity of ceftolozane/tazobactam alone and in combination with amikacin against MDR/XDR Pseudomonas aeruginosa isolates from Greece. J. Antimicrob. Chemother. 75, 2164–2172. doi: 10.1093/jac/dkaa160 [DOI] [PubMed] [Google Scholar]
- Gales A. C., Castanheira M., Jones R. N., Sader H. S. (2012). Antimicrobial resistance among gram-negative bacilli isolated from Latin America: results from SENTRY antimicrobial surveillance program (Latin America, 2008–2010). Diagn. Microbiol. Infect. Dis. 73, 354–360. doi: 10.1016/j.diagmicrobio.2012.04.007, PMID: [DOI] [PubMed] [Google Scholar]
- Gales A. C., Jones R. N., Sader H. S. (2011). Contemporary activity of colistin and polymyxin B against a worldwide collection of gram-negative pathogens: results from the SENTRY antimicrobial surveillance program (2006–09). J. Antimicrob. Chemother. 66, 2070–2074. doi: 10.1093/jac/dkr239, PMID: [DOI] [PubMed] [Google Scholar]
- Gales A. C., Menezes L. C., Silbert S., Sader H. S. (2003). Dissemination in distinct Brazilian regions of an epidemic carbapenem-resistant Pseudomonas aeruginosa producing SPM metallo-β-lactamase. J. Antimicrob. Chemother. 52, 699–702. doi: 10.1093/jac/dkg416, PMID: [DOI] [PubMed] [Google Scholar]
- Galindo-Mendez M., Navarrete-Salazar H., Pacheco-Vasquez R., Quintas-de la Paz D., Baltazar-Jimenez I., Santiago-Luna J. D., et al. (2023). Detection of plasmid-mediated resistance against Colistin in multi-drug-resistant gram-negative Bacilli isolated from a tertiary hospital. Microorganisms 11:1996. doi: 10.3390/microorganisms11081996, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwar N., Siddapur G., Sharma S. (2021). Clinical implications of culture and sensitivity data in chronic otitis media. Indian J. Otol. 27, 101–105. doi: 10.4103/indianjotol.indianjotol_8_21 [DOI] [Google Scholar]
- Gant V., Hussain A., Bain M., Longshaw C., Henriksen A. S. (2021). In vitro activity of cefiderocol and comparators against gram-negative bacterial isolates from a series of surveillance studies in England: 2014–2018. J. Global Antimicrob. Resist. 27, 1–11. doi: 10.1016/j.jgar.2021.07.014, PMID: [DOI] [PubMed] [Google Scholar]
- García-Castillo M., Del Campo R., Morosini M. I., Riera E., Cabot G., Willems R., et al. (2011). Wide dispersion of ST175 clone despite high genetic diversity of carbapenem-nonsusceptible Pseudomonas aeruginosa clinical strains in 16 Spanish hospitals. J. Clin. Microbiol. 49, 2905–2910. doi: 10.1128/JCM.00753-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fernandez S., Garcia-Castillo M., Bou G., Calvo J., Cercenado E., Delgado M., et al. (2019). Activity of ceftolozane/tazobactam against Pseudomonas aeruginosa and Enterobacterales isolates recovered from intensive care unit patients in Spain: the SUPERIOR multicentre study. Int. J. Antimicrob. Agents 53, 682–688. doi: 10.1016/j.ijantimicag.2019.02.004 [DOI] [PubMed] [Google Scholar]
- Garg A., Garg J., Kumar S., Bhattacharya A., Agarwal S., Upadhyay G. (2019). Molecular epidemiology & therapeutic options of carbapenem-resistant gram-negative bacteria. Indian J. Med. Res. 149, 285–289. doi: 10.4103/ijmr.IJMR_36_18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudereto J. J., Neto L. V. P., Leite G. C., Espinoza E. P. S., Martins R. C. R., Villas Boa Prado G., et al. (2020). Comparison of methods for the detection of in vitro synergy in multidrug-resistant gram-negative bacteria. BMC Microbiol. 20, 1–7. doi: 10.1186/s12866-020-01756-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem S. M., Gad G. F., El-Baky A., Mahmoud R., Waly N. G. (2023). Association between antibiotic resistance, biofilm formation and LASB gene in Pseudomonas Aeruginosa isolated from different clinical specimens. Bull. Pharm. Sci. Assiut Univ. 46, 421–431. doi: 10.21608/bfsa.2023.301132 [DOI] [Google Scholar]
- Ghasemian S., Karami-Zarandi M., Heidari H., Khoshnood S., Kouhsari E., Ghafourian S., et al. (2023). Molecular characterizations of antibiotic resistance, biofilm formation, and virulence determinants of Pseudomonas aeruginosa isolated from burn wound infection. J. Clin. Lab. Anal. 37:e24850. doi: 10.1002/jcla.24850, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasemshahi S., Ahmadpor M., Booskabadi A., Rezaei H., Poopak B., Hakemi-Vala M. (2022). A six-month survey of the frequency of extensively drug-resistant gram-negative bacteria by VITEK 2 system in 2020. Iranian J. Med. Microbiol. 16, 134–140. doi: 10.30699/ijmm.16.2.134 [DOI] [Google Scholar]
- Gherardi G., Linardos G., Pompilio A., Fiscarelli E., Di Bonaventura G. (2019). Evaluation of in vitro activity of ceftolozane-tazobactam compared to other antimicrobial agents against Pseudomonas aeruginosa isolates from cystic fibrosis patients. Diagn. Microbiol. Infect. Dis. 94, 297–303. doi: 10.1016/j.diagmicrobio.2019.01.012, PMID: [DOI] [PubMed] [Google Scholar]
- Ghosh S., Bornman C., Zafer M. M. (2021). Antimicrobial resistance threats in the emerging COVID-19 pandemic: where do we stand? J. Infect. Public Health 14, 555–560. doi: 10.1016/j.jiph.2021.02.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani T., Arena F., Pollini S., Di Pilato V., D’Andrea M. M., Henrici De Angelis L., et al. (2018). Italian nationwide survey on Pseudomonas aeruginosa from invasive infections: activity of ceftolozane/tazobactam and comparators, and molecular epidemiology of carbapenemase producers. J. Antimicrob. Chemother. 73, 664–671. doi: 10.1093/jac/dkx453, PMID: [DOI] [PubMed] [Google Scholar]
- Gilani M., Munir T., Latif M., Rehman S., Ansari M., Hafeez A., et al. (2015). In vitro efficacy of Doripenem against Pseudomonas aeruginosa and Acinetobacter baumannii by E-test. J. Coll. Physicians Surg. Pak. 25, 726–729. doi: 10.2015/JCPSP.726729, PMID: [DOI] [PubMed] [Google Scholar]
- Goli H. R., Nahaei M. R., Rezaee M. A., Hasani A., Kafil H. S., Aghazadeh M., et al. (2017). Prevalence and molecular characterization of class 1 integrons among clinical isolates of Pseudomonas aeruginosa in northwest of Iran. Mol. Genet. Microbiol. Virol. 32, 109–115. doi: 10.3103/S0891416817020057 [DOI] [Google Scholar]
- Goli H. R., Nahaei M. R., Rezaee M. A., Hasani A., Samadi Kafil H., Aghazadeh M., et al. (2016). Contribution of mexAB-oprM and mexXY (−oprA) efflux operons in antibiotic resistance of clinical Pseudomonas aeruginosa isolates in Tabriz, Iran. Infect. Genet. Evol. 45, 75–82. doi: 10.1016/j.meegid.2016.08.022, PMID: [DOI] [PubMed] [Google Scholar]
- Golli A. L., Cristea O. M., Zlatian O., Glodeanu A. D., Balasoiu A. T., Ionescu M., et al. (2022). Prevalence of multidrug-resistant pathogens causing bloodstream infections in an intensive care unit. Infect. Drug Resist. 15, 5981–5992. doi: 10.2147/IDR.S383285, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A. C., Ortiz T., Valenzuela A., Egoávil-Espejo R., Huerto-Huanuco R., Pinto J. A., et al. (2023). Super-infection by multiple microorganisms in COVID-19 patients. Front. Mol. Biosci. 10:1113969. doi: 10.3389/fmolb.2023.1113969, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Garcés J., Aracil B., Gil Y., Burillo A. (2009). Susceptibility of 228 non-fermenting gram-negative rods to tigecycline and six other antimicrobial drugs. J. Chemother. 21, 267–271. doi: 10.1179/joc.2009.21.3.267, PMID: [DOI] [PubMed] [Google Scholar]
- Gomila M., del Carmen G. M., Fernández-Baca V., Pareja A., Pascual M., Díaz-Antolín P., et al. (2013). Genetic diversity of clinical Pseudomonas aeruginosa isolates in a public hospital in Spain. BMC Microbiol. 13, 138–110. doi: 10.1186/1471-2180-13-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunalan A., Sarumathi D., Sastry A. S., Ramanathan V., Rajaa S., Sistla S. (2021). Effect of combined colistin and meropenem against meropenem resistant Acinetobacter baumannii and Pseudomonas aeruginosa by checkerboard method: a cross sectional analytical study. Indian J. Pharmacol. 53, 207–212. doi: 10.4103/ijp.ijp_1013_20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Song Q., Mei S., Xue Z., Li J., Ning T. (2023). Distribution of multidrug-resistant bacterial infections in diabetic foot ulcers and risk factors for drug resistance: a retrospective analysis. PeerJ. 11:e16162. doi: 10.7717/peerj.16162, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Santana J. C., Geronimo-Gallegos A., Martinez-Corona M. B., Lopez-Lopez M., Toscano-Garibay J. D., Cuevas-Schacht F., et al. (2022). High rates of extensively drug-resistant Pseudomonas aeruginosa in children with cystic fibrosis. Curr. Microbiol. 79:353. doi: 10.1007/s00284-022-03048-4, PMID: [DOI] [PubMed] [Google Scholar]
- Guzek A., Korzeniewski K., Tomaszewski D., Rybicki Z., Zwolinska E. (2017). Bacteriological assessment of pneumonia caused by gram-negative Bacteria in patients hospitalized in intensive care unit. Adv. Exp. Med. Biol. 955, 39–46. doi: 10.1007/5584_2016_163, PMID: [DOI] [PubMed] [Google Scholar]
- Guzek A., Rybicki Z., Korzeniewski K., Mackiewicz K., Saks E., Chciałowski A., et al. (2015). Etiological factors causing lower respiratory tract infections isolated from hospitalized patients. Adv. Exp. Med. Biol. 835, 37–44. doi: 10.1007/5584_2014_23, PMID: [DOI] [PubMed] [Google Scholar]
- Güzel Ç. B., Gerçeker A. A. (2008). In vitro activities of various antibiotics, alone and in combination with colistin methanesulfonate, against Pseudomonas aeruginosa strains isolated from cystic fibrosis patients. Chemotherapy 54, 147–151. doi: 10.1159/000119741, PMID: [DOI] [PubMed] [Google Scholar]
- Hackel M. A., Tsuji M., Yamano Y., Echols R., Karlowsky J. A., Sahm D. F. (2018). In Vitro activity of the Siderophore cephalosporin, Cefiderocol, against Carbapenem-nonsusceptible and multidrug-resistant isolates of gram-negative Bacilli collected worldwide in 2014 to 2016. Antimicrob. Agents Chemother. 62:e01968. doi: 10.1128/AAC.01968-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeam H. A., Askar G., Al Sulaiman K., Mansour R., Al Qahtani M. M., Abbara D., et al. (2022). Treatment of multidrug-resistant Pseudomonas aeruginosa bacteremia using ceftolozane-tazobactam-based or colistin-based antibiotic regimens: a multicenter retrospective study. J. Infect. Public Health 15, 1081–1088. doi: 10.1016/j.jiph.2022.08.020, PMID: [DOI] [PubMed] [Google Scholar]
- Hallit S., Adaime A., Hajj A. (2020). Lebanese Observatory of Pathogenic Agents (Lopa-study): a 2 year-surveillance prospective study. Lebanese Med. J. 68, 126–133. doi: 10.12816/0060171 [DOI] [Google Scholar]
- Hansen C., Pressler T., Høiby N. (2008). Early aggressive eradication therapy for intermittent Pseudomonas aeruginosa airway colonization in cystic fibrosis patients: 15 years experience. J. Cyst. Fibros. 7, 523–530. doi: 10.1016/j.jcf.2008.06.009, PMID: [DOI] [PubMed] [Google Scholar]
- Hao L., Yang X., Chen H., Wei S., Xu B., Zhao Z. (2023). Distribution and drug resistance of bacterial infection in hospitalized patients at the respiratory department before and after the COVID-19 pandemic in Guangzhou, China. Microorganisms 11:2542. doi: 10.3390/microorganisms11102542, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasman H., Hammerum A. M., Hansen F., Hendriksen R. S., Olesen B., Agersø Y., et al. (2015). Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Eur. Secur. 20:30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085 [DOI] [PubMed] [Google Scholar]
- Hassen B., Hammami S., Hassen A., Abbassi M. (2022). Molecular mechanisms and clonal lineages of colistin-resistant bacteria across the African continent: a scoping review. Lett. Appl. Microbiol. 75, 1390–1422. doi: 10.1111/lam.13818, PMID: [DOI] [PubMed] [Google Scholar]
- Hawser S., Kothari N., Jemmely N., Redder N. (2021). Surveillance of ceftobiprole against gram-positive and gram-negative clinical isolates from 2018 from different European territories. J. Glob. Antimicrob. Resist. 26, 326–329. doi: 10.1016/j.jgar.2021.07.012, PMID: [DOI] [PubMed] [Google Scholar]
- Heidari R., Farajzadeh Sheikh A., Hashemzadeh M., Farshadzadeh Z., Salmanzadeh S., Saki M. (2022). Antibiotic resistance, biofilm production ability and genetic diversity of carbapenem-resistant Pseudomonas aeruginosa strains isolated from nosocomial infections in southwestern Iran. Mol. Biol. Rep. 49, 3811–3822. doi: 10.1007/s11033-022-07225-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A., Tan E., McCarthy K., Paterson D. (2018). Activity of ceftolozane/tazobactam against a collection of Pseudomonas aeruginosa isolates from bloodstream infections in Australia. Pathology 50, 748–752. doi: 10.1016/j.pathol.2018.08.009, PMID: [DOI] [PubMed] [Google Scholar]
- Herrera-Espejo S., Cebrero-Cangueiro T., Labrador-Herrera G., Pachón J., Pachón-Ibáñez M. E., Álvarez-Marín R. (2020). In vitro activity of pentamidine alone and in combination with antibiotics against multidrug-resistant clinical Pseudomonas aeruginosa strains. Antibiotics 9:885. doi: 10.3390/antibiotics9120885, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindler J. A., Humphries R. M. (2013). Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant gram-negative bacilli. J. Clin. Microbiol. 51, 1678–1684. doi: 10.1128/JCM.03385-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishinuma T., Tada T., Kuwahara-Arai K., Yamamoto N., Shimojima M., Kirikae T. (2018). Spread of GES-5 carbapenemase-producing Pseudomonas aeruginosa clinical isolates in Japan due to clonal expansion of ST235. PLoS One 13:e0207134. doi: 10.1371/journal.pone.0207134, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishinuma T., Uchida H., Tohya M., Shimojima M., Tada T., Kirikae T. (2020). Emergence and spread of VIM-type metallo-β-lactamase-producing Pseudomonas aeruginosa clinical isolates in Japan. J. Global Antimicrob. Resist. 23, 265–268. doi: 10.1016/j.jgar.2020.09.010, PMID: [DOI] [PubMed] [Google Scholar]
- Holger D. J., El Ghali A., Bhutani N., Lev K. L., Stamper K., Kebriaei R., et al. (2023). Phage-antibiotic combinations against multidrug-resistant Pseudomonas aeruginosa in in vitro static and dynamic biofilm models. Antimicrob. Agents Chemother. 67:e0057823. doi: 10.1128/aac.00578-23, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holger D. J., Lev K. L., Kebriaei R., Morrisette T., Shah R., Alexander J., et al. (2022b). Bacteriophage-antibiotic combination therapy for multidrug-resistant Pseudomonas aeruginosa: in vitro synergy testing. J. Appl. Microbiol. 133, 1636–1649. doi: 10.1111/jam.15647, PMID: [DOI] [PubMed] [Google Scholar]
- Holger D. J., Rebold N. S., Alosaimy S., Morrisette T., Lagnf A., Belza A. C., et al. (2022a). Impact of ceftolozane–tazobactam vs. best alternative therapy on clinical outcomes in patients with multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa lower respiratory tract infections. Infect. Dis. Ther. 11, 1965–1980. doi: 10.1007/s40121-022-00687-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. S., Kim J. O., Lee H., Bae I. K., Jeong S. H., Lee K. (2015). Characteristics of Metallo-β-lactamase-producing Pseudomonas aeruginosa in Korea. Infect. Chemother. 47, 33–40. doi: 10.3947/ic.2015.47.1.33, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoşbul T., Aydogan C. N., Kaya S., Bedir O., Özcan H., Gümral R. (2022). In vitro activity of ceftazidime-avibactam and colistin against carbapenem-resistant Pseudomonas aeruginosa clinical isolates. J. Istanbul Faculty Med. 85, 355–361. doi: 10.26650/IUITFD.1092556 [DOI] [PubMed] [Google Scholar]
- Hosseininassab Nodoushan S. A., Yadegari S., Moghim S., Isfahani B. N., Fazeli H., Poursina F., et al. (2017). Distribution of the strains of multidrug-resistant, extensively drug-resistant, and Pandrug-resistant Pseudomonas aeruginosa isolates from burn patients. Adv. Biomed. Res. 6:74. doi: 10.4103/abr.abr_239_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Anderson J., Davis M., Page A. M., Bower C. W., Smith G., Jacob J. T., et al. (2022). Prevalence of colistin heteroresistance in carbapenem-resistant Pseudomonas aeruginosa and association with clinical outcomes in patients: an observational study. J. Antimicrob. Chemother. 77, 793–798. doi: 10.1093/jac/dkab461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrbacek J., Cermak P., Zachoval R. (2020). Current antibiotic resistance trends of uropathogens in Central Europe: survey from a tertiary hospital urology department 2011–2019. Antibiotics 9:630. doi: 10.3390/antibiotics9090630, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh S. C., Lee Y. J., Huang Y. T., Liao C. H., Tsuji M., Hsueh P. R. (2019). In vitro activities of cefiderocol, ceftolozane/tazobactam, ceftazidime/avibactam and other comparative drugs against imipenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii, and Stenotrophomonas maltophilia, all associated with bloodstream infections in Taiwan. J. Antimicrob. Chemother. 74, 380–386. doi: 10.1093/jac/dky425, PMID: [DOI] [PubMed] [Google Scholar]
- Hu Y., Chen J., Huang L., Liu C., Zhou H., Zhang R. (2023). Antimicrobial susceptibility study and molecular epidemiology of ceftazidime/avibactam against Pseudomonas aeruginosa collected from clinical patients in PR China (2004–2021). J. Med. Microbiol. 72:001656. doi: 10.1099/jmm.0.001656 [DOI] [PubMed] [Google Scholar]
- Hu Y., Liu C., Wang Q., Zeng Y., Sun Q., Shu L., et al. (2021a). Emergence and expansion of a Carbapenem-resistant Pseudomonas aeruginosa clone are associated with plasmid-borne Bla (KPC-2) and virulence-related genes. mSystems 6:154. doi: 10.1128/mSystems.00154-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Peng W., Wu Y., Li H., Wang Q., Yi H., et al. (2021b). A potential high-risk clone of Pseudomonas aeruginosa ST463. Front. Microbiol. 12:670202. doi: 10.3389/fmicb.2021.670202, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Sun L., Nie T., Yang Y., Wang X., Pang J., et al. (2022). Evaluation of agar dilution method in susceptibility testing of Polymyxins for Enterobacteriaceae and non-fermentative rods: advantages compared to broth microdilution and broth macrodilution. Antibiotics 11:1392. doi: 10.3390/antibiotics11101392, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-S., Wang J.-T., Tai H.-M., Chang P.-C., Huang H.-C., Yang P.-C. (2022). Metal nanoparticles and nanoparticle composites are effective against Haemophilus influenzae, Streptococcus pneumoniae, and multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 55, 708–715. doi: 10.1016/j.jmii.2022.05.003 [DOI] [PubMed] [Google Scholar]
- Huband M. D., Castanheira M., Flamm R. K., Farrell D. J., Jones R. N., Sader H. S. (2016). In vitro activity of ceftazidime-avibactam against contemporary Pseudomonas aeruginosa isolates from U.S. medical centers by census region, 2014. Antimicrob. Agents Chemother. 60, 2537–2541. doi: 10.1128/AAC.03056-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huband M. D., Lindley J. M., Mendes R. E., Fedler K. A., Benn V. J., Zhang J., et al. (2020). In vitro activity of KHP-3757 (a novel LpxC inhibitor) and comparator agents against recent and molecularly characterized Pseudomonas aeruginosa isolates from a global surveillance program (2017–2018). Diagn. Microbiol. Infect. Dis. 98:115191. doi: 10.1016/j.diagmicrobio.2020.115191, PMID: [DOI] [PubMed] [Google Scholar]
- Humphries R. M., Janssen H., Hey-Hadavi J. H., Hackel M., Sahm D. (2023). Multidrug-resistant gram-negative bacilli recovered from respiratory and blood specimens from adults: the ATLAS surveillance program in European hospitals, 2018–2020. Int. J. Antimicrob. Agents 61:106724. doi: 10.1016/j.ijantimicag.2023.106724, PMID: [DOI] [PubMed] [Google Scholar]
- Ibrahim M. E. (2018). High antimicrobial resistant rates among gram-negative pathogens in intensive care units. A retrospective study at a tertiary care hospital in Southwest Saudi Arabia. Saudi Med. J. 39, 1035–1043. doi: 10.15537/smj.2018.10.22944, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S., Al-Saryi N., Al-Kadmy I., Aziz S. N. (2021). Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol. Biol. Rep. 48, 6987–6998. doi: 10.1007/s11033-021-06690-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou P., Alexakis K., Maraki S., Kofteridis D. P. (2023). Pseudomonas bacteremia in a tertiary hospital and factors associated with mortality. Antibiotics 12:670. doi: 10.3390/antibiotics12040670, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail S. J., Mahmoud S. S. (2018). First detection of New Delhi metallo-β-lactamases variants (NDM-1, NDM-2) among Pseudomonas aeruginosa isolated from Iraqi hospitals. Iran. J. Microbiol. 10, 98–103, PMID: [PMC free article] [PubMed] [Google Scholar]
- Izadi Pour Jahromi S., Mardaneh J., Sharifi A., Pezeshkpour V., Behzad-Behbahani A., Seyyedi N., et al. (2018). Occurrence of a multidrug resistant Pseudomonas aeruginosa strains in hospitalized patients in southwest of Iran: characterization of resistance trends and virulence determinants. J. Microbiol. 11:57341. doi: 10.5812/jjm.57341 [DOI] [Google Scholar]
- Jalali Y., Sturdik I., Jalali M., Payer J. (2021). Isolated carbapenem resistant bacteria, their multidrug resistant profile, percentage of healthcare associated infection and associated mortality, in hospitalized patients in a University Hospital in Bratislava. Clin. Study 122, 379–385. doi: 10.4149/10.4149/BLL_2021_063 [DOI] [PubMed] [Google Scholar]
- Jamalifar H., Samadi N., Nowroozi J., Dezfulian M., Fazeli M. R. (2019). Down-regulatory effects of green coffee extract on las I and las R virulence-associated genes in Pseudomonas aeruginosa. DARU J. Pharm. Sci. 27, 35–42. doi: 10.1007/s40199-018-0234-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Mahrt N., Tueffers L., Barbosa C., Harjes M., Adolph G., et al. (2016). Association between clinical antibiotic resistance and susceptibility of Pseudomonas in the cystic fibrosis lung. Evol. Med. Public Health 2016, 182–194. doi: 10.1093/emph/eow016, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japoni A., Vazin A., Davarpanah M. A., Ardakani M. A., Alborzi A., Japoni S., et al. (2011). Ventilator-associated pneumonia in Iranian intensive care units. J. Infect. Dev. Countries 5, 286–293. doi: 10.3855/jidc.1212, PMID: [DOI] [PubMed] [Google Scholar]
- Jauhari S., Pal S., Goyal M., Prakash R., Juyal D. (2020). Bacteriological and antimicrobial sensitivity profile of burn wound infections in a tertiary Care Hospital of Uttarakhand. Int. J. Curr. Res. Rev. 12, 30–36. doi: 10.31782/IJCRR.2020.12127 [DOI] [Google Scholar]
- Jayarani M., Jaikumar S., Sandhya R. T. (2020). Microbiological profile and drug resistant Organism's pattern in diabetic foot ulcer patients at tertiary care hospital Puducherry. Int. J. Res. Pharmaceutical Sci. 11, 6692–6697. doi: 10.26452/ijrps.v11i4.3592 [DOI] [Google Scholar]
- Jazani N. H., Zahedi A., Garebagi N. (2012). Phenotypic detection of metallo-β-lactamase producing Pseudomonas aeruginosa isolated from Urmia hospitals. African J. Microbiol. Res. 6, 1387–1392. doi: 10.5897/AJMR11.1023 [DOI] [Google Scholar]
- Jean S.-S., Hsueh P.-R., Lee W.-S., Chang H.-T., Chou M.-Y., Chen S., et al. (2009). Nationwide surveillance of antimicrobial resistance among non-fermentative gram-negative bacteria in intensive care units in Taiwan: SMART programme data 2005. Int. J. Antimicrob. Agents 33, 266–271. doi: 10.1016/j.ijantimicag.2008.08.026 [DOI] [PubMed] [Google Scholar]
- Jean S. S., Lee Y. L., Liu P. Y., Lu M. C., Ko W. C., Hsueh P. R. (2022). Multicenter surveillance of antimicrobial susceptibilities and resistance mechanisms among Enterobacterales species and non-fermenting gram-negative bacteria from different infection sources in Taiwan from 2016 to 2018. J. Microbiol. Immunol. Infect. 55, 463–473. doi: 10.1016/j.jmii.2021.07.015, PMID: [DOI] [PubMed] [Google Scholar]
- Jean S. S., Lee W. S., Yu K. W., Liao C. H., Hsu C. W., Chang F. Y., et al. (2016). Rates of susceptibility of carbapenems, ceftobiprole, and colistin against clinically important bacteria collected from intensive care units in 2007: results from the surveillance of multicenter antimicrobial resistance in Taiwan (SMART). J. Microbiol. Immunol. Infect. 49, 969–976. doi: 10.1016/j.jmii.2014.12.008, PMID: [DOI] [PubMed] [Google Scholar]
- Jeong S., Jeon K., Lee N., Park M.-J., Song W. (2024). Changing genotypic distribution, antimicrobial susceptibilities, and risk factors of urinary tract infection caused by carbapenemase-producing pseudomonas aeruginosa. Ann. Lab. Med. 44, 38–46. doi: 10.3343/alm.2024.44.1.38, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guerra G., Heras-Canas V., Gutierrez-Soto M., Del Pilar A.-P. M., Exposito-Ruiz M., Navarro-Mari J. M., et al. (2018). Urinary tract infection by Acinetobacter baumannii and Pseudomonas aeruginosa: evolution of antimicrobial resistance and therapeutic alternatives. J. Med. Microbiol. 67, 790–797. doi: 10.1099/jmm.0.000742, PMID: [DOI] [PubMed] [Google Scholar]
- Jones R. N., Castanheira M., Hu B., Ni Y., Lin S. S., Mendes R. E., et al. (2013a). Update of contemporary antimicrobial resistance rates across China: reference testing results for 12 medical centers (2011). Diagn. Microbiol. Infect. Dis. 77, 258–266. doi: 10.1016/j.diagmicrobio.2013.07.003, PMID: [DOI] [PubMed] [Google Scholar]
- Jones R. N., Flonta M., Gurler N., Cepparulo M., Mendes R. E., Castanheira M. (2014). Resistance surveillance program report for selected European nations (2011). Diagn. Microbiol. Infect. Dis. 78, 429–436. doi: 10.1016/j.diagmicrobio.2013.10.008, PMID: [DOI] [PubMed] [Google Scholar]
- Jones R. N., Guzman-Blanco M., Gales A. C., Gallegos B., Castro A. L. L., Martino M. D. V., et al. (2013b). Susceptibility rates in Latin American nations: report from a regional resistance surveillance program (2011). Braz. J. Infect. Dis. 17, 672–681. doi: 10.1016/j.bjid.2013.07.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász E., Iván M., Pintér E., Pongrácz J., Kristóf K. (2017a). Colistin resistance among blood culture isolates at a tertiary care Centre in Hungary. J. Global Antimicrob. Resist. 11, 167–170. doi: 10.1016/j.jgar.2017.08.002, PMID: [DOI] [PubMed] [Google Scholar]
- Juhász E., Kovacs A., Pongracz J., Ivan M., Kristof K. (2017b). In vitro activity of Colistin and trimethoprim/sulfamethoxazole against consortia of multidrug resistant non-fermenting gram-negative Bacilli isolated from lower respiratory tract. Jundishapur J. Microbiol. 10:14034. doi: 10.5812/jjm.14034 [DOI] [Google Scholar]
- Jung S., Chung E. K., Jun M. S., Son E. S., Rhie S. J. (2019). Differences in Colistin administration and bacterial and treatment outcomes in critically ill patients. Sci. Rep. 9:8781. doi: 10.1038/s41598-019-44965-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabic J., Fortunato G., Vaz-Moreira I., Kekic D., Jovicevic M., Pesovic J., et al. (2023). Dissemination of metallo-β-lactamase-producing Pseudomonas aeruginosa in Serbian hospital settings: expansion of ST235 and ST654 clones. Int. J. Mol. Sci. 24:1519. doi: 10.3390/ijms24021519, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakhandki L., Takpere A. Y., Bagali S., Wavare S., Karigoudar R., Shahapur P. R. (2020). Antibiogram of gram negative bacteria isolated from the skin and soft tissue infections a guide for empirical therapy to the clinicians. J. Pure Appl. Microbiol. 14, 1353–1358. doi: 10.22207/JPAM.14.2.32 [DOI] [Google Scholar]
- Kanwar N., Harrison C. J., Pence M. A., Qin X., Selvarangan R. (2023). Comparative in vitro antipseudomonal activity of ceftolozane/tazobactam against Pseudomonas aeruginosa isolates from children with cystic fibrosis. Diagn. Microbiol. Infect. Dis. 105:115904. doi: 10.1016/j.diagmicrobio.2023.115904, PMID: [DOI] [PubMed] [Google Scholar]
- Karami P., Khaledi A., Mashoof R. Y., Yaghoobi M. H., Karami M., Dastan D., et al. (2020). The correlation between biofilm formation capability and antibiotic resistance pattern in Pseudomonas aeruginosa. Gene Rep. 18:100561. doi: 10.1016/j.genrep.2019.100561 [DOI] [Google Scholar]
- Karimzadeh I., Sadeghimanesh N., Mirzaee M., Sagheb M. M. (2017). Evaluating the resistance pattern of gram-negative bacteria during three years at the nephrology ward of a referral hospital in southwest of Iran. J. Nephropathol. 6, 210–219. doi: 10.15171/jnp.2017.35, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky J. A., Adam H. J., Baxter M. R., Lagacé-Wiens P. R., Walkty A. J., Hoban D. J., et al. (2013). In vitro activity of ceftaroline-avibactam against gram-negative and gram-positive pathogens isolated from patients in Canadian hospitals from 2010 to 2012: results from the CANWARD surveillance study. Antimicrob. Agents Chemother. 57, 5600–5611. doi: 10.1128/AAC.01485-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky J. A., Bouchillon S. K., Benaouda A., Soraa N., Zerouali K., Mohamed N., et al. (2022). Antimicrobial susceptibility testing of clinical isolates of gram-negative bacilli collected in Morocco by the ATLAS global surveillance program from 2018 to 2020. J. Global Antimicrob. Resist. 30, 23–30. doi: 10.1016/j.jgar.2022.04.011, PMID: [DOI] [PubMed] [Google Scholar]
- Karlowsky J. A., Hackel M. A., Tsuji M., Yamano Y., Echols R., Sahm D. F. (2019a). In vitro activity of cefiderocol, a siderophore cephalosporin, against gram-negative bacilli isolated by clinical laboratories in North America and Europe in 2015-2016: SIDERO-WT-2015. Int. J. Antimicrob. Agents 53, 456–466. doi: 10.1016/j.ijantimicag.2018.11.007, PMID: [DOI] [PubMed] [Google Scholar]
- Karlowsky J. A., Kazmierczak K. M., Bouchillon S. K., de Jonge B. L. M., Stone G. G., Sahm D. F. (2019b). In vitro activity of ceftazidime-avibactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa collected in Latin American countries: results from the INFORM global surveillance program, 2012 to 2015. Antimicrob. Agents Chemother. 63:e01814. doi: 10.1128/AAC.01814-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky J. A., Lob S. H., Hawser S. P., Kothari N., Siddiqui F., Alekseeva I., et al. (2023). Activity of ceftolozane/tazobactam and imipenem/relebactam against clinical isolates of Enterobacterales and Pseudomonas aeruginosa collected in central and northern Europe (Belgium, Norway, Sweden, Switzerland)—SMART 2017–21. JAC-Antimicrob. Resist. 5:dlad098. doi: 10.1093/jacamr/dlad098, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky J. A., Lob S. H., Kazmierczak K. M., Young K., Motyl M. R., Sahm D. F. (2018a). In vitro activity of imipenem-Relebactam against clinical isolates of gram-negative Bacilli isolated in Hospital Laboratories in the United States as part of the SMART 2016 program. Antimicrob. Agents Chemother. 62:62. doi: 10.1128/AAC.00169-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky J. A., Lob S. H., Young K., Motyl M. R., Sahm D. F. (2018b). Activity of imipenem/relebactam against Pseudomonas aeruginosa with antimicrobial-resistant phenotypes from seven global regions: SMART 2015–2016. J. Global Antimicrob. Resist. 15, 140–147. doi: 10.1016/j.jgar.2018.07.012, PMID: [DOI] [PubMed] [Google Scholar]
- Karlowsky J. A., Lob S. H., Young K., Motyl M. R., Sahm D. F. (2021). In vitro activity of imipenem/Relebactam against gram-negative Bacilli from pediatric patients-study for monitoring antimicrobial resistance trends (SMART) global surveillance program 2015-2017. J. Pediatric Infect. Dis. Soc. 10, 274–281. doi: 10.1093/jpids/piaa056 [DOI] [PubMed] [Google Scholar]
- Karruli A., Massa A., Bertolino L., Andini R., Sansone P., Dongiovanni S., et al. (2022). Clinical characteristics and outcome of MDR/XDR bacterial infections in a neuromuscular semi-intensive/sub-intensive care unit. Antibiotics 11:1411. doi: 10.3390/antibiotics11101411, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashfi M., Hashemi A., Eslami G., Amin M. S., Tarashi S., Taki E. (2017). The prevalence of aminoglycoside-modifying enzyme genes among Pseudomonas aeruginosa strains isolated from burn patients. Arch. Clin. Infect. Dis. 12:40896. doi: 10.5812/archcid.40896 [DOI] [Google Scholar]
- Katchanov J., Asar L., Klupp E. M., Both A., Rothe C., Konig C., et al. (2018). Carbapenem-resistant gram-negative pathogens in a German university medical center: prevalence, clinical implications and the role of novel beta-lactam/beta-lactamase inhibitor combinations. PLoS One 13:e0195757. doi: 10.1371/journal.pone.0195757, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Chauhan S., Saini A., Chauhan J., Kumar H. (2022). Assessment of colistin resistance in gram negative bacteria from clinical samples in resource-limited settings. Asian Pac J Trop Med 15, 367–373. doi: 10.4103/1995-7645.351764 [DOI] [Google Scholar]
- Kazmierczak K. M., de Jonge B. L. M., Stone G. G., Sahm D. F. (2018). In vitro activity of ceftazidime/avibactam against isolates of Pseudomonas aeruginosa collected in European countries: INFORM global surveillance 2012-15. J. Antimicrob. Chemother. 73, 2777–2781. doi: 10.1093/jac/dky267, PMID: [DOI] [PubMed] [Google Scholar]
- Kazmierczak K. M., Rabine S., Hackel M., McLaughlin R. E., Biedenbach D. J., Bouchillon S. K., et al. (2016). Multiyear, multinational survey of the incidence and global distribution of Metallo-β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 60, 1067–1078. doi: 10.1128/AAC.02379-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kc R., Adhikari S., Bastola A., Devkota L., Bhandari P., Ghimire P., et al. (2019). Opportunistic respiratory infections in HIV patients attending sukraraj tropical and infectious diseases hospital in Kathmandu, Nepal. HIV/AIDS Res. Palliative Care 11, 357–367. doi: 10.2147/HIV.S229531, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keepers T. R., Gomez M., Celeri C., Nichols W. W., Krause K. M. (2014). Bactericidal activity, absence of serum effect, and time-kill kinetics of ceftazidime-avibactam against β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 58, 5297–5305. doi: 10.1128/AAC.02894-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria A., Praharaj A. K., Kumar M., Grover N. (2013). Emergence of NDM–1 in the clinical isolates of Pseudomonas aeruginosa in India. J. Clin. Diagn. Res. 7:1328. doi: 10.7860/JCDR/2013/5509.3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F. Y., Abu-Khattab M., Almaslamani E. A., Hassan A. A., Mohamed S. F., Elbuzdi A. A., et al. (2017). Acute bacterial meningitis in Qatar: a hospital-based study from 2009 to 2013. Biomed. Res. Int. 2017, 1–8. doi: 10.1155/2017/2975610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J. Z., Ismail M., Ullah R., Ali W., Ali I. (2023). Increased burden of MDR bacterial infections; reflection from an antibiogram of ICUs of a tertiary care hospital. J. Infect. Dev. Ctries. 17, 994–998. doi: 10.3855/jidc.18142 [DOI] [PubMed] [Google Scholar]
- Khater E. (2022). Detection of carbapenem-resistant Pseudomonas aeruginosa in tertiary care hospital in Saudi Arabia. Microbes Infect. Dis. 0:1320. [Google Scholar]
- Khater E. S., AlFaki A. A. A. (2022). Detection of carbapenem-resistant Pseudomonas aeruginosa in tertiary care hospital in Saudi Arabia. Microbes Infect. Dis. 3, 693–702. doi: 10.21608/mid.2022.142569.1320 [DOI] [Google Scholar]
- Khorvash F., Yazdani M., Shabani S., Soudi A. (2017). Pseudomonas aeruginosa-producing Metallo-beta-lactamases (VIM, IMP, SME, and AIM) in the clinical isolates of intensive care units, a University Hospital in Isfahan. Iran. Adv. Biomed. Res. 6:147. doi: 10.4103/2277-9175.219412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnood S., Khosravi A. D., Jomehzadeh N., Montazeri E. A., Motahar M., Shahi F., et al. (2019). Distribution of extended-spectrum β-lactamase genes in antibiotic-resistant strains of Pseudomonas aeruginosa obtained from burn patients in Ahvaz, Iran. J. Acute Dis. 8, 53–57. doi: 10.4103/2221-6189.254426 [DOI] [Google Scholar]
- Khosravi A. D., Motahar M., Abbasi M. E. (2017). The frequency of class1 and 2 integrons in Pseudomonas aeruginosa strains isolated from burn patients in a burn center of Ahvaz, Iran. PLoS One 12:e0183061. doi: 10.1371/journal.pone.0183061, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A. D., Shafie F., Abbasi Montazeri E., Rostami S. (2016). The frequency of genes encoding exotoxin a and exoenzyme S in Pseudomonas aeruginosa strains isolated from burn patients. Burns 42, 1116–1120. doi: 10.1016/j.burns.2016.02.012, PMID: [DOI] [PubMed] [Google Scholar]
- Khudair A. N. A., Mahmood S. S. (2021). Detection of the antiseptic resistance gene among Pseudomonas aeruginosa isolates. Iraqi J. Sci., 62, 75–82. doi: 10.24996/ijs.2021.62.1.7 [DOI] [Google Scholar]
- Kidd T. J., Ramsay K. A., Hu H., Bye P. T., Elkins M. R., Grimwood K., et al. (2009). Low rates of Pseudomonas aeruginosa misidentification in isolates from cystic fibrosis patients. J. Clin. Microbiol. 47, 1503–1509. doi: 10.1128/JCM.00014-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S., Park B. K., Kim S. K., Han S. B., Lee J. W., Lee D. G., et al. (2017). Clinical characteristics and outcomes of Pseudomonas aeruginosa bacteremia in febrile neutropenic children and adolescents with the impact of antibiotic resistance: a retrospective study. BMC Infect. Dis. 17:500. doi: 10.1186/s12879-017-2597-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Yoon E.-J., Hong J. S., Choi M. H., Kim H. S., Kim Y. R., et al. (2022). Major bloodstream infection-causing bacterial pathogens and their antimicrobial resistance in South Korea, 2017–2019: phase i report from kor-glass. Front. Microbiol. 12:799084. doi: 10.3389/fmicb.2021.799084, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kırac S., Keskin D., Yarar M. (2018). Antimicrobial susceptibility pattern and multidrug resistance ındex in Pseudomonas aeruginosa among clinical isolates in Denizli, Turkey. Tanzan. J. Health Res. 20:v20i3. doi: 10.4314/thrb.v20i3.6 [DOI] [Google Scholar]
- Kiratisin P., Kazmierczak K., Stone G. G. (2021). In vitro activity of ceftazidime/avibactam and comparators against carbapenemase-producing Enterobacterales and Pseudomonas aeruginosa isolates collected globally between 2016 and 2018. J. Global Antimicrob. Resist. 27, 132–141. doi: 10.1016/j.jgar.2021.08.010, PMID: [DOI] [PubMed] [Google Scholar]
- Kiratisin P., Kempf M., Stone G., Utt E. (2023). Ceftazidime-avibactam and comparators against Pseudomonas aeruginosa isolates collected globally and in each geographical region between 2017-2020. J. Glob. Antimicrob. Resist. 34, 113–118. doi: 10.1016/j.jgar.2023.06.005, PMID: [DOI] [PubMed] [Google Scholar]
- Ko W.-C., Stone G. G. (2020). In vitro activity of ceftazidime–avibactam and comparators against gram-negative bacterial isolates collected in the Asia–Pacific region as part of the INFORM program (2015–2017). Ann. Clin. Microbiol. Antimicrob. 19, 1–12. doi: 10.1186/s12941-020-00355-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohira N., Hackel M. A., Oota M., Takemura M., Hu F., Mizuno H., et al. (2023). In vitro antibacterial activities of cefiderocol against gram-negative clinical strains isolated from China in 2020. J. Global Antimicrob. Resist. 32, 181–186. doi: 10.1016/j.jgar.2022.11.017, PMID: [DOI] [PubMed] [Google Scholar]
- Kokkayil P., Agarwal R., Mohapatra S., Bakshi S., Das B., Sood S., et al. (2018). Bacterial profile and antibiogram of blood stream infections in febrile neutropenic patients with haematological malignancies. J. Infect. Dev. Ctries. 12, 442–447. doi: 10.3855/jidc.9725 [DOI] [PubMed] [Google Scholar]
- Kombade S. P., Kaur N., Patro S. K., Nag V. L. (2021). Clinico-bacteriological and antibiotic drug resistance profile of chronic suppurative otitis media at a tertiary care hospital in Western Rajasthan. J. Family Med. Prim. Care 10, 2572–2579. doi: 10.4103/jfmpc.jfmpc_2480_20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzekwa K., Kędziora A., Stańczykiewicz B., Bugla-Płoskońska G., Wojnicz D. (2021). Benefits of usage of immobilized silver nanoparticles as Pseudomonas aeruginosa antibiofilm factors. Int. J. Mol. Sci. 23:284. doi: 10.3390/ijms23010284, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothari A., Kumar S. K., Singh V., Kumar P., Kaushal K., Pandey A., et al. (2022). Association of multidrug resistance behavior of clinical Pseudomonas aeruginosa to pigment coloration. Eur. J. Med. Res. 27:120. doi: 10.1186/s40001-022-00752-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic P., Zlojutro B., Kovacevic T., Baric G., Dragic S., Momcicevic D. (2019). Microorganisms profile and antibiotics sensitivity patterns in the only medical intensive care unit in Bosnia and Herzegovina. Microb. Drug Resist. 25, 1176–1181. doi: 10.1089/mdr.2018.0458, PMID: [DOI] [PubMed] [Google Scholar]
- Kragh K. N., Gijón D., Maruri A., Antonelli A., Coppi M., Kolpen M., et al. (2021). Effective antimicrobial combination in vivo treatment predicted with microcalorimetry screening. J. Antimicrob. Chemother. 76, 1001–1009. doi: 10.1093/jac/dkaa543, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K. M., Haglund C. M., Hebner C., Serio A. W., Lee G., Nieto V., et al. (2019). Potent LpxC inhibitors with in vitro activity against multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63:10.1128/aac, 00977–00919. doi: 10.1128/AAC.00977-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresken M., Korber-Irrgang B., Korte-Berwanger M., Pfennigwerth N., Gatermann S. G., Seifert H., et al. (2020a). Dissemination of carbapenem-resistant Pseudomonas aeruginosa isolates and their susceptibilities to ceftolozane-tazobactam in Germany. Int. J. Antimicrob. Agents 55:105959. doi: 10.1016/j.ijantimicag.2020.105959, PMID: [DOI] [PubMed] [Google Scholar]
- Kresken M., Korte-Berwanger M., Gatermann S. G., Pfeifer Y., Pfennigwerth N., Seifert H., et al. (2020b). In vitro activity of cefiderocol against aerobic gram-negative bacterial pathogens from Germany. Int. J. Antimicrob. Agents 56:106128. doi: 10.1016/j.ijantimicag.2020.106128, PMID: [DOI] [PubMed] [Google Scholar]
- Kristof K., Adamkova V., Adler A., Gospodarek-Komkowska E., Rafila A., Billova S., et al. (2021). In vitro activity of ceftazidime-avibactam and comparators against Enterobacterales and Pseudomonas aeruginosa isolates from Central Europe and Israel, 2014–2017 and 2018. Diagn. Microbiol. Infect. Dis. 101:115420. doi: 10.1016/j.diagmicrobio.2021.115420, PMID: [DOI] [PubMed] [Google Scholar]
- Kuo S.-C., Wang Y.-C., Tan M.-C., Huang W.-C., Shiau Y.-R., Wang H.-Y., et al. (2021). In vitro activity of imipenem/relebactam, meropenem/vaborbactam, ceftazidime/avibactam, cefepime/zidebactam and other novel antibiotics against imipenem-non-susceptible gram-negative bacilli from Taiwan. J. Antimicrob. Chemother. 76, 2071–2078. doi: 10.1093/jac/dkab141, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D., Matsumoto S., Kishi N., Ishii Y., Mori M. (2022). In vitro antibacterial activity of imipenem/relebactam against clinical isolates in Japan. Microbiol. Spectrum. 10, e02235–e02221. doi: 10.1128/spectrum.02235-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Chen Y.-S., Lee N.-Y., Tang H.-J., Lee S. S.-J., Lin C.-F., et al. (2019). Susceptibility rates of clinically important bacteria collected from intensive care units against colistin, carbapenems, and other comparative agents: results from surveillance of multicenter antimicrobial resistance in Taiwan (SMART). Infect. Drug Resist. 12, 627–640. doi: 10.2147/IDR.S194482, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurentiu T. A., Nicoleta M., Octav P., Gheorghe I., Marcela P., Otilia B., et al. (2017). Resistance features of pseudomonas aeruginosa strains isolated from patients with infectious complications of cardiovascular surgery. Biointerface Res. Appl. Chem. 7, 2004–2008. [Google Scholar]
- Lee S.-C., Huang S.-S., See L.-C., Tsai M.-H., Shieh W.-B. (2011). In vitro activities of nine current antibiotics against culprit bacteria in nosocomial infections in an institution in northern Taiwan. Chang Gung Med. J. 34, 580–589, PMID: [PubMed] [Google Scholar]
- Lee Y. L., Ko W. C., Hsueh P. R. (2023). In vitro activities of ceftazidime-avibactam, ceftolozane-tazobactam, meropenem-vaborbactam and other comparators against Pseudomonas aeruginosa isolates with discrepant resistance to carbapenems: data from the antimicrobial testing leadership and surveillance (ATLAS) program, 2012-2021. Int. J. Antimicrob. Agents 62:106867. doi: 10.1016/j.ijantimicag.2023.106867, PMID: [DOI] [PubMed] [Google Scholar]
- Lee J.-Y., Lim M. H., Heo S. T., Ko K. S. (2012). Repeated isolation of Pseudomonas aeruginosa isolates resistant to both polymyxins and carbapenems from 1 patient. Diagn. Microbiol. Infect. Dis. 72, 267–271. doi: 10.1016/j.diagmicrobio.2011.11.014, PMID: [DOI] [PubMed] [Google Scholar]
- Lee Y.-L., Lu M.-C., Shao P.-L., Lu P.-L., Chen Y.-H., Cheng S.-H., et al. (2019). Nationwide surveillance of antimicrobial resistance among clinically important gram-negative bacteria, with an emphasis on carbapenems and colistin: Results from the surveillance of multicenter antimicrobial resistance in Taiwan (SMART) in 2018. Int. J. Antimicrob. Agents 54, 318–328. doi: 10.1016/j.ijantimicag.2019.06.009 [DOI] [PubMed] [Google Scholar]
- Lee J.-Y., Song J.-H., Ko K. S. (2011). Identification of nonclonal Pseudomonas aeruginosa isolates with reduced colistin susceptibility in Korea. Microb. Drug Resist. 17, 299–304. doi: 10.1089/mdr.2010.0145 [DOI] [PubMed] [Google Scholar]
- Li X. Y., Liu X. G., Dong Z. L., Chai L. T., Liu Y. J., Qi J., et al. (2023). The distribution, drug susceptibility, and dynamic trends of Pseudomonas aeruginosa infection in a tertiary Hospital in China during 2016–2022. Infect. Drug Resist. 16, 3525–3533. doi: 10.2147/IDR.S408956, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. Y., Liu B., Liu L. (2021). Successfully controlling the incidence of multidrug-resistant Pseudomonas aeruginosa through antibiotic stewardship and infection control programmes at a Chinese university hospital. J. Clin. Pharm. Ther. 46, 1357–1366. doi: 10.1111/jcpt.13446, PMID: [DOI] [PubMed] [Google Scholar]
- Li X.-F., Shi H.-Q., Liang Y., Li J., Jiang B., Song G.-B. (2022). Interaction of biofilm and efflux pump in clinical isolates of carbapenem resistant P. aeruginosa. European Rev. Med. Pharmacol. Sci. 26. [DOI] [PubMed] [Google Scholar]
- Licata F., Quirino A., Pepe D., Matera G., Bianco A., Group C (2020). Antimicrobial resistance in pathogens isolated from blood cultures: a two-year multicenter hospital surveillance study in Italy. Antibiotics 10:10. doi: 10.3390/antibiotics10010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K.-Y., Lauderdale T.-L., Wang J.-T., Chang S.-C. (2016). Carbapenem-resistant Pseudomonas aeruginosa in Taiwan: prevalence, risk factors, and impact on outcome of infections. J. Microbiol. Immunol. Infect. 49, 52–59. doi: 10.1016/j.jmii.2014.01.005, PMID: [DOI] [PubMed] [Google Scholar]
- Lin J., Xu C., Fang R., Cao J., Zhang X., Zhao Y., et al. (2019). Resistance and heteroresistance to colistin in Pseudomonas aeruginosa isolates from Wenzhou, China. Antimicrob. Agents Chemother. 63, 00556–00519. doi: 10.1128/AAC.00556-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Zhao X., Huang J., Liu W., Zheng Y., Yang X., et al. (2019). Rapid screening of colistin-resistant Escherichia coli, Acinetobacter baumannii and Pseudomonas aeruginosa by the use of Raman spectroscopy and hierarchical cluster analysis. Analyst 144, 2803–2810. doi: 10.1039/C8AN02220H, PMID: [DOI] [PubMed] [Google Scholar]
- Liu P. Y., Lee Y. L., Lu M. C., Shao P. L., Lu P. L., Chen Y. H., et al. (2020). National surveillance of antimicrobial susceptibility of Bacteremic gram-negative Bacteria with emphasis on community-acquired resistant isolates: report from the 2019 surveillance of multicenter antimicrobial resistance in Taiwan (SMART). Antimicrob. Agents Chemother. 64:64. doi: 10.1128/AAC.01089-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu B., Li Y., Zhang W. (2018). Successful control of resistance in Pseudomonas aeruginosa using antibiotic stewardship and infection control programs at a Chinese university hospital: a 6-year prospective study. Infect. Drug Resist. 11, 637–646. doi: 10.2147/IDR.S163853, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-Y., Wang Y., Walsh T. R., Yi L.-X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7, PMID: [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Mushtaq S., Meunier D., Hopkins K. L., Hill R., Adkin R., et al. (2017). Activity of ceftolozane/tazobactam against surveillance and ‘problem’Enterobacteriaceae, Pseudomonas aeruginosa and non-fermenters from the British Isles. J. Antimicrob. Chemother. 72, 2278–2289. doi: 10.1093/jac/dkx136, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanes C., Pourcel C., Richardot C., Plésiat P., Fichant G., Cavallo J. D., et al. (2013). Diversity of β-lactam resistance mechanisms in cystic fibrosis isolates of Pseudomonas aeruginosa: a French multicentre study. J. Antimicrob. Chemother. 68, 1763–1771. doi: 10.1093/jac/dkt115, PMID: [DOI] [PubMed] [Google Scholar]
- Lob S. H., Hawser S. P., Siddiqui F., Alekseeva I., DeRyke C. A., Young K., et al. (2023). Activity of ceftolozane/tazobactam and imipenem/relebactam against clinical gram-negative isolates from Czech Republic, Hungary, and Poland—SMART 2017–2020. Eur. J. Clin. Microbiol. Infect. Dis. 42, 365–370. doi: 10.1007/s10096-023-04549-1, PMID: [DOI] [PubMed] [Google Scholar]
- Lockwood W., Lawson L. A. (1973). Studies on the susceptibility of 150 consecutive clinical isolates of Pseudomonas aeruginosa to tobramycin, gentamicin, colistin, carbenicillin, and five other antimicrobials. Antimicrob. Agents Chemother. 4, 281–284. doi: 10.1128/AAC.4.3.281, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longshaw C., Manissero D., Tsuji M., Echols R., Yamano Y. (2020). In vitro activity of the siderophore cephalosporin, cefiderocol, against molecularly characterized, carbapenem-non-susceptible gram-negative bacteria from Europe. JAC Antimicrob. Resist. 2:60. doi: 10.1093/jacamr/dlaa060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Montesinos I., Gómez-Zorrilla S., Palacios-Baena Z. R., Prim N., Echeverria-Esnal D., Gracia M. P., et al. (2022). Aminoglycoside or Polymyxin monotherapy for treating complicated urinary tract infections caused by extensively drug-resistant Pseudomonas aeruginosa: a propensity score-adjusted and matched cohort study. Infect. Dis. Ther. 11, 335–350. doi: 10.1007/s40121-021-00570-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Causape C., de Dios-Caballero J., Cobo M., Escribano A., Asensio O., Oliver A., et al. (2017). Antibiotic resistance and population structure of cystic fibrosis Pseudomonas aeruginosa isolates from a Spanish multi-Centre study. Int. J. Antimicrob. Agents 50, 334–341. doi: 10.1016/j.ijantimicag.2017.03.034 [DOI] [PubMed] [Google Scholar]
- Lopez-Causape C., Rojo-Molinero E., Mulet X., Cabot G., Moya B., Figuerola J., et al. (2013). Clonal dissemination, emergence of mutator lineages and antibiotic resistance evolution in Pseudomonas aeruginosa cystic fibrosis chronic lung infection. PLoS One 8:e71001. doi: 10.1371/journal.pone.0071001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly N. S., Bulitta J. B., Rao G. G., Landersdorfer C. B., Holden P. N., Forrest A., et al. (2015). Colistin and doripenem combinations against Pseudomonas aeruginosa: profiling the time course of synergistic killing and prevention of resistance. J. Antimicrob. Chemother. 70, 1434–1442. doi: 10.1093/jac/dku567, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macin S., Akyon Y. (2017). Phenotypic and genotypic virulence factors in Pseudomonas aeruginosa strains according to pigment presence. Acta Medica Mediterranea. 33, 1033–1038. doi: 10.19193/0393-6384_2017_6_163 [DOI] [Google Scholar]
- MacKenzie F., Smith S. V., Milne K., Griffiths K., Legge J., Gould I. M. (2004). Antibiograms of resistant gram-negative bacteria from Scottish CF patients. J. Cyst. Fibros. 3, 151–157. doi: 10.1016/j.jcf.2004.03.009 [DOI] [PubMed] [Google Scholar]
- Mahar U., Anwar N., Fatima N., Hassan J., Shamsi T. (2020). Emerging anti-microbial resistance in febrile neutropenia: is it high time to evaluate quality control measures? Pak J Med Sci. 36, 1246–1251. doi: 10.12669/pjms.36.6.2138, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdy M. (2022). Isolation and characterization of bacteriophages active against Pseudomonas aeruginosa strains isolated from diabetic foot infections. Archives of Razi Institute. 77:2187. doi: 10.22092/ARI.2022.359032.2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud H., Zakaria S., Kishk R., Al-Amir A. (2021). Effect of Meropenem-Colistin and Meropenem-amikacin combinations against Carbapenem-resistant Pseudomonas aeruginosa isolates in Suez Canal university hospitals. Microbes and Infectious Diseases. 0:1096. doi: 10.21608/mid.2021.53173.1096 [DOI] [Google Scholar]
- MaI M., Ma G.-C., Loza E., Ma P.-V., Baquero F., Cantón R. (2005). Breakpoints for predicting Pseudomonas aeruginosa susceptibility to inhaled tobramycin in cystic fibrosis patients: use of high-range Etest strips. J. Clin. Microbiol. 43, 4480–4485. doi: 10.1128/JCM.43.9.4480-4485.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina J. W., Onyambu F. G., Kibet P. S., Musyoki A. M. (2023). Multidrug-resistant gram-negative bacterial infections and associated factors in a Kenyan intensive care unit: a cross-sectional study. Ann. Clin. Microbiol. Antimicrob. 22:85. doi: 10.1186/s12941-023-00636-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharita R. R., El-Kholy I., Hetta H. F., Abdelaziz M. H., Hagagy F. I., Ahmed A. A., et al. (2020). Antibiogram and genetic characterization of Carbapenem-resistant gram-negative pathogens incriminated in healthcare-associated infections. Infect Drug Resist. 13, 3991–4002. doi: 10.2147/IDR.S276975, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malekzadegan Y., Abdi A., Heidari H., Moradi M., Rastegar E., Sedigh E.-S. H. (2019). In vitro activities of colistin, imipenem and ceftazidime against drug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii isolates in the south of Iran. BMC. Res. Notes 12:301. doi: 10.1186/s13104-019-4344-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallikarjuna P. V., Dhanashree B. (2023). Phenotypic and genotypic characterization of clinical Pseudomonas aeruginosa. J. Taibah Univ. Med. Sci. 18, 480–487. doi: 10.1016/j.jtumed.2022.10.012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manno G., Cruciani M., Romano L., Scapolan S., Mentasti M., Lorini R., et al. (2005). Antimicrobial use and Pseudomonas aeruginosa susceptibility profile in a cystic fibrosis Centre. Int. J. Antimicrob. Agents 25, 193–197. doi: 10.1016/j.ijantimicag.2004.11.009, PMID: [DOI] [PubMed] [Google Scholar]
- Mansour S. A., Eldaly O., Jiman-Fatani A., Mohamed M. L., Ibrahim E. M. (2013). Epidemiological characterization of P. aeruginosa isolates of intensive care units in Egypt and Saudi Arabia. East Mediterr. Health J. 19, 71–80. doi: 10.26719/2013.19.1.71, PMID: [DOI] [PubMed] [Google Scholar]
- Maraki S., Mantadakis E., Nioti E., Samonis G. (2015). Susceptibility of 2,252 Pseudomonas aeruginosa clinical isolates over 4 years to 9 antimicrobials in a tertiary Greek hospital. Chemotherapy 60, 334–341. doi: 10.1159/000437252 [DOI] [PubMed] [Google Scholar]
- Maraki S., Mavros M. N., Kofteridis D. P., Samonis G., Falagas M. E. (2012). Epidemiology and antimicrobial sensitivities of 536 multi-drug-resistant gram-negative bacilli isolated from patients treated on surgical wards. Surg. Infect. 13, 326–331. doi: 10.1089/sur.2011.115 [DOI] [PubMed] [Google Scholar]
- Marteva-Proevska Y., Velinov T., Markovska R., Dobrikova D., Pavlov I., Boyanova L., et al. (2021). High level of colistin resistant gram-negative bacteria in a university hospital in Bulgaria. Comptes Rendus de L Academie Bulgare des Sci. 74, 899–905. doi: 10.7546/CRABS.2021.06.12 [DOI] [Google Scholar]
- Martínez T., Vazquez G., Aquino E., Goering R. V., Robledo I. (2012). Two novel class I integron arrays containing IMP-18 metallo-β-lactamase gene in Pseudomonas aeruginosa clinical isolates from Puerto Rico. Antimicrob. Agents Chemother. 56, 2119–2121. doi: 10.1128/AAC.05758-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mataraci Kara E., Yilmaz M., Istanbullu Tosun A., Ozbek C. B. (2020). Synergistic activities of ceftazidime-avibactam in combination with different antibiotics against colistin-nonsusceptible clinical strains of Pseudomonas aeruginosa. Infect Dis. 52, 616–624. doi: 10.1080/23744235.2020.1767803, PMID: [DOI] [PubMed] [Google Scholar]
- Matuschek E., Åhman J., Webster C., Kahlmeter G. (2018). Antimicrobial susceptibility testing of colistin – evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin. Microbiol. Infect. 24, 865–870. doi: 10.1016/j.cmi.2017.11.020, PMID: [DOI] [PubMed] [Google Scholar]
- McCracken M. G., Adam H. J., Blondeau J. M., Walkty A. J., Karlowsky J. A., Hoban D. J., et al. (2019). Characterization of carbapenem-resistant and XDR Pseudomonas aeruginosa in Canada: results of the CANWARD 2007–16 study. J. Antimicrob. Chemother. 74, iv32–iv38. doi: 10.1093/jac/dkz285 [DOI] [PubMed] [Google Scholar]
- Medell M., Hart M., Duquesne A., Espinosa F., Valdés R. (2013). Nosocomial ventilator-associated pneumonia in Cuban intensive care units: bacterial species and antibiotic resistance. MEDICC Rev. 15, 26–29. doi: 10.37757/MR2013V15.N2.6, PMID: [DOI] [PubMed] [Google Scholar]
- Medell M., Hart M., Marrero O., Espinosa F., Oca Z. M., Valdés R. (2012). Clinical and microbiological characterization of pneumonia in mechanically ventilated patients. Braz. J. Infect. Dis. 16, 442–447. doi: 10.1016/j.bjid.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Mellouli A., Chebbi Y., El Fatmi R., Raddaoui A., Lakhal A., Torjmane L., et al. (2021). Multidrug resistant bacteremia in hematopoietic stem cell transplant recipients. La Tunisie Medicale 99, 269–276, PMID: [PMC free article] [PubMed] [Google Scholar]
- Memar M. Y., Pormehrali R., Alizadeh N., Ghotaslou R., Bannazadeh B. H. (2016). Colistin, an option for treatment of multiple drug resistant Pseudomonas aeruginosa. Physiol Pharmacol.
- Mendes R. E., Mendoza M., Banga Singh K. K., Castanheira M., Bell J. M., Turnidge J. D., et al. (2013). Regional resistance surveillance program results for 12 Asia-Pacific nations (2011). Antimicrob. Agents Chemother. 57, 5721–5726. doi: 10.1128/AAC.01121-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meradji S., Barguigua A., Bentakouk M., Nayme K., Zerouali K., Mazouz D., et al. (2016). Epidemiology and virulence of VIM-4 metallo-beta-lactamase-producing Pseudomonas aeruginosa isolated from burn patients in eastern Algeria. Burns 42, 906–918. doi: 10.1016/j.burns.2016.02.023, PMID: [DOI] [PubMed] [Google Scholar]
- Meradji S., Barguigua A., Zerouali K., Mazouz D., Chettibi H., Elmdaghri N., et al. (2015). Epidemiology of carbapenem non-susceptible Pseudomonas aeruginosa isolates in eastern Algeria. Antimicrob. Resist. Infect. Control 4:27. doi: 10.1186/s13756-015-0067-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meschiari M., Orlando G., Kaleci S., Bianco V., Sarti M., Venturelli C., et al. (2021). Combined resistance to Ceftolozane-Tazobactam and ceftazidime-avibactam in extensively drug-resistant (XDR) and multidrug-resistant (MDR) Pseudomonas aeruginosa: resistance predictors and impact on clinical outcomes besides implications for antimicrobial stewardship programs. Antibiotics 10:1224. doi: 10.3390/antibiotics10101224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickymaray S. (2019). One-step synthesis of Silver nanoparticles using Saudi Arabian Desert seasonal plant Sisymbrium irio and antibacterial activity against multidrug-resistant bacterial strains. Biomol. Ther. 9:662. doi: 10.3390/biom9110662, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikucionyte G., Zamorano L., Vitkauskiene A., López-Causapé C., Juan C., Mulet X., et al. (2016). Nosocomial dissemination of VIM-2-producing ST235 Pseudomonas aeruginosa in Lithuania. Eur. J. Clin. Microbiol. Infect. Dis. 35, 195–200. doi: 10.1007/s10096-015-2529-0, PMID: [DOI] [PubMed] [Google Scholar]
- Milczewska J., Wolkowicz T., Zacharczuk K., Mierzejewska E., Kwiatkowska M., Walicka-Serzysko K., et al. (2020). Clinical outcomes for cystic fibrosis patients with Pseudomonas aeruginosa cross-infections. Pediatr. Pulmonol. 55, 161–168. doi: 10.1002/ppul.24535, PMID: [DOI] [PubMed] [Google Scholar]
- Milojković M., Nenadović Ž., Stanković S., Božić D. D., Nedeljković N. S., Ćirković I., et al. (2020). Phenotypic and genetic properties of susceptible and multidrug-resistant Pseudomonas aeruginosa isolates in southern Serbia. Arh. Hig. Rada Toksikol. 71, 231–250. doi: 10.2478/aiht-2020-71-3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirbagheri S. Z., Meshkat Z., Naderinasab M., Rostami S., Nabavinia M. S., Rahmati M. (2015). Study on imipenem resistance and prevalence of blaVIM1 and blaVIM2 metallo-beta lactamases among clinical isolates of Pseudomonas aeruginosa from Mashhad, northeast of Iran. Iran J. Microbiol. 7, 72–78, PMID: [PMC free article] [PubMed] [Google Scholar]
- Mishra D. R., Shah D. S., Shah N., Prasad J. N., Gupta P. P., Agrawaal K. K. (2020). Study of microbiological and antibiotic sensitivity pattern of ventilator associated pneumonia (VAP) in ICU of a tertiary care hospital in Nepal. J. Family Med. Prim. Care 9, 6171–6176. doi: 10.4103/jfmpc.jfmpc_1430_20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobaraki S., Aghazadeh M., Soroush Barhaghi M. H., Yousef Memar M., Goli H. R., Gholizadeh P., et al. (2018). Prevalence of integrons 1, 2, 3 associated with antibiotic resistance in Pseudomonas aeruginosa isolates from northwest of Iran. Biomedicine 8:2. doi: 10.1051/bmdcn/2018080102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A. H., Mohamud H. A., Arslan E. (2021). Epidemiological characteristics and predisposing factors for surgical site infections caused by bacterial pathogens exhibiting multidrug-resistant patterns. Antibiotics 10:622. doi: 10.3390/antibiotics10060622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A. H., Sheikh Omar N. M., Osman M. M., Mohamud H. A., Eraslan A., Gur M. (2022). Antimicrobial resistance and predisposing factors associated with catheter-associated UTI caused by uropathogens exhibiting multidrug-resistant patterns: a 3-year retrospective study at a tertiary hospital in Mogadishu, Somalia. Trop. Med. Infect. Dis. 7:42. doi: 10.3390/tropicalmed7030042, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed N. M., Youssef A. A. (2011). In vitro activity of tigecycline and comparators against gram-negative bacteria isolated from a tertiary hospital in Alexandria, Egypt. Microb. Drug Resist. 17, 489–495. doi: 10.1089/mdr.2010.0195, PMID: [DOI] [PubMed] [Google Scholar]
- Mohammadi Barzelighi H., Bakhshi B., Daraei B., Fazeli H., Nasr E. B. (2020). Global sequence analysis and expression of Azurin gene in different clinical specimens of burn patients with Pseudomonas aeruginosa infection. Infect Drug Resist. 13, 2261–2275. doi: 10.2147/IDR.S248043, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadzadeh A., Mardaneh J., Ahmadi R., Adabi J. (2017). Evaluation of the virulence features and antibiotic resistance patterns of pathogenic Pseudomonas aeruginosa strains isolated from hospitalized patients in Gonabad, Iran. Arch. Pediatric Infect. Dis. In press. doi: 10.5812/pedinfect.41267 [DOI] [Google Scholar]
- Mohammadzamani Z., Khorshidi A., Khaledi A., Shakerimoghaddam A., Moosavi G. A., Piroozmand A. (2020). Inhibitory effects of Cinnamaldehyde, Carvacrol, and honey on the expression of exoS and ampC genes in multidrug-resistant Pseudomonas aeruginosa isolated from burn wound infections. Microb. Pathog. 140:103946. doi: 10.1016/j.micpath.2019.103946 [DOI] [PubMed] [Google Scholar]
- Mohanty S., Maurya V., Gaind R., Deb M. (2013). Phenotypic characterization and colistin susceptibilities of carbapenem-resistant of Pseudomonas aeruginosa and Acinetobacter spp. J. Infect. Dev. Ctries. 7, 880–887. doi: 10.3855/jidc.2924, PMID: [DOI] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6, e1000097–e1000096. doi: 10.1371/journal.pmed.1000097, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momenah A. M., Bakri R. A., Jalal N. A., Ashgar S. S., Felemban R. F., Bantun F., et al. (2023). Antimicrobial resistance pattern of Pseudomonas aeruginosa: An 11-year experience in a tertiary care hospital in Makkah, Saudi Arabia. Infect. Drug Resist. 16, 4113–4122. doi: 10.2147/IDR.S409726, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monogue M. L., Nicolau D. P. (2018). Antibacterial activity of ceftolozane/tazobactam alone and in combination with other antimicrobial agents against MDR Pseudomonas aeruginosa. J. Antimicrob. Chemother. 73, 942–952. doi: 10.1093/jac/dkx483, PMID: [DOI] [PubMed] [Google Scholar]
- Montero M. M., Domene Ochoa S., López-Causapé C., VanScoy B., Luque S., Sorlí L., et al. (2019). Colistin plus meropenem combination is synergistic in vitro against extensively drug-resistant Pseudomonas aeruginosa, including high-risk clones. J Glob Antimicrob Resist. 18, 37–44. doi: 10.1016/j.jgar.2019.04.012, PMID: [DOI] [PubMed] [Google Scholar]
- Montero M., Domene Ochoa S., Lopez-Causape C., VanScoy B., Luque S., Sorli L., et al. (2020). Efficacy of Ceftolozane-Tazobactam in combination with Colistin against extensively drug-resistant Pseudomonas aeruginosa, including high-risk clones, in an in vitro Pharmacodynamic model. Antimicrob. Agents Chemother. 64:e02542. doi: 10.1128/AAC.02542-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero M. M., López Montesinos I., Knobel H., Molas E., Sorlí L., Siverio-Parés A., et al. (2020). Risk factors for mortality among patients with Pseudomonas aeruginosa bloodstream infections: what is the influence of XDR phenotype on outcomes? J. Clin. Med. 9:514. doi: 10.3390/jcm9020514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi N., Kazemi N., Ghaemi M., Mirzaei B. (2021). Frequency and antimicrobial resistance pattern of bacterial isolates from patients with COVID-19 in two hospitals of Zanjan. Iranian J. Microbiol. 13, 769–778. doi: 10.18502/ijm.v13i6.8078, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata L., Cobos-Trigueros N., Martínez J. A., Soriano Á., Almela M., Marco F., et al. (2012). Influence of multidrug resistance and appropriate empirical therapy on the 30-day mortality rate of Pseudomonas aeruginosa bacteremia. Antimicrob. Agents Chemother. 56, 4833–4837. doi: 10.1128/AAC.00750-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordi R. M., Erah P. O. (2006). Susceptibility of common urinary isolates to the commonly used antibiotics in a tertiary hospital in southern Nigeria. Afr. J. Biotechnol. 5. [Google Scholar]
- Morrow B. J., Pillar C. M., Deane J., Sahm D. F., Lynch A. S., Flamm R. K., et al. (2013). Activities of carbapenem and comparator agents against contemporary US Pseudomonas aeruginosa isolates from the CAPITAL surveillance program. Diagn. Microbiol. Infect. Dis. 75, 412–416. doi: 10.1016/j.diagmicrobio.2012.12.012, PMID: [DOI] [PubMed] [Google Scholar]
- Morteza M., Roya S., Hamed H., Amir Z., Abolfazl A. (2019). Synthesis and evaluation of polymeric micelle containing piperacillin/tazobactam for enhanced antibacterial activity. Drug Deliv. 26, 1292–1299. doi: 10.1080/10717544.2019.1693708, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa S. H., Saleh S. E., Hamed S. M., Aboshanab K. M. (2022). Febrile illness of bacterial etiology in a public fever hospital in Egypt: high burden of multidrug resistance and WHO priority gram negative pathogens. Germs 12, 75–85. doi: 10.18683/germs.2022.1308, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubareck C. A., Halat D. H., Akkawi C., Nabi A., AlSharhan M. A., AlDeesi Z. O., et al. (2019). Role of outer membrane permeability, efflux mechanism, and carbapenemases in carbapenem-nonsusceptible Pseudomonas aeruginosa from Dubai hospitals: results of the first cross-sectional survey. Int. J. Infect. Dis. 84, 143–150. doi: 10.1016/j.ijid.2019.04.027, PMID: [DOI] [PubMed] [Google Scholar]
- Mulet X., Fernandez-Esgueva M., Norte C., Zamorano L., Del Barrio-Tofino E., Oliver A., et al. (2021). Validation of MALDI-TOF for the early detection of the ST175 high-risk clone of Pseudomonas aeruginosa in clinical isolates belonging to a Spanish nationwide multicenter study. Enferm Infect. Microbiol. Clin. 39, 279–282. doi: 10.1016/j.eimc.2020.05.022, PMID: [DOI] [PubMed] [Google Scholar]
- Munn Z., Moola S., Riitano D., Lisy K. (2014). The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int. J. Health Policy Manag. 3, 123–128. doi: 10.15171/ijhpm.2014.71, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa M. H., Chalhoub H., Denis O., Deplano A., Vergison A., Rodriguez-Villalobos H., et al. (2016). Antimicrobial susceptibility of Pseudomonas aeruginosa isolated from cystic fibrosis patients in northern Europe. Antimicrob. Agents Chemother. 60, 6735–6741. doi: 10.1128/AAC.01046-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Lina G., Santerre Henriksen A., Longshaw C., Jehl F. (2021). In vitro activity of cefiderocol and comparators against isolates of gram-negative pathogens from a range of infection sources: SIDERO-WT-2014-2018 studies in France. JAC Antimicrob. Resist. 3:dlab081. doi: 10.1093/jacamr/dlab081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair A., Perry A., Perry J. D., Gould F. K., Samuel J. (2020). In vitro effects of combined iron chelation, antibiotics and matrix disruption on clinical isolates of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 75, 586–592. doi: 10.1093/jac/dkz505, PMID: [DOI] [PubMed] [Google Scholar]
- Najafi K., Kafil H. S., Shokrian S., Azimi S., Asgharzadeh M., Yousefi M., et al. (2015). Virulence genes and antibiotic resistance profile of Pseudomonas aeruginosa isolates in northwest of Iran. J. Pure Appl. Microbiol. 9, 383–389. [Google Scholar]
- Narimisa N., Goodarzi F., Bavari S. (2022). Prevalence of colistin resistance of Klebsiella pneumoniae isolates in Iran: a systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 21:29. doi: 10.1186/s12941-022-00520-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedeljković N. S., Tiodorović B., Kocić B., Cirić V., Milojković M., Waisi H. (2015). Pseudomonas aeruginosa serotypes and resistance to antibiotics from wound swabs. Vojnosanit. Pregl. 72, 996–1003. doi: 10.2298/VSP131224108S [DOI] [PubMed] [Google Scholar]
- Negm E. M., Elgharabawy E. S., Badran S. G., Raafat A. O., Soliman S. T., Mahmoud H. M., et al. (2023). Analysis of cumulative antibiogram reports in intensive care units at an Egyptian university hospital. J. Infect. Public Health 16, 1220–1229. doi: 10.1016/j.jiph.2023.05.032, PMID: [DOI] [PubMed] [Google Scholar]
- Negm E. M., Mowafy S. M., Mohammed A. A., Amer M. G., Tawfik A. E., Ibrahim A. E., et al. (2021). Antibiograms of intensive care units at an Egyptian tertiary care hospital. Egyptian J. Bronchol. 15, 1–15. doi: 10.1186/s43168-021-00059-w [DOI] [Google Scholar]
- Nelson R. G., Rosowsky A. (2002). Dicyclic and tricyclic diaminopyrimidine derivatives as potent inhibitors of cryptosporidium parvum dihydrofolate reductase: structure-activity and structure-selectivity correlations. Antimicrob. Agents Chemother. 46:940. doi: 10.1128/AAC.46.3.940-940.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V. T. K., Montakantikul P., Tragulpiankit P., Houngsaitong J., Shuib M. F. (2021). Colistin dosing regimens against Pseudomonas aeruginosa in critically ill patients: An application of Monte Carlo simulation. Antibiotics 10:595. doi: 10.3390/antibiotics10050595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols W. W., de Jonge B. L., Kazmierczak K. M., Karlowsky J. A., Sahm D. F. (2016). In vitro susceptibility of global surveillance isolates of Pseudomonas aeruginosa to ceftazidime-avibactam (INFORM 2012 to 2014). Antimicrob. Agents Chemother. 60, 4743–4749. doi: 10.1128/AAC.00220-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitescu B., Pitigoi D., Talapan D., Nitescu M., Arama S. S., Pavel B., et al. (2023). Etiology and multi-drug resistant profile of bacterial infections in severe burn patients, Romania 2018-2022. Medicina (Kaunas) 59:1143. doi: 10.3390/medicina59061143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz F., de Melo B. O., da Silva L. C. N., de Souza M. A., Marques S. G., Monteiro-Neto V., et al. (2021). Molecular detection of drug-resistance genes of Bla OXA-23-Bla OXA-51 and mcr-1 in clinical isolates of Pseudomonas aeruginosa. Microorganisms 9:786. doi: 10.3390/microorganisms9040786, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojookambari N. Y., Eslami G., Hashemi A., Sadredinamin M., Tarashi S., Roshani M., et al. (2019). In vitro antimicrobial activity of Cinnamomum verum, Allium sativum, and Zingiber officinale extracts on metallo-β-lactamase-producing Pseudomonas aeruginosa: A potential therapeutic approach. Acta Microbiologica Hellenica.
- Nolan P. J., Jain R., Cohen L., Finklea J. D., Smith T. T. (2021). In vitro activity of ceftolozane-tazobactam and ceftazidime-avibactam against Pseudomonas aeruginosa isolated from patients with cystic fibrosis. Diagn. Microbiol. Infect. Dis. 99:115204. doi: 10.1016/j.diagmicrobio.2020.115204 [DOI] [PubMed] [Google Scholar]
- Nwabuisi C., Ologe F. E. (2002). Pathogenic agents of chronic suppurative otitis media in Ilorin, Nigeria. East African Med. J. 79, 202–205. doi: 10.4314/eamj.v79i4.8879, PMID: [DOI] [PubMed] [Google Scholar]
- O'Carroll M., Syrmis M., Wainwright C., Greer R., Mitchell P., Coulter C., et al. (2004). Clonal strains of Pseudomonas aeruginosa in paediatric and adult cystic fibrosis units. Eur. Respir. J. 24, 101–106. doi: 10.1183/09031936.04.00122903, PMID: [DOI] [PubMed] [Google Scholar]
- Odumosu B. T., Adeniyi B. A., Dada-AdegbolaHannah R. C. (2012). Multidrug resistant Pseudomonas aeruginosa from Southwest Nigeria hospitals. Blood 3:5. [Google Scholar]
- Olowo-okere A., Ibrahim Y. K. E., Ehinmidu J. O., Mohammed Y., Nabti L. Z., Olayinka B. O. (2020). Emergence of VIM metallo-β-lactamase among carbapenem-resistant Pseudomonas species in Northwest Nigeria. Gene Rep. 21:100877. doi: 10.1016/j.genrep.2020.100877 [DOI] [Google Scholar]
- Öncül O., Öksüz S., Acar A., Ülkür E., Turhan V., Uygur F., et al. (2014). Nosocomial infection characteristics in a burn intensive care unit: analysis of an eleven-year active surveillance. Burns 40, 835–841. doi: 10.1016/j.burns.2013.11.003, PMID: [DOI] [PubMed] [Google Scholar]
- Ouzzani M., Hammady H., Fedorowicz Z., Elmagarmid A. (2016). Rayyan—a web and mobile app for systematic reviews. Syst. Rev. 5, 1–10. doi: 10.1186/s13643-016-0384-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özhak B., Öğünç D., Yıldız E., Çolak D., Günseren F., Öngüt G. (2019). Evaluation of the BD phoenix100 system and colistin broth disk elution method for antimicrobial susceptibility testing of colistin against gram-negative bacteria. Mikrobiyoloji Bulteni 53, 254–261. doi: 10.5578/mb.68066 [DOI] [PubMed] [Google Scholar]
- Ozsurekci Y., Aykac K., Cengiz A. B., Bayhan C., Sancak B., Oncel E. K., et al. (2016). Is colistin effective in the treatment of infections caused by multidrug-resistant (MDR) or extremely drug-resistant (XDR) gram-negative microorganisms in children? Diagn. Microbiol. Infect. Dis. 85, 233–238. doi: 10.1016/j.diagmicrobio.2016.02.017, PMID: [DOI] [PubMed] [Google Scholar]
- Ozumba U. C. (2005). Antimicrobial resistance problems in a university hospital. J. Natl. Med. Assoc. 97, 1714–1718, PMID: [PMC free article] [PubMed] [Google Scholar]
- Pachori P., Gothalwal R., Gandhi P. (2019). Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 6, 109–119. doi: 10.1016/j.gendis.2019.04.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi P., Carroll K. C., Buchan B. W., Chan R. C., Dhiman N., Ford B., et al. (2018). Multicenter evaluation of the accelerate PhenoTest BC kit for rapid identification and phenotypic antimicrobial susceptibility testing using morphokinetic cellular analysis. J. Clin. Microbiol. 56, e01329–e01317. doi: 10.1128/JCM.01329-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z., Raudonis R., Glick B. R., Lin T.-J., Cheng Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013 [DOI] [PubMed] [Google Scholar]
- Pani A., Lucini V., Dugnani S., Schianchi A., Scaglione F. (2022). Effects of levofloxacin, Aztreonam, and Colistin on enzyme synthesis by P. aeruginosa isolated from cystic fibrosis patients. Antibiotics 11:1114. doi: 10.3390/antibiotics11081114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannitsis C. C., Medvecky M., Chudejova K., Skalova A., Rotova V., Spanelova P., et al. (2017). Molecular characterization of carbapenemase-producing Pseudomonas aeruginosa of Czech origin and evidence for clonal spread of extensively resistant sequence type 357 expressing IMP-7 metallo-β-lactamase. Antimicrob. Agents Chemother. 61, 01811–01817. doi: 10.1128/AAC.01811-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paprocka P., Durnas B., Mankowska A., Sklodowski K., Krol G., Zakrzewska M., et al. (2021). New beta-lactam antibiotics and Ceragenins – a study to assess their potential in treatment of infections caused by multidrug-resistant strains of Pseudomonas aeruginosa. Infect. Drug Resist. 14, 5681–5698. doi: 10.2147/IDR.S338827, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. H., Kim S. Y., Roh E. Y., Lee H. S. (2017). Difference of type 3 secretion system (T3SS) effector gene genotypes (exoU and exoS) and its implication to antibiotics resistances in isolates of Pseudomonas aeruginosa from chronic otitis media. Auris Nasus Larynx 44, 258–265. doi: 10.1016/j.anl.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Parsa P., Amirmozafari N., Nowruzi B., Bahar M. A. (2020). Molecular characterization of polymorphisms among Pseudomonas aeruginosa strains isolated from burn patients' wounds. Heliyon. 6:e05041. doi: 10.1016/j.heliyon.2020.e05041, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca M. R., Dalla Valle C., De Jesus Lopes Ribeiro A. L., Buroni S., Papaleo M. C., Bazzini S., et al. (2012). Evaluation of fluoroquinolone resistance mechanisms in Pseudomonas aeruginosa multidrug resistance clinical isolates. Microb. Drug Resist. 18, 23–32. doi: 10.1089/mdr.2011.0019 [DOI] [PubMed] [Google Scholar]
- Paterson D. L., Harris P. N. (2016). Colistin resistance: a major breach in our last line of defence. Lancet Infect. Dis. 16, 132–133. doi: 10.1016/S1473-3099(15)00463-6, PMID: [DOI] [PubMed] [Google Scholar]
- Pechorsky A., Nitzan Y., Lazarovitch T. (2009). Identification of pathogenic bacteria in blood cultures: comparison between conventional and PCR methods. J. Microbiol. Methods 78, 325–330. doi: 10.1016/j.mimet.2009.07.008, PMID: [DOI] [PubMed] [Google Scholar]
- Peña C., Gómez-Zorrilla S., Suarez C., Dominguez M., Tubau F., Arch O., et al. (2012). Extensively drug-resistant Pseudomonas aeruginosa: risk of bloodstream infection in hospitalized patients. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2791–2797. doi: 10.1007/s10096-012-1629-3 [DOI] [PubMed] [Google Scholar]
- Pérez A., Gato E., Pérez-Llarena J., Fernández-Cuenca F., Gude M. J., Oviaño M., et al. (2019). High incidence of MDR and XDR Pseudomonas aeruginosa isolates obtained from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 74, 1244–1252. doi: 10.1093/jac/dkz030, PMID: [DOI] [PubMed] [Google Scholar]
- Perez F., Hujer A. M., Marshall S. H., Ray A. J., Rather P. N., Suwantarat N., et al. (2014). Extensively drug-resistant pseudomonas aeruginosa isolates containing blaVIM-2 and elements of Salmonella genomic island 2: a new genetic resistance determinant in Northeast Ohio. Antimicrob. Agents Chemother. 58, 5929–5935. doi: 10.1128/AAC.02372-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Vázquez M., Sola-Campoy P. J., Zurita Á. M., Ávila A., Gómez-Bertomeu F., Solís S., et al. (2020). Carbapenemase-producing Pseudomonas aeruginosa in Spain: interregional dissemination of the high-risk clones ST175 and ST244 carrying blaVIM-2, blaVIM-1, blaIMP-8, blaVIM-20 and blaKPC-2. Int. J. Antimicrob. Agents 56:106026. doi: 10.1016/j.ijantimicag.2020.106026 [DOI] [PubMed] [Google Scholar]
- Perovic O., Singh-Moodley A., Lowe M. (2023). In vitro activity of ceftolozane-tazobactam against Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa obtained from blood cultures from sentinel public hospitals in South Africa. Antibiotics. 12:453. doi: 10.3390/antibiotics12030453, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petca A. (2021). Bacterial pathogens isolated from surgical site infections and their antibiotic susceptibility. Farmacia 69, 741–748. doi: 10.31925/farmacia.2021.4.15 [DOI] [Google Scholar]
- Petrova G., Strateva T., Miteva D., Lazova S., Perenovska P. (2016). Antimicrobial susceptibility of Pseudomonas aeruginosa before and after initiation of inhaled tobramycin in Bulgaria. J. Infect. Dev. Ctries. 10, 1265–1267. doi: 10.3855/jidc.7658 [DOI] [PubMed] [Google Scholar]
- Peymani A., Farivar T. N., Ghanbarlou M. M., Najafipour R. (2015). Dissemination of Pseudomonas aeruginosa producing blaIMP-1 and blaVIM-1 in Qazvin and Alborz educational hospitals, Iran. Iran J. Microbiol. 7, 302–309. [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Flamm R. K., Duncan L. R., Mendes R. E., Jones R. N., Sader H. S. (2017). Antimicrobial activity of tigecycline and cefoperazone/sulbactam tested against 18,386 gram-negative organisms from Europe and the Asia-Pacific region (2013-2014). Diagn. Microbiol. Infect. Dis. 88, 177–183. doi: 10.1016/j.diagmicrobio.2017.02.020, PMID: [DOI] [PubMed] [Google Scholar]
- Pfaller M., Shortridge D., Chen W.-T., Sader H., Castanheira M. (2022). Ceftolozane/tazobactam activity against drug-resistant Pseudomonas aeruginosa and enterobacterales causing healthcare-associated infections in eight Asian countries: report from an antimicrobial surveillance program (2016–2018). Infect. Drug Resist. 15, 6739–6753. doi: 10.2147/IDR.S387097, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Shortridge D., Sader H. S., Castanheira M., Flamm R. K. (2018). Ceftolozane/tazobactam activity against drug-resistant Enterobacteriaceae and Pseudomonas aeruginosa causing healthcare-associated infections in the Asia-Pacific region (minus China, Australia and New Zealand): report from an antimicrobial surveillance Programme (2013-2015). Int. J. Antimicrob. Agents 51, 181–189. doi: 10.1016/j.ijantimicag.2017.09.016, PMID: [DOI] [PubMed] [Google Scholar]
- Picão Renata C., Poirel L., Gales Ana C., Nordmann P. (2009). Diversity of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates causing bloodstream infections in Brazil. Antimicrob. Agents Chemother. 53, 3908–3913. doi: 10.1128/AAC.00453-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierard D., Stone G. G. (2021). In vitro antimicrobial susceptibility of clinical respiratory isolates to ceftazidime-avibactam and comparators (2016-2018). BMC Infect. Dis. 21:600. doi: 10.1186/s12879-021-06153-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt T., Sparrow M., Warner M., Stefanidou M. (2003). Survey of resistance of Pseudomonas aeruginosa from UK patients with cystic fibrosis to six commonly prescribed antimicrobial agents. Thorax 58, 794–796. doi: 10.1136/thorax.58.9.794, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourakbari B., Yaslianifard S., Yaslianifard S., Mahmoudi S., Keshavarz-Valian S., Mamishi S. (2016). Evaluation of efflux pumps gene expression in resistant Pseudomonas aeruginosa isolates in an Iranian referral hospital. Iran J Microbiol. 8, 249–256, PMID: [PMC free article] [PubMed] [Google Scholar]
- Pragsam A. K., Kumar D. T., Doss C. G. P., Iyadurai R., Satyendra S., Rodrigues C., et al. (2018). In silico and in vitro activity of ceftolozane/tazobactam against pseudomonas aeruginosa collected across Indian hospitals. Indian J. Med. Microbiol. 36, 127–130. doi: 10.4103/ijmm.IJMM_17_349 [DOI] [PubMed] [Google Scholar]
- Prasanth Manohar P. M., Thamaraiselvan Shanthini T. S., Ramankannan Ayyanar R. A., Bozdogan B., Aruni Wilson A. W., Tamhankar A., et al. (2017). The distribution of carbapenem-and colistin-resistance in gram-negative bacteria from the Tamil Nadu region in India. J Med Microbiol 66, 874–883. doi: 10.1099/jmm.0.000508 [DOI] [PubMed] [Google Scholar]
- Priyadarshi K., Dhandapani S., Sivaradjy M., Shanmugam L., Sastry A. S. (2023). Feasibility of using ceftazidime-avibactam as a therapeutic option for bloodstream infections caused by multidrug-resistant Enterobacterales and Pseudomonas aeruginosa based on its susceptibility profile. Cureus 15:37002. doi: 10.7759/cureus.37002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujji O. J. S., Nakarmi K. K., Shrestha B., Rai S. M., Jeffery S. L. A. (2019). The bacteriological profile of burn wound infections at a tertiary Burns Center in Nepal. J. Burn Care Res. 40, 838–845. doi: 10.1093/jbcr/irz096, PMID: [DOI] [PubMed] [Google Scholar]
- Qadeer A., Akhtar A., Ain Q. U., Saadat S., Mansoor S., Assad S., et al. (2016). Antibiogram of medical intensive care unit at tertiary care hospital setting of Pakistan. Cureus 8:e809. doi: 10.7759/cureus.809, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radan M., Moniri R., Khorshidi A., Gilasi H., Norouzi Z., Beigi F., et al. (2016). Emerging Carbapenem-resistant Pseudomonas aeruginosa isolates carrying Bla(IMP) among burn patients in Isfahan, Iran. Arch Trauma Res. 5:e33664. doi: 10.5812/atr.33664, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalskiy V., Pushkar D., Yakovlev S., Epstein O., Putilovskiy M., Tarasov S., et al. (2020). Distribution and antibiotic resistance profile of key gram-negative bacteria that cause community-onset urinary tract infections in the Russian Federation: RESOURCE multicentre surveillance 2017 study. J Glob Antimicrob Resist. 21, 188–194. doi: 10.1016/j.jgar.2019.09.008 [DOI] [PubMed] [Google Scholar]
- Rahimi E., Asgari A., Azimi T., Soleiman-Meigooni S. (2021). Molecular detection of carbapenemases and extended-spectrum β-lactamases-encoding genes in clinical isolates of pseudomonas aeruginosa in Iran. Jundishapur J. Microbiol. 14:115977. doi: 10.5812/jjm.115977 [DOI] [Google Scholar]
- Rajenderan S., Balaji V., Anandan S., Sahni R. D., Tansarli G. S., Falagas M. E. (2014). Determination of MIC distribution of arbekacin, cefminox, fosfomycin, biapenem and other antibiotics against gram-negative clinical isolates in South India: a prospective study. PLoS One 9:e103253. doi: 10.1371/journal.pone.0103253, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan R. A., Gebriel M. G., Kadry H. M., Mosallem A. (2018). Carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: characterization of carbapenemase genes and E-test evaluation of colistin-based combinations. Infect Drug Resist. 11, 1261–1269. doi: 10.2147/IDR.S170233, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan B., Jindal H. M., Le C. F., Gudimella R., Anwar A., Razali R., et al. (2017). Next generation sequencing reveals the antibiotic resistant variants in the genome of Pseudomonas aeruginosa. PLoS One 12:e0182524. doi: 10.1371/journal.pone.0182524, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid H., Zeb M., Jamal Q., Waqar M., Farooqi B., Majid A. (2014). Frequency and antimicrobial susceptibility pattern of Pseudomonas aeruginosa in ear swabs. World Appl. Sci. J. 30, 812–817. [Google Scholar]
- Rattanaumpawan P., Ussavasodhi P., Kiratisin P., Aswapokee N. (2013). Epidemiology of bacteremia caused by uncommon non-fermentative gram-negative bacteria. BMC Infect. Dis. 13:167. doi: 10.1186/1471-2334-13-167, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza M., Das B., Goyal V., Lodha R., Chaudhry R., Sood S., et al. (2019). Emerging multidrug resistance isolates of hospital-acquired bacterial meningitis in a tertiary care Centre in North India. J. Med. Microbiol. 68, 1585–1590. doi: 10.1099/jmm.0.001072, PMID: [DOI] [PubMed] [Google Scholar]
- Reddy R., Pathania S., Kapil A., Bakhshi S. (2014). Review of spectrum and sensitivity of bacterial bloodstream isolates in children with malignancy: a retrospective analysis from a single center. Indian J. Cancer 51, 425–427. doi: 10.4103/0019-509X.175363, PMID: [DOI] [PubMed] [Google Scholar]
- Rizek C., Fu L., Dos Santos L. C., Leite G., Ramos J., Rossi F., et al. (2014). Characterization of carbapenem-resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Ann. Clin. Microbiol. Antimicrob. 13:43. doi: 10.1186/s12941-014-0043-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodulfo H., Arcia A., Hernandez A., Michelli E., Martinez D. D. V., Guzman M., et al. (2019). Virulence factors and integrons are associated with MDR and XDR phenotypes in nosocomial strains of Pseudomonas aeruginosa in a Venezuelan university hospital. Rev. Inst. Med. Trop. 61:e20. doi: 10.1590/s1678-9946201961020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolston K. V., Gerges B., Shelburne S., Aitken S. L., Raad I., Prince R. A. (2020). Activity of cefiderocol and comparators against isolates from cancer patients. Antimicrob. Agents Chemother. 64, 01955–01919. doi: 10.1128/AAC.01955-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Reyes R., Rodriguez-Alvarado M., Lezana-Fernandez J. L., Sanchez-Lozano J. Y., Gayosso-Vazquez C., Jarillo-Quijada M. D., et al. (2020). Pseudomonas aeruginosa isolates from a cohort of Mexican children with cystic fibrosis show adaptation to a chronic phenotype. Pediatr. Infect. Dis. J. 39, 899–906. doi: 10.1097/INF.0000000000002714, PMID: [DOI] [PubMed] [Google Scholar]
- Rossolini G. M., Luzzaro F., Migliavacca R., Mugnaioli C., Pini B., De Luca F., et al. (2008). First countrywide survey of acquired metallo-β-lactamases in gram-negative pathogens in Italy. Antimicrob. Agents Chemother. 52, 4023–4029. doi: 10.1128/AAC.00707-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout B., Dash S. K., Kumar Sahu K., Behera B., Praharaj I., Otta S. (2023). Evaluation of different methods for in vitro susceptibility testing of colistin in carbapenem resistant gram-negative Bacilli. Access Microbiol. 5:595. doi: 10.1099/acmi.0.000595.v3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy Chowdhury P., Scott M., Worden P., Huntington P., Hudson B., Karagiannis T., et al. (2016). Genomic islands 1 and 2 play key roles in the evolution of extensively drug-resistant ST235 isolates of Pseudomonas aeruginosa. Open Biol. 6:150175. doi: 10.1098/rsob.150175, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruekit S., Srijan A., Serichantalergs O., Margulieux K. R., Mc Gann P., Mills E. G., et al. (2022). Molecular characterization of multidrug-resistant ESKAPEE pathogens from clinical samples in Chonburi, Thailand (2017–2018). BMC Infect. Dis. 22:695. doi: 10.1186/s12879-022-07678-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruh E., Gazi U., Güvenir M., Süer K., Çakır N. (2016). Antibiotic resistance rates of Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae isolated from a university-affiliated hospital in North Cyprus. Turk Hij Den Biyol Derg. 73, 333–344. doi: 10.5505/TurkHijyen.2016.82653 [DOI] [Google Scholar]
- Ruiz-Roldan L., Belles A., Bueno J., Azcona-Gutierrez J. M., Rojo-Bezares B., Torres C., et al. (2018). Pseudomonas aeruginosa isolates from Spanish children: occurrence in Faecal samples, antimicrobial resistance, virulence, and molecular typing. Biomed. Res. Int. 2018, 1–8. doi: 10.1155/2018/8060178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacha P., Michalska A., Ojdana D., Wieczorek P., Hauschild T., Majewski P., et al. (2015). Identification of plasmid OXA and other β-lactamase genes among carbapenem-resistant isolates of Pseudomonas aeruginosa from the clinical university hospital in northeastern Poland. New Microbiol. 38, 271–275, PMID: [PubMed] [Google Scholar]
- Sader H. S., Carvalhaes C. G., Duncan L. R., Flamm R. K., Shortridge D. (2020a). Susceptibility trends of ceftolozane/tazobactam and comparators when tested against European gram-negative bacterial surveillance isolates collected during 2012-18. J. Antimicrob. Chemother. 75, 2907–2913. doi: 10.1093/jac/dkaa278, PMID: [DOI] [PubMed] [Google Scholar]
- Sader H. S., Carvalhaes C. G., Streit J. M., Castanheira M., Flamm R. K. (2020b). Antimicrobial activity of cefoperazone-sulbactam tested against gram-negative organisms from Europe, Asia-Pacific, and Latin America. Int. J. Infect. Dis. 91, 32–37. doi: 10.1016/j.ijid.2019.11.006, PMID: [DOI] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Arends S. J. R., Goossens H., Flamm R. K. (2019). Geographical and temporal variation in the frequency and antimicrobial susceptibility of bacteria isolated from patients hospitalized with bacterial pneumonia: results from 20 years of the SENTRY antimicrobial surveillance program (1997-2016). J. Antimicrob. Chemother. 74, 1595–1606. doi: 10.1093/jac/dkz074, PMID: [DOI] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Duncan L. R., Flamm R. K. (2018a). Antimicrobial susceptibility of Enterobacteriaceae and Pseudomonas aeruginosa isolates from United States medical centers stratified by infection type: results from the international network for optimal resistance monitoring (INFORM) surveillance program, 2015-2016. Diagn. Microbiol. Infect. Dis. 92, 69–74. doi: 10.1016/j.diagmicrobio.2018.04.012, PMID: [DOI] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Farrell D. J., Flamm R. K., Jones R. N. (2015a). Ceftazidime-avibactam activity when tested against ceftazidime-nonsusceptible Citrobacter spp., Enterobacter spp., Serratia marcescens, and Pseudomonas aeruginosa from unites states medical centers (2011-2014). Diagn. Microbiol. Infect. Dis. 83, 389–394. doi: 10.1016/j.diagmicrobio.2015.06.008, PMID: [DOI] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Flamm R. K. (2017a). Antimicrobial activity of ceftazidime-avibactam against gram-negative Bacteria isolated from patients hospitalized with pneumonia in U.S. medical centers, 2011 to 2015. Antimicrob. Agents Chemother. 61:e02083. doi: 10.1128/AAC.02083-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Flamm R. K., Jones R. N. (2016). Antimicrobial activities of ceftazidime-avibactam and comparator agents against gram-negative organisms isolated from patients with urinary tract infections in U.S. medical centers, 2012 to 2014. Antimicrob. Agents Chemother. 60, 4355–4360. doi: 10.1128/AAC.00405-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Huband M., Jones R. N., Flamm R. K. (2017b). WCK 5222 (Cefepime-Zidebactam) antimicrobial activity against clinical isolates of gram-negative Bacteria collected worldwide in 2015. Antimicrob. Agents Chemother. 61:e00072. doi: 10.1128/AAC.00072-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Jones R. N., Flamm R. K. (2017c). Antimicrobial activity of ceftazidime-avibactam and comparator agents when tested against bacterial isolates causing infection in cancer patients (2013-2014). Diagn. Microbiol. Infect. Dis. 87, 261–265. doi: 10.1016/j.diagmicrobio.2016.11.019 [DOI] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Mendes R. E., Flamm R. K. (2018b). Frequency and antimicrobial susceptibility of gram-negative bacteria isolated from patients with pneumonia hospitalized in ICUs of US medical centres (2015-17). J. Antimicrob. Chemother. 73, 3053–3059. doi: 10.1093/jac/dky279, PMID: [DOI] [PubMed] [Google Scholar]
- Sader H. S., Castanheira M., Shortridge D., Mendes R. E., Flamm R. K. (2017d). Antimicrobial activity of ceftazidime-avibactam tested against multidrug-resistant Enterobacteriaceae and Pseudomonas aeruginosa isolates from U.S. medical centers, 2013 to 2016. Antimicrob. Agents Chemother. 61:e01045. doi: 10.1128/AAC.01045-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Duncan L. R., Doyle T. B., Castanheira M. (2021a). Antimicrobial activity of ceftazidime/avibactam, ceftolozane/tazobactam and comparator agents against Pseudomonas aeruginosa from cystic fibrosis patients. JAC Antimicrob. Resist. 3. doi: 10.1093/jacamr/dlab126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Farrell D. J., Castanheira M., Flamm R. K., Jones R. N. (2014a). Antimicrobial activity of ceftolozane/tazobactam tested against Pseudomonas aeruginosa and Enterobacteriaceae with various resistance patterns isolated in European hospitals (2011-12). J. Antimicrob. Chemother. 69, 2713–2722. doi: 10.1093/jac/dku184 [DOI] [PubMed] [Google Scholar]
- Sader H. S., Farrell D. J., Flamm R. K., Jones R. N. (2014b). Antimicrobial susceptibility of gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009-2011). Diagn. Microbiol. Infect. Dis. 78, 443–448. doi: 10.1016/j.diagmicrobio.2013.11.025 [DOI] [PubMed] [Google Scholar]
- Sader H. S., Farrell D. J., Flamm R. K., Jones R. N. (2014c). Ceftolozane/tazobactam activity tested against aerobic gram-negative organisms isolated from intra-abdominal and urinary tract infections in European and United States hospitals (2012). J. Infect. 69, 266–277. doi: 10.1016/j.jinf.2014.04.004, PMID: [DOI] [PubMed] [Google Scholar]
- Sader H. S., Flamm R. K., Carvalhaes C. G., Castanheira M. (2018c). Antimicrobial susceptibility of Pseudomonas aeruginosa to ceftazidime-avibactam, Ceftolozane-Tazobactam, piperacillin-Tazobactam, and Meropenem stratified by U.S. Census divisions: results from the 2017 INFORM program. Antimicrob. Agents Chemother. 62:62. doi: 10.1128/AAC.01587-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Flamm R. K., Dale G. E., Rhomberg P. R., Castanheira M. (2018d). Murepavadin activity tested against contemporary (2016-17) clinical isolates of XDR Pseudomonas aeruginosa. J. Antimicrob. Chemother. 73, 2400–2404. doi: 10.1093/jac/dky227, PMID: [DOI] [PubMed] [Google Scholar]
- Sader H. S., Flamm R. K., Pfaller M. A., Castanheira M. (2018e). 1004. Frequency of occurrence and antimicrobial susceptibility of Bacteria isolated from patients hospitalized with bloodstream infections in United States medical centers (2015–2017). Open forum infectious diseases. Oxford: Oxford University Press. [DOI] [PubMed] [Google Scholar]
- Sader H. S., Huband M. D., Castanheira M., Flamm R. K. (2017e). Pseudomonas aeruginosa antimicrobial susceptibility results from four years (2012 to 2015) of the international network for optimal resistance monitoring program in the United States. Antimicrob. Agents Chemother. 61:514. doi: 10.1128/AAC.02252-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Rhomberg P. R., Farrell D. J., Jones R. N. (2015b). Arbekacin activity against contemporary clinical bacteria isolated from patients hospitalized with pneumonia. Antimicrob. Agents Chemother. 59, 3263–3270. doi: 10.1128/AAC.04839-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader H. S., Streit J. M., Carvalhaes C. G., Huband M. D., Shortridge D., Mendes R. E., et al. (2021b). Frequency of occurrence and antimicrobial susceptibility of bacteria isolated from respiratory samples of patients hospitalized with pneumonia in Western Europe, Eastern Europe and the USA: results from the SENTRY antimicrobial surveillance program (2016–19). JAC-Antimicrob. Resist. 3:dlab117. doi: 10.1093/jacamr/dlab117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saderi H., Owlia P., Detection of Multidrug Resistant (MDR) and Extremely Drug Resistant (XDR) P (2015). aeruginosa isolated from patients in Tehran, Iran. Iran J Pathol. 10, 265–271, PMID: [PMC free article] [PubMed] [Google Scholar]
- Sadikot R. T., Blackwell T. S., Christman J. W., Prince A. S. (2005). Pathogen–host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171, 1209–1223. doi: 10.1164/rccm.200408-1044SO, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffari M., Karami S., Firoozeh F., Sehat M. (2017). Evaluation of biofilm-specific antimicrobial resistance genes in Pseudomonas aeruginosa isolates in Farabi hospital. J. Med. Microbiol. 66, 905–909. doi: 10.1099/jmm.0.000521, PMID: [DOI] [PubMed] [Google Scholar]
- Saleem S., Bokhari H. (2020). Resistance profile of genetically distinct clinical Pseudomonas aeruginosa isolates from public hospitals in Central Pakistan. J. Infect. Public Health 13, 598–605. doi: 10.1016/j.jiph.2019.08.019, PMID: [DOI] [PubMed] [Google Scholar]
- Saleem Z., Haseeb A., Abuhussain S. S. A., Moore C. E., Kamran S. H., Qamar M. U., et al. (2023). Antibiotic susceptibility surveillance in the Punjab Province of Pakistan: findings and implications. Medicina (Kaunas) 59:1215. doi: 10.3390/medicina59071215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M., Syed Khaja A. S., Hossain A., Alenazi F., Said K. B., Moursi S. A., et al. (2023). Pathogen burden among ICU patients in a tertiary care hospital in hail Saudi Arabia with particular reference to β-lactamases profile. Infect. Drug Resist. 16, 769–778. doi: 10.2147/IDR.S394777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M., Syed Khaja A. S., Hossain A., Alenazi F., Said K. B., Moursi S. A., et al. (2022). Catheter-associated urinary tract infection in intensive care unit patients at a tertiary care hospital, hail, kingdom of Saudi Arabia. Diagnostics. 12:1695. doi: 10.3390/diagnostics12071695, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimizand H., Lotfi G., Afrasiabian S., Hajibagheri K., Babahajian A., Mohammadi S. (2023). Mortality and outcomes of patients infected with extensively drug-resistant Bacteria admitted to intensive care units. Arch. Clin. Infect. Dis. 18:e132030. doi: 10.5812/archcid-132030 [DOI] [Google Scholar]
- Samonis G., Maraki S., Karageorgopoulos D., Vouloumanou E., Falagas M. (2012). Synergy of fosfomycin with carbapenems, colistin, netilmicin, and tigecycline against multidrug-resistant Klebsiella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa clinical isolates. Eur. J. Clin. Microbiol. Infect. Dis. 31, 695–701. doi: 10.1007/s10096-011-1360-5 [DOI] [PubMed] [Google Scholar]
- Samonis G., Maraki S., Rafailidis P. I., Kapaskelis A., Kastoris A. C., Falagas M. E. (2010). Antimicrobial susceptibility of gram-negative nonurinary bacteria to fosfomycin and other antimicrobials. Future Microbiol. 5, 961–970. doi: 10.2217/fmb.10.47, PMID: [DOI] [PubMed] [Google Scholar]
- Samonis G., Maraki S., Vouloumanou E., Georgantzi G., Kofteridis D., Falagas M. (2012). Antimicrobial susceptibility of non-fermenting gram-negative isolates to isepamicin in a region with high antibiotic resistance. Eur. J. Clin. Microbiol. Infect. Dis. 31, 3191–3198. doi: 10.1007/s10096-012-1684-9, PMID: [DOI] [PubMed] [Google Scholar]
- Sanchez-Lopez J., Cortes-Cuevas J. L., Diez-Aguilar M., Lopez-Causape C., Canton R., Morosini M. I. (2021). Evaluation of rapid Polymyxin Pseudomonas test in clinical Pseudomonas aeruginosa isolates with various degrees of multidrug resistance. JAC Antimicrob. Resist. 3:dlab104. doi: 10.1093/jacamr/dlab104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santella B., Serretiello E., De Filippis A., Veronica F., Iervolino D., Dell'Annunziata F., et al. (2021). Lower respiratory tract pathogens and their antimicrobial susceptibility pattern: a 5-year study. Antibiotics 10:851. doi: 10.3390/antibiotics10070851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santimaleeworagun W., Thunyaharn S., Juntanawiwat P., Thongnoy N., Harindhanavudhi S., Nakeesathit S., et al. (2020). The prevalence of colistin-resistant gram-negative bacteria isolated from hospitalized patients with bacteremia. J. Appl. Pharmaceutical Sci. 10, 56–59. doi: 10.7324/JAPS.2020.102009 [DOI] [Google Scholar]
- Sarwat T., Yousuf M., Khan A. S., Kakru D. K., Dutta R. (2021). Prevalence and antibiogram of non-fermenting gram-negative bacilli in blood stream infections: study in a tertiary care Centre, Western Uttar Pradesh, India. Trop Doct. 51, 322–325. doi: 10.1177/0049475520979298, PMID: [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Vas Nunes J., Mönnink G., Falama A.-M., Bangura J., Mathéron H., et al. (2022). Chronic wounds in Sierra Leone: pathogen spectrum and antimicrobial susceptibility. Infection 50, 907–914. doi: 10.1007/s15010-022-01762-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechner V., Gottesman T., Schwartz O., Korem M., Maor Y., Rahav G., et al. (2011). Pseudomonas aeruginosa bacteremia upon hospital admission: risk factors for mortality and influence of inadequate empirical antimicrobial therapy. Diagn. Microbiol. Infect. Dis. 71, 38–45. doi: 10.1016/j.diagmicrobio.2011.05.010 [DOI] [PubMed] [Google Scholar]
- Schülin T. (2002). In vitro activity of the aerosolized agents colistin and tobramycin and five intravenous agents against Pseudomonas aeruginosa isolated from cystic fibrosis patients in southwestern Germany. J. Antimicrob. Chemother. 49, 403–406. doi: 10.1093/jac/49.2.403 [DOI] [PubMed] [Google Scholar]
- Seale T. W., Thirkill H., Tarpay M., Flux M., Rennert O. M. (1979). Serotypes and antibiotic susceptibilities of Pseudomonas aeruginosa isolates from single sputa of cystic fibrosis patients. J. Clin. Microbiol. 9, 72–78. doi: 10.1128/jcm.9.1.72-78.1979, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefraoui I., Berrazeg M., Drissi M., Rolain J. M. (2014). Molecular epidemiology of carbapenem-resistant Pseudomonas aeruginosa clinical strains isolated from western Algeria between 2009 and 2012. Microb. Drug Resist. 20, 156–161. doi: 10.1089/mdr.2013.0161, PMID: [DOI] [PubMed] [Google Scholar]
- Seifert H., Körber-Irrgang B., Kresken M. (2018). In-vitro activity of ceftolozane/tazobactam against Pseudomonas aeruginosa and Enterobacteriaceae isolates recovered from hospitalized patients in Germany. Int. J. Antimicrob. Agents 51, 227–234. doi: 10.1016/j.ijantimicag.2017.06.024 [DOI] [PubMed] [Google Scholar]
- Şen S., Cesur S., Yilmaz N. (2016). In vitro synergistic efficacy of various antibiotic combinations against multi-drug-resistant Pseudomonas aeruginosa isolates obtained from patients in intensive care units. Acta Med. Austriaca 32:1041. [Google Scholar]
- Sendra E., López Montesinos I., Rodriguez-Alarcón A., Du J., Siverio-Parés A., Arenas-Miras M., et al. (2022). Comparative analysis of complicated urinary tract infections caused by extensively drug-resistant pseudomonas aeruginosa and extended-spectrum β-lactamase-producing klebsiella pneumoniae. Antibiotics 11:1511. doi: 10.3390/antibiotics11111511, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahri F. N., Izanloo A., Goharrizi M. A. S. B., Jamali A., Bagheri H., Hjimohammadi A., et al. (2022). Antimicrobial resistance, virulence factors, and genotypes of Pseudomonas aeruginosa clinical isolates from Gorgan, northern Iran. Int. Microbiol. 25, 709–721. doi: 10.1007/s10123-022-00256-7, PMID: [DOI] [PubMed] [Google Scholar]
- Sharan H., Katare N., Pandey A., Bhatambare G. S., Bajpai T. (2016). Emergence of hospital acquired Carbapenem resistant non-fermenters in teaching institute. J. Clin. Diagn. Res. 10, Dc20–dc3. doi: 10.7860/JCDR/2016/22607.9020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariati A., Azimi T., Ardebili A., Chirani A., Bahramian A., Pormohammad A., et al. (2018). Insertional inactivation of oprD in carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Tehran, Iran. New Microbes New Infect. 21, 75–80. doi: 10.1016/j.nmni.2017.10.013, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi H., Pouladfar G., Shakibaie M. R., Pourabbas B., Mardaneh J., Mansouri S. (2019). Prevalence of beta-lactamase genes, class 1 integrons, major virulence factors and clonal relationships of multidrug-resistant Pseudomonas aeruginosa isolated from hospitalized patients in southeast of Iran. Iran. J. Basic Med. Sci. 22, 806–812. doi: 10.22038/ijbms.2019.35063.8340, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Devkota M. D., Pokhrel B. M., Banjara M. R. (2023). Detection of Bla(NDM-1,)mcr-1 and MexB in multidrug resistant Pseudomonas aeruginosa isolated from clinical specimens in a tertiary care hospital of Nepal. BMC Microbiol. 23:153. doi: 10.1186/s12866-023-02906-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma J., Sharma D., Singh A., Sunita K. (2022). Colistin resistance and management of drug resistant infections. Can. J. Infect. Dis. Med. Microbiol. 2022, 1–10. doi: 10.1155/2022/4315030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan J. B., Dongol A., Humagain S., Joshi A., Magar S. R., Bhandari S. (2022). Antibiotic susceptibility pattern of Bacteria causing urinary tract infection. Nepal Health Research Council. [DOI] [PubMed]
- Sherchan J. B., Humagain S. (2020). Antimicrobial susceptibility pattern of gram-negative bacteria causing lower respiratory tract infections in Kathmandu university hospital. J Nepal Health Res Counc 18, 661–666. doi: 10.33314/jnhrc.v18i4.2566 [DOI] [PubMed] [Google Scholar]
- Sheth K. V., Patel T. K., Malek S. S., Tripathi C. (2012a). Antibiotic sensitivity pattern of bacterial isolates from the intensive care unit of a tertiary care hospital in India. Trop. J. Pharm. Res. 11, 991–999. [Google Scholar]
- Sheth K. V., Patel T. K., Tripathi C. (2012b). Antibiotic sensitivity pattern in neonatal intensive care unit of a tertiary care hospital of India. Asian J. Pharm. Clin. Res. 5, 46–50. [Google Scholar]
- Shiralizadeh S., Keramat F., Hashemi S. H., Majzoobi M. M., Azimzadeh M., Alikhani M. S., et al. (2023). Investigation of antimicrobial resistance patterns and molecular typing of Pseudomonas aeruginosa isolates among coronavirus disease-19 patients. BMC Microbiol. 23:84. doi: 10.1186/s12866-023-02825-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivshetty N., Kelmani C., Patil S. (2020). Imipenem resistance mechanisms in multidrug resistant Pseudomonas aeruginosa clinical isolates from South India. Res. J. Biotechnol. 15, 80–85. [Google Scholar]
- Shortridge D., Carvalhaes C., Deshpande L., Castanheira M. (2021a). Activity of meropenem/vaborbactam and comparators against gram-negative isolates from eastern and Western European patients hospitalized with pneumonia including ventilator-associated pneumonia (2014–19). J. Antimicrob. Chemother. 76, 2600–2605. doi: 10.1093/jac/dkab252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge D., Carvalhaes C. G., Streit J. M., Flamm R. K. (2021b). Susceptibility trends of ceftolozane/tazobactam and comparators when tested against U.S. gram-negative bacterial surveillance isolates (2012-2018). Diagn. Microbiol. Infect. Dis. 100:115302. doi: 10.1016/j.diagmicrobio.2020.115302, PMID: [DOI] [PubMed] [Google Scholar]
- Shortridge D., Castanheira M., Pfaller M. A., Flamm R. K. (2017). Ceftolozane-tazobactam activity against Pseudomonas aeruginosa clinical isolates from US hospitals: report from the PACTS antimicrobial surveillance program, 2012 to 2015. Antimicrob. Agents Chemother. 61, 00465–00417. doi: 10.1128/AAC.00465-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge D., Duncan L. R., Pfaller M. A., Flamm R. K. (2019a). Activity of ceftolozane-tazobactam and comparators when tested against gram-negative isolates collected from paediatric patients in the USA and Europe between 2012 and 2016 as part of a global surveillance programme. Int. J. Antimicrob. Agents 53, 637–643. doi: 10.1016/j.ijantimicag.2019.01.015, PMID: [DOI] [PubMed] [Google Scholar]
- Shortridge D., Gales A. C., Streit J. M., Huband M. D., Tsakris A., Jones R. N. (2019b). Geographic and temporal patterns of antimicrobial resistance in Pseudomonas aeruginosa over 20 years from the SENTRY antimicrobial surveillance program, 1997-2016. Open Forum Infect. Dis. 6, S63–S68. doi: 10.1093/ofid/ofy343, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge D., Pfaller M. A., Arends S. J. R., Raddatz J., DePestel D. D., Flamm R. K. (2019c). Comparison of the in vitro susceptibility of Ceftolozane-Tazobactam with the cumulative susceptibility rates of standard antibiotic combinations when tested against Pseudomonas aeruginosa from ICU patients with bloodstream infections or pneumonia. Open Forum Infect. Dis. 6:ofz240. doi: 10.1093/ofid/ofz240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge D., Pfaller M. A., Castanheira M., Flamm R. K. (2018). Antimicrobial activity of ceftolozane-tazobactam tested against Enterobacteriaceae and Pseudomonas aeruginosa collected from patients with bloodstream infections isolated in United States hospitals (2013-2015) as part of the program to assess Ceftolozane-Tazobactam susceptibility (PACTS) surveillance program. Diagn. Microbiol. Infect. Dis. 92, 158–163. doi: 10.1016/j.diagmicrobio.2018.05.011, PMID: [DOI] [PubMed] [Google Scholar]
- Shortridge D., Pfaller M. A., Streit J. M., Flamm R. K. (2020). Antimicrobial activity of ceftolozane/tazobactam tested against contemporary (2015-2017) Pseudomonas aeruginosa isolates from a global surveillance programme. J. Glob. Antimicrob. Resist. 21, 60–64. doi: 10.1016/j.jgar.2019.10.009, PMID: [DOI] [PubMed] [Google Scholar]
- Shortridge D., Streit J. M., Mendes R., Castanheira M. (2022). In vitro activity of cefiderocol against US and European gram-negative clinical isolates collected in 2020 as part of the SENTRY antimicrobial surveillance program. Microbiol. Spectrum 10, e02712–e02721. doi: 10.1128/spectrum.02712-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shravani V., Selvi G. A. S., Mantravadi H. (2023). Detection of quorum sensing virulence factor genes and its consanguinity to antibiotic sensitivity profile in the clinical isolates of Pseudomonas aeruginosa. Iran. J. Basic Med. Sci. 26, 899–905. doi: 10.22038/IJBMS.2023.67981.14992, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sid Ahmed M. A., Abdel Hadi H., Abu Jarir S., Al Khal A. L., Al-Maslamani M. A., Jass J., et al. (2020). Impact of an antimicrobial stewardship programme on antimicrobial utilization and the prevalence of MDR Pseudomonas aeruginosa in an acute care hospital in Qatar. JAC Antimicrob. Resist. 2:dlaa050. doi: 10.1093/jacamr/dlaa050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sid Ahmed M. A., Abdel Hadi H., Hassan A. A. I., Abu Jarir S., Al-Maslamani M. A., Eltai N. O., et al. (2019). Evaluation of in vitro activity of ceftazidime/avibactam and ceftolozane/tazobactam against MDR Pseudomonas aeruginosa isolates from Qatar. J. Antimicrob. Chemother. 74, 3497–3504. doi: 10.1093/jac/dkz379, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sid Ahmed M. A., Hamid J. M., Husain A. A., Hadi H. A., Skariah S., Sultan A. A., et al. (2021). Clinical outcomes, molecular epidemiology and resistance mechanisms of multidrug-resistant Pseudomonas aeruginosa isolated from bloodstream infections from Qatar. Ann. Med. 53, 2345–2353. doi: 10.1080/07853890.2021.2012588, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sid Ahmed M. A., Petkar H. M., Saleh T. M., Albirair M., Arisgado L. A., Eltayeb F. K., et al. (2023). The epidemiology and microbiological characteristics of infections caused by gram-negative bacteria in Qatar: national surveillance from the study for monitoring of antimicrobial resistance trends (SMART): 2017 to 2019. JAC Antimicrob. Resist. 5:dlad086. doi: 10.1093/jacamr/dlad086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simar S., Sibley D., Ashcraft D., Pankey G. (2017). Colistin and polymyxin B minimal inhibitory concentrations determined by Etest found unreliable for gram-negative bacilli. Ochsner J. 17, 239–242, PMID: [PMC free article] [PubMed] [Google Scholar]
- Singhal T., Shah S., Thakkar P., Naik R. (2019). The incidence, aetiology and antimicrobial susceptibility of central line-associated bloodstream infections in intensive care unit patients at a private tertiary care hospital in Mumbai, India. Indian J Med Microbiol. 37, 521–526. doi: 10.4103/ijmm.IJMM_20_3, PMID: [DOI] [PubMed] [Google Scholar]
- Singh-Moodley A., Duse A., Naicker P., Kularatne R., Nana T., Lekalakala R., et al. (2018). Laboratory based antimicrobial resistance surveillance for Pseudomonas aeruginosa blood isolates from South Africa. J. Infect. Dev. Ctries. 12, 616–624. doi: 10.3855/jidc.9539, PMID: [DOI] [PubMed] [Google Scholar]
- Sleiman A., Abdelkhalek P., Doumat G., Atallah F., Hamadeh L., Moussa P., et al. (2023). The under investigated facet of the COVID-19 pandemic: molecular analysis of secondary bacterial infections at a COVID dedicated intensive care unit within a tertiary care center in Lebanon. Front. Med. 10:1001476. doi: 10.3389/fmed.2023.1001476, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman A. M., Zarad H. O., Nariya H., Shimamoto T., Shimamoto T. (2020). Genetic analysis of carbapenemase-producing gram-negative bacteria isolated from a university teaching hospital in Egypt. Infect. Genet. Evol. 77:104065. doi: 10.1016/j.meegid.2019.104065, PMID: [DOI] [PubMed] [Google Scholar]
- Soni M., Kapoor G., Perumal N., Chaurasia D. (2023). Emergence of multidrug-resistant non-fermenting gram-negative Bacilli in a tertiary care teaching Hospital of Central India: is Colistin resistance still a distant threat? Cureus 15:e39243. doi: 10.7759/cureus.39243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone G. G., Ponce-de-Leon A. (2020). In vitro activity of ceftazidime/avibactam and comparators against gram-negative bacterial isolates collected from Latin American centres between 2015 and 2017. J. Antimicrob. Chemother. 75, 1859–1873. doi: 10.1093/jac/dkaa089, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone G. G., Smayevsky J., Kazmierczak K. (2020). Longitudinal analysis of the in vitro activity of ceftazidime-avibactam vs. Pseudomonas aeruginosa, 2012-2016. Diagn. Microbiol. Infect. Dis. 96:114835. doi: 10.1016/j.diagmicrobio.2019.05.007, PMID: [DOI] [PubMed] [Google Scholar]
- Stracquadanio S., Torti E., Longshaw C., Henriksen A. S., Stefani S. (2021). In vitro activity of cefiderocol and comparators against isolates of gram-negative pathogens from a range of infection sources: SIDERO-WT-2014-2018 studies in Italy. J. Glob. Antimicrob. Resist. 25, 390–398. doi: 10.1016/j.jgar.2021.04.019, PMID: [DOI] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- Tada T., Hishinuma T., Watanabe S., Uchida H., Tohya M., Kuwahara-Arai K., et al. (2019). Molecular characterization of multidrug-resistant Pseudomonas aeruginosa isolates in hospitals in Myanmar. Antimicrob. Agents Chemother. 63, 02397–02318. doi: 10.1128/AAC.02397-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Miyoshi-Akiyama T., Kato Y., Ohmagari N., Takeshita N., Hung N. V., et al. (2013). Emergence of 16S rRNA methylase-producing Acinetobacter baumannii and Pseudomonas aeruginosa isolates in hospitals in Vietnam. BMC Infect. Dis. 13:251. doi: 10.1186/1471-2334-13-251, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi H., Dehbashi S., Alikhani M. Y., Porbaran M., Arabestani M. R. (2020a). Prevalence and molecular typing of Metallo-β-lactamase-producing Pseudomonas aeruginosa with adhesion factors: a descriptive analysis of burn wounds isolates from Iran. Gene Reports 21:100853. doi: 10.1016/j.genrep.2020.100853 [DOI] [Google Scholar]
- Tahmasebi H., Dehbashi S., Arabestani M. R. (2020b). Prevalence and molecular typing of Colistin-resistant Pseudomonas aeruginosa (CRPA) among beta-lactamase-producing isolates: a study based on high-resolution melting curve analysis method. Infect Drug Resist. 13, 2943–2955. doi: 10.2147/IDR.S264796, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahmasebi H., Dehbashi S., Arabestani M. R. (2020c). Co-harboring of mcr-1 and β-lactamase genes in Pseudomonas aeruginosa by high-resolution melting curve analysis (HRMA): molecular typing of superbug strains in bloodstream infections (BSI). Infect. Genet. Evol. 85:104518. doi: 10.1016/j.meegid.2020.104518, PMID: [DOI] [PubMed] [Google Scholar]
- Takahashi T., Tada T., Shrestha S., Hishinuma T., Sherchan J. B., Tohya M., et al. (2021). Molecular characterisation of carbapenem-resistant Pseudomonas aeruginosa clinical isolates in Nepal. J Glob Antimicrob Resist. 26, 279–284. doi: 10.1016/j.jgar.2021.07.003, PMID: [DOI] [PubMed] [Google Scholar]
- Taleb M. H., Elmanama A. A., Taleb A. H., Tawfick M. M. (2023). Pre-and post-COVID-19 antimicrobial resistance profile of bacterial pathogens, a comparative study in a tertiary hospital. J. Infect. Dev. Ctries. 17, 597–609. doi: 10.3855/jidc.17791 [DOI] [PubMed] [Google Scholar]
- Talebi Bezmin Abadi A., Rizvanov A. A., Haertlé T., Blatt N. L. (2019). World health organization report: current crisis of antibiotic resistance. BioNanoScience 9, 778–788. doi: 10.1007/s12668-019-00658-4 [DOI] [Google Scholar]
- Talebi G., Hakemi-Vala M. (2019). Survey on some carbapenems and colistin resistance genes among Pseudomonas aeruginosa isolates from burn and cystic fibrosis patients, Tehran, Iran. Archives of. Clin. Infect. Dis. 14:e93651. doi: 10.5812/archcid.93651 [DOI] [Google Scholar]
- Tan T. Y., Ng L. S. Y. (2006). Comparison of three standardized disc susceptibility testing methods for colistin. J. Antimicrob. Chemother. 58, 864–867. doi: 10.1093/jac/dkl330, PMID: [DOI] [PubMed] [Google Scholar]
- Tantisiriwat W., Buppanharun J., Ekpanyaskul C., Onruang K., Yungyuen T., Kiratisin P., et al. (2022). In vitro activity of ceftolozane-tazobactam and other antibiotics against Pseudomonas aeruginosa infection-isolates from an academic medical center in Thailand. Antibiotics. 11:732. doi: 10.3390/antibiotics11060732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarashi S., Goudarzi H., Erfanimanesh S., Pormohammad A., Hashemi A. (2016). Phenotypic and molecular detection of metallo-beta-lactamase genes among imipenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from patients with burn injuries. Archives of. Clin. Infect. Dis. 11:e39036. doi: 10.5812/archcid.39036 [DOI] [Google Scholar]
- Taylor E., Bal A. M., Balakrishnan I., Brown N. M., Burns P., Clark M., et al. (2021). A prospective surveillance study to determine the prevalence of 16S rRNA methyltransferase-producing gram-negative bacteria in the UK. J. Antimicrob. Chemother. 76, 2428–2436. doi: 10.1093/jac/dkab186, PMID: [DOI] [PubMed] [Google Scholar]
- Tchakal-Mesbahi A., Metref M., Singh V. K., Almpani M., Rahme L. G. (2021). Characterization of antibiotic resistance profiles in Pseudomonas aeruginosa isolates from burn patients. Burns 47, 1833–1843. doi: 10.1016/j.burns.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekin R., Dal T., Pirinccioglu H., Oygucu S. E. (2013). A 4-year surveillance of device-associated nosocomial infections in a neonatal intensive care unit. Pediatr. Neonatol. 54, 303–308. doi: 10.1016/j.pedneo.2013.03.011, PMID: [DOI] [PubMed] [Google Scholar]
- Tenover F. C., Nicolau D. P., Gill C. M. (2022). Carbapenemase-producing Pseudomonas aeruginosa-an emerging challenge. Emerg. Microbes Infect. 11, 811–814. doi: 10.1080/22221751.2022.2048972, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thabet A., Ahmed S., Esmat M. (2022). Emergence of Colistin-resistant Pseudomonas aeruginasa in Sohag University hospitals, Egypt. Microbes and Infectious Diseases. 0, 958–971. doi: 10.21608/mid.2022.150919.1352 [DOI] [Google Scholar]
- Thapa T. B., Pokhrel S., Lamichhane A., Singh V. K., Shrestha O., Sapkota M., et al. (2023). Prevalence and antibiogram of bacteria causing urinary tract infection among patients with chronic kidney disease. Open Med. 18:20230824. doi: 10.1515/med-2023-0824, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen P., Henriksen A. S., Longshaw C., Yamano Y., Caldwell B., Hamprecht A. (2022). In vitro activity of cefiderocol against gram-negative bacterial pathogens in Germany. J. Global Antimicrob. Resist. 28, 12–17. doi: 10.1016/j.jgar.2021.10.029, PMID: [DOI] [PubMed] [Google Scholar]
- Tiengrim S., Mootsikapun P., Wonglakorn L., Changpradub D., Thunyaharn S., Tantisiriwat W., et al. (2017). Comparative in vitro activity of Sitafloxacin against Bacteria isolated from Thai patients with urinary tract infections and lower respiratory tract infections in 2016. J. Med. Assoc. Thail. 100. [PubMed] [Google Scholar]
- Tohamy S. T., Aboshanab K. M., El-Mahallawy H. A., El-Ansary M. R., Afifi S. S. (2018). Prevalence of multidrug-resistant gram-negative pathogens isolated from febrile neutropenic cancer patients with bloodstream infections in Egypt and new synergistic antibiotic combinations. Infect. Drug Resist. 11, 791–803. doi: 10.2147/IDR.S163293, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsopoulos P. P., Iosifidis E., Antachopoulos C., Anestis D. M., Karantani E., Karyoti A., et al. (2016). Nosocomial bloodstream infections in neurosurgery: a 10-year analysis in a center with high antimicrobial drug-resistance prevalence. Acta Neurochir. 158, 1647–1654. doi: 10.1007/s00701-016-2890-5 [DOI] [PubMed] [Google Scholar]
- Tumbarello M., De Pascale G., Trecarichi E. M., Spanu T., Antonicelli F., Maviglia R., et al. (2013). Clinical outcomes of Pseudomonas aeruginosa pneumonia in intensive care unit patients. Intensive Care Med. 39, 682–692. doi: 10.1007/s00134-013-2828-9 [DOI] [PubMed] [Google Scholar]
- Tumbarello M., Repetto E., Trecarichi E. M., Bernardini C., De Pascale G., Parisini A., et al. (2011). Multidrug-resistant Pseudomonas aeruginosa bloodstream infections: risk factors and mortality. Epidemiol. Infect. 139, 1740–1749. doi: 10.1017/S0950268810003055 [DOI] [PubMed] [Google Scholar]
- Tuon F. F., Cieslinski J., Rodrigues S. D. S., Serra F. B., Paula M. D. (2020). Evaluation of in vitro activity of ceftolozane-tazobactam against recent clinical bacterial isolates from Brazil – the EM200 study. Braz. J. Infect. Dis. 24, 96–103. doi: 10.1016/j.bjid.2020.04.004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah R., Amir M., Anjum S., Ur Rehman M., Noorul Hasan T., Sajjad Naqvi S., et al. (2023). Presence of T3SS (exoS, exoT, exoU and exoY), susceptibility pattern and MIC of MDR-Pseudomonas aeruginosa from burn wounds. J. Infect. Dev. Ctries. 17, 1130–1137. doi: 10.3855/jidc.17580, PMID: [DOI] [PubMed] [Google Scholar]
- Ullah N., Guler E., Guvenir M., Arikan A., Suer K. (2019). Isolation, identification, and antibiotic susceptibility patterns of Pseudomonas aeruginosa strains from various clinical samples in a University Hospital in Northern Cyprus. Cyprus J. Med. Sci. 4, 225–228. doi: 10.5152/cjms.2019.931 [DOI] [Google Scholar]
- Uskudar Guclu A., Altay Kocak A., Akcil Ok M., Tutluoglu B., Basustaoglu A. C., Respiratory Study G (2021). Antibacterial resistance in lower respiratory tract bacterial pathogens: a multicenter analysis from Turkey. J. Infect. Dev. Ctries. 15, 254–262. doi: 10.3855/jidc.12599 [DOI] [PubMed] [Google Scholar]
- Uzun B., Güngör S., Sezak N., Afşar İ., Şerifhan İlgün M., Demirci M. (2014). Changes in resistance percentage to antibiotics in Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from blood cultures of intensive care unit patients. Turk Hij Den Biyol Derg. 71, 1–8. doi: 10.5505/TurkHijyen.2014.68916 [DOI] [Google Scholar]
- Valenza G., Radike K., Schoen C., Horn S., Oesterlein A., Frosch M., et al. (2010). Resistance to tobramycin and colistin in isolates of Pseudomonas aeruginosa from chronically colonized patients with cystic fibrosis under antimicrobial treatment. Scand. J. Infect. Dis. 42, 885–889. doi: 10.3109/00365548.2010.509333, PMID: [DOI] [PubMed] [Google Scholar]
- Valenza G., Tappe D., Turnwald D., Frosch M., König C., Hebestreit H., et al. (2008). Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J. Cyst. Fibros. 7, 123–127. doi: 10.1016/j.jcf.2007.06.006 [DOI] [PubMed] [Google Scholar]
- Valenza G., Tuschak C., Nickel S., Krupa E., Lehner-Reindl V., Höller C. (2015). Prevalence, antimicrobial susceptibility, and genetic diversity of Pseudomonas aeruginosa as intestinal colonizer in the community. Infect Dis. 47, 654–657. doi: 10.3109/23744235.2015.1031171, PMID: [DOI] [PubMed] [Google Scholar]
- Vamsi K. S., Ramavath U. R., Reddy B., Gandhari M., Reddy Y. (2023). Efficacy of Colistin with Carbapenems combination by checkerboard assay against Carbapenem resistant non lactose fermenting gram negative Bacteria. Journal of Pure & Applied. Microbiology 17, 2104–2110. doi: 10.22207/JPAM.17.4.06 [DOI] [Google Scholar]
- Van An N., Hoang L. H., Le H. H. L., Thai Son N., Hong L. T., Viet T. T., et al. (2023). Distribution and antibiotic resistance characteristics of bacteria isolated from blood culture in a teaching hospital in Vietnam during 2014–2021. Infect. Drug Resist. 16, 1677–1692. doi: 10.2147/IDR.S402278, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Burgh S., Maghdid D. M., Ganjo A. R., Mansoor I. Y., Kok D. J., Fatah M. H., et al. (2019). PME and other ESBL-positive multiresistant Pseudomonas aeruginosa isolated from hospitalized patients in the region of Kurdistan, Iraq. Microbial Drug Resist. 25, 32–38. doi: 10.1089/mdr.2018.0036, PMID: [DOI] [PubMed] [Google Scholar]
- van der Heijden I. M., Levin A. S., De Pedri E. H., Fung L., Rossi F., Duboc G., et al. (2007). Comparison of disc diffusion, Etest and broth microdilution for testing susceptibility of carbapenem-resistant P. aeruginosa to polymyxins. Ann. Clin. Microbiol. Antimicrob. 6, 1–7. doi: 10.1186/1476-0711-6-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudeva N., Nirwan P. S., Shrivastava P. (2016). Bloodstream infections and antimicrobial sensitivity patterns in a tertiary care hospital of India. Therap. Adv. Infect. Dis. 3, 119–127. doi: 10.1177/2049936116666983, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vata A., Pruna R., Rosu F., Miftode E., Nastase E., Gina Vata L., et al. (2018). Pseudomonas Aeruginosa infections in the “Sfânta Parascheva” infectious diseases hospital of Iasi city. Romanian J. Infect. Dis. 21, 115–120. doi: 10.37897/RJID.2018.3.4 [DOI] [Google Scholar]
- Veeraraghavan B., Jesudason M. R., Prakasah J. A. J., Anandan S., Sahni R. D., Pragasam A. K., et al. (2018). Antimicrobial susceptibility profiles of gram-negative bacteria causing infections collected across India during 2014-2016: study for monitoring antimicrobial resistance trend report. Indian J. Med. Microbiol. 36, 32–36. doi: 10.4103/ijmm.IJMM_17_415, PMID: [DOI] [PubMed] [Google Scholar]
- Viasus D., Puerta-Alcalde P., Cardozo C., Suarez-Lledo M., Rodriguez-Nunez O., Morata L., et al. (2020). Predictors of multidrug-resistant Pseudomonas aeruginosa in neutropenic patients with bloodstream infection. Clin. Microbiol. Infect. 26, 345–350. doi: 10.1016/j.cmi.2019.07.002 [DOI] [PubMed] [Google Scholar]
- Viedma E., Juan C., Villa J., Barrado L., Orellana M. Á., Sanz F., et al. (2012). VIM-2–producing multidrug-resistant Pseudomonas aeruginosa ST175 clone, Spain. Emerg. Infect. Dis. 18, 1235–1241. doi: 10.3201/eid1808.111234, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosahlikova S., Drevinek P., Cinek O., Pohunek P., Maixnerova M., Urbaskova P., et al. (2007). High genotypic diversity of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis in the Czech Republic. Res. Microbiol. 158, 324–329. doi: 10.1016/j.resmic.2007.02.003, PMID: [DOI] [PubMed] [Google Scholar]
- Wadhwa R., Sharma Y., Upadhyay R. P., Bala K. (2016). Nosocomial infection by non-fermenting gram negative bacilli in tertiary care hospital: screening and cure. Int J Pharm Pharm Sci 8, 274–277. [Google Scholar]
- Waites K. B., Bade D. J., Bébéar C., Brown S. D., Davidson M. K., Duffy L. B., et al. (2011). Methods for antimicrobial susceptibility testing for human mycoplasmas; approved guideline. Europe PMC. [PubMed]
- Walkty A., Baxter M., Adam H., Karlowsky J. A., Lagacé-Wiens P., Hoban D. J., et al. (2012). Antimicrobial susceptibility of Pseudomonas aeruginosa isolates obtained from patients in Canadian hospitals: CANWARD 2008–2011. Diagn. Microbiol. Infect. Dis. 73, 361–364. doi: 10.1016/j.diagmicrobio.2012.05.007, PMID: [DOI] [PubMed] [Google Scholar]
- Walkty A., DeCorby M., Lagacé-Wiens P., Karlowsky J., Hoban D., Zhanel G. (2011). In vitro activity of ceftazidime combined with NXL104 versus Pseudomonas aeruginosa isolates obtained from patients in Canadian hospitals (CANWARD 2009 study). Antimicrob. Agents Chemother. 55, 2992–2994. doi: 10.1128/AAC.01696-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkty A., DeCorby M., Nichol K., Karlowsky J., Hoban D., Zhanel G. (2009). In vitro activity of colistin (polymyxin E) against 3,480 isolates of gram-negative bacilli obtained from patients in Canadian hospitals in the CANWARD study, 2007-2008. Antimicrob. Agents Chemother. 53, 4924–4926. doi: 10.1128/AAC.00786-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkty A., Karlowsky J. A., Adam H., Baxter M., Lagacé-Wiens P., Hoban D. J., et al. (2013). In vitro activity of ceftolozane-tazobactam against Pseudomonas aeruginosa isolates obtained from patients in Canadian hospitals in the CANWARD study, 2007 to 2012. Antimicrob. Agents Chemother. 57, 5707–5709. doi: 10.1128/AAC.01404-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkty A., Karlowsky J. A., Lagace-Wiens P., Baxter M. R., Adam H. J., Zhanel G. G. (2022). Antimicrobial resistance patterns of bacterial pathogens recovered from the urine of patients at Canadian hospitals from 2009 to 2020. JAC-Antimicrob. Resist. 4:dlac122. doi: 10.1093/jacamr/dlac122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkty A., Lagace-Wiens P., Adam H., Baxter M., Karlowsky J., Mulvey M. R., et al. (2017). Antimicrobial susceptibility of 2906 Pseudomonasaeruginosa clinical isolates obtained from patients in Canadian hospitals over a period of 8 years: results of the Canadian Ward surveillance study (CANWARD), 2008-2015. Diagn. Microbiol. Infect. Dis. 87, 60–63. doi: 10.1016/j.diagmicrobio.2016.10.003, PMID: [DOI] [PubMed] [Google Scholar]
- Walters M. S., Grass J. E., Bulens S. N., Hancock E. B., Phipps E. C., Muleta D., et al. (2019). Carbapenem-resistant Pseudomonas aeruginosa at US emerging infections program sites, 2015. Emerg. Infect. Dis. 25, 1281–1288. doi: 10.3201/eid2507.181200, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kong J., Zhang X., Liu Y., Huang Z., Yuan L., et al. (2022). Plumbagin resurrect colistin susceptible against colistin-resistant Pseudomonas aeruginosa in vitro and in vivo. Front. Microbiol. 13:1020652. doi: 10.3389/fmicb.2022.1020652, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang X. (2020). Prevalence of metallo-β-lactamase genes among Pseudomonas aeruginosa isolated from various clinical samples in China. J. Lab. Med. 44, 197–203. doi: 10.1515/labmed-2019-0162 [DOI] [Google Scholar]
- Wang Q., Wang Z., Zhang F., Zhao C., Yang B., Sun Z., et al. (2020). Long-term continuous antimicrobial resistance surveillance among nosocomial gram-negative Bacilli in China from 2010 to 2018 (CMSS). Infect Drug Resist. 13, 2617–2629. doi: 10.2147/IDR.S253104, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattal C., Goel N., Oberoi J. K., Datta S., Raveendran R. (2019). Performance of three commercial assays for colistin susceptibility in clinical isolates and mcr-1 carrying reference strain. Indian J. Med. Microbiol. 37, 488–495. doi: 10.4103/ijmm.IJMM_20_92, PMID: [DOI] [PubMed] [Google Scholar]
- Wattal C., Raveendran R., Goel N., Oberoi J. K., Rao B. K. (2014). Ecology of blood stream infection and antibiotic resistance in intensive care unit at a tertiary care hospital in North India. Braz. J. Infect. Dis. 18, 245–251. doi: 10.1016/j.bjid.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemambu S., Joshi K. (1983). Prevalence and antimicrobial sensitivity of Localpseudomonas aeruginosa at a teaching hospital. Public Health 97, 89–94. doi: 10.1016/S0033-3506(83)80004-3 [DOI] [PubMed] [Google Scholar]
- Wendel A. F., Malecki M., Mattner F., Xanthopoulou K., Wille J., Seifert H., et al. (2022). Genomic-based transmission analysis of carbapenem-resistant Pseudomonas aeruginosa at a tertiary care Centre in Cologne (Germany) from 2015 to 2020. JAC-Antimicrob. Resist. 4:dlac057. doi: 10.1093/jacamr/dlac057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wi Y. M., Choi J.-Y., Lee J.-Y., Kang C.-I., Chung D. R., Peck K. R., et al. (2017). Emergence of colistin resistance in Pseudomonas aeruginosa ST235 clone in South Korea. Int. J. Antimicrob. Agents 49, 767–769. doi: 10.1016/j.ijantimicag.2017.01.023, PMID: [DOI] [PubMed] [Google Scholar]
- Wi Y. M., Greenwood-Quaintance K. E., Schuetz A. N., Ko K. S., Peck K. R., Song J.-H., et al. (2018). Activity of ceftolozane-tazobactam against carbapenem-resistant, non-carbapenemase-producing Pseudomonas aeruginosa and associated resistance mechanisms. Antimicrob. Agents Chemother. 62, 01970–01917. doi: 10.1128/AAC.01970-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann M., Bezdan D., Zapata L., Susak H., Vogel W., Schröppel K., et al. (2015). Analysis of a long-term outbreak of XDR Pseudomonas aeruginosa: a molecular epidemiological study. J. Antimicrob. Chemother. 70, 1322–1330. doi: 10.1093/jac/dku546, PMID: [DOI] [PubMed] [Google Scholar]
- Willmann M., Kuebart I., Marschal M., Schröppel K., Vogel W., Flesch I., et al. (2013). Effect of metallo-β-lactamase production and multidrug resistance on clinical outcomes in patients with Pseudomonas aeruginosa bloodstream infection: a retrospective cohort study. BMC Infect. Dis. 13, 1–9. doi: 10.1186/1471-2334-13-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise M. G., Karlowsky J. A., Hackel M. A., Harti M. A., Ntshole B. M., Njagua E. N., et al. (2023a). In vitro activity of ceftazidime-avibactam against clinical isolates of Enterobacterales and Pseudomonas aeruginosa from sub-Saharan Africa: ATLAS global surveillance program 2017–2021. J. Global Antimicrobial Resist. 35, 93–100. doi: 10.1016/j.jgar.2023.08.022, PMID: [DOI] [PubMed] [Google Scholar]
- Wise M. G., Karlowsky J. A., Lemos-Luengas E. V., Valdez R. R., Sahm D. F. (2023b). Epidemiology and in vitro activity of ceftazidime-avibactam and comparator agents against multidrug-resistant isolates of Enterobacterales and Pseudomonas aeruginosa collected in Latin America as part of the ATLAS surveillance program in 2015–2020. Braz. J. Infect. Dis. 27:102759. doi: 10.1016/j.bjid.2023.102759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Yin D., Zhi P., Guo Y., Yang Y., Zhu D., et al. (2022). In vitro activity of MRX-8 and comparators against clinical isolated gram-negative bacilli in China. Front. Cell. Infect. Microbiol. 12:829592. doi: 10.3389/fcimb.2022.829592, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J., Jia P., Zhu Y., Yu W., Zhang J., Gao H., et al. (2022). Antimicrobial susceptibility to polymyxin B and other comparators against gram-negative bacteria isolated from bloodstream infections in China: results from CARVIS-NET program. Front. Microbiol. 13:1017488. doi: 10.3389/fmicb.2022.1017488, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S. K., Bhujel R., Mishra S. K., Sharma S., Sherchand J. B. (2020). Emergence of multidrug-resistant non-fermentative gram negative bacterial infection in hospitalized patients in a tertiary care center of Nepal. BMC. Res. Notes 13:319. doi: 10.1186/s13104-020-05163-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin M. T., Mostafa A. A.-F., Al-Askar A. A., Al-Otibi F. O. (2022). Synergistic antibacterial activity of green synthesized silver nanomaterials with colistin antibiotic against multidrug-resistant bacterial pathogens. Crystals 12:1057. doi: 10.3390/cryst12081057 [DOI] [Google Scholar]
- Yayan J., Ghebremedhin B., Rasche K. (2015). Antibiotic resistance of Pseudomonas aeruginosa in pneumonia at a single University Hospital Center in Germany over a 10-year period. PLoS One 10:e0139836. doi: 10.1371/journal.pone.0139836, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz F. N., Öksüz L., Demir E. S., Döşler S., Savage P. B., Güzel Ç. B. (2023). Efficacy of Ceragenins alone and in combinations with antibiotics against multidrug-resistant gram negative pathogens from bloodstream infections. Curr. Microbiol. 80:327. doi: 10.1007/s00284-023-03443-5, PMID: [DOI] [PubMed] [Google Scholar]
- Yilmaz G., Salyan S., Aksoy F., Koksal I. (2017). Individualized antibiotic therapy in patients with ventilator-associated pneumonia. J. Med. Microbiol. 66, 78–82. doi: 10.1099/jmm.0.000401 [DOI] [PubMed] [Google Scholar]
- Yousefi S., Nahaei M. R., Farajnia S., Aghazadeh M., Iversen A., Edquist P., et al. (2013). A multiresistant clone of Pseudomonas aeruginosa sequence type 773 spreading in a burn unit in Orumieh. Iran. Apmis. 121, 146–152. doi: 10.1111/j.1600-0463.2012.02948.x [DOI] [PubMed] [Google Scholar]
- Zerouali K., Ramdani-Bouguessa N., Boye C., Hammami A., Active G. (2016). Multicentric study in five African countries of antibiotic susceptibility for three main pathogens: Streptococcus pneumoniae, Staphylococcus aureus, and Pseudomonas aeruginosa. J. Chemother. 28, 266–272. doi: 10.1179/1973947814Y.0000000220, PMID: [DOI] [PubMed] [Google Scholar]
- Zhanel G. G., Adam H. J., Baxter M. R., Fuller J., Nichol K. A., Denisuik A. J., et al. (2019). 42936 pathogens from Canadian hospitals: 10 years of results (2007-16) from the CANWARD surveillance study. J. Antimicrob. Chemother. 74, iv5–iv21. doi: 10.1093/jac/dkz283 [DOI] [PubMed] [Google Scholar]
- Zhanel G. G., Adam H. J., Baxter M. R., Fuller J., Nichol K. A., Denisuik A. J., et al. (2013). Antimicrobial susceptibility of 22746 pathogens from Canadian hospitals: results of the CANWARD 2007–11 study. J. Antimicrob. Chemother. 68, i7–i22. doi: 10.1093/jac/dkt022 [DOI] [PubMed] [Google Scholar]
- Zhanel G. G., Adam H. J., Low D. E., Blondeau J., DeCorby M., Karlowsky J. A., et al. (2011). Antimicrobial susceptibility of 15,644 pathogens from Canadian hospitals: results of the CANWARD 2007–2009 study. Diagn. Microbiol. Infect. Dis. 69, 291–306. doi: 10.1016/j.diagmicrobio.2010.10.025, PMID: [DOI] [PubMed] [Google Scholar]
- Zhanel G. G., DeCorby M., Adam H., Mulvey M. R., McCracken M., Lagacé-Wiens P., et al. (2010). Prevalence of antimicrobial-resistant pathogens in Canadian hospitals: results of the Canadian Ward surveillance study (CANWARD 2008). Antimicrob. Agents Chemother. 54, 4684–4693. doi: 10.1128/AAC.00469-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lin Y., Zhang X., Chen L., Xu C., Liu S., et al. (2021). Combining Colistin with Furanone C-30 rescues Colistin resistance of gram-negative Bacteria in vitro and in vivo. Microbiol. Spectr. 9:e0123121. doi: 10.1128/Spectrum.01231-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chen D., Chen K., Xie M., Guo J., Chan E. W. C., et al. (2023). Epidemiological and genetic characteristics of clinical carbapenem-resistant Pseudomonas aeruginosa strains in Guangdong Province, China. Microbiol. Spectrum 11, e04261–e04222. doi: 10.1128/spectrum.04261-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Chen J., Shen H., Chen Z., Yang Q.-w., Zhu J., et al. (2021). Emergence of ceftazidime-and avibactam-resistant Klebsiella pneumoniae carbapenemase-producing Pseudomonas aeruginosa in China. Msystems 6, e00787–e00721. doi: 10.1128/mSystems.00787-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Kang Y., Zhang H., Yu W., Zhang G., Zhang J., et al. (2023). Emergence of ST463 exoU-positive, imipenem-nonsusceptible Pseudomonas aeruginosa isolates in China. Microbiol. Spectrum. 11, e00105–e00123. doi: 10.1128/spectrum.00105-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorgani A., Abofayed A., Glia A., Albarbar A., Hanish S. (2015). Prevalence of device-associated nosocomial infections caused by gram-negative bacteria in a trauma intensive care unit in Libya. Oman Med. J. 30, 270–275. doi: 10.5001/omj.2015.54, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubair K., Iregbu K. (2018). Resistance pattern and detection of metallo-beta-lactamase genes in clinical isolates of Pseudomonas aeruginosa in a Central Nigeria tertiary hospital. Niger. J. Clin. Pract. 21, 176–182. doi: 10.4103/njcp.njcp_229_17, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.