Abstract
Background
Emerging evidence reveals a complex relationship between cardiovascular disease (CVD) and cancer, which share common risk factors and biological pathways.
Objectives
The aim of this study was to evaluate common epigenetic signatures for CVD and cancer incidence in 3 ethnically diverse cohorts: Native Americans from the SHS (Strong Heart Study), European Americans from the FHS (Framingham Heart Study), and European Americans and African Americans from the ARIC (Atherosclerosis Risk In Communities) study.
Methods
A 2-stage strategy was used that included first conducting untargeted epigenome-wide association studies for each cohort and then running targeted models in the union set of identified differentially methylated positions (DMPs). We also explored potential molecular pathways by conducting a bioinformatics analysis.
Results
Common DMPs were identified across all populations. In a subsequent meta-analysis, 3 and 1 of those DMPs were statistically significant for CVD only and both cancer and CVD, respectively. No meta-analyzed DMPs were statistically significant for cancer only. The enrichment analysis pointed to interconnected biological pathways involved in cancer and CVD. In the DrugBank database, elements related to 1-carbon metabolism and cancer and CVD medications were identified as potential drugs for target gene products. In an additional analysis restricted to the 950 SHS participants who developed incident CVD, the C index for incident cancer increased from 0.618 (95% CI: 0.570-0.672) to 0.971 (95% CI: 0.963-0.978) when adjusting the models for the combined cancer and CVD DMPs identified in the other cohorts.
Conclusions
These results point to molecular pathways and potential treatments for precision prevention of CVD and cancer. Screening based on common epigenetic signatures of incident CVD and cancer may help identify patients with newly diagnosed CVD at increased cancer risk.
Key Words: cancer, cardio-oncology, cardiovascular disease, DNA methylation, multicohort
Central Illustration
Cardiovascular disease (CVD) and cancer are the leading causes of death worldwide.1 In the United States, CVD and cancer were the first and second most common causes of death in both 2021 and 2022.2 Both diseases share risk factors, including aging, cigarette smoking, physical inactivity, an unhealthy diet, excess adiposity, and genetic and environmental factors.3 They also share biological pathways, including inflammation, oxidative stress, apoptosis, and angiogenesis.3,4 Cancer treatments, including chemotherapy, radiotherapy, targeted cancer therapies, and immunotherapy, can contribute to cardiotoxicity.4, 5, 6 While CVD and cancer have traditionally been studied separately, the field of cardio-oncology has grown in recent years.1
Epigenetic marks are able to regulate gene expression, and their dysregulation might lead to the disruption of essential biological processes, potentially leading to disease.7 To our knowledge, however, epigenetic changes associated with CVD and cancer have only been studied separately.8,9 Also, DNA methylation (DNAm), the most well studied epigenetic mark, has not yet been jointly evaluated for CVD and cancer outcomes in population-based studies.
The objective of this study was to evaluate whether common epigenetic marks for incident cancer and CVD exist across different racial/ethnic groups in the United States. We conducted a multicohort epigenome-wide association study (EWAS) of CVD only, cancer only, and both cancer and CVD (Ca-CVD) using data from 4 ethnically diverse cohorts: the SHS (Strong Heart Study) (Native Americans), the FHS (Framingham Heart Study) (European Americans), and European and African American subsamples of the ARIC (Atherosclerosis Risk In Communities) study. As a secondary objective, we aimed to explore potential molecular pathways that link the etiologies of CVD and cancer by conducting bioinformatic analyses. These analyses included an integrative analysis of protein-protein interaction networks among proteins encoded by genes annotated to identified CVD, cancer, and Ca-CVD differentially methylated positions (DMPs), network enrichment analyses, and identification of potential drugs in the DrugBank database for target genes from the most relevant nodes observed in the protein interaction network. In addition, to explore if DNAm might contribute to the identification of candidates for cancer screening in patients with newly diagnosed CVD, we evaluated the predictive ability of DNAm for cancer incidence in participants who had CVD incident events.
Methods
Study populations
The SHS protocol was reviewed and approved by the Institutional Review Boards of the participating institutions, the 12 participating tribes, and the Indian Health Service Institutional Review Boards for the 3 geographic areas where the tribes reside. The FHS was approved by the Institutional Review Board of Boston University Medical Center. ARIC received approval from the Institutional Review Boards of the participating institutions.
The SHS
The SHS is a prospective cohort study funded by the National Heart, Lung, and Blood Institute to investigate CVD and its risk factors in American Indian adults.10 From 1989 to 1991, a total of 4,549 men and women aged 45 to 75 years, members of 12 tribes based in the Northern Plains (North Dakota and South Dakota), the Southern Plains (Oklahoma), and the Southwest (Arizona), agreed to participate. After the exclusion of participants with prevalent CVD, prevalent cancer and missing data for relevant covariates, 2,186 participants were included in this study. DNAm was measured using the Illumina MethylationEPIC BeadChip array (details and preprocessing are described in Supplemental Methods).
The FHS
The FHS is a population-based cohort that started in 1948.11 A total of 5,209 men and women of European ancestry between the ages of 30 and 62 years from the town of Framingham, Massachusetts, were recruited. The FHS Offspring Cohort, founded in 1971, is a second-generation study in which children of the FHS Original Cohort were recruited. After excluding individuals with prevalent CVD and cancer at examination 8, as well as individuals with missing data for other relevant covariates, 1,474 Offspring Cohort participants were included in this study. DNAm was measured using the Illumina Infinium HumanMethylation450 BeadChip array (details and preprocessing are described in Supplemental Methods).
The ARIC
The ARIC study is a prospective cohort study that was initiated in 1987 to investigate CVD risk.12 The study involved 15,792 participants sampled from 4 U.S. communities, including Jackson, Mississippi; Washington County, Maryland; suburban Minneapolis, Minnesota; and Forsyth County, North Carolina. The baseline cohort consisted of individuals aged 45 to 64 years, with the Jackson sample comprising exclusively Black residents. All analyses were conducted separately in the 2 ARIC subgroups, African American participants of the ARIC study (ARICb) and European American participants of the ARIC study (ARICw). For this study, individuals with prevalent CVD or cancer at the time of blood draw for each visit were excluded, along with those who had missing data for relevant covariates. The final analysis included a total of 2,212 African American participants and 903 European American participants. DNAm was measured using the Infinium HumanMethylation450 BeadChip array (details and preprocessing are described in Supplemental Methods).
Cancer and CVD incidence ascertainment and follow-up definition
Details of cancer and CVD assessment for each cohort are described in Supplemental Methods. Briefly, in the SHS, CVD was assessed using annual morbidity surveillance reviews of hospitalization and death records. Incident CVD was defined as the first occurrence of fatal or nonfatal coronary heart disease, stroke or heart failure, or other nonfatal CVD.10 Cancer incidence was assessed using interviews, death certificates, and/or chart reviews.
In the FHS, CVD was assessed using medical histories, physical examinations during study visits, hospitalization records, and personal physician records. Incident CVD was defined as a composite of coronary heart disease, cerebrovascular events, peripheral artery disease, and heart failure.13 Cancer was assessed by interviews, death certificates, and/or chart reviews that included pathology reports.14
In ARIC, CVD was assessed through active surveillance of hospitalizations and cohort follow-up. Incident CVD included incident heart failure, myocardial infarction, ischemic stroke, and atrial fibrillation.15 Cancer incidence was identified linking the cohort with state cancer registries and through active surveillance of the cohort involving recording hospital discharge codes for all participants.16
For all cohorts, we classified the primary endpoints into 3 groups: incident cancer in participants who did not develop CVD, incident CVD in participants who did not develop cancer, and Ca-CVD for participants who developed both cancer and CVD during follow-up (in either order). We calculated follow-up from the date of baseline examination to the date of the cancer or CVD diagnosis, respectively, or administrative censoring (2017 in the SHS, 2018 for CVD and 2019 for cancer in the FHS, and 2015 in ARIC), whichever occurred first. For the combined Ca-CVD endpoint, we considered the date of the event that happened second. For participants who died of any cause of death other than cancer or CVD, we censored the follow-up at date of death.
Statistical analysis
Multicohort 2-stage epigenome-wide association approach and meta-analysis
Traditionally, EWAS have been conducted evaluating each CpG (cytosine followed by a guanine with a phosphate link) site individually in separate models. However, epigenetic marks are interrelated and are more likely to influence biological processes all together. Thus, considering them jointly in a model that appropriately accounts for the particular challenges of epigenomic data (eg, high dimensions and correlations) is a more realistic approach.17
To evaluate the consistency of CVD, cancer, and Ca-CVD DMPs across populations, we implemented a 2-stage EWAS approach. In the first stage, we conducted an untargeted EWAS for each cohort, using GLMnet penalized Cox regression (Cox elastic-net, R package glmnet),18 which simultaneously considers all CpG sites as independent variables in the same model (see Supplemental Methods). We fit an elastic-net Cox model using cancer, CVD, and Ca-CVD time-to-event outcomes in separate models.
In the second stage, we conducted a targeted EWAS. We defined the union set of DMPs identified across all cohorts from the untargeted elastic-net in the previous step (Central Illustration). Subsequently, we ran elastic-net models in each cohort separately for that union set list of DMPs. We calculated individual HRs and 95% CIs using Cox proportional hazards models for the DMPs that were identified in the targeted EWAS in common for all cohorts. We used Cox proportional hazards models instead of elastic-net for the calculation of effect estimates, given that elastic-net applies shrinkage and therefore might lead to biased effect estimates. In those DMPs, we subsequently conducted a meta-analysis of Cox regression coefficients using the R package meta.19 The 2-stage approach was implemented for each of the 3 endpoints: CVD, cancer, and Ca-CVD. All models were adjusted for age, sex, body mass index, smoking status (never, former, or current), DNAm-based smoking score,20 and estimated cell counts. CVD and Ca-CVD models were additionally adjusted for low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, diabetes (yes or no), hypertension treatment (yes or no) and systolic blood pressure. SHS models were additionally adjusted for study center (Arizona, Oklahoma, or North Dakota and South Dakota), 5 genetic principal components (PCs) that accounted for population stratification, and, for CVD and Ca-CVD models, albuminuria (<30, 30-300, and ≥300 mg/g). FHS models were additionally adjusted for batch, as DNAm data were processed in 2 different batches. ARIC models were adjusted for 5 PCs to correct batch effects in each cohort. For White participants, ARIC models were additionally adjusted for field center (Washington County, suburban Minneapolis, and Forsyth County). In addition to effect sizes and P values from the meta-analysis, we reported the I2 statistic, a measure of heterogeneity among studies.21 As a sensitivity analysis, we ran models for the CpGs included in the meta-analysis that were statistically significant in the SHS among never smokers.
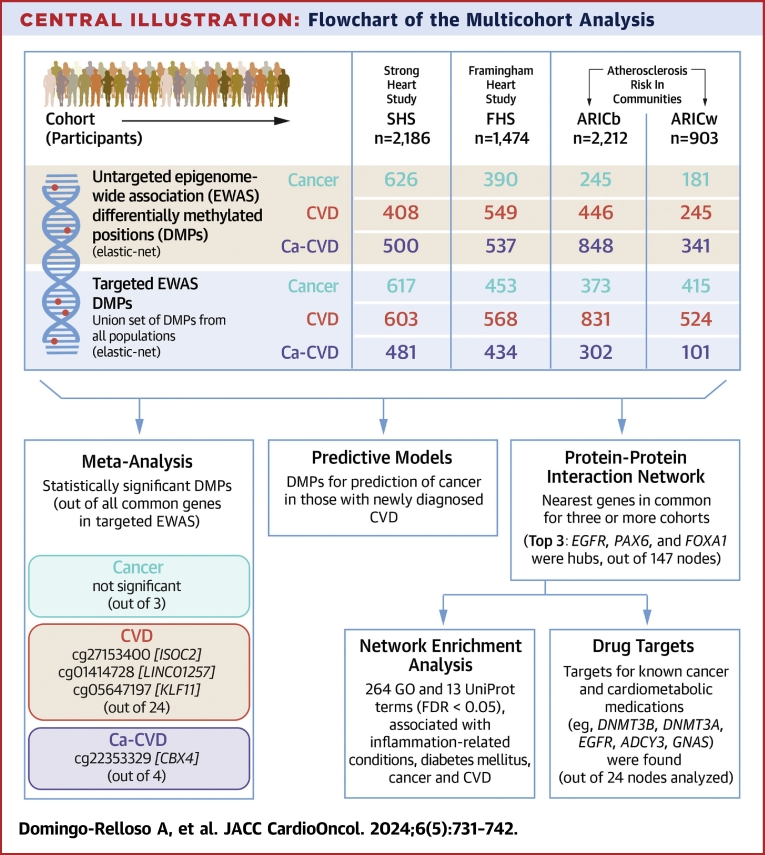
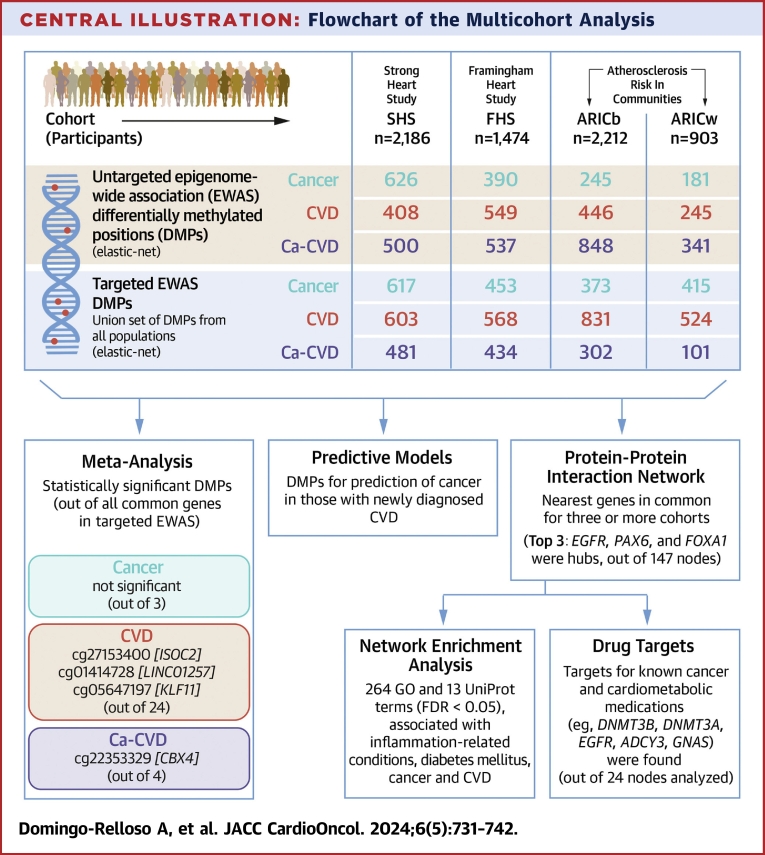
Central Illustration.
Flowchart of the Multicohort Analysis
Differentially methylated positions (DMPs) in the untargeted epigenome-wide association study (EWAS) were obtained by fitting separate elastic-net models for each cohort. Targeted EWAS DMPs were obtained by fitting an elastic-net model, separately for each cohort, to the union set of DMPs identified in the untargeted EWAS for all cohorts. DMPs in the targeted EWAS were annotated to their closest gene to perform a protein-protein interaction network in common nodes for at least 3 cohorts (highlighted in black lines in Venn diagram; Supplemental Figure 2). Subsequently, a gene-set analysis was carried out on the network. ARICb = African American participants of the Atherosclerosis Risk In Communities study; ARICw = European American participants of the Atherosclerosis Risk In Communities study; Ca-CVD = cancer and cardiovascular disease; CVD = cardiovascular disease; FDR = false discovery rate; FHS = Framingham Heart Study; GO = Gene Ontology; SHS = Strong Heart Study.
Molecular pathway analyses
We conducted a protein-protein interaction analysis of protein-coding genes annotated to DMPs identified in the second stage of the 2-stage approach described in the previous subsection (targeted EWAS DMPs) (Central Illustration). We obtained reported biological interactions among the protein nodes from the STRING database version 11.5 (see Supplemental Methods).22 A network enrichment analysis was performed by incorporating available information for the relationships between nodes based on Gene Ontology (GO) and UniProt databases. We also attempted to identify common relevant biological mechanisms for cancer and CVD by evaluating which nodes are classified as drug targets within the DrugBank database version 5.1. DrugBank combines detailed data annotations with comprehensive drug target information,23 which enables the assessment of whether gene products are potential drug targets of specific compounds.
Predictive models of incident cancer in participants who developed CVD first
To explore if measuring DNAm in blood, which is an accessible tissue in the clinical setting, might contribute to the identification of candidates for risk stratification for cancer screening in patients with newly diagnosed CVD, we evaluated in the SHS the predictive ability of baseline blood DNAm signatures identified in FHS, ARICb, and ARICw for cancer incidence. Time to incident cancer was evaluated starting from the time of CVD diagnosis. Predictive ability was measured by the concordance index (C index) obtained from elastic-net models; values close to 1 indicate good predictive performance. Ninety-five percent CIs for the C indexes were calculated using quantile bootstrap. We also calculated the Brier score for each predictive model, which measures calibration.24 In this case, values close to 0 indicate good calibration. We restricted this analysis to participants who were free of cancer at the time of their first CVD event (n = 950). We compared the predictive ability of a model adjusted for clinical risk factors (age, sex, smoking status, body mass index, and study center) with the predictive ability of these additional models: 1) adjusted for clinical risk factors, estimated cell counts, genetic PCs, and the epigenetic smoking score; 2) additionally adjusted for cancer DMPs identified in FHS, ARICb, and ARICw; and 3) additionally adjusted for Ca-CVD DMPs identified in FHS, ARICb, and ARICw. We were unable to conduct parallel analysis of incident CVD among individuals newly diagnosed with cancer because of the small number of cases (n = 15 cancer cases happening before CVD).
Results
Descriptive analysis
In the SHS (n = 2,186), we observed 277, 823, and 142 incident cancer, CVD, and Ca-CVD cases, respectively (Table 1). In the FHS (n = 154), we observed 179, 294, and 50 incident CVD, cancer, and Ca-CVD cases, respectively. In ARICb (n = 2,212), we observed 389, 622, and 244 incident cancer, CVD, and Ca-CVD cases, respectively. In ARICw (n = 903), there were 193, 222, and 84 incident cancer, CVD, and Ca-CVD cases, respectively. Numbers of specific cancer and CVD endpoints in each cohort are shown in Supplemental Table 1. In all cohorts, participants with incident CVD were older and more likely to have baseline hypertension and diabetes compared with those without incident CVD. Baseline current smoking was more common in participants with incident cancer, CVD, and Ca-CVD (Table 1) compared with those without.
Table 1.
Participant Characteristics in the Participating Cohorts
| No Cancer and No CVD | Cancer | CVD | Cancer and CVD | |
|---|---|---|---|---|
| Strong Heart Study | (n = 944) | (n = 277) | (n = 823) | (n = 142) |
| Follow-up, y | 26.1 (12.4-27.3) | 14.9 (8.7-20.5) | 9.9 (5.4-16.7) | 17.7 (10.8-22.8) |
| Age, y | 53 (48-60) | 56 (50-64) | 57 (51-64) | 57 (51-63) |
| Female | 61.1 | 54.2 | 56.5 | 49.3 |
| Current smoking | 34.7 | 46.3 | 40.1 | 47.2 |
| BMI, kg/m2 | 29.2 (25.6-33.1) | 29.2 (25.6-33.9) | 30.1 (27.1-34.2) | 30.0 (27.3-34.5) |
| Diabetes | 32.8 | 37.7 | 52.6 | 45.8 |
| LDL cholesterol, mg/dL | 115 (94-137) | 118 (101-138) | 123 (102-144) | 120 (103-140) |
| HDL cholesterol, mg/dL | 45 (38-54) | 43 (37-53) | 42 (36-50) | 42 (36-49) |
| Hypertension | 14.5 | 16.5 | 27.1 | 21.1 |
| SBP, mm Hg | 121 (111-132) | 124 (111-136) | 127 (116-140) | 125 (111-138) |
| Framingham Heart Study | (n = 1,017) | (n = 294) | (n = 179) | (n = 50) |
| Follow-up | 11.5 (10.5-12.4) | 4.8 (2.5-7.3) | 5.9 (3.2-9.1) | 11.4 (10.1-12.4) |
| Age, y | 62 (57-68) | 65 (60-71) | 70 (63-77) | 72.5 (65-78) |
| Female | 39.5 | 45.1 | 48.5 | 40 |
| Current smoking | 50.9 | 55.8 | 55.9 | 56 |
| BMI, kg/m2 | 27.0 (24.3-30.6) | 27.9 (24.8-31.3) | 28.9 (25.4-32.5) | 30.4 (26.4-34.2) |
| Diabetes | 8.8 | 11.9 | 18.3 | 22 |
| LDL cholesterol, mg/dL | 109 (88-131) | 108 (88-125.25) | 101 (81-127) | 102.5 (80.25-125) |
| HDL cholesterol, mg/dL | 57 (46-70) | 58 (47-69) | 52 (42-65) | 53.5 (43.25-69.25) |
| Hypertension | 36.3 | 45.3 | 59.8 | 64 |
| SBP, mm Hg | 124 (114-135) | 127 (116-138.25) | 131 (122-145) | 127 (118-142) |
| Atherosclerosis Risk In Communities study (African American) | (n = 957) | (n = 389) | (n = 622) | (n = 244) |
| Follow-up | 23.8 (22.4-24.8) | 10.7 (6.2-15.4) | 12.5 (6.7-18.7) | 12.2 (6.4-17.9) |
| Age, y | 54 (51-59) | 56 (51-61) | 57 (52-62) | 58 (53-62) |
| Female | 71.7 | 49.4 | 62.4 | 51.2 |
| Current smoking | 19.6 | 29.3 | 24.8 | 35.7 |
| BMI, kg/m2 | 28.6 (25.7-32.5) | 28.6 (25.3-32.5) | 29.8 (26.6-34.0) | 29.6 (25.7-33.7) |
| Diabetes | 12.7 | 15.9 | 32.0 | 23.0 |
| LDL cholesterol, mg/dL | 130.8 (106.1-155.1) | 129.9 (106.4-157.2) | 135.9 (109.0-162.6) | 132.9 (108.2-158.4) |
| HDL cholesterol, mg/dL | 55 (44-67) | 50 (42-62) | 48 (39-59) | 47 (39-56) |
| Hypertension | 37.8 | 35.2 | 57.1 | 54.9 |
| SBP, mm Hg | 120 (110-132) | 121 (110-132) | 128 (116-143) | 129 (119-141) |
| Atherosclerosis Risk In Communities study (European American) | (n = 404) | (n = 193) | (n = 222) | (n = 84) |
| Follow-up | 24.2 (22.0-25.0) | 11.5 (7.2-17.3) | 14.5 (8.6-21.2) | 14.0 (9.0-18.9) |
| Age, y | 58 (54-63) | 59 (56-64) | 61 (57-65) | 61 (58-64) |
| Female | 68.3 | 51.8 | 51.8 | 41.7 |
| Current smoking | 15.1 | 19.7 | 22.5 | 22.6 |
| BMI, kg/m2 | 25.3 (22.8-27.7) | 25.7 (23.4-28.4) | 26.3 (23.6-29.5) | 26.5 (24.5-28.2) |
| Diabetes | 3.0 | 5.2 | 7.2 | 3.6 |
| LDL cholesterol, mg/dL | 126.1 (106.2-147.8) | 127.2 (107.8-145.3) | 129.4 (107.0-154.4) | 127.2 (108.2-145.8) |
| HDL cholesterol, mg/dL | 52 (42-65) | 47 (40-61) | 46 (37-59) | 46 (39-59) |
| Hypertension | 13.6 | 14.5 | 21.6 | 14.3 |
| SBP, mm Hg | 111 (104-122) | 112 (105-124) | 118 (110-133) | 115 (107-129) |
Values are median (Q1-Q3) or %.
BMI = body mass index; CVD = cardiovascular disease; HDL = high-density lipoprotein; LDL = low-density lipoprotein; SBP = systolic blood pressure.
Multicohort 2-stage approach and meta-analysis
The Central Illustration shows a summary of the number of DMPs identified for each cohort and each endpoint in the untargeted and targeted EWAS, as well as the number of statistically significant DMPs identified in the meta-analysis of common DMPs from the targeted EWAS (see more detailed version of this figure in Supplemental FIgure 1). The overlap between DMPs from the untargeted EWAS across cohorts for each endpoint can be found in Supplemental Figure 1. In the subsequent targeted EWAS, 24 DMPs were common across all populations for CVD, 3 for cancer and 4 for Ca-CVD. Of these common DMPs, with a cutoff P value of <0.05, 3 DMPs were statistically significant in the meta-analysis for CVD—annotated to genes ISOC2 (cg27153400), LINC01257 (cg01414728), and KLF11 (cg05647197)—and 1 was statistically significant for Ca-CVD, annotated to CBX4 (Table 2). With a less stringent cutoff P value of 0.1, 2 additional DMPs were statistically significant for CVD, annotated to HLA-L and ATXN1, and 1 for Ca-CVD, annotated to SIX1. No DMPs were statistically significant in the meta-analysis for the cancer only endpoint. In the sensitivity analysis among never smokers, point estimates were mostly directionally consistent, although CIs were wider, as expected given the reduction in sample size. Tenth and 90th percentiles of DNAm in statistically significant DMPs in the meta-analysis are shown in Supplemental Table 2.
Table 2.
Meta-Analyzed HRs for Differentially Methylated Positions in Common for All Cohorts
| DMP | Gene | HR (95% CI)a | P Value | I2 |
|---|---|---|---|---|
| CVD | ||||
| cg27153400 | ISOC2 | 1.05 (1.01-1.10) | 0.015 | 0.00 |
| cg05647197 | KLF11 | 0.88 (0.79-0.97) | 0.014 | 0.00 |
| cg01414728 | LINC01257 | 0.82 (0.69-0.98) | 0.029 | 0.53 |
| cg16055914 | HLA-L | 0.88 (0.77-1.01) | 0.063 | 0.54 |
| cg00151370 | ATXN1 | 1.25 (0.97-1.59) | 0.080 | 0.68 |
| Cancer and CVD | ||||
| cg22353329 | CBX4 | 1.44 (1.12-1.84) | 0.004 | 0.00 |
| cg03032497 | SIX1 | 1.11 (0.99-1.24) | 0.073 | 0.00 |
All models were adjusted for age, sex, body mass index, smoking status (never, former, or current), DNA methylation–based smoking score, and estimated cell counts. CVD and Ca-CVD models were additionally adjusted for low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, diabetes (yes or no), hypertension treatment (yes or no) and systolic blood pressure. SHS (Strong Heart Study) models were additionally adjusted for study center (Arizona, Oklahoma, or North Dakota and South Dakota), 5 genetic PCs and, for CVD and Ca-CVD models, albuminuria (normal albumin levels, microalbuminuria, and macroalbuminuria). FHS (Framingham Heart Study) models were additionally adjusted for batch, as DNA methylation data were processed in two different batches. ARIC (Atherosclerosis Risk in Communities) models were additionally adjusted for five PCs to correct batch effects, and models for European American participants of ARIC were additionally adjusted for field center (Jackson, Mississippi; Washington County, Maryland; suburban Minneapolis, Minnesota; and Forsyth County, North Carolina). DMPs in common for all cohorts in the targeted epigenome-wide association study (see Central Illustration) were included in the meta-analysis. No cytosines followed by a guanine with a phosphate link were significant in the meta-analysis for the cancer only endpoint. P values adjusted for false discovery rate (using the Benjamini-Hochberg approach) were 0.18 for cg27153400, 0.18 for cg05647197, 0.23 for cg01414728, 0.37 for cg16055914, 0.38 for cg00151370, 0.019 for cg22353329, and 0.15 for cg03032497.
Ca-CVD = cancer and cardiovascular disease; CVD = cardiovascular disease; DMP = differentially methylated position; PC = principal component.
HRs (95% CIs) comparing the 90th and 10th percentiles of DNA methylation proportions, calculated as the mean percentile across all cohorts.
Molecular pathways analyses
We conducted a protein-protein interaction network analysis of protein-coding genes annotated to DMPs that were identified in elastic-net analysis of at least 3 cohorts from the targeted EWAS (Supplemental Figure 2). The resulting protein interaction network contained 147 nodes and 217 edges (Figure 1, Supplemental File, sheets M and N). For CVD only, the most connected node in the network was EGFR, with 19 edges (statistically significant in ARICw, FHS, and SHS). PAX6 was the most connected node associated only with Ca-CVD DMPs in the network (14 interactions). For only cancer, FOXA1 was the most connected node (10 interactions). The most connected node shared by cancer and CVD was NKX2-5, with 10 interactions. PTPRN2 was the only node associated to Ca-CVD, cancer, and CVD DMPs in at least 3 cohorts. Additionally, an unrestricted approach was applied to analyze the 358 protein-coding genes (confidence score 0.0 or greater), resulting in a network of 352 nodes and 3,734 edges (Supplemental Figure 3, Supplemental File, sheets Q and R). The main findings were similar for both networks.
Figure 1.
Protein-Protein Interaction Network for Protein-Coding Genes Associated With at Least 3 Cohorts
The network was analyzed using Cytoscape and included 147 nodes and 217 interactions. The sizes of the nodes are proportional to the number of connections. Increasingly darker solid edge lines indicate protein interactions with increasing confidence scores. The interactions and their confidence scores (0.5 or greater) were obtained from the STRING database. The circle shape indicates that the node was common for 3 cohorts, whereas the triangle shape indicates that it was common for all 4 cohorts. Different colors are used for each outcome. CpG = cytosine followed by a guanine with a phosphate link; CVD = cardiovascular disease.
Network enrichment analysis
A total of 264 GO and 13 UniProt terms were statistically significant at a false discovery rate–corrected (calculated using the Benjamini-Hochberg approach) P value <0.05. The top 5 GO terms for the biological process category were animal organ development, tissue development, cell differentiation, anatomical structure development, and developmental process. For the molecular function category, the top 5 terms were all related to regulatory region sequence-specific DNA binding. For the cellular component category, the top terms were chromatin, chromosome, and membrane-bounded organelle related. The UniProt top terms were DNA binding, nucleus, transcription regulation, repressor, and homeobox (Supplemental File, sheet O). In the DrugBank search, among the 24 overlapping nodes between at least 2 endpoints in 3 or more cohorts and the most connected nodes (Supplemental File, sheet P), we identified 8 drug target proteins (UniProt ID) for treatments related to the 1-carbon metabolism and epigenetic functions, including antioxidant elements (MGMT and HDAC4), cancer (DNMT3B, DNMT3A, and EGFR), and cardiometabolic diseases (ADCY3 and GNAS) treatments (Supplemental Table 3).
Incident cancer predictive accuracy among individuals with prior cardiovascular events
Of 950 SHS participants who were free of cancer at the time of incident CVD, 127 subsequently developed cancer (accumulated follow-up time was 9,185.2 person-years). The C indexes with 95% CIs are shown in Table 3. The baseline predictive model (including clinical risk factors but not blood DNAm) had a C index of 0.618 (95% CI: 0.570-0.672) for all cancer, with a Brier score of 0.698. The C index when including the union set of cancer DMPs identified in FHS, ARICb, and ARICw (858 CpGs) improved to 0.727 (95% CI: 0.651-0.789), with a Brier score of 0.116. When including the union set of Ca-CVD DMPs identified in FHS, ARICb, and ARICw (642 CpGs), it improved to 0.971, (95% CI: 0.963-0.978), with a Brier score of 0.113.
Table 3.
C Indexes for Cancer in Participants With Cardiovascular Disease in the Strong Heart Study
| Adjustment Variable | Number of CpGs | C Index (95% CI) |
|---|---|---|
| Clinical risk factorsa | 0 | 0.618 (0.570-0.672) |
| Clinical risk factors + estimated cell counts, genetic PCs, and smoking epigenetic score | 0 | 0.669 (0.621-0.714) |
| Clinical risk factors + estimated cell counts, genetic PCs, and smoking epigenetic score + cancer DMPs in FHS, ARICw, and ARICb | 858 | 0.727 (0.651-0.789) |
| Clinical risk factors + estimated cell counts, genetic PCs, and smoking epigenetic score + Ca-CVD DMPs in FHS, ARICw, and ARICb | 642 | 0.971 (0.963-0.978) |
ARICb = African American participants of the Atherosclerosis Risk In Communities study; ARICw = European American participants of the Atherosclerosis Risk In Communities study; CpG = cytosine followed by a guanine with a phosphate link; FHS = Framingham Heart Study; PC = principal component; other abbreviations as in Table 2.
Age, sex, smoking status, body mass index, and study center.
Discussion
In this multicohort study, we found common epigenetic signatures for CVD and cancer across ethnically diverse populations, supporting the hypothesis of a common biological background for both diseases. Many of the proteins encoded by those genes were involved in known molecular pathways for cancer and CVD. Elements related to 1-carbon metabolism and cancer and CVD medications were identified as potential drugs for target gene products from most relevant nodes in the protein-interaction network. Many DMPs identified in the cohort-specific EWAS, however, were not common across populations, possibly indicating that population-specific environments might also induce differential epigenetic footprints. We also found a substantial increase in predictive accuracy for incident cancer among participants with newly diagnosed CVD using blood DNAm at 642 CpG sites.
Some of the genes annotated to the DMPs that were statistically significant in our meta-analysis have biological functions relevant for CVD and cancer. The KLF11 gene plays a role in the regulation of pancreatic beta cells, and variants of this gene might contribute to the development of diabetes, one of the main risk factors for CVD.25 Also, the CBX4 gene has been proposed as an oncogene26 and has been reported to promote tumor progression and proliferation in both lung27 and gastric28 cancer. In addition, blocking CBX4 expression showed protective effects against drug resistance for hepatocellular carcinoma.29
Our protein-protein interaction network further supports the interconnection of mechanisms in common for CVD and cancer. The most connected nodes of the network include the NKX2-5, PTPRN2, and EGFR genes (Figure 1), among others. NKX2-5 encodes a homeobox-containing transcription factor with functions in heart formation and development. Genetic variants of this gene have been associated with congenital heart disease,30 and transcriptomic and epigenomic data indicate that this gene contributes to electrocardiographic phenotypes.31 In addition, NKX2-5 has been associated with acute lymphoblastic leukemia32 and colorectal cancer.33 DNAm in positions annotated to PTPRN2 was associated with vascular and cardiac disease,34 sarcomas,35 and breast cancer.36 Last, EGFR is a transmembrane glycoprotein receptor for members of the epidermal growth factor family, with functions widely associated with cancer development and treatment37 as well as CVD.38 Regarding the most connected nodes for cancer and Ca-CVD endpoints, the FOXA1 gene encodes a member of the forkhead class of DNA-binding proteins, related to several types of cancer39 and also to heart failure.40 The protein product of PAX6 gene is a regulator of gene transcription, and has been associated to cancer41 and CVD.42 DNMT3A is one of the most commonly somatically mutated gene in clonal hematopoiesis.43 These mutations increase steeply with age. Therefore, we conducted a post hoc sensitivity analysis for the Ca-CVD endpoint evaluating the potential interaction between DNAm at cg23009818 (annotated to DNMT3A) by age, with no evidence of a differential association (P value for interaction = 0.78).
In addition, the network enrichment analysis identified general pathways associated with inflammation-related conditions and diabetes mellitus, and also with cancer and CVD. Alternatively, the DrugBank search pointed to proteins encoded by genes annotated to our identified DMPs as targets for known cancer and cardiometabolic medications (genes DNMT3B, DNMT3A, EGFR, ADCY3, and GNAS), as well as proteins with zinc binding sites (genes MGMT and HDAC4), which have a known role in the redox balance and epigenetic regulation.44 The most relevant drug target was EGFR, which has been previously identified as a central target in cardiotoxicity induced by targeted therapy in cancer treatment.38 Interestingly, cysteine dietary supplementation was associated with expression of the human MGMT gene. Cysteine is a nonessential amino acid needed to form glutathione, which also has redox regulatory functions.45 Oxidative stress and glutathione play a key role in 1-carbon metabolism, which provides methyl groups that constitute the substrate for DNAm.46
In this study, we were also able to evaluate the epigenetic susceptibility of patients with CVD to develop cancer. Experimental data in mice and humans support that CVD can predispose to cancer development.47 Our results support that DNAm signatures might be used to identify individuals at risk for cancer after newly developing CVD, as we found a substantial increase in predictive accuracy after including Ca-CVD DMPs in our model. Conversely, we could not evaluate the predictive ability of DNAm profiles in individuals who develop CVD after cancer. Thus, additional studies to guide treatment are needed to prevent cardiovascular complication of cancer therapies in patients with cancer on the basis of their epigenetic profiles.
Study limitations
Of note, we found many more common significant DMPs across cohorts for CVD than for cancer, and no DMPs were statistically significant in the meta-analysis for cancer only. This might be related to statistical power, as many more cases of CVD than cancer were present. Another explanation could be the heterogeneity of the cancer endpoint, as epigenetic changes might be specific to different types of cancer, which constitutes one of the limitations of our study. We were not able, however, to conduct separate analysis for each type of specific cancer because of the small number of cases. Additional cardio-oncology studies that focus on specific CVD and cancer endpoints leveraging multiple cohorts are needed. In addition, nonfatal cancer data in the SHS might be incomplete, as the SHS is not linked to cancer registry data. However, FHS and ARIC do have linkage to the cancer registry data, which adds robustness to the identified DMPs. On the other hand although elastic-net can deal with multicollinearity and has shown good performance in high-dimensional data,48 shrinkage might lead to biased effect estimates and therefore to loss of relevant DMPs selection, which might challenge causal discovery. Nevertheless, the DMPs identified in our study for each of the considered endpoints are robust, given that they were identified in 4 ethnically diverse populations.
Strengths of our study include the multicohort approach and the ethnic diversity of the participants included in this study, the large sample size, the elastic-net approach that alleviates the need to apply multiple-comparisons correction, the ability to account for a wide range of confounders, the prospective nature and long follow-up duration of the studies, and the combination of an EWAS with subsequent in silico analyses leveraging the mechanistic evidence available from well-established bioinformatic datasets to assess the potential biological plausibility of the findings.
Conclusions
We reported epigenetic signatures in common for cancer and CVD. Our bioinformatic analyses additionally support that underlying common molecular pathways are related to cancer and CVD onset. Future studies that experimentally evaluate the role of the identified target genes in CVD and cancer are needed. The epigenetic signatures reported in this study could potentially help identify individuals with newly diagnosed CVD at increased cancer risk, thus enabling the prevention and control of cancer and CVD.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: We conducted an ethnically diverse multicohort study to identify epigenetic marks, which point to common underlying molecular mechanisms for CVD and cancer and might aid in the early detection of cancer in patients with newly diagnosed CVD.
TRANSLATIONAL OUTLOOK: Future research and experimental studies are needed to confirm the role of the identified epigenetic marks as treatment targets in cardio-oncology. Measuring DNAm might enable the identification of candidates for intensified cancer prevention and screening in newly diagnosed clinical CVD.
Funding Support and Author Disclosures
This work was supported by grants from the National Heart, Lung, and Blood Institute (under contracts 75N92019D00027, 75N92019D00028, 75N92019D00029, and 75N92019D00030) and previous grants (R01HL090863, R01HL109315, R01HL109301, R01HL109284, R01HL109282, and R01HL109319 and cooperative agreements U01HL41642, U01HL41652, U01HL41654, U01HL65520, and U01HL65521); by the National Institute of Health Sciences (R01ES021367, R01ES025216, P42ES033719, and P30ES009089); by the Spanish Funds for Research in Health Sciences, Instituto de Salud Carlos III, cofunded by European Regional Development Funds (PI22CIII/00029, PI15/00071 and CP12/03080); and the by Spanish Agency for Research (PID2023-147163OB-C22, PID2019-108973RB-C21 and PID2020-117114GB-I00 from Ministerio de Ciencia e Innovación) by “Ministerio de Ciencia e Innovación,” Maria Zambrano grant ZA21-063, funded by the Ministry of Universities of the Government of Spain, financed by the European Union, NextGeneration EU, to Dr Riffo-Campos; ANID–Millennium Science Initiative Program—NCS2021_013 and ANID FONDAP 152220002 (CECAN) and a fellowship from the “La Caixa” Foundation (ID 100010434), code LCF/BQ/DR19/11740016. The ARIC study has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services, under contracts 75N92022D00001, 75N92022D00002, 75N92022D00003, 75N92022D00004, and 75N92022D00005. Funding was also supported by grants 5RC2HL102419 and R01NS087541. Studies on cancer in ARIC are also supported by the National Cancer Institute (U01 CA164975). Cancer data were provided by the Maryland Cancer Registry, Center for Cancer Prevention and Control, Maryland Department of Health, with funding from the State of Maryland and the Maryland Cigarette Restitution Fund. The collection and availability of cancer registry data are also supported by cooperative agreement NU58DP007114, funded by the Centers for Disease Control and Prevention. The FHS is funded by National Institutes of Health contract N01-HC-25195. The laboratory work for this investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, and a National Institutes of Health Director’s Challenge Award (Daniel Levy, principal investigator). Dr Belsky is a fellow of the CIFAR CBD Network. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the Centers for Disease Control and Prevention, the U.S. Department of Health and Human Services, or Instituto de Salud Carlos III. The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors thank the staff members and participants of the SHS, ARIC, and FHS for their important contributions.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental methods, references, tables, and figures, please see the online version of this paper.
Contributor Information
Arce Domingo-Relloso, Email: ad3531@cumc.columbia.edu.
Maria Tellez-Plaza, Email: m.tellez@isciii.es.
Appendix
References
- 1.Michel L., Schadendorf D., Rassaf T. Oncocardiology: new challenges, new opportunities. Herz. 2020;45:619–625. doi: 10.1007/s00059-020-04951-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad F.B., Cisewski J.A., Xu J., Anderson R.N. Provisional mortality data—United States, 2022. MMWR Morb Mortal Wkly Rep. 2023;72:488–492. doi: 10.15585/mmwr.mm7218a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koene R.J., Prizment A.E., Blaes A., Konety S.H. Shared risk factors in cardiovascular disease and cancer. Circulation. 2016;133:1104–1114. doi: 10.1161/CIRCULATIONAHA.115.020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Kindi S.G., Oliveira G.H. Onco-cardiology: a tale of interplay between 2 families of diseases. Mayo Clin Proc. 2016;91:1675–1677. doi: 10.1016/j.mayocp.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Hong R.A., Iimura T., Sumida K.N., Eager R.M. Cardio-oncology/onco-cardiology. Clin Cardiol. 2010;33:733–737. doi: 10.1002/clc.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aleman B.M.P., Moser E.C., Nuver J., et al. Cardiovascular disease after cancer therapy. EJC Suppl. 2014;12:18–28. doi: 10.1016/j.ejcsup.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zoghbi H.Y., Beaudet A.L. Epigenetics and human disease. Cold Spring Harb Perspect Biol. 2016;8:1–28. doi: 10.1101/cshperspect.a019497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinnaird A., Zhao S., Wellen K.E., Michelakis E.D. Metabolic control of epigenetics in cancer. Nat Rev Cancer. 2016;16:694–707. doi: 10.1038/nrc.2016.82. [DOI] [PubMed] [Google Scholar]
- 9.van der Harst P., de Windt L.J., Chambers J.C. Translational perspective on epigenetics in cardiovascular disease. J Am Coll Cardiol. 2017;70:590–606. doi: 10.1016/j.jacc.2017.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee E.T., Welty T.K., Fabsitz R., et al. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132:1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- 11.Tsao C.W., Vasan R.S. Cohort Profile: The Framingham Heart Study (FHS): overview of milestones in cardiovascular epidemiology. Int J Epidemiol. 2015;44:1800–1813. doi: 10.1093/ije/dyv337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright J.D., Folsom A.R., Coresh J., et al. The Atherosclerosis Risk in Communities (ARIC) study: JACC focus seminar 3/8. J Am Coll Cardiol. 2021;77:2939. doi: 10.1016/j.jacc.2021.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Agostino R.B., Vasan R.S., Pencina M.J., et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation. 2008;117:743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 14.Kreger B.E., Splansky G.L., Schatzkin A. The cancer experience in the Framingham Heart Study cohort. Cancer. 1991;67(1):1–6. doi: 10.1002/1097-0142(19910101)67:1<1::aid-cncr2820670102>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 15.Almuwaqqat Z., Jokhadar M., Norby F.L., et al. Association of antidepressant medication type with the incidence of cardiovascular disease in the ARIC study. J Am Heart Assoc. 2019;8(11) doi: 10.1161/JAHA.119.012503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshu C.E., Barber J.R., Coresh J., et al. Enhancing the infrastructure of the Atherosclerosis Risk in Communities (ARIC) study for cancer epidemiology research: ARIC Cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:295. doi: 10.1158/1055-9965.EPI-17-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domingo-Relloso A., Feng Y., Rodriguez-Hernandez Z., et al. Omics feature selection with the extended SIS R package: identification of a body mass index epigenetic multi-marker in the Strong Heart Study. Am J Epidemiol. 2024;193(7):1010–1018. doi: 10.1093/aje/kwae006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon N., Friedman J., Hastie T., Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarzer G., Carpenter J.R., Rücker G. Springer; Cham, Switzerland: 2015. Meta-Analysis With R. [Google Scholar]
- 20.Joehanes R., Just A.C., Marioni R.E., et al. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet. 2016;9:436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Lissa C. Heterogeneity statistics. In: Doing Meta-Analysis in R and Exploring Heterogeneity Using metaforest. Accessed April 30, 2023. https://cjvanlissa.github.io/Doing-Meta-Analysis-in-R/heterogeneity-statistics.html.
- 22.Szklarczyk D., Gable A.L., Lyon D., et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knox C., Law V., Jewison T., et al. DrugBank 3.0: a comprehensive resource for “omics” research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C., Li S.X., Caraballo C., et al. Performance metrics for the comparative analysis of clinical risk prediction models employing machine learning. Circ Cardiovasc Qual Outcomes. 2021;14 doi: 10.1161/CIRCOUTCOMES.120.007526. [DOI] [PubMed] [Google Scholar]
- 25.Neve B., Fernandez-Zapico M.E., Ashkenazi-Katalan V., et al. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc Natl Acad Sci U S A. 2005;102:4807–4812. doi: 10.1073/pnas.0409177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang N., Niu G., Pan Y.H., et al. CBX4 transcriptionally suppresses KLF6 via interaction with HDAC1 to exert oncogenic activities in clear cell renal cell carcinoma. EBioMedicine. 2020;53 doi: 10.1016/j.ebiom.2020.102692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu C., Zhang Q., Tang Q., et al. CBX4 promotes the proliferation and metastasis via regulating BMI-1 in lung cancer. J Cell Mol Med. 2020;24:618–631. doi: 10.1111/jcmm.14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W., Chen H., Wang Z., Liu J., Lei X., Chen W. Chromobox 4 (CBX4) promotes tumor progression and stemness via activating CDC20 in gastric cancer. J Gastrointest Oncol. 2022;13:1058–1072. doi: 10.21037/jgo-22-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao W., Ma B., Tian Z., et al. Inhibiting CBX4 efficiently protects hepatocellular carcinoma cells against sorafenib resistance. Br J Cancer. 2021;124:1237. doi: 10.1038/s41416-020-01240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González-Castro T.B., Tovilla-Zárate C.A., López-Narvaez M.L., et al. Association between congenital heart disease and NKX2.5 gene polymorphisms: systematic review and meta-analysis. Biomark Med. 2020;14:1747–1757. doi: 10.2217/bmm-2020-0190. [DOI] [PubMed] [Google Scholar]
- 31.Benaglio P., D’Antonio-Chronowska A., Ma W., et al. Allele-specific NKX2-5 binding underlies multiple genetic associations with human electrocardiographic traits. Nat Genet. 2019;51:1506–1517. doi: 10.1038/s41588-019-0499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagel S., Scherr M., Kel A., et al. Activation of TLX3 and NKX2-5 in t(5;14)(q35;q32) T-cell acute lymphoblastic leukemia by remote 3′-BCL11B enhancers and coregulation by PU.1 and HMGA1. Cancer Res. 2007;67:1461–1471. doi: 10.1158/0008-5472.CAN-06-2615. [DOI] [PubMed] [Google Scholar]
- 33.Li H., Wang J., Huang K., et al. Nkx2.5 functions as a conditional tumor suppressor gene in colorectal cancer cells via acting as a transcriptional coactivator in p53-mediated p21 expression. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.648045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krolevets M., ten Cate V., Prochaska J.H., et al. DNA methylation and cardiovascular disease in humans: a systematic review and database of known CpG methylation sites. Clin Epigenet. 2023;15:1–16. doi: 10.1186/s13148-023-01468-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vargas A.C., Gray L.A., White C.L., et al. Genome wide methylation profiling of selected matched soft tissue sarcomas identifies methylation changes in metastatic and recurrent disease. Sci Rep. 2021;11:1–17. doi: 10.1038/s41598-020-79648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sengelaub C.A., Navrazhina K., Ross J.B., Halberg N., Tavazoie S.F. PTPRN2 and PLCβ1 promote metastatic breast cancer cell migration through PI(4,5)P2-dependent actin remodeling. EMBO J. 2016;35:62–76. doi: 10.15252/embj.201591973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uribe M.L., Marrocco I., Yarden Y. EGFR in cancer: signaling mechanisms, drugs, and acquired resistance. Cancers (Basel) 2021;13:2748. doi: 10.3390/cancers13112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chitturi K.R., Burns E.A., Muhsen I.N., Anand K., Trachtenberg B.H. Cardiovascular risks with epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors and monoclonal antibody therapy. Curr Oncol Rep. 2022;24:475–491. doi: 10.1007/s11912-022-01215-1. [DOI] [PubMed] [Google Scholar]
- 39.Augello M.A., Hickey T.E., Knudsen K.E. FOXA1: master of steroid receptor function in cancer. EMBO J. 2011;30:3885–3894. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hannenhalli S., Putt M.E., Gilmore J.M., et al. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 41.Birknerová N., Kova?íková H., Baranová I., et al. DNA hypermethylation of CADM1, PAX5, WT1, RARβ, and PAX6 genes in oropharyngeal cancer associated with human papillomavirus. Epigenetics. 2022;17:1301–1310. doi: 10.1080/15592294.2021.2018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D.J., Lee M.J., Cho H.B., et al. Differential expression of Pax6 following bilateral common carotid artery occlusion. In Vivo. 2023;37:655–660. doi: 10.21873/invivo.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheller M., Ludwig A.K., Göllner S., et al. Hotspot DNMT3A mutations in clonal hematopoiesis and acute myeloid leukemia sensitize cells to azacytidine via viral mimicry response. Nat Cancer. 2021;2:527–544. doi: 10.1038/s43018-021-00213-9. [DOI] [PubMed] [Google Scholar]
- 44.Oteiza P.I. Zinc and the modulation of redox homeostasis. Free Radic Biol Med. 2012;53:1748–1759. doi: 10.1016/j.freeradbiomed.2012.08.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fra A., Yoboue E.D., Sitia R. Cysteines as redox molecular switches and targets of disease. Front Mol Neurosci. 2017;10:167. doi: 10.3389/fnmol.2017.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lionaki E., Ploumi C., Tavernarakis N. One-carbon metabolism: pulling the strings behind aging and neurodegeneration. Cells. 2022;11(2):214. doi: 10.3390/cells11020214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meijers W.C., Maglione M., Bakker S.J.L., et al. Heart failure stimulates tumor growth by circulating factors. Circulation. 2018;138:678–691. doi: 10.1161/CIRCULATIONAHA.117.030816. [DOI] [PubMed] [Google Scholar]
- 48.Zou H., Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc B Stat Methodol. 2005;67(2):301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.